Abstract
Topoisomerase II resolves intrinsic topological problems of double-stranded DNA. As part of its essential cellular functions, the enzyme generates DNA breaks, but the regulation of this potentially dangerous process is not well understood. Here we report single-molecule fluorescence experiments that reveal a previously uncharacterized sequence of events during DNA cleavage by topoisomerase II: nonspecific DNA binding, sequence-specific DNA bending, and stochastic cleavage of DNA. We have identified unexpected structural roles of Mg2+ ions coordinated in the TOPRIM (topoisomerase-primase) domain in inducing cleavage-competent DNA bending. A break at one scissile bond dramatically stabilized DNA bending, explaining how two scission events in opposing strands can be coordinated to achieve a high probability of double-stranded cleavage. Clamping of the protein N-gate greatly enhanced the rate and degree of DNA bending, resulting in a significant stimulation of the DNA cleavage and opening reactions. Our data strongly suggest that the accurate cleavage of DNA by topoisomerase II is regulated through a tight coordination with DNA bending.
Keywords: N-gate clamping, single-molecule FRET, type II topoisomerase, G-segment selection, indirect readout
The double helical nature of DNA imposes intrinsic topological problems during replication, repair, and transcription (1–3). Additionally, the topological state of the genetic material needs to be tightly regulated in order to promote proper biochemical interactions between DNA and a variety of proteins (1–4). Topoisomerases are enzymes that resolve topological problems within the double helix by repeated cycles of DNA cleavage and ligation (1–3, 5, 6).
As a subclass of the topoisomerase family, type II topoisomerases are found in all organisms from bacteria to human, and even in some viruses (1–3, 5, 6). The essential roles of type II topoisomerases in cell metabolism, differences between bacterial and human homologues, and hyperactivation of these enzymes in cancer cells have been utilized for clinical treatments of bacterial infections and numerous cancers (3, 7–9).
Extensive studies for more than twenty years have established that type II topoisomerases use a “two gate” mechanism for DNA strand passage (3, 10, 11), in which a DNA duplex (the transport or T-segment) is transported through an enzyme-mediated transient opening in a separate DNA duplex (the gate or G-segment). The directionality of strand passage is the N-terminal gate of the enzyme to the C-terminal gate. As a result of the double-stranded DNA passage mechanism, each catalytic event changes the linking number of DNA by two. The transport of the T-segment through the G-segment is thought to be initiated by the N-gate clamping motion induced by the binding of ATP to the enzyme (3, 6, 10, 12).
Although the double-stranded DNA breaks generated by type II topoisomerases are essential for the cellular functions of these enzymes, it is a dangerous process in which an aberrant operation can damage chromosomal integrity. In fact, widely prescribed anticancer and antibacterial drugs initiate cell death by increasing the cellular concentration of topoisomerase II-mediated DNA strand breaks (3, 7–9). Thus, the DNA cleavage reaction of type II topoisomerases needs to be tightly regulated.
Despite the paramount importance of the cleavage reaction of type II topoisomerases in biology and practical medicine, many fundamental questions concerning the regulation of the cleavage reaction remain unanswered. For instance, decades of biochemical studies revealed that the DNA cleavage reaction occurs only at specific sequences (13), but it is not understood how the cleavage sites are selected. In most in vitro assays, the cleavage efficiency of type II topoisomerases is very low (3). However, a mechanism to increase the enzymatic cleavage efficiency of type II topoisomerases has not yet been identified. In contrast to the low cleavage efficiency of type II topoisomerases, the probability of a double-stranded break is extremely high, suggesting that cleavage of the two separate strands is efficiently communicated. However, the communication mechanism has not yet been delineated.
Here we report single-molecule fluorescence resonance energy transfer (FRET) assays (14) that monitor the individual reaction steps of the interaction between human topoisomerase IIα (hTopoIIα) and DNA. Similar methodologies using different labeling schemes recently have been used to monitor opening of the G-segment DNA and narrowing of the N-gate in Drosophila topoisomerase II and Escherichia coli gyrase, respectively (15–17). Results of the present study strongly suggest the following: (i) a sharp bend in DNA is dynamically induced by the enzyme as a prerequisite to the cleavage reaction; (ii) although the DNA binding step appears to be nonspecific with regard to DNA sequence, bending was observed only with cleavable sequences, indicating that the deformability of DNA sequences is an important determinant of G-segment selection. (iii) the interaction of divalent metal ions with the highly conserved acidic residues in the TOPRIM (topoisomerase-primase) domain is critical for DNA bending, revealing an unexpected structual role for divalent ions in the enzymatic function of type II topoisomerases; (iv) a break at one scissile bond dramatically increases the bending lifetime, providing a physical mechanism for the coordination of double-stranded breaks; (v) the rate and degree of DNA bending are greatly enhanced by clamping of the protein N-gate, resulting in a great stimulation of DNA cleavage and opening, which is disfavored under normal conditions. Overall, our work provides evidence that Mg2+-mediated DNA bending by topoisomerase II is the basis for both the cleavage site selection and the regulation of double-stranded DNA cleavage by the enzyme.
Results
Observation of Single-Enzyme Binding Events.
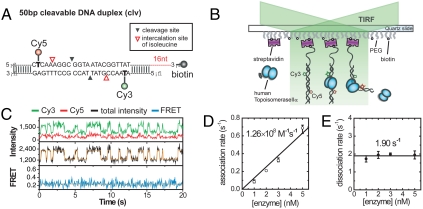
A partial DNA duplex containing a central cleavage site for hTopoIIα (18) and flanking FRET probes (Cy3 and Cy5) was prepared [clv (DNA construct) in SI Appendix, Fig. S1; Fig. 1A]. The duplex has a biotinylated single-stranded overhang to avoid any potential steric hindrance caused by surface immobilization. After immobilizing DNA molecules on a polymer-coated quartz surface, hTopoIIα was delivered into a detection chamber, and the fluorescence signals of single DNA molecules were monitored using a total-internal-reflection fluorescence microscope (Fig. 1B).
Fig. 1.
Observation of single-enzyme binding events. (A) A DNA construct (clv) used for the experiments. (B) Schematic diagram of single-molecule FRET experiments. (C) Representative time traces of Cy3 fluorescence (green, top), Cy5 fluorescence (red, top), the sum of Cy3 and Cy5 fluorescences (black with orange, middle), and corresponding FRET efficiency (blue with orange, bottom) in the presence of 5 nM hTopoIIα. To make transitions clearer, orange lines are added as an eye guide. The same color convention is used throughout the paper. These experiments were performed without divalent ions. (D) Association rates at varying enzyme concentrations. The association rate constant (1.26 × 108 M-1 s-1) was obtained from a linear fit of the data. (E) Dissociation rates at varying enzyme concentrations, whose average is 1.90 s-1.
In the absence of enzyme, fluorescence intensities showed a single-state behavior with small fluctuations limited by shot-noise (SI Appendix, Fig. S2). Upon addition of hTopoIIα, however, large intensity jumps with appreciable dwell times were observed (Fig. 1C). The frequency of the intensity jumps increased linearly with enzyme concentration (Fig. 1D, SI Appendix, Fig. S3A), while the lifetime of the high intensity state did not show any appreciable change over the examined enzyme concentration range (Fig. 1E; SI Appendix, Fig. S3B). Thus, we infer that the intensity jumps correspond to the binding events of single enzymes (19, 20). The association rate constant obtained from the enzyme titration experiments in Fig. 1D (1.26 × 108 M-1 s-1) shows that the association step is diffusion limited (21, 22). Thus, hindrance of DNA-enzyme interactions by dye-labeling or surface immobilization is negligible. This conclusion is further supported by the fact that similar dissociation constants were obtained in single-molecule and bulk studies (SI Appendix, Fig. S4). As expected from a diffusion-limited binding event, the association rate was similar at varying salt concentrations (SI Appendix, Fig. S5A). The dissociation rate, however, rapidly decreased at lower salt concentrations (SI Appendix, Fig. S5B), indicating that the dissociation constant is very sensitive to ionic strength (23).
Mg2+-Induced DNA Bending as a Prerequisite for the Cleavage Reaction.
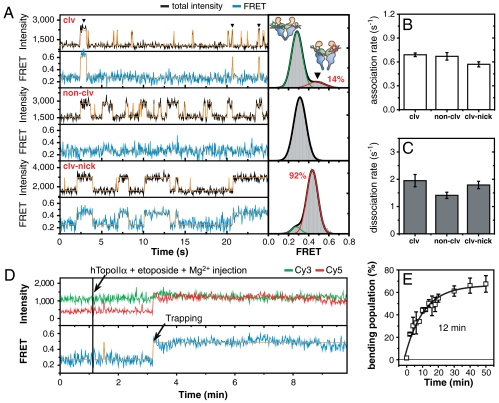
Remarkably, in the above experiments, which were performed in the absence of divalent ions, FRET efficiencies remained constant (E = 0.27) during the repeated association/dissociation events of the enzyme (Fig. 1C). In the presence of 5 mM Mg2+, however, the situation changed dramatically. Large FRET jumps (from E = 0.27 to E = 0.50) were observed in some of the enzyme-DNA binding events (Fig. 2A, top), indicating that a substantial amount of DNA deformation was stochastically induced by hTopoIIα. Similar FRET jumps were observed in the presence Ca2+ (SI Appendix, Fig. S6). The FRET jump, however, was not observed when a noncleavable DNA sequence was used as the substrate (24) (non-clv in SI Appendix, Fig. S1; Fig. 2A, middle), but became more frequent and stable when a nick was introduced in one of the two scissile bonds of the cleavage sequence (clv-nick in SI Appendix, Fig. S1; Fig. 2A, bottom; SI Appendix, Fig. S7B). Introduction of a nick in non-clv, however, did not induce any FRET jump during enzyme binding events (SI Appendix, Fig. S8). Combining the above observation with the knowledge that a nick in one strand greatly increases the cleavage efficiency of the opposite strand (25, 26), we conclude that DNA deformation induced by hTopoIIα occurs in a sequence-specific fashion with a strong correlation between the cleavage efficiency and the population of the deformed state (Fig. 2A, right).
Fig. 2.
Divalent ion-induced DNA bending as a precleavage complex. (A) Representative intensity and FRET time traces for three DNA duplexes (clv, top; non-clv, middle; clv-nick, bottom) in the presence of 5 mM Mg2+ (left), and corresponding FRET histograms of DNA duplexes with bound enzyme (right). Bending population of clv (14%) was obtained by fitting the FRET histogram to sum of two Gaussian functions. (B, C) Comparison of association rates (B) and dissociation rates (C) of hTopoIIα (5 nM) on three DNA duplexes in the absence of Mg2+. (D) Representative intensity and FRET time traces of clv after the addition of hTopoIIα (5 nM), and etoposide (500 μM) in the presence of 5 mM Mg2+. (E) Relative population of cleavage complex as a function of incubation time. The trapping time (12 min) was obtained by fitting the data to a single-exponential function. Error bars represent standard errors of two datasets.
Sharp bending of the G-segment DNA is supported by a number of experiments including recent high-resolution X-ray structures, atomic force microscopy, and FRET (27–31). With this context in mind, the FRET jumps specifically observed only in cleavable sequences in Fig. 2A are interpreted as reflecting the sharp bending of G-segment DNA induced by hTopoIIα. In contrast to the favorable correlations of enzyme-induced DNA bending and straightening rates with the cleavage efficiencies of the three DNA duplexes (SI Appendix, Fig. S7B), such a correlation was not observed with either the association or dissociation steps regardless of the presence of Mg2+ ions (SI Appendix, Fig. S7A; Fig. 2 B and C; SI Appendix, Fig. S9).
The requirement of Mg2+ ions for DNA bending is reminiscent of the critical role of divalent ions in G-segment cleavage (32). This requirement raises a question as to whether the bending conformation observed in the previous experiments represents a precleavage complex or a product of the cleavage reaction. To address this issue, we investigated how fast DNA duplexes were trapped in the cleavage state by an anticancer drug such as etoposide at saturating concentrations (SI Appendix, Fig. S10). A recent high-resolution X-ray structure (33) shows that etoposide stabilizes the cleavage complex by intercalating between the bases of the cleaved scissile bond (3, 7, 34), providing an indirect way to monitor DNA cleavage events. Fig. 2D shows representative time traces of a real-time buffer exchange experiment—hTopoIIα and etoposide were added to the clv-immobilized detection chamber (black line) while single-molecule FRET images were being taken. With the 1-s time resolution used in the experiment, individual binding and bending events could not be detected clearly. Instead, irreversible trapping events to the high FRET state occurred very slowly, with an average trapping time of 12 min (Fig. 2E; SI Appendix, Fig. S11). This finding indicates that most of the bending population (Fig. 2A, top right) is not cleaved, suggesting that it represents a precleavage complex. This conclusion is supported by several lines of evidence. First, previous studies discovered that only a small fraction of the topoisomerase II-DNA complex (less than 1% for clv) is trapped in the cleavage state (3, 18, 35). Second, significant bending of clv was observed in single-molecule FRET studies that employed a nonreactive mutant hTopoIIα, Y805F, in which the catalytic tyrosine was replaced with a phenylalanine (SI Appendix, Fig. S12). Thus, DNA cleavage is not required for bending. Consistent with this conclusion, the existence of a precleavage complex with a sharp bend but no cleavage of the G-segment has been reported in two recent X-ray crystal studies (29, 36). On the basis of these observations, we conclude that the cleavage reaction of type II topoisomerases goes through three distinct and well ordered reaction steps: nonspecific enzyme-DNA binding, sequence-specific DNA bending, and finally, cleavage. This conclusion is consistent with the previous observation that the DNA site-selection step of topoisomerase II appears to occur after initial binding but before cleavage (24, 37). Taken together, we propose that the cleavage efficiency of a DNA sequence is not related to its binding affinity (24, 37), but rather to its deformability.
Roles of Divalent Ions and Conserved Acidic Residues in DNA Bending.
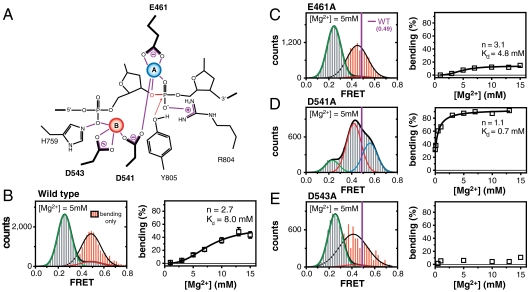
The above observation revealed an unexpected role of Mg2+ ions in the cleavage reaction of topoisomerase II; they are critical for inducing the large-scale structural change of G-segment DNA into a cleavage-competent form. Because the traditional view has been that divalent ions coordinated at the reaction center by interacting with acidic residues in the region play mainly a catalytic or structural role (Fig. 3A) (29, 30, 36, 38–40), we asked whether those interactions also are involved in DNA bending.
Fig. 3.
Roles of acidic residues in DNA bending. (A) A two-metal-ion model of DNA cleavage by human topoisomerase IIα (30). Highly conserved amino acids in the TOPRIM region are indicated by thick lines. The blue (A site) and red (B site) spheres are coordinated metal ions. Noncovalent interactions are indicated by violet lines. (B–E) FRET histograms (left) of DNA duplexes with bound enzyme in the presence of 5 mM Mg2+ and bending populations (right) at varying Mg2+ concentrations of WT (Wild Type) (B), E461A (C), D541A (D), and D543A (E). The high cooperativity of Mg2+ titration experiments of (B) is consistent with the two-metal-ion model of (A). To more clearly visualize the difference of FRET values between the bending conformations of the mutants and the WT, the exaggerated histograms of bending conformations of the mutants (red) and the FRET position of the WT bending conformation (purple line) are overlaid in (B–E). In (B), error bars were obtained from three independent experiments. The data in (B–D, right) were fit to the Hill equation.
To address the question, we performed Mg2+ titration experiments with mutant hTopoIIα proteins in which the highly conserved acidic residues in the TOPRIM domain that bind the active site metal ions (E461A, D541A, or D543A) were replaced with alanine. In bulk DNA cleavage assays, the cleavage reaction of hTopoIIα was greatly hampered by these mutations (32).
Single-molecule FRET studies revealed that all of these mutations had significant consequences for DNA bending (Fig. 3 B–E). In E461A, the bending population was mildly reduced, but the FRET value corresponding to the bent conformation was clearly distinguished from that of the wild-type enzyme (red in Fig. 3C). In D543A, both the DNA bending population and the FRET value corresponding to the bending conformation were greatly reduced (Fig. 3E). In 541A, two distinct bending conformations were observed whose FRET values were clearly distinguished from those induced by wild type, hTopoIIα, and the overall population of those bent states was much more populated (Fig. 3D). Qualitatively similar results were observed in the corresponding cysteine mutants (SI Appendix, Fig. S13). Collectively, these results illustrate that the conserved acidic residues in the TOPRIM region (E461, D541, and D543), which are responsible for binding the active site metal ions (30, 32), play critical roles in inducing cleavage-competent DNA bending. The results also indicate that global bending of DNA, per se, is not sufficient for DNA cleavage, but a more detailed coordination of protein, DNA, and divalent ions is required.
Coordination of Double-Stranded DNA Breaks Through DNA Bending.
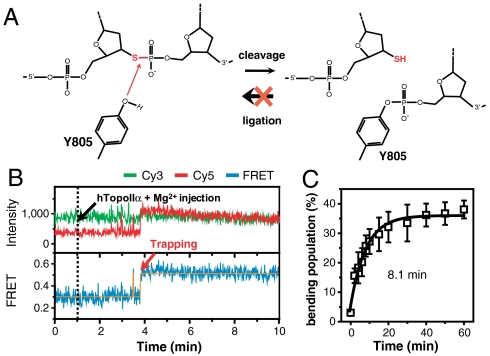
Next, we addressed what happens to the bending conformation when a single-stranded break is generated at the scissile bond on one of the DNA strands. To trap rare cleavage events, we prepared a DNA duplex whose 3′-bridging oxygen at one of the scissile bonds was replaced with a sulfur atom (clv-S in SI Appendix, Fig. S1). The sulfur atom at the cleavage site stabilizes the cleavage complex by blocking (or greatly inhibiting) the ligation process (Fig. 4A) (25). Thus, we expected that irreversible cleavage events in the modified strand might be kinetically distinguished from transient bending events without cleavage. Fig. 4B shows representative time traces of a real-time buffer exchange experiment—10 nM hTopoIIα was added to the clv-S-immobilized detection chamber (black dashed line) while single-molecule FRET images were being taken. As in Fig. 2D, irreversible trapping events to the high FRET state were observed, resulting in the accumulation of the high FRET population with the time constant of 8.1 min (Fig. 4C; SI Appendix, Fig. S14). This kind of irreversible trapping event was not observed in clv, and it was interpreted as the cleavage events in the sulfur-modified strand. Because ligation of the unmodified strand is not affected by the phosphorothiolation in the opposite strand (26), it is expected that DNA cleavage and ligation are continuously taking place in the unmodified strand even after the sulfur-modified strand is irreversibly cleaved. Therefore, the above observation provides a clue for an intriguing question as to how cleavage reactions in both strands are coordinated (25, 26, 41). Once one strand is cut, the cleavage probability of the other strand is greatly enhanced through an elongated dwell time of the cleavage-competent bending complex. Consistently, long-lived bent states of clv were observed with a rare but sufficiently higher probability than expected from the existence of single bending conformation with 0.27-s lifetime (SI Appendix, Fig. S15). With this finding in mind, the dramatic increase of the bending lifetime of a nicked sample (SI Appendix, Fig. S16), and the resulting increase of cleavage efficiency can be understood in the same context.
Fig. 4.
Single-stranded break greatly increases the bending lifetime. (A) Schematic drawing showing the effect of replacing the 3′-bridging oxygen of the scissile bond with sulfur on cleavage/ligation reaction of topoisomerase II. (B) Representative intensity and FRET time traces of clv-S after the addition of hTopoIIα (10 nM) in the presence of 5 mM Mg2+. (C) Relative population of cleavage complex as a function of incubation time. The cleavage time (8.1 min) was obtained by fitting the data to a single-exponential function. Error bars represent standard deviations that were obtained from three independent experiments.
Coordination of Cleavage and Opening of Gate-DNA with the N-Gate Clamping Motion of the Enzyme.
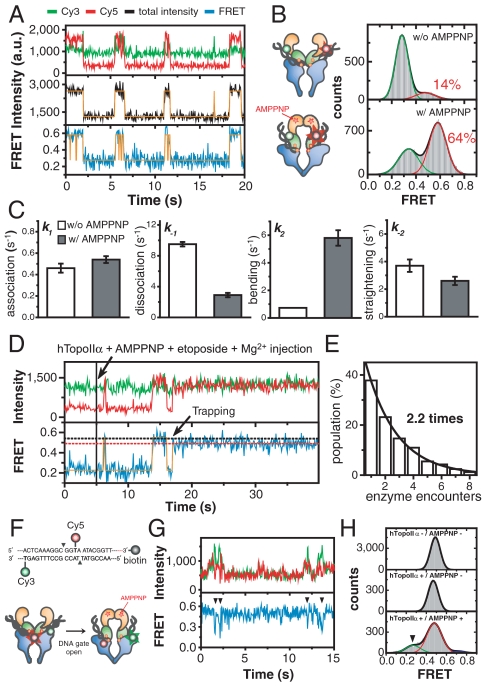
Our single-molecule observations and previous bulk cleavage assays consistently indicate that the DNA cleavage reaction of hTopoIIα is tightly down-regulated under normal reaction conditions (3, 18, 35). From a biological perspective, it is imperative that the genetic material should be protected from aberrant cleavage until the reaction is absolutely required for enzymatic function. Therefore, what is the regulation mechansim that triggers the DNA cleavage reaction of type II topoisomerases? There have been a number of reports indicating a coupling between N-gate clamping of the protein and DNA cleavage (41–45). To understand the coupling mechanism, we investigated how N-gate clamping affects the DNA bending, cleavage, and opening events of the G-segment. To this end, we carried out injection experiments with AMPPNP, a nonhydrolyzable analogue of ATP (42). Compared to Fig. 2A, AMPPNP dramatically affected the DNA-enzyme interaction (Fig. 5A). The population of bent DNA increased more than fourfold, and the FRET value corresponding to the bent conformation shifted to higher value (Fig. 5B). This finding suggests that N-gate clamping induces a greater deformation of the gate-DNA. Analysis of association/dissociation and bending/straightening kinetics revealed that the most dramatic change occurred in the bending and dissociation steps (Fig. 5C; SI Appendix, Fig. S17), resulting in numerous bending events of DNA duplexes during single encounter with an enzyme.
Fig. 5.
Effects of N-gate clamping on gate-DNA bending/cleavage/opening. (A) Representative intensity and FRET time traces of clv in the presence of AMPPNP (1 mM). (B) Comparison of FRET histograms of clv with (bottom) and without (top) AMPPNP. The histograms were fit to the sum of two Gaussian functions. (C) Comparison of kinetic parameters (k1: association rate, k-1; dissociation rate, k2: bending rate, k-2: straightening rate) of clv with (solid bars) and without (open bars) AMPPNP. (D) Representative intensity and FRET time traces of clv after the addition of hTopoIIα (5 nM), AMPPNP (1 mM) and etoposide (500 μM). The cleavage complex was trapped by etoposide. Simultaneous with the trapping event, the FRET efficiency of the bent state drops to the original FRET value (red dashed line) corresponding to the bent state in the absence of AMPPNP. (E) Relative populations of DNA cleavage trapping events as a function of enzyme encounter number after the addition of hTopoIIα (5 nM), AMPPNP (1 mM) and etoposide (500 μM). The average number of encounters per a cleavage event was obtained by fitting the data to a single-exponential function. (F) A DNA construct (clv-open in SI Appendix, Fig. S1; top) and experimental scheme (bottom) to directly observe DNA gate opening. (G) Representative intensity and FRET time traces of clv-open in the presence of AMPPNP (1 mM). Transitions to low FRET states are denoted by solid arrowheads. (H) Comparison of FRET histograms (top: without the enzyme and AMPPNP, middle: with the enzyme and without AMPPNP, bottom: with the enzyme and AMPPNP) of clv-open. Histograms were fitted to Gaussian functions. The FRET state corresponding to DNA gate opening was denoted by a solid arrowhead.
Next, we investigated the effects of N-gate clamping on the cleavage reaction of hTopoIIα. Compared to the slow rate of DNA cleavage observed in the absence of AMPPNP (Fig. 2E) (12 min trapping time on average, which corresponds to 330 encounters between DNA and the enzyme), trapping events occurred very rapidly (Fig. 5D), requiring an average of just 2.2 DNA encounters with the enzyme (Fig. 5E). Therefore, we conclude that N-gate clamping of hTopoIIα enhances the cleavage of gate-DNA as well as its bending, which is confirmed by the ensemble cleavage assay (SI Appendix, Fig. S18).
Finally, we investigated whether N-gate clamping induced an opening of the DNA gate. Because the previous labeling scheme was not sensitive to DNA gate opening, we designed a different labeling scheme, in which opening of DNA gate is expected to induce a transition to a low FRET state (Fig. 5F). With the new labeling scheme, transitions to low FRET states were observed only in the presence of AMPPNP (Fig. 5 G and H). This finding suggests that opening of DNA gate is tightly coupled with the clamping motion of the N-gate. Intriguingly, an apparent change in DNA bending and opening was not observed in the presence of ATP (SI Appendix, Fig. S19).
Discussion
The transient nature of key steps of the catalytic cycle of type II topoisomerases has hindered a deeper understanding of the reaction mechanisms of these enzymes. Therefore, we developed single-molecule fluorescence methods that were capable of visualizing the individual steps of the topoisomerase II-mediated DNA cleavage reaction. These methods allowed us to dissect the operational and regulatory mechanisms of the DNA cleavage reaction, which is summarized in SI Appendix, Fig. S20 and below.
It has long been known that type II topoisomerases cleave DNA in a sequence-dependent manner (3, 5, 24), but the mechanistic basis for G-segment selection was not apparent. We found that the cleavage reaction of type II topoisomerases proceeds through three ordered steps: DNA binding, bending, and cleavage. DNA bending was observed only in specific sequences that could be cleaved by the enzyme, while binding was not (Fig. 2 A–C). This finding suggests that the G-segment is selected by topoisomerase II via an “indirect readout” mechanism—by the physical properties, or deformability, of a DNA sequence, as opposed to the chemical information stored in the DNA sequence (46, 47). Consistent with this conclusion, recent high-resolution crystal structures indicated few direct interactions between DNA bases and eukaryotic topoisomerase II (27, 30).
The use of divalent metal cations by type II topoisomerases for their ATPase, cleavage, and ligation activities has been known for decades (3, 5, 32, 48). Our observations revealed an unexpected role of divalent metal ions in the operation of type II topoisomerases. DNA bending by hTopoIIα required the active site divalent cations, and this metal ion-dependent global conformational change was an intermediate reaction step en route to DNA cleavage. Furthermore, we showed that the highly conserved acidic residues in the TOPRIM domain are critical for the proper bending of gate-DNA.
Topoisomerase II coordinates cleavage of the two scissile bonds of the gate-DNA, but the mechanism underlying this coordination was not clear (25, 26). We observed that a single-stranded break increased the bending state lifetime dramatically, suggesting a unique biophysical mechanism by which the two independent cleavage events in the cleavage site are coordinated: cleavage of the first strand increases the flexibility of DNA, and as a result enhances the probability of the second strand cleavage by maintaining the DNA substrate in a cleavage-competent bent conformation (26).
There have been intriguing questions about the coupling mechanism of DNA cleavage and N-gate clamping. We found that the rate and degree of gate-DNA bending are greatly enhanced in the presence of AMPPNP, resulting in stimulated cleavage and opening of gate-DNA. Therefore, the cleavage and opening reactions of gate-DNA are well coordinated with the N-gate clamping motion of the enzyme through modulation of DNA bending. Through this tight control of DNA cleavage/opening processes, the probability of an accidental double-stranded breaks can be minimized, resulting in a protection of the genome from cytotoxic lesions caused by a misoperation of type II topoisomerases.
Materials and Methods
Detailed information on single-molecule measurements and determination of kinetic rates are available in SI Appendix, Materials and Methods.
Wild-type and mutant human topoisomerase IIα proteins were expressed in Saccharomyces cerevisiae JEL1Δtop1 cells and purified as described previously (49, 50). HPLC-purified DNA strands were purchased from Integrated DNA Technologies except the phosphorothiolate modified DNA strand, which was purchased from Purimex. DNA oligonucleotides (SI Appendix, Fig. S1) were labeled at the amine group of an internal amino modifier (dTC6) with either Cy3 or Cy5 (51). DNA duplexes were annealed by slowly cooling the mixture of the biotinylated strand and nonbiotinylated strand in 2∶3 molar ratio at 10 μM concentration in a buffer containing 10 mM Tris-HCl (pH 8.0) and 50 mM NaCl.
Supplementary Material
Acknowledgments.
We thank all the members of our labs for their kind discussion and help, and I.K. Chung (Yonsei University) for the contribution to this work at early stage. This work was supported by Creative Research Initiatives (Physical Genetics Laboratory, 2009-0081562), and by World Class University (WCU) Program of National Research Foundation of Korea to S.H., and by National Institutes of Health (NIH) research Grant GM033944 to N.O.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1115704109/-/DCSupplemental.
References
- 1.Wang JC. Cellular roles of DNA topoisomerases: a molecular perspective. Nat Rev Mol Cell Biol. 2002;3:430–440. doi: 10.1038/nrm831. [DOI] [PubMed] [Google Scholar]
- 2.Nitiss JL. DNA topoisomerase II and its growing repertoire of biological functions. Nat Rev Cancer. 2009;9:327–337. doi: 10.1038/nrc2608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Deweese JE, Osheroff N. The DNA cleavage reaction of topoisomerase II: wolf in sheep’s clothing. Nucleic Acids Res. 2009;37:738–748. doi: 10.1093/nar/gkn937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Koster DA, Crut A, Shuman S, Bjornsti MA, Dekker NH. Cellular strategies for regulating DNA supercoiling: a single-molecule perspective. Cell. 2010;142:519–530. doi: 10.1016/j.cell.2010.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Champoux JJ. DNA topoisomerases: structure, function, and mechanism. Annu Rev Biochem. 2001;70:369–413. doi: 10.1146/annurev.biochem.70.1.369. [DOI] [PubMed] [Google Scholar]
- 6.Corbett KD, Berger JM. Structure, molecular mechanisms, and evolutionary relationships in DNA topoisomerases. Annu Rev Biophys Biomol Struct. 2004;33:95–118. doi: 10.1146/annurev.biophys.33.110502.140357. [DOI] [PubMed] [Google Scholar]
- 7.Pommier Y, Leo E, Zhang H, Marchand C. DNA topoisomerases and their poisoning by anticancer and antibacterial drugs. Chem Biol. 2010;17:421–433. doi: 10.1016/j.chembiol.2010.04.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Liu LF. DNA topoisomerase poisons as antitumor drugs. Annu Rev Biochem. 1989;58:351–375. doi: 10.1146/annurev.bi.58.070189.002031. [DOI] [PubMed] [Google Scholar]
- 9.Nitiss JL. Targeting DNA topoisomerase II in cancer chemotherapy. Nat Rev Cancer. 2009;9:338–350. doi: 10.1038/nrc2607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Schoeffler AJ, Berger JM. DNA topoisomerases: harnessing and constraining energy to govern chromosome topology. Q Rev Biophys. 2008;41:41–101. doi: 10.1017/S003358350800468X. [DOI] [PubMed] [Google Scholar]
- 11.Roca J, Wang JC. DNA transport by a type II DNA topoisomerase: evidence in favor of a two-gate mechanism. Cell. 1994;77:609–616. doi: 10.1016/0092-8674(94)90222-4. [DOI] [PubMed] [Google Scholar]
- 12.Roca J, Wang JC. The capture of a DNA double helix by an ATP-dependent protein clamp: a key step in DNA transport by type II DNA topoisomerases. Cell. 1992;71:833–840. doi: 10.1016/0092-8674(92)90558-t. [DOI] [PubMed] [Google Scholar]
- 13.Capranico G, Binaschi M. DNA sequence selectivity of topoisomerases and topoisomerase poisons. Biochim Biophys Acta. 1998;1400:185–194. doi: 10.1016/s0167-4781(98)00135-3. [DOI] [PubMed] [Google Scholar]
- 14.Ha T, et al. Probing the interaction between two single molecules: fluorescence resonance energy transfer between a single donor and a single acceptor. Proc Natl Acad Sci USA. 1996;93:6264–6268. doi: 10.1073/pnas.93.13.6264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Smiley RD, Collins TR, Hammes GG, Hsieh TS. Single-molecule measurements of the opening and closing of the DNA gate by eukaryotic topoisomerase II. Proc Natl Acad Sci USA. 2007;104:4840–4845. doi: 10.1073/pnas.0700342104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gubaev A, Hilbert M, Klostermeier D. The DNA-gate of Bacillus subtilis gyrase is predominantly in the closed conformation during the DNA supercoiling reaction. Proc Natl Acad Sci USA. 2009;106:13278–13283. doi: 10.1073/pnas.0902493106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gubaev A, Klostermeier D. DNA-induced narrowing of the gyrase N-gate coordinates T-segment capture and strand passage. Proc Natl Acad Sci USA. 2011;108:14085–14090. doi: 10.1073/pnas.1102100108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fortune JM, et al. Site-specific DNA cleavage by Chlorella virus topoisomerase II. Biochemistry. 2002;41:11761–11769. doi: 10.1021/bi025802g. [DOI] [PubMed] [Google Scholar]
- 19.Luo G, Wang M, Konigsberg WH, Xie XS. Single-molecule and ensemble fluorescence assays for a functionally important conformational change in T7 DNA polymerase. Proc Natl Acad Sci USA. 2007;104:12610–12615. doi: 10.1073/pnas.0700920104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hwang H, Kim H, Myong S. Protein induced fluorescence enhancement as a single molecule assay with short distance sensitivity. Proc Natl Acad Sci USA. 2011;108:7414–7418. doi: 10.1073/pnas.1017672108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Berg OG, von Hippel PH. Diffusion-controlled macromolecular interactions. Annu Rev Biophys Biophys Chem. 1985;14:131–160. doi: 10.1146/annurev.bb.14.060185.001023. [DOI] [PubMed] [Google Scholar]
- 22.Halford SE, Marko JF. How do site-specific DNA-binding proteins find their targets? Nucleic Acids Res. 2004;32:3040–3052. doi: 10.1093/nar/gkh624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Leontiou C, Lightowlers R, Lakey JH, Austin CA. Kinetic analysis of human topoisomerase IIalpha and beta DNA binding by surface plasmon resonance. FEBS Lett. 2003;554:206–210. doi: 10.1016/s0014-5793(03)01172-4. [DOI] [PubMed] [Google Scholar]
- 24.Mueller-Planitz F, Herschlag D. DNA topoisomerase II selects DNA cleavage sites based on reactivity rather than binding affinity. Nucleic Acids Res. 2007;35:3764–3773. doi: 10.1093/nar/gkm335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Deweese JE, Burgin AB, Osheroff N. Using 3′-bridging phosphorothiolates to isolate the forward DNA cleavage reaction of human topoisomerase IIalpha. Biochemistry. 2008;47:4129–4140. doi: 10.1021/bi702194x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Deweese JE, Osheroff N. Coordinating the two protomer active sites of human topoisomerase IIalpha: nicks as topoisomerase II poisons. Biochemistry. 2009;48:1439–1441. doi: 10.1021/bi8021679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Dong KC, Berger JM. Structural basis for gate-DNA recognition and bending by type IIA topoisomerases. Nature. 2007;450:1201–1205. doi: 10.1038/nature06396. [DOI] [PubMed] [Google Scholar]
- 28.Laponogov I, et al. Structural insight into the quinolone-DNA cleavage complex of type IIA topoisomerases. Nat Struct Mol Biol. 2009;16:667–669. doi: 10.1038/nsmb.1604. [DOI] [PubMed] [Google Scholar]
- 29.Bax BD, et al. Type IIA topoisomerase inhibition by a new class of antibacterial agents. Nature. 2010;466:935–940. doi: 10.1038/nature09197. [DOI] [PubMed] [Google Scholar]
- 30.Schmidt BH, Burgin AB, Deweese JE, Osheroff N, Berger JM. A novel and unified two-metal mechanism for DNA cleavage by type II and IA topoisomerases. Nature. 2010;465:641–644. doi: 10.1038/nature08974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hardin AH, et al. Direct measurement of DNA bending by type IIA topoisomerases: implications for non-equilibrium topology simplification. Nucleic Acids Res. 2011;39:5729–5743. doi: 10.1093/nar/gkr109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Deweese JE, Osheroff N. The use of divalent metal ions by type II topoisomerases. Metallomics. 2010;2:450–459. doi: 10.1039/c003759a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wu CC, et al. Structural basis of type II topoisomerase inhibition by the anticancer drug etoposide. Science. 2011;333:459–462. doi: 10.1126/science.1204117. [DOI] [PubMed] [Google Scholar]
- 34.Bromberg KD, Burgin AB, Osheroff N. A two-drug model for etoposide action against human topoisomerase IIalpha. J Biol Chem. 2003;278:7406–7412. doi: 10.1074/jbc.M212056200. [DOI] [PubMed] [Google Scholar]
- 35.Wang JC. Unlocking and opening a DNA gate. Proc Natl Acad Sci USA. 2007;104:4773–4774. doi: 10.1073/pnas.0701070104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Laponogov I, et al. Structural basis of gate-DNA breakage and resealing by type II topoisomerases. PLoS One. 2010;5:e11338. doi: 10.1371/journal.pone.0011338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bromberg KD, Hendricks C, Burgin AB, Osheroff N. Human topoisomerase IIalpha possesses an intrinsic nucleic acid specificity for DNA ligation Use of 5′ covalently activated oligonucleotide substrates to study enzyme mechanism. J Biol Chem. 2002;277:31201–31206. doi: 10.1074/jbc.M204741200. [DOI] [PubMed] [Google Scholar]
- 38.Deweese JE, Burgin AB, Osheroff N. Human topoisomerase IIalpha uses a two-metal-ion mechanism for DNA cleavage. Nucleic Acids Res. 2008;36:4883–4893. doi: 10.1093/nar/gkn466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Pitts SL, et al. Use of divalent metal ions in the DNA cleavage reaction of topoisomerase IV. Nucleic Acids Res. 2011;39:4808–4817. doi: 10.1093/nar/gkr018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Noble CG, Maxwell A. The role of GyrB in the DNA cleavage-religation reaction of DNA gyrase: a proposed two metal-ion mechanism. J Mol Biol. 2002;318:361–371. doi: 10.1016/S0022-2836(02)00049-9. [DOI] [PubMed] [Google Scholar]
- 41.Mueller-Planitz F, Herschlag D. Coupling between ATP binding and DNA cleavage by DNA topoisomerase II: a unifying kinetic and structural mechanism. J Biol Chem. 2008;283:17463–17476. doi: 10.1074/jbc.M710014200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Osheroff N. Eukaryotic topoisomerase II. Characterization of enzyme turnover. J Biol Chem. 1986;261:9944–9950. [PubMed] [Google Scholar]
- 43.Lindsley JE, Wang JC. On the coupling between ATP usage and DNA transport by yeast DNA topoisomerase II. J Biol Chem. 1993;268:8096–8104. [PubMed] [Google Scholar]
- 44.Robinson MJ, et al. Effects of quinolone derivatives on eukaryotic topoisomerase II. A novel mechanism for enhancement of enzyme-mediated DNA cleavage. J Biol Chem. 1991;266:14585–14592. [PubMed] [Google Scholar]
- 45.Mueller-Planitz F, Herschlag D. Interdomain communication in DNA topoisomerase II. DNA binding and enzyme activation. J Biol Chem. 2006;281:23395–23404. doi: 10.1074/jbc.M604119200. [DOI] [PubMed] [Google Scholar]
- 46.Rohs R, et al. Origins of specificity in protein-DNA recognition. Annu Rev Biochem. 2010;79:233–269. doi: 10.1146/annurev-biochem-060408-091030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Garvie CW, Wolberger C. Recognition of specific DNA sequences. Mol Cell. 2001;8:937–946. doi: 10.1016/s1097-2765(01)00392-6. [DOI] [PubMed] [Google Scholar]
- 48.Sissi C, Palumbo M. Effects of magnesium and related divalent metal ions in topoisomerase structure and function. Nucleic Acids Res. 2009;37:702–711. doi: 10.1093/nar/gkp024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Worland ST, Wang JC. Inducible overexpression, purification, and active site mapping of DNA topoisomerase II from the yeast Saccharomyces cerevisiae. J Biol Chem. 1989;264:4412–4416. [PubMed] [Google Scholar]
- 50.Kingma PS, Greider CA, Osheroff N. Spontaneous DNA lesions poison human topoisomerase IIalpha and stimulate cleavage proximal to leukemic 11q23 chromosomal breakpoints. Biochemistry. 1997;36:5934–5939. doi: 10.1021/bi970507v. [DOI] [PubMed] [Google Scholar]
- 51.Roy R, Hohng S, Ha T. A practical guide to single-molecule FRET. Nat Methods. 2008;5:507–516. doi: 10.1038/nmeth.1208. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.