Abstract
Mechanical cues affect many important biological processes in metazoan cells, such as migration, proliferation, and differentiation. Such cues are thought to be detected by specialized mechanosensing molecules linked to the cytoskeleton, an intracellular network of protein filaments that provide mechanical rigidity to the cell and drive cellular shape change. The most abundant such filament, actin, forms branched networks nucleated by the actin-related protein (Arp) 2/3 complex that support or induce membrane protrusions and display adaptive behavior in response to compressive forces. Here we show that filamentous actin serves in a mechanosensitive capacity itself, by biasing the location of actin branch nucleation in response to filament bending. Using an in vitro assay to measure branching from curved sections of immobilized actin filaments, we observed preferential branch formation by the Arp2/3 complex on the convex face of the curved filament. To explain this behavior, we propose a fluctuation gating model in which filament binding or branch nucleation by Arp2/3 occur only when a sufficiently large, transient, local curvature fluctuation causes a favorable conformational change in the filament, and we show with Monte Carlo simulations that this model can quantitatively account for our experimental data. We also show how the branching bias can reinforce actin networks in response to compressive forces. These results demonstrate how filament curvature can alter the interaction of cytoskeletal filaments with regulatory proteins, suggesting that direct mechanotransduction by actin may serve as a general mechanism for organizing the cytoskeleton in response to force.
Keywords: actin-based motility, autocatalytic branching, bending fluctuations, worm-like chain, force sensing
Mechanical forces from a metazoan cell’s environment are transduced into biochemical signals during many biological processes, such as the differentiation, proliferation, and migration of cells, to regulate processes ranging from cytoskeletal remodeling to gene expression (1). Mechanotransduction has been thought to occur primarily via specialized mechanosensing molecules, which stretch or unfold in response to applied forces (2, 3), whereas the filament networks that make up the bulk of the cytoskeleton have been studied primarily as materials, whose mechanical properties determine how they transmit or absorb forces (4, 5). We asked whether filamentous actin (F-actin), a major part of the cytoskeleton, can act as a mechanosensor in its own right.
The actin cytoskeleton consists of an organized network of filaments that bear both tensile and compressive forces and largely determine the shape and rigidity of metazoan cells (4). Growth of one specialized cytoskeletal structure, the branched actin network (6–8), produces forces that act on cellular membranes to help them protrude or change shape (9–12) and plays an important role in cell motility, the trafficking of cellular membranes including endocytosis, and the motility of intracellular pathogens (13, 14). When this protrusive growth is opposed by resistance from the surrounding cytoskeleton or plasma membrane, the actin network compresses, and filaments in the network bend (15–18). In vitro studies have shown that compressive forces applied to branched networks cannot only reversibly deform them (15) but can also alter their density (19) and growth velocity (10, 11), suggesting that their architecture may respond actively to mechanical forces. Although the binding of many actin binding proteins (ABPs) to the side of an actin filament has been characterized (20) and, in some cases, shown to depend on the filament’s twist (21, 22) or its bound nucleotide (23, 24), the response of most F-actin–ABP interactions to filament bending is unknown (5). The only such response that has been documented is an increased frequency of severing by actophorin or its homolog actin depolymerizing factor (ADF)/cofilin at highly curved sections of actin filaments (23, 25).
Bending of F-actin is particularly relevant to its interaction with the actin-related protein (Arp) 2/3 complex because of the complex’s central regulatory and structural roles in the formation of branched actin networks (6). Upon activation by two molecules of nucleation-promoting factor (NPF) localized at or near a membrane, the Arp2/3 complex nucleates a new, “daughter” filament from the side of a preexisting “mother” filament, forming a Y-shaped branch that serves as the basic structural unit of these networks (26–28) (Fig. S1). Importantly, the Arp2/3 binding site on F-actin spans three actin monomers along F-actin’s long-pitch helix, suggesting that its binding may be affected by changes in both monomer conformation and intermonomer distance induced by bending stresses (29).
The mechanism of Arp2/3 branch nucleation (Fig. S1) is understood to involve conformational changes in the Arp2/3 complex induced by the binding of NPFs (26, 27, 28, 30, 31). Additional conformational changes in both the Arp2/3 complex and several monomers in the mother filament probably occur upon the binding of the ternary complex of NPFs, Arp2/3, and G-actin to the mother filament or during a subsequent activation step that is necessary to allow branch nucleation (26, 31), because the bound NPFs appear to partially overlap the F-actin binding surface of the Arp2/3 complex (32). Once formed, the branch can then survive for minutes in vitro before dissociating (a process called “debranching”) (33). The rate of debranching has been shown to depend on the nucleotide bound to the mother filament (ATP, ADP-Pi or ADP) (33), and on the presence of the actin stabilizing drug phalloidin (34, 35). Branch nucleation appears to happen most readily on actin in the ATP-bound state (36), although it is not yet fully determined whether this is due to enhanced nucleation or stability against fast debranching. Experiments that used saturating amounts of phosphate to stabilize actin in the ADP-Pi-bound state showed a rate of barbed end creation similar to that on unstabilized actin (33). The regulation of both branch nucleation and branch stability by direct mechanical factors has not yet been studied. We asked whether filament bending by externally imposed geometric constraints plays a regulatory role at any point in this actin branch nucleation pathway.
Results and Discussion
Surface-Based Branching Assay Reveals That Actin Filament Curvature Biases Branching Direction.
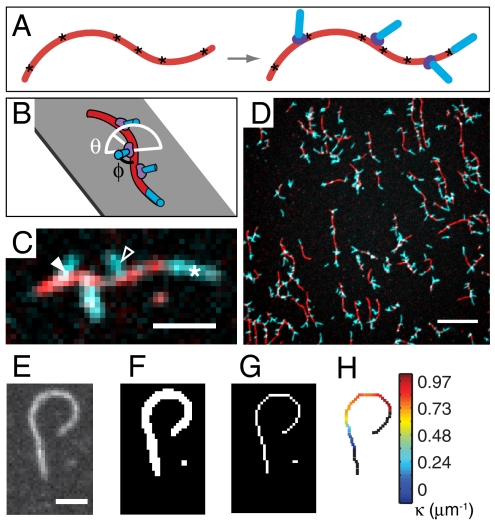
To examine whether and how the bending of filaments affects their interaction with the Arp2/3 complex, we imaged branch nucleation from fluorescently labeled F-actin that was preimmobilized on a surface before incubation with Arp2/3 complex, an NPF, and monomeric actin (G-actin) (Fig. 1 A–C). From the total of 403 images (Fig. 1D) acquired in five independent experiments, we measured the distribution of curvature along the immobilized mother filaments (37) (Fig. 1 E–H). Curvature varied smoothly as observed by fluorescence microscopy and could be measured on filaments spanning at least 3 μm, with a spatial resolution of approximately 1.1 μm. We were able to infer the location of Arp2/3 complex binding on the mother filament, with a spatial resolution of approximately 500 nm, from the location and direction of the short and stiff actin branches it nucleated, which were imaged separately from mother filaments using a two-color fluorescent labeling strategy (34, 36). Filament curvature at branch points and the direction of branch growth (Fig. 1C) determined the sign of the curvature value assigned to each branch. Branches on the convex side of the filament curve were assigned negative curvature, and branches on the concave side were assigned positive curvature.
Fig. 1.
Branching from curved filaments was observed in vitro. (A) Mother filaments (red) immobilized via biotin-streptavidin tethers (asterisks) before nucleation of branches (cyan blue) by Arp2/3 complex (violet). (B) Actin branches grow at a branch angle φ ∼ 70° to the mother filament (black line) with an azimuthal angle θ from 0° to 180° (white line). (C) Fluorescence image of actin growth at mother filament ends (white asterisk) and on branches on concave (open arrowhead) and convex (filled arrowhead) sides of mother filament curves. (Scale bar: 2 μm.) (D) Sample field of view. (Scale bar: 10 μm.) (E–G) Filament image thresholded and skeletonized to an 8-connected digital curve. (Scale bar: 2 μm.) (H) Mother filament curvature measured with the tangent angle method.
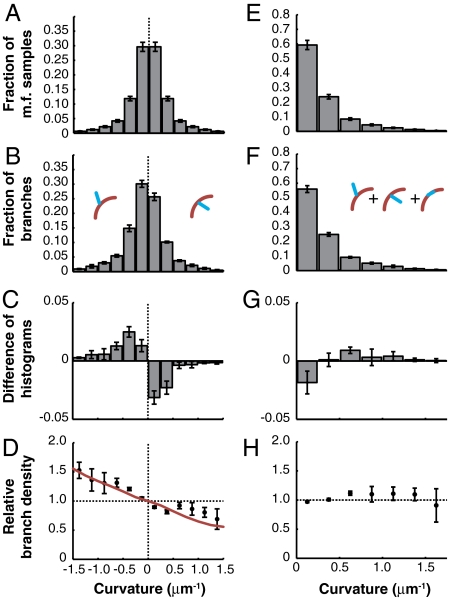
Interestingly, we observed that branches were more likely to be found on the convex surface of a curved filament than on the concave surface. We compared the distribution of curvatures measured at equally spaced points 182-nm apart along a total of 27.4 mm of mother filaments where branches could have formed (Fig. 2A), to the distribution of curvatures observed at 10,443 branch points, where branches actually formed (Fig. 2B). If branch density were independent of mother filament curvature, the two distributions would be identical after normalization. Instead, we found that the distributions were different (Fig. 2C and Table S1) and calculated their ratio, which we call the relative branch density (Fig. 2D).
Fig. 2.
Filament curvature biases branching direction. (A) Mother filament curvature distribution and (B) the distribution of mother filament curvature at branch points measured with the tangent angle method. (C) The difference and (D) the ratio of the histograms in B and A. The latter is called the relative branch density. The red curve represents the best fit (by least squares) by the fluctuation gating model with a 5 μm-1 threshold curvature. C, D, G, and H were normalized using a simulated control (see Materials and Methods). (E–H) The unsigned curvature distributions corresponding to A–D. Error bars: SEM, n = 5 independent experiments.
The relative branch density increased with negative curvature, indicating that extensional strain on the Arp2/3-binding surface of F-actin makes branch nucleation more likely, whereas compressional strain makes it less likely (Fig. 2D). We quantified the trend with a weighted least-squares linear fit to the relative branch density calculated from a subset of mother filament curvature samples selected randomly, one per filament to strictly satisfy the assumptions underlying linear regression. The weights were the number of samples of mother filament curvature in each curvature bin. The relative linear branch density decreased with a slope of -33% per μm-1 of curvature [95% C.I.: (-40, -26%), R2 = 0.56] (Fig. S2A and Table S2). Thus, the probability of finding a branch on the convex side of a filament with a curvature of 1 μm-1 is 99% higher than finding it on the concave side. Linear regression against the full dataset containing multiple curvature measurements from each mother filament yielded similar results (Table S2).
To quantify the effect of filament bending on total linear branch density, we carried out the same analysis as above with unsigned branch curvatures (Fig. 2 E and F) and found that the likelihood of branching per unit length shows a weak dependence on curvature (Fig. 2 G and H and Fig. S2B) with a slope of 13% per μm-1 [95% CI: (3.3, 23%)]. However, the linear fit does not describe the unsigned curvature data very well (Fig. S2B and Table S2, R2 = 0.17), and the size of the deviation from a flat curve is comparable to the size of systematic errors in digital curvature estimation (Materials and Methods, Fig. S2D, and SI Materials and Methods). In addition, fitting the data with a higher-order polynomial did not significantly improve the fit (p = 0.06, ANOVA). We conclude that total linear branch density depends weakly on absolute curvature, and we focus on studying the predominant effect of mother filament curvature on branch direction.
To confirm that the existence of a branching bias due to curvature was robust to the analysis method, we applied an alternative spline-based curvature estimation algorithm (37, 38) (Fig. S3). The exact value of the slope depended on the curvature estimation method, but the observation of branch direction bias due to curvature [-14% per μm-1 of curvature, 95% C.I.: (-17, -10%)] was unchanged (Table S2). We also checked how our estimate of bias in the direction of branching was affected by changes in image magnification and found only a weak effect (Fig. S4 and Table S3, p = 0.076).
As biochemical controls, we verified that our results do not depend on the mode of actin labeling (Fig. S5 A, C, and E, and Table S3) (p = 0.69). We also tested whether stabilizing the mother filament in the ADP-Pi-bound state by adding 25 mM phosphate affected the observed branching bias and did not observe an effect (Fig. S5 C–E and Table S3) (p = 0.998), nor did we observe a significant change in slope due to phalloidin stabilization of F-actin (Fig. S5 A, B, and E, and Table S3) (p = 0.10).
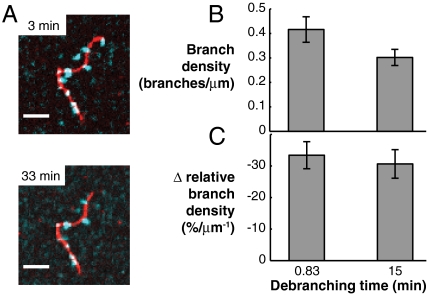
The Observed Bias in Branch Direction Is Not Caused by Debranching.
Mother filament curvature may influence one or several of the steps in the branch nucleation pathway. Because we imaged the end products of this branching pathway, we could not address the effect of mother filament curvature on Arp2/3 binding separately from branch nucleation. To address the role of debranching, we incubated samples in which branching had occurred for 2 min for an additional 33 min in the absence of Arp2/3 complex. We did not observe debranching during the additional incubation time (Fig. 3A), even with a high concentration of blocking protein (2 mg/mL BSA) included in solution to prevent nonspecific adsorption of branches onto the coverslip surface. To quantify branch density and its dependence on curvature, we incubated different samples for either 50 s or 15 min before stabilization with phalloidin. In these experiments, the branch density decreased, but not to a statistically significant extent (Fig. 3B), and there was not a statistically significant difference in the slope of the relative branch density as a function of curvature (Fig. 3C and Table S3). These results indicate that mother filament curvature primarily acts on branch nucleation.
Fig. 3.
Branch stability does not affect the branching bias. (A) Actin branches (cyan blue) grown from unstabilized mother filaments (red) and incubated in buffer with unlabeled actin but without phalloidin stabilization for times shown exhibited little to no debranching when the same sample was imaged at the two time points. (Scale bars: 5 μm.). (B and C) To obtain enough images for curvature analysis, identical but separate samples were prepared with incubation times of 0.83 or 15 min. in KMEI buffer (see SI Materials and Methods) with unlabeled actin before stabilization with phalloidin and imaging. (B) We found a decrease in overall branch density between short and long incubation samples, but it was not statistically significant (p = 0.12, Welch’s t test, n = 4). (C) There was no significant difference in the slope of relative branch density with respect to curvature (Table S3, p = 0.66). Error bars: SEM.
It is also possible that curvature acts on the stability of very short branches that were proposed by Mahaffy and Pollard to dissociate before microscopy-based methods can detect them (33). However, the lack of dependence on phosphate added at a concentration similar to that used by Mahaffy and Pollard suggests that curvature most likely acts on nucleation rather than dissociation. Overall, we do not exclude the possibility that curvature may affect fast debranching that we do not detect, but we favor the interpretation that curvature primarily affects branch nucleation. Itchetovkin et al. observed an enhanced branch density on filaments stabilized in the ATP-bound state (36), suggesting that the presence of ATP may have an effect on the nucleation process and may also affect sensitivity to curvature. However in our experiments, freshly polymerized actin containing ATP was only present on filament ends, where curvature could not be accurately measured.
Monte Carlo Simulations Revealed the Nanometer-Scale Curvature Fluctuations of Constrained Filaments.
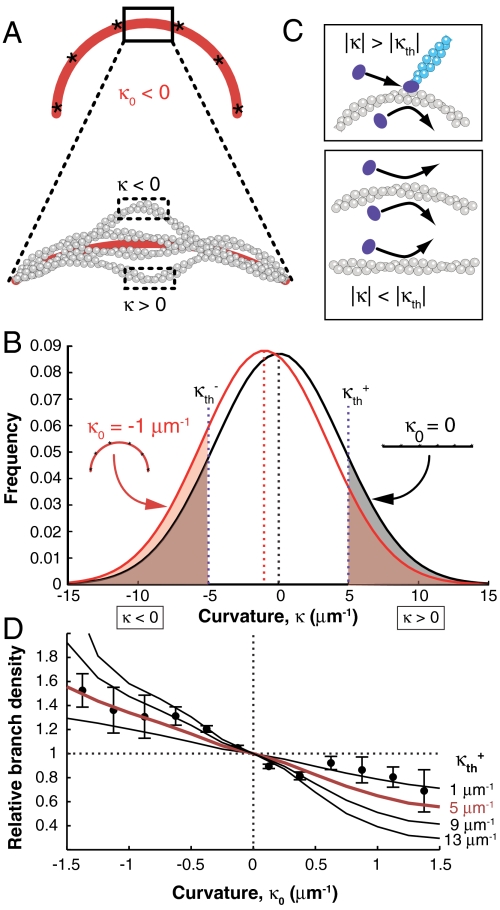
Because the length scale relevant to Arp2/3 binding and branch nucleation is 5–10 nm, well below the length scale at which fluorescence microscopy can measure curvature and also below the micrometer length scale at which curvature can be externally imposed, we used Monte Carlo simulations of a discretized worm-like chain (WLC) polymer (Fig. S6 A and B, and SI Materials and Methods) to assess the nanometer-scale implications of the micrometer-scale curvature. The validity of the WLC model to F-actin elasticity has been demonstrated for filament curvatures as high as 5 μm-1 (39). In our work, the WLC polymer, with the persistence length of actin (Lp = 9 μm) (40), was pinned to a plane with imposed curvature, κ0, mimicking the experiment (Fig. 4A).
Fig. 4.
The branching bias can be explained by a shift in the curvature fluctuations of a WLC filament tethered to a curved path. (A) Schematic of the WLC polymer tethered at six points (asterisks) to a curve with imposed curvature κ0 < 0. Fluctuations with local curvature κ < 0 and κ > 0 are possible. Curvature was calculated from the section between the middle two tethered particles, in order to avoid end effects (SI Materials and Methods). (B) Distribution of local curvature fluctuations for a filament tethered to a straight (black) or curved (red) path. Shaded areas indicate probability of branching. (C) The fluctuation gating model predicts a threshold convex local curvature beyond which stable binding and branching by the Arp2/3 complex (violet) can occur. (D) Relative branch density calculated from the ratio of the red- and black-shaded areas in B for several values of κth plotted with experimental data (also shown in Fig. 2D, with the red line corresponding to the same value of κth). Error bars: SEM.
Despite being constrained to an average curvature of κ0, the simulated filament exhibits large thermal fluctuations in nanometer-length-scale local curvature about that average (Fig. 4 A and B, and Fig. S6E). The breadth of the local curvature distribution is large in comparison to the range of experimentally accessible imposed curvatures. Therefore, for the side of a filament with convex average curvature of -1 μm-1, locally concave fluctuations occur almost as often as locally convex ones (Fig. S6 C–E). This small, 10% asymmetry is inconsistent with the larger, 99% asymmetry in branch density we observed between the two sides of filaments with -1 μm-1 average curvature (Fig. 2D and Fig. S2A). Strong differences between the sides of the filament with convex and concave average curvature only occur in the extreme tails of the corresponding curvature distributions. Therefore, we conclude that branching must be sensitive to local curvature fluctuations that are far from the average. In addition, because such extreme local curvature fluctuations occur rarely, making the system slow to reach chemical equilibrium, we discuss the effect of curvature on branch nucleation by Arp2/3 in kinetic terms.
A Fluctuation Gating Model for Branching by the Arp2/3 Complex Is Consistent with the Experimental Data.
Two lines of evidence support the hypothesis that curvature regulates branch nucleation kinetics. First, the Arp2/3 complex binds F-actin in solution with a slow on-rate, perhaps because it must wait for a favorable structural fluctuation of the filament (31). Second, a structural model of the Arp2/3-actin branch shows a local distortion involving subdomain 2 of an actin monomer at the Arp2/3 binding site (29). Extensional strain could weaken longitudinal intermonomer contacts in F-actin, helping to stabilize a transition state with high local curvature and increasing the kinetic rate of either Arp2/3 binding or branch nucleation.
Based on this evidence, we developed a filament fluctuation gating model, conceptually similar to fluctuation-gated binding of ligands to proteins (41). In our model, stable Arp2/3 ternary complex binding and branch nucleation occur only when the local curvature of the filament fluctuates beyond a threshold value κth (Fig. 4C). A sharp threshold is chosen because in the extreme wings of the local curvature distribution, probability attenuates so rapidly that the only pertinent model parameter is the lowest curvature value where branching is greatly enhanced, in effect, κth. Thus, the probability that a branch forms on either side of the curved filament under our model is the net probability of respective curvature values in excess of +κth or -κth (Fig. 4 B and C). Our calculations assess how extreme this curvature threshold needs to be in order to account for the curvature preference we observe experimentally.
With these simple assumptions, the fluctuation gating model captures the shape of the curvature-dependent branching bias and agrees quantitatively with our data over the entire experimental range for a value of κth = 5 μm-1 (red curve, Figs. 2D and 4D). These results are consistent with a mechanism in which F-actin bending fluctuations play a role in regulating branch formation by the Arp2/3 complex, suggesting that branching can be regulated by alterations of bending fluctuations of filaments due to constraints on actin network architecture or by binding of other ABPs. However, because of its coarse-grained resolution, this model cannot make predictions about conformational changes of the actin monomer caused by bending in the Arp2/3 binding site on the scale of individual amino acid residues. This model is presented in the simplest form that is consistent with our data and experimental parameters, but it could be extended to include details about the dependence of branch direction on curvature based on future findings. For example, we currently have little data in the very high convex curvature regime, where the branch density may decrease as the curvature distorts the mother filament to such an extent that it can no longer accommodate branch nucleation.
Autocatalytic Branching Amplifies the Branching Bias.
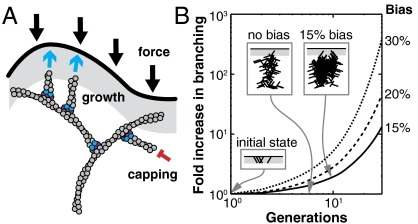
Directionally biased branching has important implications for branched actin assembly in vivo, where autocatalytic nucleation amplifies small effects (26, 42). A large fraction of filaments in a branched actin network adopt an approximately -35° orientation (43), and the side of a bent filament experiencing extensional strain is the same side that typically faces the bending force. Excess growth on the convex side of the curved filament would therefore create more branches oriented toward the bending force, reinforcing the branched network (Fig. 5A). The excess of branches on the most convex side of the mother filament may also define a preferred plane for branching that coincides with the plane of filament curvature, possibly contributing to the flat and thin shape of lamellipodia. It would also lead to more filaments growing into the membrane-adjacent zone where new branches can be nucleated, increasing total branch density. We studied this effect with a different, stochastic simulation of branching in two dimensions (Fig. 5B) and found that, for a 15% bias toward the membrane, the total number of filaments is double that of the zero bias case after only 10 branch generations (Fig. 5B). Based on our experimental data (Fig. 2D), a 15% curvature bias corresponds to a radius of curvature of 2.3 μm and a bending energy of 0.6 kBT per μm of ATP-bound filament (40). This amount of curvature could result from a lateral force of 1 pN applied perpendicularly to the end of a 0.05-μm-long filament fixed at the other end (44), which reflects the average force per filament due to membrane tension and rigidity (45, 46) and the approximate length of free F-actin (47) at the leading edge of the cell. If the length of free F-actin is longer at the leading edge (48, 49), the filaments require even less force to bend. Therefore, even modest filament curvature that is caused by the normal force balance of branched actin growth against a membrane can generate a significant bias in the direction of actin branch nucleation.
Fig. 5.
A bias in the direction of branching can increase the total amount of actin in a branched network. (A) In a branched network, compressive forces bend filaments away from the membrane (black). Excess branching on the convex side of a bent filament creates more branches pointing toward the membrane, increasing the number of filaments pushing against the membrane (cyan blue arrows). Capping (red) can occur anywhere, but filaments can only branch in the branching zone (gray). (B) Results of a stochastic branching simulation (Materials and Methods) in which rigid branches with angles of ± 36° and -108° grow with a given bias (right column) toward the membrane. (Insets) Schematic snapshots of branching with 0% and 15% bias (gray, branching zone).
Conclusions
We have shown that F-actin curvature regulates Arp2/3 complex activity, providing the cell with a distributed, filament-dependent mechanism for sensing and responding to the compressive stress on branched actin networks. Our results suggest the possibility that mechanical stress on cytoskeletal filaments can modulate how they interact with their binding partners. The actin filament takes on a diversity of structural states as it grows, interacts with binding proteins, encounters physical constraints, and fluctuates due to thermal motion (50, 51). It is likely that other side-binding ABPs besides the Arp2/3 complex exhibit similar sensitivity to local actin curvature, providing a direct mechanism for altering organization of the actin cytoskeleton in response to force. For example, filament severing by the ADF/cofilin homolog actophorin occurs more readily at points of high curvature (23), consistent with a recently elucidated mechanism for severing by cofilin that depends on a mechanism that takes advantage of the mechanical instability at the border between two structural states of F-actin (25). However, it is not yet known whether cofilin binding or the cooperativity of cofilin binding is affected by local filament curvature, although it has been shown that its binding lowers the persistence length of actin (52) and that its binding is enhanced by tension on F-actin (53). Nor have other proteins that modify the persistence length of actin, such as drebrin (54) or tropomyosin (40), been tested for sensitivity to F-actin curvature. The methods we have developed can be used as a platform to investigate the curvature dependence of other ABP–filament interactions and the role of actin filament bending in mechanotransduction and cytoskeletal reorganization.
Materials and Methods
Branching Assays.
A flow channel was assembled with a plasma-cleaned (ca. 300 mTorr oxygen; Plasmod; March Instruments) coverslip and incubated with 10 mg/mL biotinylated BSA (A6043; Sigma-Aldrich) in 80 mM piperazine-N-N′-bis(2-ethanesulfonic acid) pH 6.9, 1 mM EGTA, and 4 mM MgCl2 for 180 min at 4 °C, and 0.2 mg/mL streptavidin (S4762; Sigma-Aldrich) in 1× calcium and magnesium free PBS (Cellgro; Mediatech) for 30–60 min at 23 °C. The channel was washed with buffer composed of nine parts G buffer (5 mM Tris, 0.2 mM ATP, 0.1 mM CaCl2, 0.5 mM DTT, 1 mM NaN3, pH 8.0 at 4 °C) and one part either 10× KMEI buffer (500 mM KCl, 10 mM MgCl2, 10 mM EGTA, 100 mM imidazole, pH 7.0) or 10× PKMEI buffer (92 mM KH2PO4, 158 mM K2HPO4, 92 mM KCl, 10 mM MgCl2, 10 mM EGTA, 100 mM imidazole, pH 7.0). In both cases, the final pH was 7.0 at 23 °C. We refer to the compound buffers as KMEI and PKMEI, respectively.
F-actin (10–20% biotinylated and 30% AF546-labeled) was polymerized 1 h at 23 °C in KMEI or PKMEI and 0.5 mM ATP (Roche Applied Science). F-actin (50 nM) and 1 μM of G-actin containing similar ratios of unlabeled, biotinylated, and AF546- or Cy3- labeled monomers was incubated in the channel for 5 min. We estimated that approximately 10% of biotin moieties on a filament bind streptavidin and thus the spacing between surface attachment points was 140–270 nm. The channel was washed with KMEI or PKMEI and incubated with branching mix containing 0.5 mM ATP, 100 nM of the N-terminal WCA domain of neural Wiskott–Aldrich syndrome protein, 100 nM Arp2/3 complex, 2 mg/mL BSA (A0281; Sigma-Aldrich), and 0.6–1 uM actin (50% AF488 labeled), for 70–120 s at 23 °C. The concentrations and incubation times were chosen such that individual branches were sparse enough to be individually identified. Finally, the channel was washed with KMEI or PKMEI containing 1 μM phalloidin (Sigma-Aldrich) and in the case of cysteine-labeled mother filaments, an oxygen scavenger system consisting of 250 μg/mL glucose oxidase (Sigma-Aldrich), 4.5 mg/mL glucose, and 30 μg/mL catalase (Roche Applied Science). Modified branching assays for measuring the effect of phalloidin and for measuring branch stability as a function of curvature are described in SI Materials and Methods.
Curvature Estimation, Tangent Angle Method.
We estimated filament curvature from skeletonized digital filament images (Fig. 1 E–H) by implementing the tangent angle-based method (method II) described previously (37) with MATLAB (The Mathworks) using a Gaussian differentiating kernel with σ = 3 pixels and a filter size of m = 9 pixels, chosen empirically by estimating the curvature of digital circles of known radius. The accuracy of this and similar local methods, compared to other curvature estimation strategies such as the global spline fit method (SI Materials and Methods), has been previously demonstrated (37, 38). Curvature error as evaluated using the tangent angle method was 0.08 μm-1, which is less than the bin width we used for histograms. End effects made curvature unreliable m points in from each end, so these end segments and any branches growing on them were excluded from further analysis. The resulting curves were sampled in two ways. First, to maximize the number of data points collected, curvature was measured every 1.1 pixels (182 nm), accepting the fact that curvature is correlated between nearby points on the same filament. Secondly, to remove any potential correlations between the curvature values sampled and thus satisfy the assumptions of the Kolmogorov–Smirnov test and weighted linear regression procedures, one point was randomly chosen on each filament, in a region away from the ends, and its curvature was measured in an identical way. Both curvature samples were used to create histograms of the curvature distribution of mother filaments for each experiment, which were normalized to sum to one.
Branch Analysis.
Branches were manually identified using two criteria: (i) overlap between one end of the green branch and the red mother filament, and (ii) lack of collinearity with the end of any other filament. Long branches that had one blurred end due to thermal fluctuations over the 1- to 1.5-s exposure were counted if their blurred end pointed away from the mother filament. Branch points were assigned the average curvature of the three closest pixels on the nearest mother filament skeleton; any branch point more than three pixels away from a skeleton was ignored. Branch orientation was determined by taking the cross-product of the branch vector and the tangent vector to the mother filament curve, which was calculated by smoothing the digitized tangent vectors to the same curve with a Gaussian filter (37) in the tangent angle-based analysis, or from the cubic spline in the spline-based analysis. Branches with indeterminate direction were assigned the absolute value of curvature and only used in the analysis of total linear branch density. Histogram bins with zero branches or curvature measurements were ignored.
Calculation of Relative Branch Density.
Controls with randomly located and oriented simulated branches were created for each image to quantify the artifacts of digital curvature estimation. They were analyzed in parallel with real data to create control histograms for each dataset. The linear branch density as a function of curvature was calculated by taking a ratio of the branch curvature histogram to the mother filament curvature histogram. The control density was subtracted from the real density to remove digitization artifacts (Fig. S2 C and D). We quantified the trend with a least-squares linear fit to the relative branch density (Fig. S2 A and B) weighted by the number of samples of mother filament curvature in each curvature bin. Statistical calculations are described in SI Materials and Methods.
The branching bias we observe is most likely an underestimate of the true directional bias because our experiment averages over branches growing horizontally along the plane of the surface, where bending strain is highest (θ = 0° or 180° in Fig. 1B) and those growing at other angles, because the fluorescence images are 2D projections of 3D branches (Fig. 1 B and C). Large-amplitude bending of mother filaments in three dimensions was not observed except at untethered ends, due to the stiffness of F-actin (40), but small-amplitude, out-of-plane local curvature fluctuations over length scales of nanometers were underestimated by construction in simulations because curvature in experiments was also measured in projection (see SI Materials and Methods).
Stochastic Model of Bias Propagation During Branching.
To quantify how the directional branching bias is amplified by branched actin growth, we created a simple 2D model in which actin filaments are represented as rigid rods of constant length, corresponding to the average filament length that can grow before capping occurs, and growing at ± 36° and ± 108° relative to the direction of network growth. Starting with 30 filaments at each of the four angles, the filaments were allowed to grow two branches, each pointing either away from or toward the direction of network growth, for 30 generations. The “branching zone” was defined such that only filaments at ± 36°, grown in the last generation could give rise to new branches. Each branch had a 0.5 + b probability of growing into the branching zone (and toward the direction of growth) and a 0.5 - b probability of growing out of the branching zone (and away from the direction of growth), where 2b is the directional branching bias.
Supplementary Material
Acknowledgments.
We thank S. Parekh and A. Liu for proteins, S. Pronk, D. Sept, B. Smith, and D. Richmond for discussions, L. Nilsson and E. Schmid for critical reading of the manuscript, M. Valentine and P. Fordyce for technical advice, J. Chung for statistics consulting, and L. Jacob for statistics advice. V.I.R. was supported by a University of California Biotechnology Research and Education Program Graduate Research and Education in Adaptive Biotechnology Training Grant, The Paul and Daisy Soros Fellowship for New Americans, and a National Science Foundation (NSF) Graduate Research Fellowship Program (GRFP) Fellowship. E.B.W. was supported by a Department of Defense National Defense Science and Engineering Graduate Fellowship. O.C. was supported by an NSF GRFP fellowship. Support for this work was provided by an National Institutes of Health R01 grant from the National Institute of General Medial Sciences (D.A.F.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission. A.M. is a guest editor invited by the Editorial Board.
See Commentary on page 2693.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1114292109/-/DCSupplemental.
References
- 1.Farge E. Mechanotransduction in development. Curr Top Dev Biol. 2011;95:243–265. doi: 10.1016/B978-0-12-385065-2.00008-6. [DOI] [PubMed] [Google Scholar]
- 2.Vogel V, Sheetz M. Local force and geometry sensing regulate cell functions. Nat Rev Mol Cell Biol. 2006;7:265–275. doi: 10.1038/nrm1890. [DOI] [PubMed] [Google Scholar]
- 3.Hoffman BD, Grashoff C, Schwartz MA. Dynamic molecular processes mediate cellular mechanotransduction. Nature. 2011;475:316–323. doi: 10.1038/nature10316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Janmey PA, McCulloch CA. Cell mechanics: Integrating cell responses to mechanical stimuli. Annu Rev Biomed Eng. 2007;9:1–34. doi: 10.1146/annurev.bioeng.9.060906.151927. [DOI] [PubMed] [Google Scholar]
- 5.Fletcher DA, Mullins RD. Cell mechanics and the cytoskeleton. Nature. 2010;463:485–492. doi: 10.1038/nature08908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Goley ED, Welch MD. The Arp2/3 complex: An actin nucleator comes of age. Nat Rev Mol Cell Biol. 2006;7:713–726. doi: 10.1038/nrm2026. [DOI] [PubMed] [Google Scholar]
- 7.Pollard TD, Borisy GG. Cellular motility driven by assembly and disassembly of actin filaments. Cell. 2003;112:453–465. doi: 10.1016/s0092-8674(03)00120-x. [DOI] [PubMed] [Google Scholar]
- 8.Welch MD, Mullins RD. Cellular control of actin nucleation. Annu Rev Cell Dev Biol. 2002;18:247–288. doi: 10.1146/annurev.cellbio.18.040202.112133. [DOI] [PubMed] [Google Scholar]
- 9.Giardini PA, Fletcher DA, Theriot JA. Compression forces generated by actin comet tails on lipid vesicles. Proc Natl Acad Sci USA. 2003;100:6493–6498. doi: 10.1073/pnas.1031670100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Marcy Y, Prost J, Carlier M, Sykes C. Forces generated during actin-based propulsion: A direct measurement by micromanipulation. Proc Natl Acad Sci USA. 2004;101:5992–5997. doi: 10.1073/pnas.0307704101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Parekh SH, Chaudhuri O, Theriot JA, Fletcher DA. Loading history determines the velocity of actin-network growth. Nat Cell Biol. 2005;7:1219–1223. doi: 10.1038/ncb1336. [DOI] [PubMed] [Google Scholar]
- 12.Prass M, Jacobson K, Mogilner A, Radmacher M. Direct measurement of the lamellipodial protrusive force in a migrating cell. J Cell Biol. 2006;174:767–772. doi: 10.1083/jcb.200601159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Firat-Karalar EN, Welch MD. New mechanisms and functions of actin nucleation. Curr Opin Cell Biol. 2011;23:4–13. doi: 10.1016/j.ceb.2010.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gouin E, Welch MD, Cossart P. Actin-based motility of intracellular pathogens. Curr Opin Microbiol. 2005;8:35–45. doi: 10.1016/j.mib.2004.12.013. [DOI] [PubMed] [Google Scholar]
- 15.Chaudhuri O, Parekh SH, Fletcher DA. Reversible stress softening of actin networks. Nature. 2007;445:295–298. doi: 10.1038/nature05459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gardel ML, et al. Elastic behavior of cross-linked and bundled actin networks. Science. 2004;304:1301–1305. doi: 10.1126/science.1095087. [DOI] [PubMed] [Google Scholar]
- 17.Kim T, Hwang W, Lee H, Kamm RD. Computational analysis of viscoelastic properties of crosslinked actin networks. PLoS Comput Biol. 2009;5:e1000439. doi: 10.1371/journal.pcbi.1000439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lieleg O, Kayser J, Brambilla G, Cipelletti L, Bausch AR. Slow dynamics and internal stress relaxation in bundled cytoskeletal networks. Nat Mater. 2011;10:236–242. doi: 10.1038/nmat2939. [DOI] [PubMed] [Google Scholar]
- 19.Soo FS, Theriot JA. Large-scale quantitative analysis of sources of variation in the actin polymerization-based movement of Listeria monocytogenes. Biophys J. 2005;89:703–723. doi: 10.1529/biophysj.104.051219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.McGough A. F-actin-binding proteins. Curr Opin Struct Biol. 1998;8:166–176. doi: 10.1016/s0959-440x(98)80034-1. [DOI] [PubMed] [Google Scholar]
- 21.De La Cruz EM. Cofilin binding to muscle and non-muscle actin filaments: Isoform-dependent cooperative interactions. J Mol Biol. 2005;346:557–64. doi: 10.1016/j.jmb.2004.11.065. [DOI] [PubMed] [Google Scholar]
- 22.Galkin VE, Orlova A, Lukoyanova N, Wriggers W, Egelman EH. Actin depolymerizing factor stabilizes an existing state of F-actin and can change the tilt of F-actin subunits. J Cell Biol. 2001;153:75–86. doi: 10.1083/jcb.153.1.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Maciver SK, Zot HG, Pollard TD. Characterization of actin filament severing by actophorin from Acanthamoeba castellanii. J Cell Biol. 1991;115:1611–1620. doi: 10.1083/jcb.115.6.1611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Okreglak V, Drubin DG. Cofilin recruitment and function during actin-mediated endocytosis dictated by actin nucleotide state. J Cell Biol. 2007;178:1251–1264. doi: 10.1083/jcb.200703092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.McCullough BR, et al. Cofilin-linked changes in actin filament flexibility promote severing. Biophys J. 2011;101:151–159. doi: 10.1016/j.bpj.2011.05.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pollard TD. Regulation of actin filament assembly by Arp2/3 complex and formins. Annu Rev Biophys Biomol Struct. 2007;36:451–477. doi: 10.1146/annurev.biophys.35.040405.101936. [DOI] [PubMed] [Google Scholar]
- 27.Padrick SB, Doolittle LK, Brautigam CA, King DS, Rosen MK. Arp2/3 complex is bound and activated by two WASP proteins. Proc Natl Acad Sci USA. 2011;108:E472–E479. doi: 10.1073/pnas.1100236108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ti S, Jurgenson CT, Nolen BJ, Pollard TD. Structural and biochemical characterization of two binding sites for nucleation-promoting factor WASp-VCA on Arp2/3 complex. Proc Natl Acad Sci USA. 2011;108:E463–E471. doi: 10.1073/pnas.1100125108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rouiller I, et al. The structural basis of actin filament branching by the Arp2/3 complex. J Cell Biol. 2008;180:887–895. doi: 10.1083/jcb.200709092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Goley ED, Rodenbusch SE, Martin AC, Welch MD. Critical conformational changes in the Arp2/3 complex are induced by nucleotide and nucleation promoting factor. Mol Cell. 2004;16:269–279. doi: 10.1016/j.molcel.2004.09.018. [DOI] [PubMed] [Google Scholar]
- 31.Beltzner CC, Pollard TD. Pathway of actin filament branch formation by Arp2/3 complex. J Biol Chem. 2008;283:7135–7144. doi: 10.1074/jbc.M705894200. [DOI] [PubMed] [Google Scholar]
- 32.Xu X, et al. Three-dimensional reconstructions of Arp2/3 complex with bound nucleation promoting factors. EMBO J. 2011:236–247. doi: 10.1038/emboj.2011.343. 10.1038/emboj.2011.343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mahaffy RE, Pollard TD. Kinetics of the formation and dissociation of actin filament branches mediated by Arp2/3 complex. Biophys J. 2006;91:3519–3528. doi: 10.1529/biophysj.106.080937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Blanchoin L, et al. Direct observation of dendritic actin filament networks nucleated by Arp2/3 complex and WASP/Scar proteins. Nature. 2000;404:1007–1011. doi: 10.1038/35010008. [DOI] [PubMed] [Google Scholar]
- 35.Mahaffy RE, Pollard TD. Influence of phalloidin on the formation of actin filament branches by Arp2/3 complex. Biochemistry. 2008;47:6460–6467. doi: 10.1021/bi702484h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ichetovkin I, Grant W, Condeelis J. Cofilin produces newly polymerized actin filaments that are preferred for dendritic nucleation by the Arp2/3 complex. Curr Biol. 2002;12:79–84. doi: 10.1016/s0960-9822(01)00629-7. [DOI] [PubMed] [Google Scholar]
- 37.Worring M, Smeulders AWM. Digital curvature estimation. CVGIP Imag Understan. 1993;58:366–382. [Google Scholar]
- 38.Bicek AD, Tüzel E, Kroll DM, Odde DJ. Analysis of microtubule curvature. Methods Cell Biol. 2007;83:237–268. doi: 10.1016/S0091-679X(07)83010-X. [DOI] [PubMed] [Google Scholar]
- 39.Arai Y, et al. Tying a molecular knot with optical tweezers. Nature. 1999;399:446–448. doi: 10.1038/20894. [DOI] [PubMed] [Google Scholar]
- 40.Isambert H, et al. Flexibility of actin filaments derived from thermal fluctuations. Effect of bound nucleotide, phalloidin, and muscle regulatory proteins. J Biol Chem. 1995;270:11437–11444. doi: 10.1074/jbc.270.19.11437. [DOI] [PubMed] [Google Scholar]
- 41.McCammon JA, Northrup SH. Gated binding of ligands to proteins. Nature. 1981;293:316–317. doi: 10.1038/293316a0. [DOI] [PubMed] [Google Scholar]
- 42.Carlsson AE. Growth velocities of branched actin networks. Biophys J. 2003;84:2907–2918. doi: 10.1016/S0006-3495(03)70018-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Verkhovsky AB, et al. Orientational order of the lamellipodial actin network as demonstrated in living motile cells. Mol Biol Cell. 2003;14:4667–4675. doi: 10.1091/mbc.E02-10-0630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Belendez T, Neipp C, Belendez A. Large and small deflections of a cantilever beam. Eur J Phys. 2002;23:371–379. [Google Scholar]
- 45.Abraham VC, Krishnamurthi V, Taylor DL, Lanni F. The actin-based nanomachine at the leading edge of migrating cells. Biophys J. 1999;77:1721–1732. doi: 10.1016/S0006-3495(99)77018-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Mogilner A, Edelstein-Keshet L. Regulation of actin dynamics in rapidly moving cells: A quantitative analysis. Biophys J. 2002;83:1237–1258. doi: 10.1016/S0006-3495(02)73897-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Svitkina TM, Borisy GG. Arp2/3 complex and actin depolymerizing factor/cofilin in dendritic organization and treadmilling of actin filament array in lamellipodia. J Cell Biol. 1999;145:1009–1026. doi: 10.1083/jcb.145.5.1009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Urban E, Jacob S, Nemethova M, Resch GP, Small JV. Electron tomography reveals unbranched networks of actin filaments in lamellipodia. Nat Cell Biol. 2010;12:429–435. doi: 10.1038/ncb2044. [DOI] [PubMed] [Google Scholar]
- 49.Small JV, Winkler C, Vinzenz M, Schmeiser C. Reply: Visualizing branched actin filaments in lamellipodia by electron tomography. Nat Cell Biol. 2011;13:1013–1014. doi: 10.1038/ncb2321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kueh HY, Mitchison TJ. Structural plasticity in actin and tubulin polymer dynamics. Science. 2009;325:960–963. doi: 10.1126/science.1168823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Reisler E, Egelman EH. Actin structure and function: What we still do not understand. J Biol Chem. 2007;282:36133–36137. doi: 10.1074/jbc.R700030200. [DOI] [PubMed] [Google Scholar]
- 52.McCullough BR, Blanchoin L, Martiel J, Dela Cruz EM. Cofilin increases the bending flexibility of actin filaments: Implications for severing and cell mechanics. J Mol Biol. 2008;381:550–558. doi: 10.1016/j.jmb.2008.05.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Hayakawa K, Tatsumi H, Sokabe M. Actin filaments function as a tension sensor by tension-dependent binding of cofilin to the filament. J Cell Biol. 2011;195:721–727. doi: 10.1083/jcb.201102039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Sharma S, Grintsevich EE, Phillips ML, Reisler E, Gimzewski JK. Atomic force microscopy reveals drebrin induced remodeling of F-actin with subnanometer resolution. Nano Lett. 2011;11:825–827. doi: 10.1021/nl104159v. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.