Abstract
We use an integrated approach to understand breast cancer heterogeneity by modeling mRNA, copy number alterations, microRNAs, and methylation in a pathway context utilizing the pathway recognition algorithm using data integration on genomic models (PARADIGM). We demonstrate that combining mRNA expression and DNA copy number classified the patients in groups that provide the best predictive value with respect to prognosis and identified key molecular and stromal signatures. A chronic inflammatory signature, which promotes the development and/or progression of various epithelial tumors, is uniformly present in all breast cancers. We further demonstrate that within the adaptive immune lineage, the strongest predictor of good outcome is the acquisition of a gene signature that favors a high T-helper 1 (Th1)/cytotoxic T-lymphocyte response at the expense of Th2-driven humoral immunity. Patients who have breast cancer with a basal HER2-negative molecular profile (PDGM2) are characterized by high expression of protumorigenic Th2/humoral-related genes (24–38%) and a low Th1/Th2 ratio. The luminal molecular subtypes are again differentiated by low or high FOXM1 and ERBB4 signaling. We show that the interleukin signaling profiles observed in invasive cancers are absent or weakly expressed in healthy tissue but already prominent in ductal carcinoma in situ, together with ECM and cell-cell adhesion regulating pathways. The most prominent difference between low and high mammographic density in healthy breast tissue by PARADIGM was that of STAT4 signaling. In conclusion, by means of a pathway-based modeling methodology (PARADIGM) integrating different layers of molecular data from whole-tumor samples, we demonstrate that we can stratify immune signatures that predict patient survival.
Keywords: functional genomics, integrated molecular data, omics, perturbated pathway
Progression of malignant tumors as well as response to chemotherapy and targeted therapy is increasingly appreciated to be dependent on the immunological makeup of the host as well as modulation of the immune system by therapeutic antibodies. Cancer cells are surrounded by stroma consisting of nonmalignant hematopoietic and mesenchymal cells in the ECM. It has been shown for many tumor types that the presence of a lymphocytic infiltrate in different types of cancers is a positive predictor of clinical outcome and that response to neoadjuvant chemotherapy is increased in tumors with a prominent immune infiltrate (1). Newly developed targeted therapies in breast cancer, such as trastuzumab, as well as in hematological malignancies, such as rituximab and alemtuzumab, have been shown to interact with immunological pathways, having an impact on response and clinical outcome (2). It has been shown that not only the presence but the composition of the lymphocytic infiltrate is of prognostic significance (3). Although chronic inflammation is clearly associated with a protumorigenic phenotype, the adaptive immune response exerts more paradoxical roles during tumor progression. Both CD8+ cytotoxic T cells (CTLs) and natural killer (NK) cells harbor antitumorigenic properties and are the main regulators of tumor immune surveillance (3, 4). Moreover, T-helper 1 (Th1)-differentiated cells also contribute to tumor cell clearance by favoring the differentiation of M1-polarized macrophages, which have tumoricidal properties (5). In contrast, although B cells contribute to antitumor immunity in the acute phase, chronic Th2-driven B-cell signaling is also protumorigenic through its ability to recruit myeloid cells, including monocytes and mast cells, and the polarization of macrophages to an M2 phenotype (6, 7). Based on these studies, the combination of high levels of tumor-associated macrophages, robust Th2 responses, and low CTL/NK cell infiltration was used to create an immune cell signature in breast cancer to predict patient survival (reviewed in 8). Regulatory T cells also promote tumorigenesis and metastasis through their ability to suppress the tumor cytotoxic activity of CTL and NK cells (9).
Whole-genome expression studies have been applied to clinical specimens representing crude tumor and stromal mixtures (10–12) as well as to microdissected fractions of each (13). Laser capture microdissection studies of normal epithelium and stroma from breast reduction mammoplasty or specimens obtained after surgical treatment of patients with breast cancer allowed the creation of an expression-based stromal-derived prognostic predictor (SDPP) that stratifies disease outcome independent of standard clinical prognostic factors (13). The SDPP revealed differential immune responses as well as angiogenic and hypoxic responses, highlighting the importance of stromal biology in tumor progression. In this study, we show that by applying a pathway-based methodology to model the interactions among different layers of molecular data, we can identify tumors with a variety of immune responses by means of the spectrum of cytokines expressed. These profiles can be obtained from whole tumor samples and mirror the leukocyte complexity in the tumors. To characterize each gene's activity level in a tumor in the context of a pathway and associated clinical data, single genes as well as known interactions among genes are studied. We use each gene's integrated pathway levels to identify and classify the tumors directly according to the most deregulated pathways (across molecular data types) and then investigate the relationship of the newly detected clusters with the previously described subclasses of breast cancer at various molecular levels. Furthermore, we show that these cytokine signatures are associated with angiopoietin receptor Tie2-mediated signaling, regulating angiogenesis and the FOXM1 pathway.
Results
Pathway Recognition Algorithm Using Data Integration on Genomic Models for Classification of Invasive Ductal Carcinoma with Prognostic Significance.
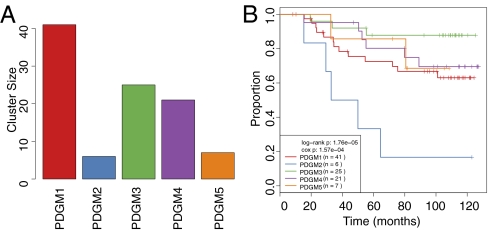
Pathway recognition algorithm using data integration on genomic models (PARADIGM) analyses based on mRNA expression and copy number alterations (CNAs) of the MicroMetastases Project (MicMa) cohort identified the existence of five different clusters (Fig. 1A) and showed that combining mRNA expression and DNA copy number (Fig. 1B) leads to better discrimination of patients with respect to prognosis than any of the molecular levels studied separately (Fig. S1). No overall improvement of the prognostic value of the PARADIGM clusters was observed by adding microRNA expression and DNA methylation to the analysis, although when looking into the specific pathways, both microRNA and methylated genes clearly contributed to the observed pathway aberrations. The pathways whose perturbations most strongly contributed to this classification were those of Tie2 angiopoietin receptor-mediated signaling and, most notably, tumor immunity (T-cell) and interleukin signaling, where nearly every gene or complex in the pathway deviated from normal breast tissue. Most prominently seen were IL-4, IL-6, IL-12, and IL-23 signaling. Other prominent pathways involve endothelins; FoxM1 transcription, which is also deregulated in the ovarian and glioblastoma TCGA (The Cancer Genome Atlas) datasets; and ERBB4, which was also previously found to be deregulated in breast and ovarian cancers (14). Based on this analysis, we have identified the following patient groups with significantly different prognoses, which can be roughly characterized as follows (Figs. 1–3):
PDGM1 = high FOXM1, high Th1/Th2 ratio, basal/ERBB2
PDGM2 = high FOXM1, low Th1/Th2 ratio, basal
PDGM3 = high FOXM1, innate immune genes, macrophage-dominated, luminal
PDGM4 = high ERBB4, low angiopoietin signaling, luminal
PDGM5 = low FOXM1, low macrophage signature, luminal A
Fig. 1.
Distribution of identified PARADIGM clusters and survival in the discovery (MicMa) dataset. (A) Each bar represents the size of each cluster. (B) Survival curves of the MicMa PARADIGM clusters after mapping to the Chin dataset.
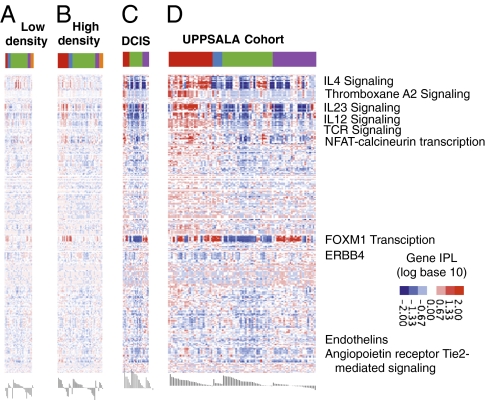
Fig. 3.
Normal to cancer. Heat maps of PARADIGM integrated pathway levels (IPLs) for each dataset. Normal breast, low mammographic density (A); normal breast, high mammographic density (B); DCIS (C); and invasive breast cancer (D; UPPSALA cohort). Each row shows the IPL of a gene or complex across all three cohorts. The colored bar across the top shows the MicMa-derived PARADIGM clusters, as in Fig. 1. Members of pathways of interest are labeled by their pathway. Red represents an activated IPL, and blue represents a deactivated IPL.
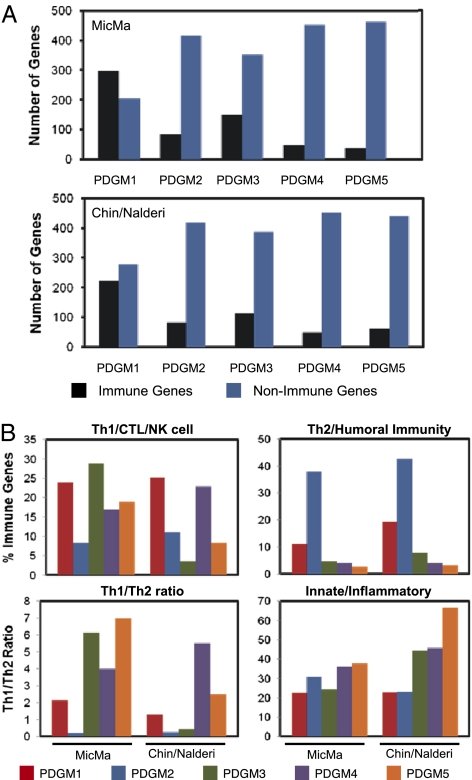
The most significantly differentially expressed pathways and genes that comprise the different clusters are summarized in Dataset S1 for both the discovery dataset (MicMa) and the validation datasets [cohort of Chin et al. (20) and Uppsala University Hospital (UPPSALA) cohort]. To gain more insight into the key pathways that contribute to the differences in patient outcome between the five groups (PDGM1–5), we stratified the top 500 genes and/or pathways identified within each group based on their biological function. We observed that tumors within the PDGM1 group displayed a significantly higher percentage of differentially regulated genes within the immune lineage (44–60%) compared with the PDGM2 group (17%) (Fig. 2A and Dataset S2). This is consistent with the significantly poorer prognosis observed within the PDGM2 patient group relative to PDGM1 patients. However, the remaining groups (PDGM3–5) also exhibit prolonged survival rates despite the fact that the immune response is underrepresented in these tumors (7–30% of all differentially expressed genes) (Fig. 2A). This suggests that both the nature of the immune infiltrate and the frequency with which the immune response is deregulated in tumors may contribute to the differential ability of each signature to predict patient outcome. This is particularly important, given the paradoxical role of the immune response during cancer development as reviewed above. Therefore, we next categorized each differentially expressed immune gene and/or pathway into five specific functional groups: (i) antitumor immunity (Th1 cell, NK cell, CTL, and M1 macrophage), (ii) Th2/humoral immunity, (iii) innate immunity/inflammatory response, (iv) pan-T cell, and (v) pan-leukocyte genes. We observed that all clusters associated with good outcome (PDGM1, PDGM3, PDGM4, and PDGM5) were significantly enriched in genes associated with antitumor immunity (8–29%) at the expense of the Th2/humoral immune response (3–20%) (Fig. 2B and Dataset S3). This is consistent with the higher ratio of Th1/Th2 genes in these groups (1.3- to 7-fold) (Fig. 2B). Conversely, the PDGM2 group, which is associated with poorer outcome, is significantly enriched in Th2/humoral-related genes (24–38%) and only displays a Th1/Th2 ratio of 0.2 (Fig. 2B and Dataset S3). Surprisingly, in the Chin dataset, the PDGM3 cluster was not enriched in an antitumor immune signature as in the MicMa dataset but, instead, displayed elevated expression of innate immune genes, including those specific to macrophages. Interestingly, the innate/inflammatory immune response, which confers protumorigenic properties, is not significantly altered between clusters (Fig. 2B and Dataset S2). Taken together, these analyses suggest that the ratio between Th1- vs. Th2-driven immunity is the best predictor of overall patient survival. Indeed, the cluster of patients presenting with the worst outcome exhibit the highest percentage of Th2 signaling, despite the fact that the overall immune response is significantly underrepresented in this group.
Fig. 2.
(A) High Th1/Th2 ratio distinguishes a good outcome in the PDGM clusters. The top 500 genes within each cluster (PDGM1–5) were classified based on their biological function. The percentage of immune and nonimmune genes in each cluster is shown within the discovery and validation datasets. (B) Immune genes identified in A were divided into functional groups: (i) Th1/CTL/NK cell, (ii) Th2/humoral immunity, and (iii) innate/inflammatory. The percentage of genes in each group, both within the discovery and validation datasets, is shown. The Th1/Th2 ratio for each dataset is also represented.
Within the nonimmune genes, the most differentially expressed ones are related to cell signaling, including tyrosine kinase, PI3K, PKC, and TGF-β pathways (Dataset S2). We also observe an elevated apoptotic signature, particularly within PDGM1 and PDGM3 tumors. Despite the large number of nonimmune genes within these groups, reaching up to 80% of all differentially expressed genes, the immune-related genes are able to discriminate patients based on outcome.
PARADIGM Analysis of Normal Breast Tissue with High and Low Mammographic Density Compared with Breast Carcinoma.
To verify that the observed immune signaling clusters do not exist in normal breast tissue, we examined a cohort of healthy women from the Norwegian breast cancer screening program using the same experimental procedure [PARADIGM analysis of mRNA expression data and array-comparative genomic hybridization (CGH) data]. The cohort was stratified by low and high mammographic density, shown as the top- and bottom-quartile % density (Fig. 3 A and B). This analysis shows that the observed interleukin signaling profiles are indeed very weak or even absent in healthy tissue, although prominently present already in ductal carcinoma in situ (DCIS) (Fig. 3C) and invasive cancers (Fig. 3D), where the PARADIGM signatures showed distorted Tie2 angiopoietin receptor-mediated signaling, tumor immunity (T-cell) and interleukin signaling, endothelins, and FoxM1 and ERBB4 pathways. The most significant difference by a t statistic between low- and high-density breast tissue assessed using PARADIGM was observed for STAT4 signaling (P < 7.678e-05, 2% false discovery rate) (Dataset S4).
PARADIGM for Classification in DCIS.
Given the involvement of the immune response in premalignant hyperplastic glands in mouse models (15) and the massive differences in cytokine signaling in invasive ductal carcinoma (IDC) observed here, we analyzed a dataset of DCIS cases to find whether this strong immune response and interleukin signaling in invasive tumors are present in premalignant (preinvasive) stages as well. DCIS is a noninvasive form of breast cancer in which some lesions are believed to transit rapidly to IDC, whereas others remain unchanged. We have previously studied gene expression patterns of 31 cases of pure DCIS, 36 cases of pure invasive cancer, and 42 cases of mixed diagnosis (invasive cancer with an in situ component) (16) and observed heterogeneity in the transcriptomes in DCIS of high histological grade, identifying a distinct subgroup of DCIS with gene expression characteristics more similar to those of advanced tumors. These groups correlated significantly to the PARADIGM clusters described here (Dataset S4). The heat map of the PARADIGM results for the patients with pure DCIS is shown in Fig. 3C, and that for the entire UPPSALA cohort [including patients with invasive ductal carcinoma (IDC) and invasive lobular carcinoma (ILC)] is shown in Fig. 3D. None of the pure DCIS tumors were of the PDGM2 type, characterized by signaling typical for high macrophage activity (Fig. 3). In agreement, experimental studies have demonstrated that macrophages in primary mammary adenocarcinomas regulate late-stage carcinogenesis as a result of their proangiogenic properties (17, 18), as well as fostering pulmonary metastasis by providing EGF to malignant mammary epithelial cells. Again, among the top deregulated pathways identified by the PARADIGM analysis in DCIS were those involving IL-2, IL-4, IL-6, IL-12, and IL-23 signaling (Dataset S2). In both datasets (DCIS and UPPSALA), as well as in the MicMa cohort (Figs. 3 and 4), cytotoxic T-cell signaling predominated, along with a large number of chemokines that are known to recruit CD8+ T cells. For example, IL-12 is produced by antigen-presenting cells and stimulates IFN-γ production from NK and T cells. IFN-γ signaling, which is elevated in the PDGM1 cluster of the DCIS dataset, is produced from the Th1, NK, and CTL cells and initiates an antitumor immune response through its ability to polarize macrophages to the M1 phenotype. In DCIS, another strong contributor (Dataset S1) was NOX4, an oxygen-sensing NADPH oxidase that resembles the protein responsible for the production of reactive oxygen species (ROS) in neutrophils and granulocytes during a primary immune response. Also, fibronectin 1 (FN1) and PDGF receptor B (PDGFRB) appeared repeatedly together specifically in DCIS, along with COL1A2, IL-12/IL-12 receptor/TYK2/JAK2/SPHK2, ESR1, and KRT14. Phase I clinical trials have shown that the clinical effect of trastuzumab (Herceptin) is potentiated by the coadministration of IL-12 to patients with HER2-overexpressing tumors, and this effect is mediated by the stimulation of IFN-γ production in the NK cells (19).
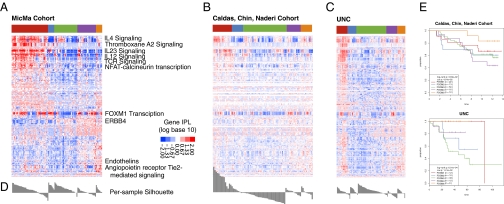
Fig. 4.
Validation datasets. Heat maps of PARADIGM integrated pathway levels (IPLs) for each dataset. Discovery dataset (A; MicMa), validation set 1 (B; Chin), and validation set 2 (C; UNC). Each row shows the IPL of a gene or complex across all three cohorts. The colored bar across the top shows the MicMa-derived PARADIGM clusters, as in Fig. 1. Members of pathways of interest are labeled by their pathway. Red represents an activated IPL, and blue represents a deactivated IPL. (D) Under each heat map, a silhouette plot illustrates the ratio between distance to centroid of belonging cluster vs. distance to all other members. (E) Survival curves.
Validation of the Classification of the PARADIGM Clusters in Three Independent Datasets.
The identification of the PARADIGM clusters (Fig. 4A) was validated in three previously published datasets: (i) the dataset of Chin et al. (20) (Fig. 4B), which had a higher frequency of estrogen receptor (ER)-negative and high-grade tumors compared with the MicMa dataset; (ii) a dataset from the University of North Carolina, Chapel Hill (UNC) (Fig. 4C); and (iii) the dataset of the UPPSALA cohort (16) (Fig. 4D). The validity of the clusters was estimated in two ways: (a) functional, comparing the type of pathways and their general proportions in each cluster in both validation and discovery datasets as described above and shown in Dataset S1, and (b) testing for consistency of clusters across datasets. These approaches showed that the PARADIGM subtypes were found to be reproducible in the other datasets (P = 3e-13, P = 2e-19, P = 5e-9, for the Chin, UNC, and UPPSALA datasets, respectively). Additionally, in each dataset, the native PARADIGM subtypes correlated better to the MicMa mapped types than to any other clinical feature or subtyping derived from the dataset. All clusters were consistent across datasets, except for cluster 3, which was different in both silhouette plot and gene composition across the cohorts. In conclusion, these analyses show that the PARADIGM clusters are largely stable across different breast cancer datasets, except for PDGM 3, which was also characterized by a different immune specter in the discovery and validation datasets.
PARADIGM Clusters and Survival.
The cluster with poor survival in the discovery dataset (MicMa), PDGM2, had poorer survival in the Chin and UNC cohorts (Fig. 4E). The cluster (PDGM5) enriched in luminal A breast cancer had the next best survival in the MicMa dataset and the best survival in the other two validation datasets. PDGM4 had a relative lower survival, which is interesting because it also contains a lot of luminal A samples. The most incoherent cluster was PDGM3, which was characterized by good prognosis in the MicMa series but bad prognosis in the Chin and UNC datasets. This is consistent with the high Th1/Th2 ratio for PDGM3 in the MicMa dataset but the extremely low Th1/Th2 ratio for this cluster in the validation datasets (Fig. 2). In evaluation of the consistency of the clusters, PDGM3 was also demonstrated to be unstable, as shown by the silhouette graph (Fig. 4D). The PDGM3 cluster had a significantly higher proportion of antitumor immune signature in the MicMa dataset and a lower proportion in the Chin dataset, which displayed elevated expression of innate immune genes, including those specific to macrophages, which corresponds to the observed differences in survival. These results suggest that the PARADIGM profiles, although concordant with the molecular profiles derived by mRNA expression alone, may carry additional prognostic information. The results, however, need to be combined with information about treatment, for which there may be differences between Norway, the United Kingdom, and the United States, because the patient's immune constitution has been shown to be critical for the outcome of chemotherapy and targeted treatment.
PARADIGM Clusters and Clinical and Molecular Parameters in DCIS and IDC.
There was a considerable concordance between the immune signaling classification proposed here and the well-established classification by mRNA expression (luminal A, luminal B, basal, ERBB2, normal-like) for both the MicMa and UPPSALA cohorts (Dataset S5). Samples belonging to the basal/ERBB2 clusters were predominantly seen in the PDGM1 and pure basal in PDGM2 (bad prognosis) clusters [i.e., identification of a subset of basal tumors with a very bad prognosis (PDGM2)]. Luminal A samples were predominantly seen in the PDGM3 and PDGM4 (best prognosis in MicMa) clusters. The distribution of the correlations to the centroids for each molecular subtype (luminal A, luminal B, basal, ERBB2, and normal-like) is shown for each PDGM subtype in Fig. S2. The correlations to the basal centroid are positive for PDGM1 and PDGM2 and negative for the other three PDGM clusters. The correlations to the centroids in ERBB2 and to the luminal cluster in PDGM2 are also negative, suggesting that these tumors are of the triple negative-like type. PARADIGM clustering offers a rather significant distinction between two clusters within luminal A breast cancer (PDGM3 and PDGM4) and luminal B breast cancer (PDGM4) clusters. Accordingly, association with the ER status and TP53 mutation status was observed in both datasets. No associations with stage, grade, Ki67, or Her2 staining were observed in DCIS, but an association with Ki67 was seen in IDC (Dataset S5).
PARADIGM Clusters and Fraction of Nontumoral Infiltrating Cells.
To exclude the possibility that the observed classification may be attributable to different proportions of infiltrating noncancerous cells, we tested the degree of nonaberrant cell infiltration in the separate PARADIGM clusters in three of the datasets (MicMa, UNC, and UPPSALA). To obtain a more precise estimation of the nonaberrant cell admixture, we applied our recently developed allele-specific copy number analysis of tumors (ASCAT) tool (21) to obtain nonaberrant cell infiltration estimates from SNP-CGH data of the MicMa, UNC, and UPPSALA datasets (Fig. S3). By means of aggregation of ASCAT profiles across our series, we were able to see a deviation only for the PDGM1 cluster in all three datasets (Fig. S2 and Dataset S6). This cluster had a significantly lower fraction of tumor cells, which may represent a higher proportion of infiltrating lymphocytes, in concordance with the PARADIGM signatures. This cluster is also enriched for ERBB2-like tumors, which are known to be characterized by large leukocyte infiltration. For the remaining PARADIGM clusters, no such association with the amount of infiltrating cells was observed (Dataset S6). This confirms that the PARADIGM clustering is not merely following the amount of infiltrating cells but, instead, is driven by the differences in quality rather than quantity of the infiltrating lymphocytes, as detailed above.
Discussion
To understand how genomic changes disturb distinct biological functions that can explain tumor phenotypes and make tumors vulnerable to targeted treatment, we need an understanding of perturbations at a pathway level. PARADIGM (14) identifies active pathways in subsets of patients that are indistinguishable if genes are studied at a single level. The method uses techniques from probabilistic graphical models to integrate functional genomics data onto a known pathway structure. It has previously been applied to analysis of copy number and mRNA expression data from the TCGA glioblastoma and ovarian datasets (14). PARADIGM analysis can also be used to connect genomic alterations at multiple levels, such as DNA methylation or copy number and mRNA or microRNA expression, and can thus integrate any number of omics layers of data in each individual sample. Although DNA methylation and microRNA expression contribute to the deregulated pathways and seem to have a distinct contribution to the prognosis and molecular profiles of breast cancer in the MicMa cohort (22–24), we did not find improvement in the prognostic value of the PARADIGM clusters by adding these two molecular profile types. One explanation for this is that the prognostic value of microRNA and DNA methylation analyses is recapitulated by mRNA expression because of their high correlation. However, this hypothesis requires further analysis, because the choice of analysis platforms (e.g., limited Illumina 1505 CpG cancer panel for methylation) and our limited knowledge of true microRNA targets may be the factors limiting our ability to measure and effectively model microRNA and DNA methylation information comprehensively.
The integrated analysis of deregulated pathways described here points to the fact that patients with breast cancer who have a basal HER2-negative molecular profile (PDGM2) are characterized by high expression of protumorigenic Th2/humoral-related genes (24–38%) and a low Th1/Th2 ratio. The luminal molecular subtypes are again differentiated by low or high FOXM1 and ERBB4 signaling. In DCIS, the deregulated genes/pathways seem to contribute to functions in the ECM, cell-cell interaction, fibrosis, and keratinization. For instance, FN1 belongs to a family of high-molecular-weight glycoproteins present on cell surfaces, in extracellular fluids, in connective tissues, and in basement membranes. Fibronectins interact with other ECM proteins and cellular ligands, such as collagen, fibrin, and integrins. Fibronectins are involved in adhesive and migratory processes of cells. PDGFR, together with the EGF receptor (EGFR), signals through EGFRs and PDGFRs, which are important receptor tyrosine kinases in breast cancer. Importantly, PDGFR, found here to be overexpressed in certain DCIS cases, is a target of sunitinib (25) and a secondary target of imatinib mesylate (Gleevec) (26). Contrary to the immunostimulatory role of trastuzumab (Herceptin), which is mediated by increased IFN-γ production, imatinib was shown to inhibit IFN-γ production by TCR-activated CD4+ T cells (2). These observations are of interest for our argument to the degree that they illuminate the interaction between growth factor receptors presented on the surface of DCIS and malignant cells and immune constitution. It was shown that stimulatory autoantibodies to PDGFR appeared to trigger an intracellular loop involving Ras, ERK1/ERK2, and ROS that leads to increased type I collagen expression (12). This is in line with COL1A2 expression, also observed as deregulated in DCIS in our study.
In the cluster with the worst prognosis in IDC, PDGM2, IL-4 signaling is strongly deregulated in conjunction with STAT6, which has been shown to prevent growth inhibition in human breast cancer cells (27). Also analyzing the top 500 genes, the IL-4 signature is the highest in PDGM2, which fits with the increased number of B-cell genes. IL-4 signaling has also been shown to promote mast cell activation, which can support greater tumor growth (28). Conversely, in PDGM5, macrophage activation is decreased and NK cell activity is increased because of IL-23 signaling. A cancer-dependent polarization of the immune response toward Th2- and B-cell recruitment on one side and Th1-cell proliferation on the other has been discussed (15). IL-4 is a Th2-derived cytokine that stimulates B-cell differentiation and chronic inflammation in cancer cells. Th2 cells also secrete IL-10, which mediates immunosuppression in these cancers. This immunosuppression was shown to occur predominantly in basal and ERBB2 breast cancers.
FOXM1 Transcription.
FOXM1 is a key regulator of cell cycle progression, and its endogenous FOXM1 expression oscillates according to the phases of the cell cycle. FOXM1, confirmed as a human protooncogene, is found up-regulated in the majority of solid human cancers, including liver, breast, lung, prostate, cervix, uterus, colon, pancreas, brain, and basal cell carcinoma, which is the most common human cancer. FOXM1 is thought to promote oncogenesis through its multiple roles in cell cycle and chromosomal/genomic maintenance (29). Aberrant up-regulation of FOXM1 in primary human skin keratinocytes can directly induce genomic instability in the form of loss of heterozygosity and CNAs (30). A recent report showed that aberrant up-regulation of FOXM1 in adult human epithelial stem cells induces a precancer phenotype in a 3D-organotypic tissue regeneration system, a condition similar to human hyperplasia (31). FoxM1 affects both G1/S and G2/M by regulating the G1/S transition to diminish nuclear levels of the inhibitory cell cycle regulators p21Cip1 and p27Kip1 and by activating Cdc25A transcription. During the G2/M transition, FoxM1 activates transcription of Cdc25B, which is essential for activation of the Cdk1–cyclin-B complex 4, and FoxM1 is also required for the expression of Aurora B kinase and Polo-like kinase (Plk1 and Plk2). In addition, Laoukili et al. (32) identified a transcriptional target of FoxM1, centromere protein F (CENP-F), that is required for proper mitotic spindle checkpoint function and chromosome stability. We see clearly two groups of patients with breast cancer who have high and low activity of this pathway broken mainly according to interleukin signaling activity. Fig. S4 illustrates the opposite activation modus of this pathway (red represents activated vs. blue represents inactivated) for cluster PDGM3, as opposed to the rest of the clusters with worse survival, and the molecular levels that contribute to it (mRNA, CNA, microRNA, or DNA methylation according to the shape of the figures). Down-regulation of MMP2 in PDGM3 is attributable to DNA methylation, whereas in the rest of the tumors, it is attributable to DNA deletion. Of the microRNAs, hsa-let7-b was up-regulated in PDGM3 and down-regulated in the rest of the tumors, complementary to its target, the aurora kinase B (AURKB). Both DNA amplification and mRNA expression were seen as causes of deregulation of expression.
Angiopoietin Receptor Tie2-Mediated Signaling.
The angiopoietin family plays an important role in angiogenesis during the development and growth of human cancers. Ang2 functions in angiogenesis to antagonize Ang1-mediated Tie2 signaling, which is critical for blood vessel maturation and stabilization. Ang2 modulates angiogenesis in a cooperative manner with another important angiogenic factor, VEGF A (33). Recent data suggest more complicated roles for Ang2 in angiogenesis in invasive phenotypes of cancer cells during progression of human cancers (33, 34). Certain angiopoietin family members can activate Tie1 (e.g., Ang1 induces Tie1 phosphorylation in endothelial cells) (34). Tie1 phosphorylation is Tie2-dependent (34). Ang1-mediated AKT and ERK phosphorylation is predominantly Tie2-mediated, and Tie1 down-regulates this pathway. Thus, the main role for Tie1 is to modulate blood vessel morphogenesis because of its ability to down-regulate Tie2-driven signaling and endothelial survival. Both Tie2-mediated signaling as well as VEGF receptor 1 (VEGFR1)- and VEGFR2-mediated signaling and specific signals were observed in this dataset.
ERBB4 contributes to proliferation and cell movement in mammary morphogenesis and the directional cell movements of Erbb4-expressing mammary primordial epithelia while promoting mammary cell fate. Most reports are consistent with a role for ErbB4 in reversing growth stimuli triggered by other ErbB family members during puberty; however, significant association of survival with ERBB4 expression has not been confirmed (35).
Conclusion
Breast cancer development is characterized by significant increases in the presence of both innate and adaptive immune cells, with B cells, T cells, and macrophages representing the most abundant leukocytes present in neoplastic stroma (3). High Ig levels in tumor stroma (and serum) and increased presence of extrafollicular B cells and T-regulatory cells as well as high ratios of CD4/CD8 or Th2/Th1 T lymphocytes in primary tumors or in lymph nodes have been shown to correlate with tumor grade, stage, and overall patient survival (36). Some leukocytes exhibit antitumor activity, including CTLs and NK cells (37). Other leukocytes, such as mast cells, B cells, dendritic cells, granulocytes, and macrophages, exhibit more bipolar roles, through their capacity to either hamper or potentiate tumor progression (38). In this study, we show that the perturbation in the immune response (TCR) and interleukin signaling, including IL-4, IL-6, IL-12, and IL-23, can lead to classification of subclasses with prognostic value. We provide evidence that these events are mirrored in high-throughput molecular data and interfere strongly with molecular subclassification of breast tumors. Given the increasing importance of immune constitution for the success of chemotherapy and targeted treatment, this additional information may prove useful in the clinic in the future.
Materials and Methods
MicMa Molecular Profiling.
The analysis was conducted on approximately 110 breast carcinomas with mRNA expression from an Agilent Technologies whole human genome, 4 × 44-K, one-color oligo array, CNAs from an Illumina Human-1 109-K BeadChip array (21), microRNA from an Agilent Technologies Human miRNA Microarray Kit (V2), and DNA methylation profiling from a GoldenGate Methylation Cancer Panel I (Illumina). Data were analyzed according to the manufacturers’ protocols and as detailed in SI Materials and Methods.
Data Preprocessing, PARADIGM, Clustering, and Survival Analysis.
Briefly, pathway files were from the Pathway Interaction Database (40), and PARADIGM preprocessing and parameters were as previously described. Clustering was performed by HOPACH 2.10 (41) and in R version 2.12 (www.r-project.org/), as were survival analysis and cluster enrichments. Full details are provided in SI Materials and Methods.
Supplementary Material
Acknowledgments
This study was supported by Norwegian Research Council Grants 183621/S10 and 175240/S10 (to V.N.K. and A.-L.B.-D.), Norwegian Cancer Society Grants PK80108002 and PK60287003, and the Radium Hospital Foundation. J.U.-S. acknowledges funding from Canadian Institutes of Health Research Operating Grant MOP-111143 and is also the recipient of a Canadian Institutes of Health Research New Investigator Salary support award. P.V.L. is a postdoctoral researcher of the Research Foundation-Flanders. C.M.P. was supported by the National Cancer Institute Breast Specialized Program of Research Excellence (SPORE) program (Grant P50-CA58223), by National Institutes of Health Grant RO1-CA138255, and by the Breast Cancer Research Foundation.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission. K.P. is a guest editor invited by the Editorial Board.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1108781108/-/DCSupplemental.
References
- 1.Ladoire S, et al. In situ immune response after neoadjuvant chemotherapy for breast cancer predicts survival. J Pathol. 2011;224:389–400. doi: 10.1002/path.2866. [DOI] [PubMed] [Google Scholar]
- 2.Stagg J, et al. Anti-ErbB-2 mAb therapy requires type I and II interferons and synergizes with anti-PD-1 or anti-CD137 mAb therapy. Proc Natl Acad Sci USA. 2011;108:7142–7147. doi: 10.1073/pnas.1016569108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.DeNardo DG, Coussens LM. Inflammation and breast cancer. Balancing immune response: Crosstalk between adaptive and innate immune cells during breast cancer progression. Breast Cancer Res. 2007;9:212. doi: 10.1186/bcr1746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bannard O, Kraman M, Fearon DT. Secondary replicative function of CD8+ T cells that had developed an effector phenotype. Science. 2009;323:505–509. doi: 10.1126/science.1166831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mellor AL, Munn DH. Creating immune privilege: Active local suppression that benefits friends, but protects foes. Nat Rev Immunol. 2008;8:74–80. doi: 10.1038/nri2233. [DOI] [PubMed] [Google Scholar]
- 6.de Visser KE, Korets LV, Coussens LM. De novo carcinogenesis promoted by chronic inflammation is B lymphocyte dependent. Cancer Cell. 2005;7:411–423. doi: 10.1016/j.ccr.2005.04.014. [DOI] [PubMed] [Google Scholar]
- 7.DeNardo DG, et al. CD4(+) T cells regulate pulmonary metastasis of mammary carcinomas by enhancing protumor properties of macrophages. Cancer Cell. 2009;16:91–102. doi: 10.1016/j.ccr.2009.06.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.De Palma M, Lewis CE. Cancer: Macrophages limit chemotherapy. Nature. 2011;472:303–304. doi: 10.1038/472303a. [DOI] [PubMed] [Google Scholar]
- 9.Grivennikov SI, Greten FR, Karin M. Immunity, inflammation, and cancer. Cell. 2010;140:883–899. doi: 10.1016/j.cell.2010.01.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Perou CM, et al. Molecular portraits of human breast tumours. Nature. 2000;406:747–752. doi: 10.1038/35021093. [DOI] [PubMed] [Google Scholar]
- 11.Sørlie T, et al. Gene expression patterns of breast carcinomas distinguish tumor subclasses with clinical implications. Proc Natl Acad Sci USA. 2001;98:10869–10874. doi: 10.1073/pnas.191367098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bergamaschi A, et al. Extracellular matrix signature identifies breast cancer subgroups with different clinical outcome. J Pathol. 2008;214:357–367. doi: 10.1002/path.2278. [DOI] [PubMed] [Google Scholar]
- 13.Finak G, et al. Stromal gene expression predicts clinical outcome in breast cancer. Nat Med. 2008;14:518–527. doi: 10.1038/nm1764. [DOI] [PubMed] [Google Scholar]
- 14.Vaske CJ, et al. Inference of patient-specific pathway activities from multi-dimensional cancer genomics data using PARADIGM. Bioinformatics. 2010;26:i237–i245. doi: 10.1093/bioinformatics/btq182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ursini-Siegel J, et al. Receptor tyrosine kinase signaling favors a protumorigenic state in breast cancer cells by inhibiting the adaptive immune response. Cancer Res. 2010;70:7776–7787. doi: 10.1158/0008-5472.CAN-10-2229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Muggerud AA, et al. Molecular diversity in ductal carcinoma in situ (DCIS) and early invasive breast cancer. Mol Oncol. 2010;4:357–368. doi: 10.1016/j.molonc.2010.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lin EY, Pollard JW. Tumor-associated macrophages press the angiogenic switch in breast cancer. Cancer Res. 2007;67:5064–5066. doi: 10.1158/0008-5472.CAN-07-0912. [DOI] [PubMed] [Google Scholar]
- 18.Lin EY, et al. Vascular endothelial growth factor restores delayed tumor progression in tumors depleted of macrophages. Mol Oncol. 2007;1:288–302. doi: 10.1016/j.molonc.2007.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bekaii-Saab TS, et al. A phase I trial of paclitaxel and trastuzumab in combination with interleukin-12 in patients with HER2/neu-expressing malignancies. Mol Cancer Ther. 2009;8:2983–2991. doi: 10.1158/1535-7163.MCT-09-0820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chin SF, et al. Using array-comparative genomic hybridization to define molecular portraits of primary breast cancers. Oncogene. 2007;26:1959–1970. doi: 10.1038/sj.onc.1209985. [DOI] [PubMed] [Google Scholar]
- 21.Van Loo P, et al. Allele-specific copy number analysis of tumors. Proc Natl Acad Sci USA. 2010;107:16910–16915. doi: 10.1073/pnas.1009843107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Naume B, et al. Presence of bone marrow micrometastasis is associated with different recurrence risk within molecular subtypes of breast cancer. Mol Oncol. 2007;1:160–171. doi: 10.1016/j.molonc.2007.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Enerly E, et al. miRNA-mRNA integrated analysis reveals roles for miRNAs in primary breast tumors. PLoS ONE. 2011;6:e16915. doi: 10.1371/journal.pone.0016915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rønneberg JA, et al. Methylation profiling with a panel of cancer related genes: Association with estrogen receptor, TP53 mutation status and expression subtypes in sporadic breast cancer. Mol Oncol. 2011;5:61–76. doi: 10.1016/j.molonc.2010.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Fratto ME, et al. New perspectives: Role of sunitinib in breast cancer. Clin Ter. 2010;161:475–482. [PubMed] [Google Scholar]
- 26.Weigel MT, et al. In vitro effects of imatinib mesylate on radiosensitivity and chemosensitivity of breast cancer cells. BMC Cancer. 2010;10:412. doi: 10.1186/1471-2407-10-412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gooch JL, Christy B, Yee D. STAT6 mediates interleukin-4 growth inhibition in human breast cancer cells. Neoplasia. 2002;4:324–331. doi: 10.1038/sj.neo.7900248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.de Visser KE, Eichten A, Coussens LM. Paradoxical roles of the immune system during cancer development. Nat Rev Cancer. 2006;6:24–37. doi: 10.1038/nrc1782. [DOI] [PubMed] [Google Scholar]
- 29.Wonsey DR, Follettie MT. Loss of the forkhead transcription factor FoxM1 causes centrosome amplification and mitotic catastrophe. Cancer Res. 2005;65:5181–5189. doi: 10.1158/0008-5472.CAN-04-4059. [DOI] [PubMed] [Google Scholar]
- 30.Teh MT, Gemenetzidis E, Chaplin T, Young BD, Philpot MP. Upregulation of FOXM1 induces genomic instability in human epidermal keratinocytes. Mol Cancer. 2010;9:45. doi: 10.1186/1476-4598-9-45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gemenetzidis E, et al. Induction of human epithelial stem/progenitor expansion by FOXM1. Cancer Res. 2010;70:9515–9526. doi: 10.1158/0008-5472.CAN-10-2173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Laoukili J, et al. FoxM1 is required for execution of the mitotic programme and chromosome stability. Nat Cell Biol. 2005;7:126–136. doi: 10.1038/ncb1217. [DOI] [PubMed] [Google Scholar]
- 33.Hashizume H, et al. Complementary actions of inhibitors of angiopoietin-2 and VEGF on tumor angiogenesis and growth. Cancer Res. 2010;70:2213–2223. doi: 10.1158/0008-5472.CAN-09-1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yuan HT, et al. Activation of the orphan endothelial receptor Tie1 modifies Tie2-mediated intracellular signaling and cell survival. FASEB J. 2007;21:3171–3183. doi: 10.1096/fj.07-8487com. [DOI] [PubMed] [Google Scholar]
- 35.Sundvall M, et al. Role of ErbB4 in breast cancer. J Mammary Gland Biol Neoplasia. 2008;13:259–268. doi: 10.1007/s10911-008-9079-3. [DOI] [PubMed] [Google Scholar]
- 36.Bates GJ, et al. Quantification of regulatory T cells enables the identification of high-risk breast cancer patients and those at risk of late relapse. J Clin Oncol. 2006;24:5373–5380. doi: 10.1200/JCO.2006.05.9584. [DOI] [PubMed] [Google Scholar]
- 37.Dunn GP, Koebel CM, Schreiber RD. Interferons, immunity and cancer immunoediting. Nat Rev Immunol. 2006;6:836–848. doi: 10.1038/nri1961. [DOI] [PubMed] [Google Scholar]
- 38.de Visser KE, Coussens LM. The inflammatory tumor microenvironment and its impact on cancer development. Contrib Microbiol. 2006;13:118–137. doi: 10.1159/000092969. [DOI] [PubMed] [Google Scholar]
- 39.Wiedswang G, et al. Detection of isolated tumor cells in bone marrow is an independent prognostic factor in breast cancer. J Clin Oncol. 2003;21:3469–3478. doi: 10.1200/JCO.2003.02.009. [DOI] [PubMed] [Google Scholar]
- 40.Schaefer CF, et al. PID: the Pathway Interaction Database. Nucleic Acids Res. 2009;37(Database issue):D674–D679. doi: 10.1093/nar/gkn653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.van der Laan MJ, Pollard KS. A new algorithm for hybrid hierarchical clustering with visualization and the bootstrap. J Stat Plan Inference. 2003;117:275–303. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.