Abstract
Dramatic changes in chromatin structure and histone modification occur during oocyte growth, as well as a global cessation of transcription. The role of histone modifications in these processes is poorly understood. We report the effect of conditionally deleting Hdac1 and Hdac2 on oocyte development. Deleting either gene has little or no effect on oocyte development, whereas deleting both genes results in follicle development arrest at the secondary follicle stage. This developmental arrest is accompanied by substantial perturbation of the transcriptome and a global reduction in transcription even though histone acetylation is markedly increased. There is no apparent change in histone repressive marks, but there is a pronounced decrease in histone H3K4 methylation, an activating mark. The decrease in H3K4 methylation is likely a result of increased expression of Kdm5b because RNAi-mediated targeting of Kdm5b in double-mutant oocytes results in an increase in H3K4 methylation. An increase in TRP53 acetylation also occurs in mutant oocytes and may contribute to the observed increased incidence of apoptosis. Taken together, these results suggest seminal roles of acetylation of histone and nonhistone proteins in oocyte development.
Oogenesis is a protracted process that encompasses meiosis and oocyte growth, and results in the only cell that, following fertilization, can develop into an organism (1). In mice, at approximately day 13.5 of gestation, oogonia undergo a final round of DNA replication and enter the first meiotic prophase, at which point they are called oocytes. By the time of birth, oocytes are arrested in diplotene of the first meiotic prophase, are approximately 15 to 20 μm in diameter, and reside in primordial follicles in which the oocyte is surrounded by a single layer of flattened follicle cells. Oocyte growth is coordinated with follicle cell proliferation; the diameter of full-grown oocytes is approximately 80 μm. During the growth phase, oocytes acquire the ability to resume meiosis (i.e., acquisition of meiotic competence) and support development to term (i.e., acquisition of developmental competence). In vivo, release of gonadotropins triggers resumption of meiosis of full-grown oocytes present in preovulatory follicles with oocytes maturing to and arresting at metaphase II; fertilization triggers resumption and completion of meiosis (2).
Oocyte growth is accompanied by dramatic changes in gene expression, but starting at approximately midgrowth phase, transcription decreases such that full-grown oocytes are essentially transcriptionally inactive (3), and transcriptional quiescence appears critical for acquisition of developmental competence (4). Transcriptional quiescence is associated with chromatin condensation as well as changes in histone posttranslational modifications (PTMs) (5); histone PTMs such as phosphorylation, methylation, ubiquitination, and acetylation are intimately linked to transcriptional regulation and required for many biological processes (6, 7), with histone acetylation typically associated with transcriptionally permissive chromatin (8). The role of histone PTMs that occur during oocyte development is poorly understood.
Histone acetylation is controlled by two opposing enzymes: histone acetyltransferases and histone deacetylases (HDACs). HDACs catalyze deacetylation of histones and many other nonhistone proteins such as tubulin (9) and transcription factors, e.g., TRP53 (P53) (10), E2F1 (11), and CREB (12). In mammals, 18 HDACs have been identified to date, and are grouped into four classes according to their homology (13). Class I HDACs (HDAC1, 2, 3, and 8) show homology to the yeast protein RPD3, are usually detected in the nucleus, and show ubiquitous expression in various mammalian cell lines and tissues. Class II HDACs (4–7, 9, 10) have a high degree of homology to Hda1 protein and can shuttle between the nucleus and the cytoplasm. Class III HDACs are homologous to the yeast Sir2 HDAC, and HDAC11 is the sole member of the class IV HDACs (14). HDACs are implicated in development of cancer, regulation of cell proliferation, apoptosis, and cell cycle (15, 16).
Among class I HDACs, HDAC1 and HDAC2 share 83% amino acid homology and are found together in almost all repressive transcriptional complexes (17), which suggests a high degree functional redundancy should exist between the two proteins. Consistent with this proposal is that deleting either HDAC1 or HDAC2 in cardiomyocytes (18), neuron precursors (19), B cells (20), and embryonic epidermis (21) does not evoke an obvious phenotype. HDAC1 and HDAC2, however, may have distinct functions. For example, germ-line deletion of HDAC1 causes mouse embryo lethality before embryonic day 10.5 even though HDAC2 is up-regulated (22), and HDAC2 specifically regulates synaptic plasticity and memory formation, a function that is not compensated by overexpressing HDAC1 (23). A unique role of HDAC1 is also observed in embryonic stem cells (24). In addition, we have shown that HDAC1 is the major HDAC that is critical for preimplantation development in mouse (25). Essentially nothing is known about the role(s) of HDAC1 and HDAC2 in oocyte development.
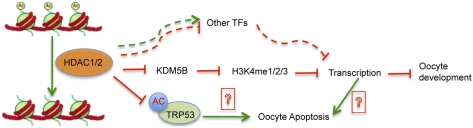
We report here that deleting both Hdac1 and Hdac2, but not individually, in mouse oocytes results in arrest of oocyte development at the midgrowth stage before competence to undergo germinal vesicle (GV) breakdown (GVBD) was acquired, a stage of oocyte development normally found in secondary follicles. This developmental arrest is accompanied by a precocious decrease in global transcription that occurs in setting of a marked increase in histone acetylation and is likely a result of increased expression of lysine demethylase 5B (KDM5B) that in turn results in a decrease in histone H3K4 methylation, a transcription activation mark. In addition, acetylation of TRP53 is elevated in mutant oocytes and may contribute to the observed increased incidence of apoptosis observed in mutant oocytes.
Results
Conditional Targeting of Hdac1 and Hdac2 Results in Infertility, Decreased Ovary Size, and Defects in Oocyte Development.
Both HDAC1 and HDAC2 are present in full-grown oocytes, but the temporal and spatial pattern of expression were not assessed (25). Accordingly, we first established the temporal and spatial pattern of HDAC1 and HDAC2 expression during oocyte growth. Immunocytochemical analysis revealed that, as anticipated, HDAC1 and HDAC2 were concentrated in the nucleus throughout the growth phase (Fig. 1). The intensity of HDAC1 nuclear staining displayed a progressive decrease during the course of oocyte growth and following GVBD colocalized with chromosomes. In contrast, nuclear HDAC2 staining increased between days 5 and 12 postpartum and then remained relatively constant for the duration of the growth phase. Following GVBD, HDAC2 was uniformly dispersed throughout the cytoplasm (Fig. 1).
Fig. 1.
HDAC1 and HDAC2 protein expression patterns during oocyte development. Immunocytochemical analysis of HDAC1 or HDAC2 expression during oogenesis. All samples for a given HDAC were processed for immunocytochemistry together, and all images were taken at the same laser power. The experiment was conducted three times (at least three mice per experiment), and at least 25 oocytes were analyzed for each sample. Shown are representative examples. 5d, 5 d postpartum; 12d, 12 d postpartum; GV, full-grown oocyte; MI, metaphase I; MII, metaphase II. DNA was stained with SYTOX green or DAPI (blue). (Scale bar: 35 μm.)
To establish the in vivo role(s) of HDAC1 and HDAC2 during oocyte growth, we generated conditional mutant oocytes using Zp3-Cre transgenic mice (26, 27) in combination with conditional loss of function alleles of Hdac1 or/and Hdac2 (18). Hdac1 or Hdac2 mutants are referred to as Hdac1−/− or Hdac2−/−, respectively. Hdac1-Hdac2 mutants (i.e., double mutant) are referred to as Hdac1:2−/−. Hdac1 heterozygotes–Hdac2-null oocytes are referred to as Hdac1−/+/Hdac2−/− and Hdac1-null–Hdac2 heterozygote oocytes are referred to as Hdac1−/−/Hdac2−/+. Mutant female mice were bred to WT males for at least 6 mo to test for fertility (Table 1). Hdac1−/− and Hdac1−/−/Hdac2−/+ mice were fully fertile, Hdac2−/− mice were subfertile, and Hdac1−/+/Hdac2−/− and Hdac1:2−/− were infertile. Taken together, these results suggest that HDAC1 and HDAC2 can compensate for loss of function of the other and that HDAC2 is the major HDAC implicated in oocyte development.
Table 1.
Hdac1:2−/− female mice are infertile
| Germline genotype | No. of females | Average litter size |
| Hdac1(floxed/ floxed)Hdac2(floxed/ floxed); WT | 12 | 9.6 ± 2.1 |
| Zp3-Cre Hdac1(+/floxed)Hdac2(+/floxed); Hdac1−/+/Hdac2−/+ | 10 | 9.6 ± 1.5 |
| Zp3-Cre Hdac1(floxed/-)Hdac2(+/+); Hdac1−/− | 15 | 9.1 ± 1.6 |
| Zp3-Cre Hdac1(floxed/-)Hdac2(+/floxed); Hdac1−/−/Hdac2−/+ | 7 | 9.2 ± 0.9 |
| Zp3-Cre Hdac1(+/+)Hdac2(floxed/-); Hdac2−/− | 10 | 7.8 ± 1.3* |
| Zp3-Cre Hdac1(+/floxed)Hdac2(floxed/-); Hdac1−/+/Hdac2−/− | 5 | 0 |
| Zp3-Cre Hdac1(floxed/-)Hdac2(floxed/-); Hdac1:2−/− | 15 | 0 |
The abbreviations for genotypes used in the text are in bold.
*P < 0.05.
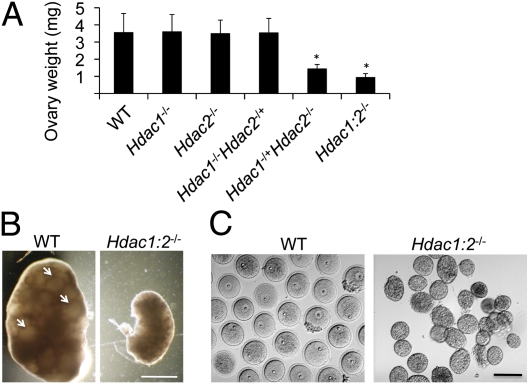
To determine the molecular basis for the infertility of Hdac1−/+/Hdac2−/− and Hdac1:2−/− mice, we first examined ovary development (Fig. 2 A and B). The weight of ovaries obtained from Hdac1−/+/Hdac2−/− or Hdac1:2−/− mice 6 wk of age was significantly less than that obtained from WT, Hdac1−/−, Hdac2−/−, and Hdac1−/−/Hdac2−/+ mice, whose weights were all similar. Mature follicles were clearly visible in WT ovaries but absent in ovaries obtained from Hdac1:2−/− mice (Fig. 2B). Consistent with the absence of mature follicles was that only secondary follicles, and no full-grown oocytes, were obtained from ovaries of Hdac1:2−/− mice (Fig. 2C), a finding that clearly accounts for their infertility. Hdac1−/+/Hdac2−/− mice were also infertile, and phenotypic characterization of these mice will be described elsewhere.
Fig. 2.
Conditional targeting of Hdac1 and Hdac2 in oocytes results in decreased ovary size and defective oogenesis. (A) Ovaries from mice 6 wk of age from the different genotypes were collected, and their weights measured. Data are expressed as mean ± SEM, and six ovaries (one ovary per mouse) per genotype were measured. The other genotypes are described in the text (*P < 0.0001). (B) Ovary morphology from WT and Hdac1:2−/− mice 6 wk of age. WT ovaries show presence of mature follicles (arrows), whereas such follicles are absent in ovaries from Hdac1:2−/− mice. (Scale bar: 1 mm.) (C). Full-grown oocytes are recovered from WT mice, whereas only secondary follicles are recovered from Hdac1:2−/− mice. (Scale bar: 100 μm.)
Deletion of both Hdac1 and Hdac2 in Oocytes Leads to Developmental Arrest at Secondary Follicle Stage.
Histological analysis of ovarian sections derived from ovaries of 18-d-old mice revealed that, compared with WT, virtually no small antral follicles were observed in Hdac1:2−/− mice (Fig. 3 A and B). Moreover, the number of secondary follicles was significantly reduced in Hdac1:2−/− mice. The increased number of primordial follicles observed in Hdac1:2−/− mice suggested that initiation of follicular development was also affected. We do not have an explanation for this unexpected finding, but it could possibly reflect that it was easier to identify primordial follicles in ovaries of mutant mice.
Fig. 3.
Developmental block at secondary follicle stage in Hdac1:2−/− mice. (A) Histological analysis of ovaries obtained from WT and Hdac1:2−/− mice 18 d of age. In WT ovaries, primary (PF), secondary (SF), and antral follicles (AF) are indicated. In ovaries from Hdac1:2−/− mice, no antral follicles are found, and there is an increase in the number of degenerating follicles (white arrow). (Scale bars: low-magnification image, 0.5 mm; higher-magnification image, 0.17 mm.) (B) Follicle counts from ovaries obtained from WT and mutant mice 18 d of age: primordial (PmF), primary (PF), secondary (SF), antral follicles (AF), and dead/degenerating follicles (DF). Numbers are mean ± SEM of counts of three sequential sections from serially sectioned ovaries. Data are from four biological replicates (*P < 0.05 and **P < 0.01). (C) Ovaries were collected from WT and Hdac1:2−/− mice 10 d of age to 6 wk of age and weighed. The data are expressed as mean ± SEM of n ≥ 8 mice (*P < 0.05 and **P < 0.01). (D) Histological analysis of ovaries obtained from WT and Hdac1:2−/− 12 d of age. In ovaries from mutant mice, there is an increase in the number of degenerating follicles (white arrow). (Scale bar: 0.5 mm.) (E) Follicle counts from ovaries obtained from WT and Hdac1:2−/− mice 12 d of age. Numbers are mean ± SEM of counts of three sequential sections from serially sectioned ovaries. Data are from four biological replicates (*P < 0.05).
Examination of ovarian sections indicated that follicular development arrested at the secondary follicle stage in Hdac1:2−/− mice and the increased periodic acid/Schiff-positive staining (Fig. 3A) in the sections indicated oocyte degeneration. Ovary weight was similar in WT and mutants until day 12 postpartum, after which time the weight of ovaries obtained from Hdac1:2−/− mice failed to increase (Fig. 3C). Histological analysis of ovarian sections from 12-d-old WT and Hdac1:2−/− mice revealed fewer oocytes present in secondary follicles (Fig. 3 D and E). Consistent with this finding is that fewer oocytes were obtained from mutant ovaries [268 ± 12/mouse (WT, 16 mice analyzed) vs. 165 ± 10/mouse (mutant, 20 mice analyzed); P < 0.001]. In addition, WT oocytes obtained from 12-d-old mice were slightly larger than those obtained from Hdac1:2−/− mice (54.3 ± 0.5 μm vs. 51.4 ± 0.7 μm, respectively; P < 0.03). Furthermore, the incidence of apoptosis was greater in Hdac1:2−/− oocytes than WT oocytes by using a TUNEL assay [5.8 ± 1.8% (WT) vs. 15.8 ± 2.2% (mutant) in which 250 and 137 WT and mutant oocytes were analyzed, respectively; P < 0.05], a finding consistent with the increased number of degenerating oocytes present at day 18 in double-mutant oocytes (Fig. 3A). There was no apparent increase in the incidence of apoptosis in oocytes when ovaries from Hdac1−/−, Hdac2−/−, Hdac1−/+/Hdac2−/−, or Hdac1−/−/Hdac2−/+ were analyzed.
Effect of Deleting Hdac1 and Hdac2 on Expression of Class I HDACs.
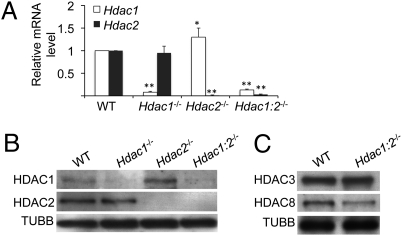
Targeted disruption of Hdac1 resulted in a dramatic decrease in the amount of Hdac1 mRNA with no apparent effect on the amount of Hdac2 mRNA (Fig. 4A). In contrast, targeted disruption of Hdac2 resulted not only in a decrease in the amount of Hdac2 mRNA but also a compensatory increase in the amount of Hdac1 mRNA. The amount of both transcripts was reduced in Hdac1:2−/− oocytes. Whereas Hdac2 transcripts were essentially undetectable in Hdac2−/− and Hdac1:2−/− oocytes, detectable amounts of Hdac1 transcripts were present in Hdac1−/− and Hdac1:2−/− oocytes and may reflect inherent differences in their stability and/or efficiency in Cre-mediated excision.
Fig. 4.
Effect of deleting Hdac1 or Hdac2 or both on Hdac1 and Hdac2 transcript and protein abundance. (A) Relative abundance of Hdac1 and Hdac2 transcripts in oocytes obtained from mice 12 d of age. Data are expressed relative to that in WT oocytes. The experiment was performed four times and the data expressed as mean ± SEM (*P < 0.05 and **P < 0.001). (B) Total protein was extracted from oocytes (n = 300 for HDAC1 and n = 100 for HDAC2) obtained from mice 12 d of age for immunoblotting; at least five mice were used. The amount of HDAC1 and HDAC2 in WT oocytes was set as 100%. Quantification of the band intensities revealed that the amounts of HDAC1 relative to WT were 32%, 223%, and 36% in Hdac1−/−, Hdac2−/−, and Hdac1:2−/− oocytes, respectively, and the amounts of HDAC2 relative to WT were 97%, 0%, and 0% in Hdac1−/−, Hdac2−/−, and Hdac1:2−/− oocytes, respectively. The experiment was performed two times. (C) Immunoblot analysis of HDAC3 and HDAC8 expression in WT and mutant oocytes obtained from mice 12 d of age; 200 oocytes were used and collected from at least five mice. The experiment was conducted two times, and similar results were obtained in each case. In B and C, β-tubulin (TUBB) was used as a loading control.
Although the amount of Hdac1 mRNA was effectively reduced in Hdac1−/− oocytes, a modest amount of HDAC1 protein (∼30%) was present in oocytes obtained from mice 12 d of age compared with WT, and there was no effect on the amount of HDAC2 protein (Fig. 4B). The amount of HDAC1 protein present likely reflected the stability of HDAC1 protein present in primordial and primary oocytes and that the Zp3 promoter becomes active at 5 d postpartum, i.e., after initiation of oocyte growth with its highest activity at approximately days 10 to 12 postpartum (28, 29). Targeting Hdac2 resulted in undetectable amounts of HDAC2 protein and an increase in the amount of HDAC1 protein (Fig. 4B), which was consistent with the compensatory increase in the amount of Hdac1 transcript in these oocytes. The amount of HDAC1 and HDAC2 in Hdac1:2−/− oocytes was also consistent with their amounts in their single respective knockouts. HDAC1 and HDAC2 nuclear staining were also reduced significantly as assessed by immunocytochemistry of Hdac1:2−/− oocytes (see Fig. 8B). We also determined the amount of HDAC3 and HDAC8, the other two Class I HDACs, in Hdac1:2−/− oocytes (Fig. 4C). Although the amount of HDAC3 protein was not changed in Hdac1:2−/− oocytes, the amount of HDAC8 protein was reduced by approximately 50%. It was unlikely that this change accounted for infertility in Hdac1:2−/− mice because Hdac8−/+ females are fertile (30).
Fig. 8.
Deletion of both Hdac1 and Hdac2 leads to apoptosis and TRP53 acetylation. (A) The relative abundance of antiapoptotic transcripts is decreased and that of proapoptotic transcripts increased in Hdac1:2−/− oocytes isolated from mice 12 d of age. The experiment was performed three times using at least three mice per group, and the data are expressed as mean ± SEM (*P < 0.05). (B) TRP53K379 acetylation is increased in mutant oocytes. Immunocytochemical detection of TRP53K379 acetylation was performed in WT, Hdac1 and Hdac2 single mutants, and Hdac1:2−/− oocytes obtained from mice 12 d of age; only the nucleus is shown. For each protein, at least 20 oocytes for each genotype were analyzed, and the experiment was conducted three times using at least three mice of each genotype per experiment. (Scale bar: 10 μm.)
Loss of both Hdac1 and Hdac2 Effects Expression of Other Components of HDAC1/2-Containing Complexes and Increases Histone Acetylation.
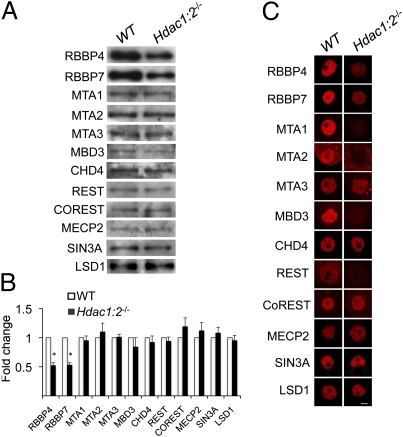
HDAC1 and HDAC2 exert their function within large multiprotein complexes, with NURD, SIN3A, COREST, and NODE being the best characterized to date (14, 31). We sought to determine if expression of other components contained in these complexes was perturbed in Hdac1:2−/− oocytes by immunoblotting (Fig. 5 A and B). The amount of RBBP4 and RBBP7 protein was significantly reduced and the amount of MBD3 and REST protein was modestly reduced in mutant oocytes, whereas there was no apparent change in the amount of CHD4, MTA1, MTA2, MTA3, COREST, MECP2, LSD1, and SIN3A. Immunocytochemical analyses revealed that the amount of nuclear-associated MTA1, MTA2, MBD3, and REST was significantly reduced (Fig. 5C), suggesting that these proteins were now located in the cytoplasm. Cytoplasmic localization of MTA1 was also observed in MTA2-depleted embryos (32). Although the small amounts of biological material precluded biochemical characterization of these four HDAC1/2-containing complexes, the findings suggest that their function was likely compromised in Hdac1:2−/− oocytes.
Fig. 5.
Expression of components of HDAC1- and HDAC2-containing complexes in Hdac1:2−/− oocytes. (A) Relative amount of NuRD, SIN3A, and COREST complex components RBBP4, RBBP7, MTA1, MTA2, MTA3, CHD4, REST, COREST, MECP2, SIN3A, and LSD1 was determined by immunoblot analysis by using total protein extracts from WT and mutant oocytes obtained from at least five mice 12 d of age. Equal numbers of oocytes were loaded per lane. The TUBB loading control is not shown because the immunoblot is a composite of several experiments for which β-tubulin (TUBB) was used as a loading control for each experiment. The experiment was performed three with two times, and similar results were obtained. (B) Quantification of the data shown in A. Data are expressed as mean ± SEM (*P < 0.05). (C) Immunocytochemical detection of RBBP4, RBBP7, MTA1, MTA2, MTA3, CHD4, REST, COREST, MECP2, SIN3A, and LSD1 in WT and mutant oocytes obtained from mice 12 d of age. At least 20 oocytes for each genotype were analyzed, and the experiment was conducted three times with at least three mice used for each experiment. Shown are representative images, and only the nucleus is shown. (Scale bar: 10 μm.)
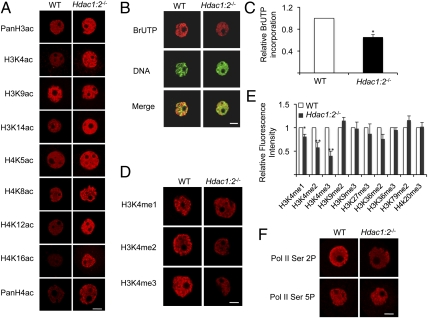
As anticipated, histone acetylation was increased in Hdac1:2−/− oocytes (Fig. 6A). We detected a significant increase in bulk acetylation of histone H3 and H4, as well as acetylation of specific lysine residues, e.g., H3K4/14 and H4K5/8/12/16. Nevertheless, there was no apparent change in the extent of histone H3K9 acetylation. Hdac1 and Hdac2 single mutant oocytes did not exhibit any change in histone acetylation. Histone H4K16 acetylation, however, showed a slight increase in Hdac2−/− oocytes (Fig. S1). That deletion of either Hdac1 or Hdac2 did not affect histone H3 and H4 acetylation suggests compensatory functions of HDAC1 and HDAC2 in histone acetylation in growing oocytes. Last, because no apparent changes of H3K9 acetylation were observed in Hdac1:2−/− oocytes, other HDACs may be responsible for deacetylating H3K9.
Fig. 6.
Increased histone acetylation and decreased global transcription and histone H3K4 methylation in Hdac1:2−/− oocytes. (A) Different acetylated histones were analyzed by immunocytochemistry using oocytes obtained from WT and mutant mice 12 d of age. For each histone variant, at least 20 oocytes from each genotype were analyzed, and the experiment was conducted three times with at least three mice used for each experiment. Shown are representative images, and only the nucleus is shown. (Scale bar: 10 μm.) (B) Global transcription was assayed by BrUTP incorporation by WT and mutant oocytes isolated from at least three mice 12 d of age. (Scale bar: 10 μm.) (C) Quantification of data shown in A. The relative fluorescence intensities of the nuclei were determined and the average value for WT oocytes was set as 1. The experiment was performed three times, with at least three mice used for each experiment, and data are expressed as mean ± SEM, with at least 60 oocytes analyzed for each group (*P < 0.001). (D) Immunocytochemical detection of different H3K4 methylated species on WT and mutant oocytes obtained from mice 12 d of age; only the nucleus is shown, and at least three mice were used for each experiment. (Scale bar: 10 μm.) Signal intensities relative to WT for H3K4me1–3 are 81 ± 1% (n = 4), 58 ± 12% (n = 5), and 40 ± 8% (n = 5), respectively. (E) Quantification of the data shown in D as well as in Fig. S2 shows staining of other methylated histone species. Nuclear staining intensity of specific methylated lysine in WT oocytes was set to 1, and the data are expressed as mean ± SEM. At least 20 oocytes for each genotype and for each modified histone were analyzed; the experiment was conducted three times and at least three mice were used for each experiment (*P < 0.05 and **P < 0.001). (F) Immunocytochemical detection of CTD phosphorylated on S2 or S5 in WT and mutant oocytes obtained from mice 12 d of age; only the nucleus is shown, and at least three mice were used for each experiment. At least 50 oocytes were analyzed, and representative images are shown. The signal intensity for S2 phosphorylation in mutant oocytes is less than that of WT (mean ± SEM, 64 ± 2%; P < 0.01). (Scale bar: 10 μm.)
Reduced Transcription Is Accompanied by Histone H3K4 Demethylation in Hdac1:2−/− Oocytes.
Histone acetylation is associated with transcriptionally permissive chromatin (7) and HDACs typically function as transcriptional corepressors (33). To ascertain whether an increase in global transcription was observed in Hdac1:2−/− oocytes, transcription run-on assays were conducted by monitoring 5-bromouridine 5′-triphosphate (BrUTP) incorporation using mutant oocytes obtained from mice at 12 d of age. BrUTP incorporation by mutant oocytes, however, was reduced by approximately 35% compared with WT oocytes (Fig. 6 B and C), and the amount of poly(A)-containing RNA was reduced by approximately 20% from 16.9 ± 1.6 pg/oocyte in WT oocytes to 13.8 ± 2.2 pg/oocyte in mutant oocytes (mean ± SEM, n = 9; P < 0.01).
The decrease in transcription prompted us to examine whether other histone marks that govern transcription were altered in Hdac1:2−/− oocytes (34, 35). We focused our attention on histone methylation marks because they are linked with both transcription activation and repression. In general, active promoters are marked by trimethylated H3K4 (H3K4me3), whereas dimethylated H3K4 (H3K4me2) is often found in the coding region. H3K4 methylation follows assembly of a preinitiation complex, suggesting that it is involved in processes that maintain rather than initiate transcription (36). Methylation of H3K36 and H3K79 is also correlated with transcriptional activation, whereas methylation of H3K9 and H3K27 are hallmarks of transcriptional repression, and H4K20me3 is an integral part of heterochromatin-mediated silencing (37).
A significant decrease in the intensity of nuclear staining was observed for each of the methylated species of H3K4 (Fig. 6D). No changes were noted for the other methylated histones (i.e., H3K9me3, H3K36me2, H3K36me3, H3K27me3, H4K20me3, and H3K79me2; Fig. 6E and Fig. S2). Moreover, we did not observe any changes of H3K4 di- and trimethylation in Hdac1−/−or Hdac2−/− oocytes. These results suggest that reduced methylation of H3K4 is responsible for the global decrease in transcription, noting that the 35% decrease in BrUTP incorporation is similar to the decrease in H3K4me2/3. Also consistent with the 35% global decrease in transcription in mutant oocytes was that the amount of carboxy-terminal domain (CTD) S2 phosphorylation was reduced approximately 40% in mutant oocytes, with little effect on the amount of CTD S5 phosphorylation (Fig. 6F). Phosphorylation of the heptapeptide (YSPTSPS) repeats located in the CTD of the largest subunit of RNA polymerase II (Pol II) is linked to Pol II function; during elongation, S2 is phosphorylated by the CTD kinase PTEFB (38), whereas initiation is coupled with phosphorylation of S5 by TFIIH and mediators (39, 40).
HDAC1 and HDAC2 Regulate H3K4 Methylation Through KDM5B.
The decrease in methylation of H3K4 could result from increased acetylation in Hdac1:2−/− oocytes because acetylation and methylation are mutually exclusive modifications. Alternatively, changes in expression of genes involved in H3K4 methylation or demethylation that result in a decrease in the steady-state amount of H3K4 methylated species could be the primary contributing factor to decreased H3K4 methylation, with the increase in H3K4 acetylation being a secondary effect.
It was unlikely that a decrease in methylases that methylate H3K4 was responsible for the observed decrease in H3K4 methylation. The abundance of Mll2 transcripts, which encode the methylase largely responsible for H3K4 methylation in mouse oocytes (41), was not decreased in mutant oocytes (Table S1). Moreover, the amount of the core components WDR5, RBBP5, and ASH2L that comprise MLL complexes (42), as well as the amount of MLL2 (and MLL1), was not changed in Hdac1:2−/− oocytes (Fig. S3). In contrast, the relative abundance of Kdm5b, which encodes the only histone lysine demethylase that can demethylate each methylated form of H3K4 (43), was increased by 17 fold (Table S1). This increase, which was confirmed by quantitative RT-PCR (qRT-PCR) that revealed a 43-fold increase (average of two experiments) in the abundance of Kdm5b transcripts in mutant oocytes, provides an explanation for the decrease in H3K4 methylation. We were not able to find an anti-KDM5B antibody that was suitable for either immunoblotting or immunocytochemistry. To address whether an increase in the amount of KDM5B could contribute to the decrease in H3K4 methylation, we found that siRNA-targeting of Kdm5b transcripts, which resulted in a 90% decrease in Kdm5b mRNA, resulted in an increase in the amount of H3K4me3 relative to controls (Fig. 7 A and B). Reciprocally, overexpressing KDM5B in WT oocytes resulted in a decrease in H3K4me3 relative to controls (Fig. 7 C and D).
Fig. 7.
RNAi-mediated targeting of Kdm5b in Hdac1:2−/− oocytes leads to an increase in histone H3K4 methylation. (A) Hdac1:2−/− oocytes obtained from mice 12 d of age were injected with control siRNA or Kdm5b siRNA, and the amount of Kdm5b mRNA relative to that present in the control siRNA-injected oocytes was determined by qRT-PCR 52 h following injection. The experiment was performed three times, with at least three mice used for each experiment, and the data are presented as mean ± SEM (*P < 0.001). (B) Immunocytochemical detection of H3K4me3 was performed on oocytes injected as described in A, and only the nucleus is shown. (Scale bar: 10 μm.) The experiment was performed two times (50 oocytes were analyzed in each group and collected from at least three mice), and quantification of the data revealed a 23 ± 6% increase in intensity of the H3H4me3 signal. (C) WT oocytes obtained from mice 12 d of age were injected with Gfp cRNA (control) or Kdm5b cRNA (Kdm5b-O), and the amount of Kdm5b transcript relative to that present in the control oocytes was determined by qRT-PCR 24 h following injection. The experiment was performed twice and the data are presented as mean ± SEM (*P < 0.05). (D) Immunocytochemical detection of H3K4me3 was performed 50 h after injection; only the nucleus is shown. (Scale bar: 10 μm.) The experiment was performed two times (24 oocytes analyzed in each group), and quantification of the data revealed a 28 ± 5% decrease in intensity of the H3H4me3 signal.
To determine whether an increase in global transcription occurred in these treated oocytes, we attempted to assay BrUTP incorporation, but the prolonged period in culture resulted in oocytes being extremely fragile and not suitable for such assays. The oocytes were suitable, however, for immunocytochemistry, but we did not observe an increase in CTD S2 phosphorylation. Although this finding may indicate that the decrease in H3K4 methylation was not the primary cause for the decrease in BrUTP incorporation in Hdac1:2−/− oocytes, it was also possible that (i) not enough time had elapsed to observe a rescue effect or (ii), more likely, that an irreversible decrease in transcription occurred, perhaps because of initiation of apoptosis (as detailed later).
Transcriptome Analysis of Hdac1 and Hdac2 Mutant Oocytes.
Transcript profiling experiments were conducted by using mutant oocytes to ascertain whether mis-regulation of specific genes would shed more light on the observed phenotype. Hierarchical clustering revealed that Hdac1:2−/− oocytes clustered separately from their single mutant brethren and WT, which clustered together (Fig. S4A). These findings are consistent with infertility being observed in only Hdac1:2−/− mice. The paucity of affected transcripts in Hdac1−/− oocytes [two increased (Eo30030I06Rik by 5.8 fold and Mt1 by 25.4 fold) and four decreased, not including Hdac1 (Olfr49 by 1.4 fold, Rnase6 by 20 fold, Slc7A8 by 2.7-fold, and a gene with ID no. 10339666 by 2.7 fold)] may reflect the residual amount of HDAC1 protein in these oocytes. The number of affected transcripts in Hdac2−/− oocytes was much higher (307 increased and 150 decreased; Table S2), and a synergistic effect was clearly observed in the double KO, in which the relative abundance of 2,666 transcripts were increased and 2,182 were decreased (Table S1). A Venn diagram displaying the intersection of the affected transcripts is shown in Fig. S4B, and the shared transcripts are provided in Table S3. The differences in transcript abundance detected by microarrays were confirmed for a set of randomly selected genes that showed no difference or an increase or decrease (Fig. S4C).
Analysis of what pathways were affected in Hdac1:2−/− oocytes revealed that, for transcripts exhibiting a decrease in abundance, the top four most enriched functional categories were transcription, regulation of transcription, DNA-dependent regulation of transcription, and regulation of RNA metabolic process (Table S4), which is consistent with a reduced level of global transcription. For transcripts that exhibited an increase in relative abundance, negative regulation of gene expression was an enriched category—consistent with the global decrease in transcription—as was the category of genes involved in regulating cell death and apoptosis (Table S4). As described elsewhere in the present study, Hdac1:2−/− oocytes undergo apoptosis.
Hdac1:2−/− Oocytes Undergo Apoptosis Initiated by TRP53 Hyperacetylation.
As previously described, a striking feature noted in Hdac1:2−/− mice was that, not only did follicle development arrest at the secondary follicle stage, but the oocytes were undergoing apoptosis. Consistent with Hdac1:2−/− oocytes undergoing apoptosis was an increased abundance of proapoptotic transcripts Bim (44) and Bad (45) and decreased abundance of antiapoptotic transcripts Bcl-2 (46) and Bcl2l1 (also called Bcl-XL) (47) (Fig. 8A). Furthermore, there was a small but significant increase in Trp53 transcript abundance and a pronounced increase in p21 (Cdkn1a) transcript abundance (Fig. 8A); p21 is a direct target of TRP53 (48) and TRP53 can play a central role in apoptosis (49).
We explored whether Hdac1 and Hdac2 modulated TRP53 function in oocytes because TRP53 activity is repressed in an HDAC1-dependent manner through deacetylation (50, 51) and activated by ING2 (inhibitor of growth protein 2) via enhancing TRP53 acetylation (52); Ing2 expression was increased in Hdac1:2−/− oocytes (Table S1). Accordingly, we ascertained whether TRP53 was hyperacetylated in Hdac1:2−/− oocytes (Fig. 8B). Immunocytochemistry revealed a significant increase in the nuclear intensity of TRP53 acetylated on K379. No apparent increase in TRP53K379 acetylation was noted for Hdac1−/− or Hdac2−/− oocytes, a finding consistent with female mice harboring these oocytes being fully fertile. These results suggest that HDAC1 and HDAC2 can compensate for the function of the other regarding deacetylating TRP53 and that hyperacetylation of TRP53 may contribute to the increased incidence of apoptosis in Hdac1:2−/− oocytes. Consistent with the latter proposal is that incubating WT oocytes in medium containing the HDAC inhibitor trichostatin A (TSA) not only resulted in an increase in TP53K379 acetylation (Fig. 9A), but also in an increase in apoptosis as assayed by TUNEL (5.3% in control oocytes vs. 13.5% in TSA-treated oocytes in which 150 and 85 control and TSA-treated oocytes were analyzed, respectively; P < 0.05, Fisher exact test). The relative abundance of p21 transcripts was also increased (Fig. 9B).
Fig. 9.
TSA induces TRP53 acetylation in WT oocytes and increased expression of proapoptotic genes. (A) Immunocytochemical detection of acetylated TRP53 after TSA treatment of WT oocytes for 24 h and 48 h, and only the nucleus is shown. At least 20 oocytes were analyzed, and the experiment was performed two times using at least three mice per experiment. Shown are representative images. (B) Relative abundance of anti- and proapoptotic transcripts determined by qRT-PCR following TSA treatment for 72 h. The experiment was performed three times using at least three mice per experiment, and the data are expressed as mean ± SEM (*P < 0.05).
Discussion
We report here that deletion of both Hdac1 and Hdac2 in mouse oocytes, but not either alone, results in infertility. Follicle development arrests at the secondary follicle stage and the oocyte transcriptome is dramatically perturbed. Moreover, Hdac1:2−/− oocytes, which display a global decrease in transcription and histone H3K4 methylation, and likely undergo apoptosis induced by hyperacetylation of TRP53. This combination of factors serves as the basis for the observed infertility and indicates that chromatin-associated factors with HDAC activity are essential for oocyte development.
Deletion of both Hdac1 and Hdac2 in mouse oocytes results in decreased ovary size, arrest of follicle development at the secondary follicle stage, and infertility. In contrast, there was no loss of fertility in females harboring Hdac1-deficient oocytes, and only a small decrease in fertility is observed following loss of Hdac2 in oocytes. The residual HDAC1 protein in Hdac1:2−/− oocytes—a similar amount of HDAC1 protein is also present in Hdac1−/− oocytes—is not sufficient to maintain fertility in female mice harboring Hdac1:2−/− oocytes. Taken together, these results indicate that HDAC1 and HDAC2 provide compensatory functions during oocyte development, a finding consistent with loss of both HDAC1 and HDAC2 function being required to observe phenotypic consequences in many cell types (18–21). Last, because (i) Hdac8−/+ mice are fertile (the amount of HDAC8 is reduced by approximately 50% in Hdac1:2−/− oocytes), (ii) the amount of HDAC3 protein is unaffected in Hdac1:2−/− oocytes, and (iii) Hdac1−/+/Hdac2−/− mice are sterile even though there is residual HDAC1 protein in the oocytes, it appears HDAC2 is the major class I HDAC essential for oocyte development. The increased amount of HDAC1 in Hdac2−/− oocytes, relative to the amount of HDAC1 in WT oocytes, may account for the observation that only a mild subfertility is observed for Hdac2−/− females.
Bidirectional communication exists between oocytes and the surrounding granulosa/cumulus cells in which the oocyte drives the conversation (53); such communication is essential for oocyte growth and follicle cell proliferation. Perturbation of the oocyte transcriptome in Hdac1:2−/− oocytes could disrupt such communication and result in the absence of follicle cell proliferation. Of note is that, although the microarrays did not detect any change in expression of the oocyte-specific genes Bmp15 and Gdf9, which are involved in this communication pathway and stimulate granulosa cell proliferation (54), we observed that qRT-PCR revealed an approximate 50% decrease in transcript abundance for each.
The primary role of HDAC2 in oocyte development contrasts with HDAC1 serving as the HDAC critical for preimplantation development (25), i.e., the oocyte-to-embryo transition entails a switch from HDAC2 as the major HDAC for oocyte development to HDAC1 being critical for preimplantation embryo development. HDAC1 serves as the major HDAC in mouse ES cells because loss of HDAC1 protein, but not HDAC2 protein, results in a substantial decrease in total HDAC activity (24) as well as in several HDAC1/2-containing complexes (20, 22, 24), despite up-regulation in the amount of HDAC2 and HDAC3 (22, 24). Up-regulation of both HDAC2 and HDAC3 is observed in preimplantation mouse embryos following RNAi-mediated targeting of Hdac1 transcripts (25). Loss of Hdac2 does not result in an increase in the amount of HDAC1 protein in ES cells (24), a situation similar to that observed in preimplantation embryos (25). The dominant role of HDAC1 function in preimplantation mouse embryos is consistent with transformation of oocytes into totipotent blastomeres, which are more closely related to pluripotent ES cells than highly differentiated oocytes.
A developmental switch in HDAC1 and 2 function was unanticipated in light of the apparent redundancy of HDAC1 and HDAC2 function in many somatic cell types and because HDAC1 and HDAC2, which can form heterodimers, often reside within the same chromatin-remodeling complex that retains activity—albeit reduced—in the absence of the other HDAC (22). Of interest is that 40% to 60% of HDAC1 and HDAC2 are not present in chromatin-remodeling complexes and are not associated with each other, raising the possibility that these free forms exert specific distinct functions underlying the oocyte-to-embryo transition (20).
The decrease in global transcription in Hdac1:2−/− oocytes appears linked to a decrease in methylated H3K4 that is likely a consequence of overexpression of Kdm5b; KDM5B functions as a strong transcriptional repressor in different systems (55, 56). There is a very tight correlation between the extent of the decrease in global transcription, decrease in methylated H3K4—in particular di- and trimethylated species—and S2 phosphorylation, but not S5 phosphorylation, of Pol II's CTD. Unfortunately, causality could not be established linking the decrease in global transcription to a decrease in H3K4 methylation, because even though siRNA targeting of Kdm5b results in a partial increase in H3K4 methylation, no increase in transcription was observed. As described earlier, failure to restore transcription could be a consequence that not enough time had elapsed to observe a rescue effect or that an irreversible decrease in transcription occurred.
The small perturbations in the transcriptome observed for Hdac1−/− or Hdac2−/−mutant oocytes, which sharply contrast with the dramatic perturbations in Hdac1:2−/− oocytes, is consistent with compensatory roles for HDAC1 and HDAC2 in oocyte development. That virtually no genes are mis-expressed in Hdac1−/− oocytes is also consistent with HDAC2 serving as the major HDAC in oocyte development. The marked transcriptome perturbation observed in Hdac1:2−/− oocytes is unlikely a secondary consequence of apoptosis because only a small fraction of the oocytes used for the microarray analyses were apoptotic.
Female mice deficient in the oocyte-specific TATA-binding protein 2 (TBP2) (57) display phenotypes very similar to that observed for Hdac1:2−/− mice, including infertility, small ovaries, arrest of follicle growth before antrum formation, a decrease in Pol II CTD S2 phosphorylation and histone H3K4 methylation, and a perturbed transcriptome (57); an increase in apoptosis, however, was not observed in Tbp2-deficient oocytes. Interestingly, the relative abundance of approximately 50% of the perturbed transcripts is increased in Tbp2-deficient oocytes. It should be noted, however, that total ovarian tissue was used to conduct the microarray analyses, and therefore the assumption is that the affected transcripts are those present in the oocytes and not somatic tissue.
Oocyte development is characterized by progressive changes in gene expression with dramatic changes occurring during the primordial-to-primary follicle stage and then again during the secondary follicle-to-small antral follicle transition (58), which is accompanied by the acquisition of meiotic competence (59). The similarity in phenotypes with failure to undergo this transition suggests that Hdac1 and Hdac2 double mutants and Tbp2 mutants share a common mechanism that is linked to a decrease in transcription and a perturbed transcriptome. Indeed, there are mis-expressed genes shared by the two mutants (Table S5), but note that, for some genes, a gene that is up-regulated in one mutant may be down-regulated in the other mutant, e.g., Zfp92 is up-regulated in Tbp2−/− oocytes but down-regulated in Hdac1:2−/− oocytes. Oocyte development likely requires constant surveillance of the transcriptome—perhaps most vigilant during a major transition such as the secondary follicle-to-small antral follicle transition—such that oocytes with a highly perturbed transcriptome fail to develop. Such a mechanism would increase the likelihood that only oocytes capable of supporting development are generated and thereby enhance reproductive fitness.
One of TRP53's many functions is to play a role in the apoptotic pathway, and indeed, expression of antiapoptotic genes is decreased and proapoptotic genes increased in Hdac1:2−/− oocytes. TRP53 activity is maintained at low levels in unstressed cells by MDM2, which is a key negative regulator (60). TRP53 also undergoes a host of PTMs in response to stress that include phosphorylation, sumoylation, and acetylation, with acetylation of lysine residues located near the carboxyl terminus required for full activity. TRP53 is hyperacetylated in Hdac1:2−/− oocytes, but not in either single mutant, i.e., acetylated TRP53 is presumably a substrate for either HDAC1 or HDAC2 in oocytes. Hyperacetylation of TRP53 in Hdac1:2−/− oocytes may contribute to the observed increased incidence of apoptosis in mutant oocytes. Consistent with this proposal is that inhibiting HDAC function with TSA in WT oocytes results in TRP53 hyperacetylation and induces apoptosis in the treated oocytes. We cannot exclude, however, that the increased incidence of apoptosis in mutant oocytes is a consequence of the dramatic perturbation in the transcriptome.
A model that emerges is that, in the absence of both Hdac1 and Hdac2, reduced methylation of H3K4 contributes to a global reduction in transcription as well as mis-expression of many genes, which prevents development beyond the secondary follicle stage. The increased incidence of apoptosis, a likely consequence of a dramatically perturbed transcriptome and hyperacetylation of TRP53, would eliminate these oocytes over a period of several days. The outcome of these events would result in female infertility.
Materials and Methods
Generation of Mouse Lines.
Female mice carrying Hdac1 and Hdac2 floxed alleles (18) were crossed with Zp3-Cre males (27), and genotyping was performed as previously described (18, 26). Mice were maintained in according with institutional guidelines established by the University of Pennsylvania Institutional Animal Care and Use Committee, and all experiments were conducted in accordance with the National Institutes of Health Guide for the Care and Use of Laboratory Animals.
Histological Analysis of Ovaries.
Ovaries from 12- and 18-d-old mice were fixed for 2 to 3 h at room temperature in Bouin solution, dehydrated in ethanol and toluene, and embedded in paraffin. Follicles were categorized and counted by examining serial 5-μm sections through the entire ovary and stained with periodic acid/Schiff reagent and Lillie–Mayer hematoxylin (57, 61).
Oocyte and Embryo Collection, Culture, and siRNA Microinjection.
Cumulus cell-free, GV-intact oocytes were obtained from equine CG-primed females as previously described (25). The collection medium for oocytes was bicarbonate-free MEM (Earle salts) containing 25 mM Hepes (pH 7.3), 3 mg/mL polyvinylpyrrolidone, and 2.5 μM milrinone to prevent GV breakdown (62). Oocytes were matured in vitro in CZB medium (63) containing 0.01% polyvinyl alcohol.
Meiotically incompetent oocytes (i.e., oocytes that do not spontaneously resume meiosis when placed in a suitable culture medium) were obtained from 12-d-old female mice as described previously (64). Incompetent oocytes were injected with 10 pL of the appropriate siRNA using a Picoliter Injector Microinjection System (Harvard Apparatus); the culture medium was bicarbonate-free Whitten medium (65) containing 0.01% polyvinyl alcohol and 25 mM Hepes, pH 7.3. Following microinjection, the oocytes were then cultured in CZB medium at 37 °C in 5% CO2 in an atmosphere of air. A predesigned siRNA for Kdm5b and a scrambled siRNA (Am4611; Ambion) served as the control. The Kdm5b siRNA (s93703; sense sequence, 5′-CUCCGAUACAUGAUUGAAAtt-3′; antisense sequence, 5′-UUUCAAUCAUGUAUCGGAGtg-3′) targets nucleotides 3915 to 3933 of the Kdm5b mRNA (GenBank accession no. NM152895). Both siRNAs were diluted in Milli-Q water to a final concentration of 5 μM for microinjection.
Quantification of Total Amount of poly(A)-Containing RNA.
Fifty growing oocytes were collected from 12-d-old mice and the total amount of total poly(A) mRNA was determined using the Poly(A) mRNA Detection System (Promega) according to the manufacturer's instructions.
In Vitro Transcription Assay.
BrUTP incorporation assays were performed as previously described (64). Fluorescence was detected on a Leica TCS SP laser-scanning confocal microscope. The intensity of fluorescence was quantified using ImageJ software (National Institutes of Health) as previously described (66).
TUNEL Labeling Assay.
TUNEL assays were performed with an In Situ Cell Death Detection Kit (Roche Diagnostics) according to the manufacturer's instructions.
RNA Extraction, Microarray Analysis, and Real-Time RT-PCR.
Total RNA was extracted from 80 oocytes isolated from mice 12 d of age by using the PicoPure RNA kit (Arcturus), amplified with the Ovation Pico WTA system (NuGen), and then fragmented and labeled with the FL-Ovation cDNA Biotin Module V2 (NuGen). Four independent biological replicates were hybridized to GeneChip Mouse 1.1 ST microarrays (Affymetrix). Raw microarray data were analyzed as previously described by using MAS5, GeneSpring (version 7), SAM, and EASE software (58). A 1.4-fold cutoff was used for EASE analysis; four biological replicates provide sufficient statistical power and confidence to detect a 1.4-fold change in transcript abundance (67). Microarray data were validated in at least two independent biological replicates by real-time quantitative PCR. Quantitative PCR analysis was performed with the ABI TaqMan Assay-on-demand probe/primer sets as previously described (25), and probes are listed in SI Materials and Methods. Two incompetent oocyte equivalents of cDNA was used for each real-time PCR with a minimum of three replicates as well as minus RT and minus template controls for each gene. Unless otherwise stated, quantification was normalized to Ubf or histone 2A mRNA.
Antibodies.
Sources and dilution of antibodies used for experiments reported is provided in SI Materials and Methods.
Immunostaining of Oocytes/Eggs and Quantification of Fluorescence Intensity.
Oocytes or embryos were fixed in 2% paraformaldehyde in PBS solution for 20 min at room temperature, and then permeabilized with 0.2% Triton X-100 in PBS solution for 10 min. Immunocytochemical staining was performed by incubating the fixed samples with primary antibodies overnight at 4 °C, followed by secondary antibodies conjugated with Cy5 or FITC for 60 min; omission of the primary antibody served as control in which case no signal was observed (Fig. S1 shows a representative image). The DNA was stained with DAPI or 1 μM SYTOX Green (Molecular Probes). The cells were then washed and mounted under a coverslip with gentle compression in VectaShield antibleaching solution (Vector Labs). Fluorescence was detected on a Leica TCS SP laser-scanning confocal microscope. For each experiment, all samples were processed in parallel and the intensity of fluorescence was quantified using ImageJ software (National Institutes of Health).
Immunoblot Analysis.
Protein samples were solubilized in Laemmli sample buffer (68), resolved by SDS/PAGE (5–15% gel), and transferred to a nitrocellulose membrane. The membrane was blocked by soaking in Blotto [Tris-buffered saline solution with 0.1% Tween-20 (TBST) and 5% nonfat dried milk] for 1.5 h and incubated overnight with the primary antibody in blocking solution. The membrane was then washed three times with TBST, incubated with a secondary antibody conjugated with horseradish peroxidase for 45 min, and washed five times with TBST. The signal was detected with ECL Advance Western blotting detection reagents (Amersham), following the manufacturer's recommendations. The primary antibodies were diluted as described earlier, and secondary antibodies (Amersham ECL-HRP Linked Secondary Antibody; GE Healthcare) were diluted 1:20,000.
Statistics.
All proportional data were subjected to an arcsine transformation before statistical analysis. A P value lower than 0.05 was considered to be statistically significant.
Supplementary Material
Acknowledgments
We thank John Eppig for providing slides of serially sectioned ovaries for histological analyses and Paula Stein for critically reading the manuscript. This research was supported by National Institutes of Health Grant HD022681 (to R.M.S.).
Footnotes
The authors declare no conflict of interest.
Data deposition: The microarray data reported in this paper have been deposited in the Gene Expression Omnibus (GEO) database, www.ncbi.nlm.nih.gov/geo (accession no. GSE34786).
This article is a PNAS Direct Submission.
See Author Summary on page 2704 (volume 109, number 8).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1118403109/-/DCSupplemental.
References
- 1.Rodrigues P, Limback D, McGinnis LK, Plancha CE, Albertini DF. Oogenesis: Prospects and challenges for the future. J Cell Physiol. 2008;216:355–365. doi: 10.1002/jcp.21473. [DOI] [PubMed] [Google Scholar]
- 2.van den Hurk R, Zhao J. Formation of mammalian oocytes and their growth, differentiation and maturation within ovarian follicles. Theriogenology. 2005;63:1717–1751. doi: 10.1016/j.theriogenology.2004.08.005. [DOI] [PubMed] [Google Scholar]
- 3.Moore GP, Lintern-Moore S. Transcription of the mouse oocyte genome. Biol Reprod. 1978;18:865–870. doi: 10.1095/biolreprod18.5.865. [DOI] [PubMed] [Google Scholar]
- 4.De La Fuente R, Eppig JJ. Transcriptional activity of the mouse oocyte genome: Companion granulosa cells modulate transcription and chromatin remodeling. Dev Biol. 2001;229:224–236. doi: 10.1006/dbio.2000.9947. [DOI] [PubMed] [Google Scholar]
- 5.Kageyama S, et al. Alterations in epigenetic modifications during oocyte growth in mice. Reproduction. 2007;133:85–94. doi: 10.1530/REP-06-0025. [DOI] [PubMed] [Google Scholar]
- 6.Bhaumik SR, Smith E, Shilatifard A. Covalent modifications of histones during development and disease pathogenesis. Nat Struct Mol Biol. 2007;14:1008–1016. doi: 10.1038/nsmb1337. [DOI] [PubMed] [Google Scholar]
- 7.Jenuwein T, Allis CD. Translating the histone code. Science. 2001;293:1074–1080. doi: 10.1126/science.1063127. [DOI] [PubMed] [Google Scholar]
- 8.Grunstein M. Histone acetylation in chromatin structure and transcription. Nature. 1997;389:349–352. doi: 10.1038/38664. [DOI] [PubMed] [Google Scholar]
- 9.Hubbert C, et al. HDAC6 is a microtubule-associated deacetylase. Nature. 2002;417:455–458. doi: 10.1038/417455a. [DOI] [PubMed] [Google Scholar]
- 10.Luo J, Su F, Chen D, Shiloh A, Gu W. Deacetylation of p53 modulates its effect on cell growth and apoptosis. Nature. 2000;408:377–381. doi: 10.1038/35042612. [DOI] [PubMed] [Google Scholar]
- 11.Martínez-Balbás MA, Bauer UM, Nielsen SJ, Brehm A, Kouzarides T. Regulation of E2F1 activity by acetylation. EMBO J. 2000;19:662–671. doi: 10.1093/emboj/19.4.662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Minucci S, Pelicci PG. Histone deacetylase inhibitors and the promise of epigenetic (and more) treatments for cancer. Nat Rev Cancer. 2006;6:38–51. doi: 10.1038/nrc1779. [DOI] [PubMed] [Google Scholar]
- 13.Bolden JE, Peart MJ, Johnstone RW. Anticancer activities of histone deacetylase inhibitors. Nat Rev Drug Discov. 2006;5:769–784. doi: 10.1038/nrd2133. [DOI] [PubMed] [Google Scholar]
- 14.Brunmeir R, Lagger S, Seiser C. Histone deacetylase HDAC1/HDAC2-controlled embryonic development and cell differentiation. Int J Dev Biol. 2009;53:275–289. doi: 10.1387/ijdb.082649rb. [DOI] [PubMed] [Google Scholar]
- 15.Mehnert JM, Kelly WK. Histone deacetylase inhibitors: Biology and mechanism of action. Cancer J. 2007;13:23–29. doi: 10.1097/PPO.0b013e31803c72ba. [DOI] [PubMed] [Google Scholar]
- 16.Haberland M, Montgomery RL, Olson EN. The many roles of histone deacetylases in development and physiology: Implications for disease and therapy. Nat Rev Genet. 2009;10:32–42. doi: 10.1038/nrg2485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Grozinger CM, Schreiber SL. Deacetylase enzymes: Biological functions and the use of small-molecule inhibitors. Chem Biol. 2002;9:3–16. doi: 10.1016/s1074-5521(02)00092-3. [DOI] [PubMed] [Google Scholar]
- 18.Montgomery RL, et al. Histone deacetylases 1 and 2 redundantly regulate cardiac morphogenesis, growth, and contractility. Genes Dev. 2007;21:1790–1802. doi: 10.1101/gad.1563807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Montgomery RL, Hsieh J, Barbosa AC, Richardson JA, Olson EN. Histone deacetylases 1 and 2 control the progression of neural precursors to neurons during brain development. Proc Natl Acad Sci USA. 2009;106:7876–7881. doi: 10.1073/pnas.0902750106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yamaguchi T, et al. Histone deacetylases 1 and 2 act in concert to promote the G1-to-S progression. Genes Dev. 2010;24:455–469. doi: 10.1101/gad.552310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.LeBoeuf M, et al. Hdac1 and Hdac2 act redundantly to control p63 and p53 functions in epidermal progenitor cells. Dev Cell. 2010;19:807–818. doi: 10.1016/j.devcel.2010.10.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lagger G, et al. Essential function of histone deacetylase 1 in proliferation control and CDK inhibitor repression. EMBO J. 2002;21:2672–2681. doi: 10.1093/emboj/21.11.2672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Guan JS, et al. HDAC2 negatively regulates memory formation and synaptic plasticity. Nature. 2009;459:55–60. doi: 10.1038/nature07925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Dovey OM, Foster CT, Cowley SM. Histone deacetylase 1 (HDAC1), but not HDAC2, controls embryonic stem cell differentiation. Proc Natl Acad Sci USA. 2010;107:8242–8247. doi: 10.1073/pnas.1000478107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ma P, Schultz RM. Histone deacetylase 1 (HDAC1) regulates histone acetylation, development, and gene expression in preimplantation mouse embryos. Dev Biol. 2008;319:110–120. doi: 10.1016/j.ydbio.2008.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lan ZJ, et al. GCNF-dependent repression of BMP-15 and GDF-9 mediates gamete regulation of female fertility. EMBO J. 2003;22:4070–4081. doi: 10.1093/emboj/cdg405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lewandoski M, Wassarman KM, Martin GR. Zp3-cre, a transgenic mouse line for the activation or inactivation of loxP-flanked target genes specifically in the female germ line. Curr Biol. 1997;7:148–151. doi: 10.1016/s0960-9822(06)00059-5. [DOI] [PubMed] [Google Scholar]
- 28.Epifano O, Liang L-F, Familari M, Moos MC, Jr, Dean J. Coordinate expression of the three zona pellucida genes during mouse oogenesis. Development. 1995;121:1947–1956. doi: 10.1242/dev.121.7.1947. [DOI] [PubMed] [Google Scholar]
- 29.Lira SA, Kinloch RA, Mortillo S, Wassarman PM. An upstream region of the mouse ZP3 gene directs expression of firefly luciferase specifically to growing oocytes in transgenic mice. Proc Natl Acad Sci USA. 1990;87:7215–7219. doi: 10.1073/pnas.87.18.7215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Haberland M, Mokalled MH, Montgomery RL, Olson EN. Epigenetic control of skull morphogenesis by histone deacetylase 8. Genes Dev. 2009;23:1625–1630. doi: 10.1101/gad.1809209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yang XJ, Seto E. Lysine acetylation: codified crosstalk with other posttranslational modifications. Mol Cell. 2008;31:449–461. doi: 10.1016/j.molcel.2008.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ma P, Lin S, Bartolomei MS, Schultz RM. Metastasis tumor antigen 2 (MTA2) is involved in proper imprinted expression of H19 and Peg3 during mouse preimplantation development. Biol Reprod. 2010;83:1027–1035. doi: 10.1095/biolreprod.110.086397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Taunton J, Hassig CA, Schreiber SL. A mammalian histone deacetylase related to the yeast transcriptional regulator Rpd3p. Science. 1996;272:408–411. doi: 10.1126/science.272.5260.408. [DOI] [PubMed] [Google Scholar]
- 34.Berger SL. The complex language of chromatin regulation during transcription. Nature. 2007;447:407–412. doi: 10.1038/nature05915. [DOI] [PubMed] [Google Scholar]
- 35.Strahl BD, Allis CD. The language of covalent histone modifications. Nature. 2000;403:41–45. doi: 10.1038/47412. [DOI] [PubMed] [Google Scholar]
- 36.Shilatifard A. Chromatin modifications by methylation and ubiquitination: Implications in the regulation of gene expression. Annu Rev Biochem. 2006;75:243–269. doi: 10.1146/annurev.biochem.75.103004.142422. [DOI] [PubMed] [Google Scholar]
- 37.Hublitz P, Albert M, Peters AH. Mechanisms of transcriptional repression by histone lysine methylation. Int J Dev Biol. 2009;53:335–354. doi: 10.1387/ijdb.082717ph. [DOI] [PubMed] [Google Scholar]
- 38.Ni Z, Schwartz BE, Werner J, Suarez JR, Lis JT. Coordination of transcription, RNA processing, and surveillance by P-TEFb kinase on heat shock genes. Mol Cell. 2004;13:55–65. doi: 10.1016/s1097-2765(03)00526-4. [DOI] [PubMed] [Google Scholar]
- 39.Hengartner CJ, et al. Temporal regulation of RNA polymerase II by Srb10 and Kin28 cyclin-dependent kinases. Mol Cell. 1998;2:43–53. doi: 10.1016/s1097-2765(00)80112-4. [DOI] [PubMed] [Google Scholar]
- 40.Komarnitsky P, Cho EJ, Buratowski S. Different phosphorylated forms of RNA polymerase II and associated mRNA processing factors during transcription. Genes Dev. 2000;14:2452–2460. doi: 10.1101/gad.824700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Andreu-Vieyra CV, et al. MLL2 is required in oocytes for bulk histone 3 lysine 4 trimethylation and transcriptional silencing. PLoS Biol. 2010;8:e1000453. doi: 10.1371/journal.pbio.1000453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wysocka J, et al. WDR5 associates with histone H3 methylated at K4 and is essential for H3 K4 methylation and vertebrate development. Cell. 2005;121:859–872. doi: 10.1016/j.cell.2005.03.036. [DOI] [PubMed] [Google Scholar]
- 43.Yamane K, et al. PLU-1 is an H3K4 demethylase involved in transcriptional repression and breast cancer cell proliferation. Mol Cell. 2007;25:801–812. doi: 10.1016/j.molcel.2007.03.001. [DOI] [PubMed] [Google Scholar]
- 44.O'Connor L, et al. Bim: A novel member of the Bcl-2 family that promotes apoptosis. EMBO J. 1998;17:384–395. doi: 10.1093/emboj/17.2.384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Chao DT, Korsmeyer SJ. BCL-2 family: Regulators of cell death. Annu Rev Immunol. 1998;16:395–419. doi: 10.1146/annurev.immunol.16.1.395. [DOI] [PubMed] [Google Scholar]
- 46.Vaux DL. Immunopathology of apoptosis—introduction and overview. Springer Semin Immunopathol. 1998;19:271–278. doi: 10.1007/BF00787224. [DOI] [PubMed] [Google Scholar]
- 47.Janumyan YM, et al. Bcl-xL/Bcl-2 coordinately regulates apoptosis, cell cycle arrest and cell cycle entry. EMBO J. 2003;22:5459–5470. doi: 10.1093/emboj/cdg533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.el-Deiry WS, et al. WAF1, a potential mediator of p53 tumor suppression. Cell. 1993;75:817–825. doi: 10.1016/0092-8674(93)90500-p. [DOI] [PubMed] [Google Scholar]
- 49.Haupt S, Berger M, Goldberg Z, Haupt Y. Apoptosis - the p53 network. J Cell Sci. 2003;116:4077–4085. doi: 10.1242/jcs.00739. [DOI] [PubMed] [Google Scholar]
- 50.Ito A, et al. MDM2-HDAC1-mediated deacetylation of p53 is required for its degradation. EMBO J. 2002;21:6236–6245. doi: 10.1093/emboj/cdf616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Tang Y, Zhao W, Chen Y, Zhao Y, Gu W. Acetylation is indispensable for p53 activation. Cell. 2008;133:612–626. doi: 10.1016/j.cell.2008.03.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Nagashima M, et al. DNA damage-inducible gene p33ING2 negatively regulates cell proliferation through acetylation of p53. Proc Natl Acad Sci USA. 2001;98:9671–9676. doi: 10.1073/pnas.161151798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Su YQ, Sugiura K, Eppig JJ. Mouse oocyte control of granulosa cell development and function: Paracrine regulation of cumulus cell metabolism. Semin Reprod Med. 2009;27:32–42. doi: 10.1055/s-0028-1108008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.McNatty KP, et al. Bone morphogenetic protein 15 and growth differentiation factor 9 co-operate to regulate granulosa cell function in ruminants. Reproduction. 2005;129:481–487. doi: 10.1530/rep.1.00517. [DOI] [PubMed] [Google Scholar]
- 55.Scibetta AG, et al. Functional analysis of the transcription repressor PLU-1/JARID1B. Mol Cell Biol. 2007;27:7220–7235. doi: 10.1128/MCB.00274-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Tan K, et al. Human PLU-1 Has transcriptional repression properties and interacts with the developmental transcription factors BF-1 and PAX9. J Biol Chem. 2003;278:20507–20513. doi: 10.1074/jbc.M301994200. [DOI] [PubMed] [Google Scholar]
- 57.Gazdag E, et al. TBP2 is essential for germ cell development by regulating transcription and chromatin condensation in the oocyte. Genes Dev. 2009;23:2210–2223. doi: 10.1101/gad.535209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Pan H, O'brien MJ, Wigglesworth K, Eppig JJ, Schultz RM. Transcript profiling during mouse oocyte development and the effect of gonadotropin priming and development in vitro. Dev Biol. 2005;286:493–506. doi: 10.1016/j.ydbio.2005.08.023. [DOI] [PubMed] [Google Scholar]
- 59.Sorensen RA, Wassarman PM. Relationship between growth and meiotic maturation of the mouse oocyte. Dev Biol. 1976;50:531–536. doi: 10.1016/0012-1606(76)90172-x. [DOI] [PubMed] [Google Scholar]
- 60.Freedman DA, Wu L, Levine AJ. Functions of the MDM2 oncoprotein. Cell Mol Life Sci. 1999;55:96–107. doi: 10.1007/s000180050273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Zhang M, Su YQ, Sugiura K, Xia G, Eppig JJ. Granulosa cell ligand NPPC and its receptor NPR2 maintain meiotic arrest in mouse oocytes. Science. 2010;330:366–369. doi: 10.1126/science.1193573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Tsafriri A, Chun S-Y, Zhang R, Hsueh AJW, Conti M. Oocyte maturation involves compartmentalization and opposing changes of cAMP levels in follicular somatic and germ cells: Studies using selective phosphodiesterase inhibitors. Dev Biol. 1996;178:393–402. doi: 10.1006/dbio.1996.0226. [DOI] [PubMed] [Google Scholar]
- 63.Chatot CL, Ziomek CA, Bavister BD, Lewis JL, Torres I. An improved culture medium supports development of random-bred 1-cell mouse embryos in vitro. J Reprod Fertil. 1989;86:679–688. doi: 10.1530/jrf.0.0860679. [DOI] [PubMed] [Google Scholar]
- 64.Svoboda P, Stein P, Schultz RM. RNAi in mouse oocytes and preimplantation embryos: Effectiveness of hairpin dsRNA. Biochem Biophys Res Commun. 2001;287:1099–1104. doi: 10.1006/bbrc.2001.5707. [DOI] [PubMed] [Google Scholar]
- 65.Whitten WK. Nutrient requirements for the culture of preimplantation mouse embryo in vitro. Adv Biosci. 1971;6:129–139. [Google Scholar]
- 66.Aoki F, Worrad DM, Schultz RM. Regulation of transcriptional activity during the first and second cell cycles in the preimplantation mouse embryo. Dev Biol. 1997;181:296–307. doi: 10.1006/dbio.1996.8466. [DOI] [PubMed] [Google Scholar]
- 67.Zeng F, Baldwin DA, Schultz RM. Transcript profiling during preimplantation mouse development. Dev Biol. 2004;272:483–496. doi: 10.1016/j.ydbio.2004.05.018. [DOI] [PubMed] [Google Scholar]
- 68.Laemmli UK. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]