Abstract
Luminal breast cancers express estrogen (ER) and/or progesterone (PR) receptors and respond to hormone therapies. Basal-like “triple negative” cancers lack steroid receptors but are cytokeratin (CK) 5-positive and require chemotherapy. Here we show that more than half of primary ER+PR+ breast cancers contain an ER−PR−CK5+ “luminobasal” subpopulation exceeding 1% of cells. Starting from ER+PR+ luminal cell lines, we generated lines with varying luminal to luminobasal cell ratios and studied their molecular and biological properties. In luminal disease, luminobasal cells expand in response to antiestrogen or estrogen withdrawal therapies. The phenotype and gene signature of the hormone-resistant cells matches that of clinical triple negative basal-like and claudin-low disease. Luminobasal cell expansion in response to hormone therapies is regulated by Notch1 signaling and can be blocked by γ-secretase inhibitors. Our data establish a previously unrecognized plasticity of ER+PR+ luminal breast cancers that, without genetic manipulation, mobilizes outgrowth of hormone-resistant basal-like disease in response to treatment. This undesirable outcome can be prevented by combining endocrine therapies with Notch inhibition.
Keywords: basal breast cancer, endocrine therapy, estrogen receptor, Slug
Gene expression profiling distinguishes among intrinsic breast cancer subtypes (1). “Luminal” subtypes account for >70% of tumors (2) and are characterized by presence of estrogen (ER) and/or progesterone (PR) receptors, expression of cytokeratin (CK) 8/18, low or no epidermal growth factor receptors 1 (EGFR) or 2 (HER2), and absence of CK5. Although most luminal cancers have a favorable prognosis and respond to antiestrogens or aromatase inhibitors, development of hormone resistance associated with tumor recurrence is common (2). The “basal-like” cancers are classified as “triple negative” (TN) if they lack ER, PR, and HER2 but retain EGFR and/or CK5, or “5 negative phenotype” if they lack all five markers (3). A recently identified “claudin-low” subtype is enriched for mesenchymal and stem cell markers (4). Basal-like and claudin-low cancers are hormone-independent, characterized by brief disease-free survival, a high proliferative index and poor histologic grade (4, 5), and require aggressive chemotherapy.
Molecular profiling of whole-tumor extracts is clinically and biologically informative. However, most tumors contain heterogeneous cell types (6), in which case subtype classification can be biased toward the most abundant subpopulations. That minor populations matter is evident from studies demonstrating the tumor-initiating potential of rare cells (7, 8). How this intratumoral cellular heterogeneity arises continues to be debated, although both genomic mutations and epigenetic changes seem to define the ultimate mosaic (9). Further, recent evidence suggests that the molecular subtypes reflect the cellular hierarchy of the normal breast. Thus, the claudin-low signature matches a mammary stem cell profile (4); the basal-like signature is consistent with that of committed luminal progenitor cells; and luminal signatures resemble those of differentiated luminal epithelial cells (4, 10). Given this hierarchy, it is reasonable to speculate that differentiated ER+PR+ luminal tumors might contain cell subpopulations with undifferentiated or progenitor properties.
The latest guidelines for immunohistochemical (IHC) quantitation of steroid receptors in luminal breast cancers recommend that ER and PR assays be considered positive if at least 1% of nuclei are stained (11). This raises questions about the properties and function of the remaining ER−PR− cells in luminal disease. We recently identified an ER−PR−CK5+ cell subpopulation in luminal breast cancer models that express basal-like markers (12). These cells, referred to here as “luminobasal” cells, have tumor-initiating potential. Similar cells are up-regulated in patients whose luminal tumors develop resistance to chemo- and hormone therapies (13).
We now show in an analysis of 72 primary breast cancers that more than half of ER+PR+ tumors contain an ER−PR−CK5+ luminobasal subpopulation exceeding 1% of cells. Starting from ER+PR+ luminal cell lines, we generated lines with varying luminal/luminobasal cell ratios and studied their molecular and biological properties. In these models of luminal disease, luminobasal cells expand in response to estrogen withdrawal or antiestrogen therapies. The phenotype and gene signature of the luminobasal cells matches that of TN basal-like and claudin-low tumors of patients. We show that luminobasal cell expansion in response to hormone therapies is regulated by Notch1 signaling and can be prevented by γ-secretase inhibitors (GSIs) of Notch. Our data establish a previously unrecognized plasticity of ER+PR+ luminal breast cancers that, without genetic manipulation, mobilizes outgrowth of hormone-resistant basal-like disease in response to common endocrine therapies. We propose that this undesirable outcome can be avoided by combining GSIs with endocrine therapies.
Results
ER−PR−CK5+ Luminobasal Cell Subpopulation in ER+PR+ Luminal Breast Cancers.
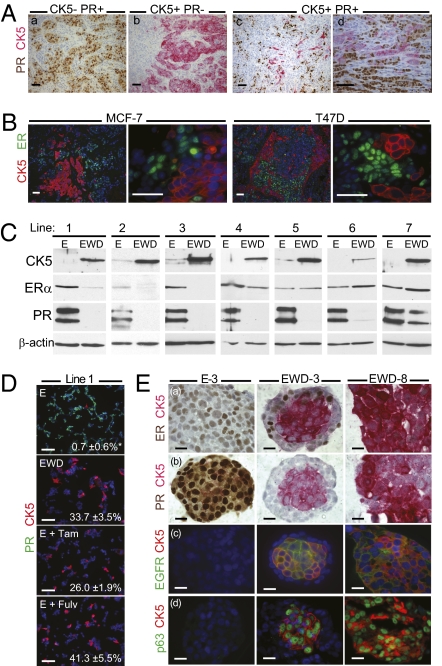
Although >70% of breast cancers are ER+ and/or PR+ and classified as luminal, such tumors often harbor poorly characterized ER−PR− cells. We previously identified a luminal tumor cell subpopulation that lacks steroid receptors but expresses CK5 (12). To quantify these “luminobasal” cells in luminal disease, 72 primary tumors of known ER status (14) were subjected to dual PR and CK5 analysis by IHC (Fig. 1A). A cutoff of ≥1% PR+ nuclei or ≥1% CK5+ cytoplasm was considered positive in accord with accepted thresholds (11). Seventy-six percent of the tumors were ER+ and/or PR+ (Fig. 1Aa); the rest were receptor-negative, of which ≈45% were CK5+ (Fig. 1Ab). Among the 55 luminal tumors, 44.7% were CK5− by the above definition. The remaining 55.3% were PR+ but contained at least 1% CK5+ luminobasal cells (Fig. 1 Ac and Ad), >97% of which lacked PR. In some tumors, luminobasal cells constituted at least one-third of the population. Thus, more than half of all luminal breast cancers contain a substantial luminobasal cell subpopulation lacking steroid receptors that would be resistant to hormone therapies (13) and could usurp the tumor in response to such treatments. We therefore sought to develop luminobasal cell models to study their regulation and implication for development of hormone resistance.
Fig. 1.
Luminobasal cells in clinical breast cancers and derivation of models with varying luminal/luminobasal ratios. (A) Primary tumors from 72 patients (14) were stained by IHC for CK5 (pink) and PR (brown). Four representative tumors are (a) pure luminal, (b) pure basal, and (c and d) mixed luminal/luminobasal. (Scale bars, 50 μm.) (B) CK5 (red) and ER (green) dual IHC in xenografts grown from MCF-7 or T47D luminal breast cancer cells. Cells (106) were injected into mammary glands of ovx'd mice supplemented with E. (Scale bars, 20 μm.) (C) Western blots of CK5, ER, and PRA or PRB isoforms in seven cell lines grown independently from T47D xenografts. Cells were cultured >45 d in 1 nM E or EWD media (Table S1). (D) Line 1 cells stained by ICC for CK5 (red) and PR (green) after culturing for >45 d in E or EWD media, or in E plus 100 nM Tam or Fulv. Percentage CK5+ luminobasal content is shown. *P < 0.01. (Scale bars, 20 μm.) (E) 3D colonies of lines E-3 (pure luminal), EWD-3 (mixed), and EWD-8 (pure luminobasal) were sectioned and stained by dual IHC for CK5 (pink) and luminal markers ER (a), PR (b, brown); or basal markers EGFR (c), p63 (d, green). (Scale bars, 20 μm.)
Estrogen Deprivation Expands a Latent Luminobasal-Cell Subpopulation in Luminal Tumor Xenografts.
Under estrogen (E)-treated or E-withdrawn (EWD) conditions in vitro, ER+PR+CK5− breast cancer cell lines (MCF-7, BT-474, and T47D) rarely or never contain ER−PR−CK5+ luminobasal cells (Fig. S1 A, B, and C). However, when grown as orthotopic solid tumors in immune compromised mice, clusters of CK5+ luminobasal cells are found interspersed among the expected ER+ luminal cells (Fig. 1B). Analogous to the clinical samples (Fig. 1A), the CK5+ luminobasal cell nuclei are ER−PR−. We used the solid tumors to generate cell lines that enable study of luminobasal cells in vitro. For this, cells passaged in mice were returned to culture in the presence of E or under EWD conditions. Specifically, eight T47D sublines were generated (Table S1) and designated for the in vitro hormone condition and xenograft of origin (i.e., xenograft line 2 yielded lines E-2 and EWD-2).
Lines 1–7, exposed to hormones in vivo, had <1% luminobasal cells in vitro in the presence of E, which rose to 20–50% in response to EWD (Fig. 1C and Table S1). The T47D tumor-derived lines grew well in E with the luminobasal subpopulation at <1%. For example, dual CK5/PR immunocytochemistry (ICC) (Fig. 1D) illustrates that line 1 E-treated cells contained 0.7% CK5+ cells. However, under EWD conditions, or if antiestrogens tamoxifen (Tam) or fulvestrant (Fulv) were added to E, the cells underwent crisis. When proliferation resumed, 33.7% (EWD), 26% (E plus Tam), or 41.3% (E plus Fulv) of the surviving cells were luminobasal (Fig. 1D). Thus, both EWD and antiestrogens expand the luminobasal subpopulation. The regulatory role for E in this transition was confirmed by switching line 2 between E and EWD conditions (Fig. S1D). Line 8, grown in the mouse without hormone, was enriched for luminobasal cells (90–100%) in vitro with E or under EWD conditions (EWD-8; Fig. 1E). EWD also promoted luminobasal-cell expansion from tumor-derived BT474 cells, but not MCF-7 cells (Fig. S1 B and C). These results reflect development of hormone resistance in patients (13) and attest to the fidelity of the models.
In 3D cultures, line EWD-3, a mixture of luminal and luminobasal cells, developed remarkable colonies with the luminobasal CK5+ cells clustered in a central core, surrounded by the ER+ luminal subpopulation (Fig. 1Ea). Despite ER positivity, the EWD-3 luminal outer ring cells were PR−, apparently due to absence of E (Fig. 1Eb), confirming the E-dependence of this important marker of ER activity in breast cancers (15). The E-3, EWD-3, and EWD-8 colonies were further evaluated for established markers of basal and luminal breast epithelium (16, 17). The basal markers CK5, EGFR, and p63 colocalized to the luminobasal EWD-3 core and EWD-8 colonies (Fig. 1E) and were absent in the luminal E-3 and EWD-3 ring cells. Luminal markers MUC1, GATA3, FOXA1, and CK18 (Fig. S2A) were restricted to E-3 cells and EWD-3 ring cells and absent in CK5+ luminobasal cells. Unlike PR (Fig. 1E), these luminal markers are E-independent. Luminobasal cells lack myoepithelial lineage markers CK14, α-smooth muscle actin, and vimentin (17) and are HER2− (Fig. S2B). We conclude that despite their luminal origin, luminobasal cells resemble TN basal-like breast cancer cells and expand within luminal tumors if E signaling is prevented.
Despite Their Luminal Cell Derivation, the Gene Signature of Luminobasal Cells Matches TN Basal-Like and Claudin-Low Breast Cancer Subtypes.
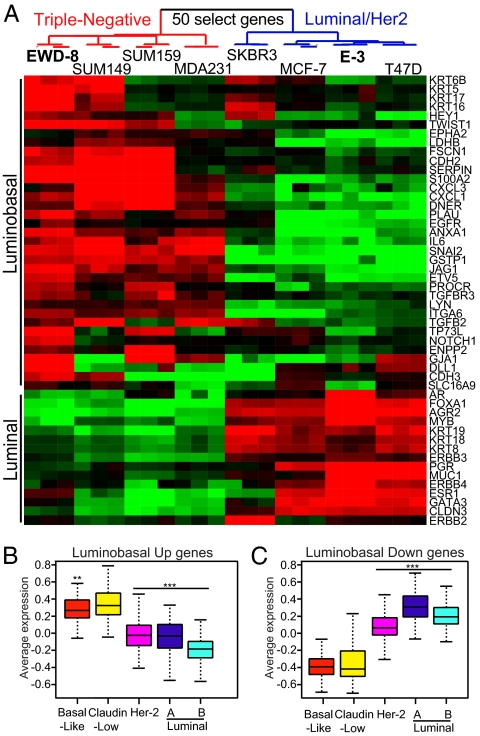
Expression of basal-like markers suggested that luminobasal cells had launched a gene expression program associated with basal-like TN disease. To verify this, pure luminal (E-3) and luminobasal (EWD-8) lines were expression profiled, and gene sets were compared with those of established breast cancer cell lines and primary breast cancers. Analysis of 16,934 genes showed that EWD-8 cells clustered with the parental T47D and E-3 lines (Fig. S3A), confirming the luminal and T47D origin of the cells. Their T47D origin and genetic similarity were also demonstrated by short tandem repeat analysis, karyotyping, and genotype array analysis (SI Materials and Methods). Using a two-class significance analysis of microarrays and a false discovery rate of 0 (SI Materials and Methods), 1,298 genes were identified that defined a “luminobasal signature” (Dataset S1). Using a 50-gene subset (Fig. 2A) or all 1,298 signature genes (Fig. S3B), cluster analysis demonstrated that EWD-8 clusters with TN MDA231, SUM159, and SUM149 cells, whereas E-3 clusters with luminal MCF-7 and parental T47D breast cancer cells. Both were distinct from the HER2+ SKBR3 cell line. These results were confirmed (Fig. S3C) using the larger 51-cell-line database of Neve et al. (18). We conclude that the T47D-derived, EWD-8 line has acquired a basal-like TN profile, while retaining the broader signature of its luminal origins.
Fig. 2.
Despite their luminal derivation, luminobasal-rich EWD-8 cells cluster with TN cell lines and TN basal-like and claudin-low breast cancers. (A) Cluster analysis of 50 “luminobasal signature” genes up-regulated (red) or down-regulated (green) in EWD-8 cells compared with other breast cancer cell lines. (B and C) ANOVA box plots of the average gene expression values for 511 up-regulated (B) or 453 down-regulated (C) genes in the luminobasal signature, compared with genes in the 516 breast tumor dataset. Sixty-six percent of tumors fall within the interquartile range (IQR; colored box); the bar indicates the median value; whiskers show the range within subtype and are 1.5*IQR. **P < 0.001, ***P < 0.0001.
We next asked how the luminobasal signature of EWD-8 relates to subtype classification of clinical breast cancers. Using a combined dataset of 516 primary tumors (Materials and Methods), we find that overexpressed (Fig. 2B) or underexpressed (Fig. 2C) luminobasal genes matched those of basal-like or claudin-low tumors and differed from luminal or HER2+ tumors. This pattern was confirmed using the independent NKI265 dataset (Fig. S3D). In related analyses, the luminobasal signature genes correctly clustered breast tumors according to their luminal, HER2, or basal-like TN status in the 516 and NKI265 datasets (Fig. S4). The similarity of gene expression patterns between our lines and primary tumors of patients suggests that malignant breast cells follow a conserved genetic path to establish a luminal vs. basal-like fate that we can now model.
Luminobasal Cells Exhibit Aggressive, E-Independent Growth in vivo.
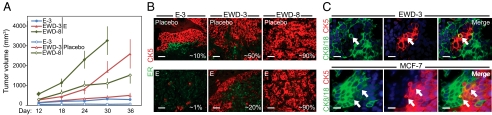
Compared with luminal breast cancers, TN basal-like tumors are more aggressive (3). To compare the aggressiveness of luminal and luminobasal cells, we injected our cell lines into mammary glands of immune-compromised ovariectomized (ovx'd) mice supplemented with placebo or E-releasing pellets (Fig. 3A). The study was terminated early because EWD-3 and EWD-8 tumors were highly aggressive, reaching allowable size limits in less than half the time of parental T47D cells. Tumor growth was proportional to the luminobasal population, with EWD-8 the fastest and capable of rapid and extensive E-independent growth (Fig. 3A). Nonetheless, E further stimulated growth of all three lines, perhaps reflecting ER expression below the detection threshold of our standard IHC assays.
Fig. 3.
Luminobasal-enriched xenografts grow without E in vivo and contain rare cells dual positive for luminal and luminobasal markers. (A) Lines E-3, EWD-3, and EWD-8 were implanted into mammary glands of ovx'd nude mice supplemented with placebo or E. Tumor size was measured weekly and volumes calculated. n = 4 or 5 mice per line per treatment. (B) Tumors from A were paraffin-embedded and stained by dual immunofluorescence for CK5 (red) and ER (green). Percentage CK5+ luminobasal content is shown. (Scale bars, 50 μm.) (C) E-treated xenografts grown from EWD-3 cells and MCF-7 cells were stained for CK8/18 (green) and CK5 (red). Arrows show rare cells (yellow) dual positive for CK18 and CK5. (Scale bars, 20 μm.)
Luminobasal cells were rare (≈1%) in E-treated E-3 tumors but represented ≈10% of cells in EWD control tumors (Fig. 3B). E-treated EWD-3 tumors contained ≈20% luminobasal cells, which increased to ≈50% in EWD mice. EWD-8 tumors were >90% luminobasal regardless of treatment (Fig. 3B). These studies confirm in vitro (Fig. 1 C and D) and patient data (13) reporting luminobasal cell expansion in EWD conditions. Analysis of T47D and MCF-7 tumors confirmed that most tumor cells exhibit either basal or luminal features (Fig. 3C and Fig. S5). However, rare cells (<1%) failed this clear-cut distinction and instead were dual (yellow) CK8/18+CK5+ (Fig. 3C, arrows) or GATA3+CK5+ (Fig. S5); a pattern attributed to luminal progenitor cells (10). We conclude that our models exhibit the extensive cellular heterogeneity observed in clinical samples of luminal breast cancers (Fig. 1A), and that the luminobasal cell subpopulation behaves aggressively in vivo and survives and expands under prolonged E-suppressive conditions.
Targeting the Luminobasal Population Through Notch.
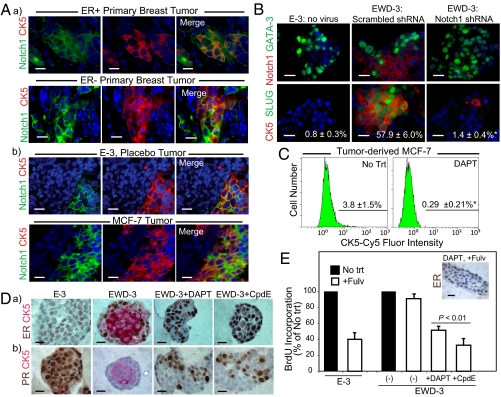
The Notch pathway has been implicated in regulating lineage and differentiation decisions in the normal breast (19, 20). We noted many Notch pathway genes in the luminobasal gene signature, including Notch1 receptor and multiple Notch ligands (Fig. S6A). To analyze this, sections of primary breast tumors (Fig. 4Aa) and xenografted luminal tumors (Fig. 4Ab) were costained for CK5 (red) and Notch1 (green). Notch1 was specifically coexpressed with CK5 in the luminobasal cell subpopulation of both patient samples and xenografts (Fig. 4A) and 3D colonies (Fig. S6B). Consistent with these results, Notch1 transcripts were elevated in basal-like/claudin-low, ESR1− tumors (Fig. S6C), and strong constitutive Notch-dependent transcriptional activity was detected in the Notch1 expressing EWD-8 luminobasal cells (Fig. S6D). This transcriptional activity could be specifically suppressed by three different GSIs: DAPT, compound E (CpdE), and XIX (all defined in Table S1 legend) (Fig. S6D). Notch1 also plays a functional role in the expansion of CK5+Slug+GATA3− luminobasal cells during EWD. The low number (0.8%) of luminobasal cells in the E-3 line rose to 57.9% under EWD conditions plus a scrambled shRNA, but this expansion was significantly suppressed (1.4%) by a Notch1-targeted shRNA (Fig. 4B).
Fig. 4.
Expansion of luminobasal cells by EWD involves Notch signaling. (A) Paraffin sections of (a) primary breast cancers or (b) E-3 and MCF-7 xenografts were dual-stained for Notch1 (green) and CK5 (red). (Scale bars, 20 μm.) (B) EWD-3 cells were transduced with lentivirus encoding an shRNA targeting Notch1 or a scrambled control shRNA and propagated for >45 d under EWD conditions. Paraffin sections of 3D colonies were stained by dual IHC for CK5, Notch1, Slug, and GATA-3 for comparison with luminal E-3 colonies. Numbers are percentage luminobasal content. *P < 0.01. (C) Flow analysis of CK5 expression in tumor-isolated MCF-7 cells treated 14 d with 2 μM DAPT. *P < 0.01. (D) Line 3 was grown >45 d in E or EWD media, or EWD supplemented with the GSIs DAPT or CpdE. 3D colonies were stained by IHC for CK5 (red) and (a) ER or (b) PR (brown). (E) Colonies grown as in D were treated 7 d with 100 nM Fulv. Cell proliferation was assessed by IHC staining for BrdU-positive nuclei. Inset: IHC for ER. Receptors are degraded by Fulv.
GSIs Maintain a Luminal Cell Phenotype Responsive to Endocrine Therapies.
GSIs are in early clinical trials as monotherapy for breast cancers because of the putative role of Notch in breast stem cells (21). We speculated that additionally, GSIs could target the Notch1-expressing luminobasal subpopulation. To test this, tumor-derived MCF-7 cells were treated 2 wk with DAPT, and the CK5+ fraction was assessed by flow cytometry (Fig. 4C) and ICC (Fig. S7A). DAPT significantly decreased MCF-7 luminobasal cell number from 3.8% to 0.29% (Fig. 4C). To show whether GSIs prevent expansion of the CK5+ luminobasal cell subpopulation in response to endocrine therapies, line 3 was propagated directly from tumors in the presence of E or EWD, or under EWD conditions supplemented with DAPT or CpdE (Fig. 4D). Notably, the GSIs had no quantifiable impact on growth of the EWD cultures, which entered and exited crisis synchronously. In the presence of E (E-3), pure ER+PR+ luminal colonies were observed. EWD yielded mixed colonies (EWD-3), with expansion of luminobasal core cells readily apparent. However, under the same EWD conditions, DAPT and CpdE suppressed outgrowth of the luminobasal subpopulation, allowing maintenance of ER (Fig. 4Da). Interestingly, E-independent, heterogeneous PR expression was also observed, demonstrating PR up-regulation by E-independent pathways (Fig. 4Db). Similar results were observed in line 7 cells (Fig. S7B).
Preservation of the ER+ luminal phenotype by Notch1-shRNA or GSIs under EWD conditions was confirmed by absence of luminobasal markers (CK5, Notch1, Slug, EGFR) and maintenance of luminal markers (GATA3, FOXA1; Fig. 4B and Fig. S7C). To show whether the ER+ cells were responsive to hormone therapies under these conditions, E-3, EWD-3, and EWD-3/GSI lines were treated with Fulv to degrade the ER protein (22) (Fig. 4E, Inset). Proliferation was quantified by BrdU incorporation. In E-dependent, pure luminal E-3 controls, Fulv suppressed growth, as expected. The mixed luminal/luminobasal EWD-3 line was relatively insensitive to Fulv, but Fulv sensitivity was maintained by DAPT or CpdE cotreatment (Fig. 4E). We propose that for patients afflicted with luminal disease, hormone therapies in combination with a Notch inhibitor to maintain the ER+ luminal state could provide greater benefit than E/ER-targeted therapies alone.
Discussion
Luminal breast cancers, the majority of tumors diagnosed in women, are now defined by presence of at least 1% ER+ or PR+ cells (11). This raises questions about the biology and origin of the remaining, often the bulk, receptor-negative cells in luminal disease. We used established ER+PR+ breast cancer lines to develop models of luminal tumor heterogeneity that address these questions. These models generate not only the classic ER+PR+CK5− luminal cell subpopulation, but also a subset of ER+PR− cells (Fig. 4Db) that often puzzle clinicians, as well as the ER−PR−CK5+ luminobasal cells (Fig. 1 Ac and Ad) resembling TN basal-like disease we describe here. That established luminal cell lines (23) retain the ability to yield such extensive cellular heterogeneity without genetic manipulation is evidence of their plasticity. Here we characterize the ER−PR−CK5+ luminobasal subpopulation found in more than half of luminal breast tumors.
Luminobasal Cells and Hormone Resistance.
We demonstrate that ER− luminobasal cells survive and expand in the absence of E (Fig. 3B), the principal therapeutic modality for patients with luminal disease. EWD clearly favors outgrowth of cells with TN profiles, consistent with observations in primary tumors (24), including patients treated with neoadjuvant Tam or aromatase inhibitors (13). We propose that therapies targeting the proliferative actions of E can have the unintended consequence of promoting outgrowth of an ER− luminobasal cell subpopulation. Other studies support a role for ER− cells in hormone resistance. Creighton et al. (24) assessed expression profiles of ER+ breast cancers before and after letrozole therapy. In 18 tumors defined as ER+ by IHC and candidates for aromatase inhibition, only 9 had pretreatment gene expression profiles consistent with luminal disease, demonstrating that ER alone is insufficient to define this tumor subtype (4). After letrozole, four of the nine luminal tumors changed to a claudin-low profile; a switch also observed in patients treated with neoadjuvant docetaxel (24).
The global gene signature of luminobasal-rich EWD-8 cells clusters with the parental luminal T47D cells (Fig. S3A), attesting to the overall cytogenetic stability and identity of the two lines. However, the luminobasal-specific signature clusters with genes expressed in TN basal-like cell lines, and with claudin-low or basal-like breast cancers (Fig. 2A), suggesting that under therapeutic pressures a restricted (≈7.7%), highly conserved gene expression program transitions luminal cells to the more aggressive basal-like/claudin-low state. We speculate that this is epigenetically driven because the same plasticity is demonstrable in multiple independent lines and primary tumors (Fig. 1A) and because the cell types show significant genetic similarity by karyotype and SNP profiling (SI Materials and Methods). Without complete genome sequencing information, however, positive selection for cells with de novo mutations cannot be entirely excluded. Because they have not been genetically manipulated, these lines should be important models for defining mechanistically, the pathways taken by breast cancer cells as they transition among various intrinsic subtypes.
Origins of the Luminobasal Phenotype and Notch Signaling.
It has been suggested (4) that the breast cancer subtypes segregate along a mammary epithelial cell differentiation hierarchy, with claudin-low tumors resembling mammary “stem cells” (MaSC), basal-like tumors resembling “luminal progenitors,” and luminal A and B tumors resembling “mature” luminal cells. Accordingly, we find that the luminobasal signature is most similar to that of the MaSC fraction identified in the normal breast by Lim et al. (10). Claudin-low tumors exhibit features (4) analogous to cells that have undergone dedifferentiation through an epithelial to mesenchymal transition (EMT) (25). Two prominent mesenchymal transcription factors, Slug (SNAI2; 100.2-fold) and Twist1 (10.6-fold), are up-regulated in the luminobasal signature. Expression levels of Slug and Twist1 are highest in claudin-low tumors of the 516-tumor dataset, and both genes identify letrozole-resistant disease (24). Slug overexpression (Fig. 4B) is particularly interesting considering recent reports of Slug accumulation in basal-like tumors associated with BRCA1 mutations (26). Notch signaling may also foster Slug accumulation, because coexpression of Notch1 and Jag1 leads to Slug overexpression in breast cancer cells (27), and Notch1 depletion prevented Slug accumulation in our studies (Fig. 4B).
Our model describes a Notch-dependent mechanism enabling the balance between luminal and basal-like disease. That luminobasal cells group together both clinically (Fig. 1A and Fig. 4A) and experimentally (Fig. 1E) could be explained by feedback loops activated between adjacent cells expressing Notch receptors and ligands (Fig. S6A). Other indicators that the luminal/luminobasal cell ratio is controlled by Notch signaling are as follows. (i) Notch1 receptor is detectable in cells expressing basal markers (Fig. 4 and Fig. S6). (ii) Notch transcriptional activity is elevated under EWD conditions in parallel with expansion of luminobasal cell number (Fig. S6D). Analogous activation of Notch in basal-like breast cancers (28, 29) and E suppression of Notch in ER+ luminal breast cancer cells (30) have been reported. (iii) GSIs or anti-Notch1 shRNA maintains a luminal state (Fig. 4D and Fig. S7) despite E deprivation. Thus, an ER+ luminal phenotype is preserved in the face of EWD if Notch remains suppressed.
The origin of luminobasal cells in luminal tumors may be analogous to the hierarchy in the epithelial compartment of the normal breast, where cells that express basal features coexist with committed luminal cells (17). Recent reports on BRCA1-related basal-like disease conclude that basal tumors originate from a luminal, not a basal, progenitor cell (10, 26, 31). Luminobasal cells could also emerge from direct conversion or reprogramming of the luminal cell state, a plasticity reminiscent of the EMT (26). Our ability to derive a cell line (EWD-8) that fits the core basal description (ER−PR−CK5+EGFR+; Fig. 1E) and gene expression profile, exhibits E-independent, aggressive tumor growth (Fig. 3) but is syngeneic with a classic, established luminal ER+PR+CK5− parental cell line, supports a luminal origin of basal-like cells. Rare cells in both T47D and MCF-7 tumors that are double positive for luminal and basal markers (Fig. 3C and Fig. S5) are interesting in that regard. We speculate that luminobasal cells sit at the nexus of the transition between luminal and basal-like cancers. In luminal disease, the balance between luminal and luminobasal cells is reversible and regulatable by E and Notch signaling. However, once transition to the basal-like/claudin-low state is complete (EWD-8 line) we find the phenotype to be irreversible. Neither exposure to E nor GSIs can restore the luminal state under these conditions (Fig. 3B), analogous to failed attempts to restore a luminal phenotype to TN cells by targeting MAPK (32).
Conclusions
The implications of our data are grave for the development of resistance to ER-targeted endocrine therapies. They predict that antiestrogens or aromatase inhibitors will raise the number of ER− cells in resistant or recurrent disease, as reported in a small neoadjuvant study (13). We suggest that outgrowth of the luminobasal cell subpopulation is undesirable and demonstrate that combination therapies targeting Notch with GSIs to maintain cells in an ER+ luminal state, while targeting ER or E with endocrine therapies, could be highly effective. With regard to Notch, combination therapy is essential because GSI monotherapy would not suppress tumor growth or kill cells. Additionally, better outcomes could be achieved if patients with ER+ tumors that contain luminobasal cell subpopulations were prospectively identified. Considering our initial data (Fig. 1A), more than half of patients with luminal disease fit into that category, but ER and PR IHC is insufficient to detect these tumors.
Materials and Methods
Experimental methods are detailed in SI Materials and Methods. Methods include xenografts and generation of tumor-derived lines, gene expression profiling and genetic analyses, primary breast cancer data, and statistical analyses. A complete list of reagents and antibodies is provided in Table S2.
Supplementary Material
Acknowledgments
We thank the University of Colorado Cancer Center's Core facilities; Jessica Rice, B.A., and Dr. Christopher D. Coldren for help with the genotyping array analysis; and Dr. Marileila Garcia for karyotype analysis. This study was supported by National Research Service Award F32 CA142096 (to J.M.H.); US Department of Defense Grant BC085270 (to J.C.H.); National Institutes of Health Grant RO1 CA026869-31, the National Foundation for Cancer Research, the Breast Cancer Research Foundation, and the Avon Foundation for Women (to K.B.H.); and the Helsinki University Central Hospital Research Foundation.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission. K.P. is a guest editor invited by the Editorial Board.
Data deposition: The gene expression microarray reported in this paper has been deposited with the Gene Expression Omnibus (GEO) database, www.ncbi.nlm.nih.gov/geo (accession no. GSE31870).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1106509108/-/DCSupplemental.
References
- 1.Perou CM, et al. Molecular portraits of human breast tumours. Nature. 2000;406:747–752. doi: 10.1038/35021093. [DOI] [PubMed] [Google Scholar]
- 2.Dunnwald LK, Rossing MA, Li CI. Hormone receptor status, tumor characteristics, and prognosis: A prospective cohort of breast cancer patients. Breast Cancer Res. 2007;9:R6. doi: 10.1186/bcr1639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cheang MC, et al. Basal-like breast cancer defined by five biomarkers has superior prognostic value than triple-negative phenotype. Clin Cancer Res. 2008;14:1368–1376. doi: 10.1158/1078-0432.CCR-07-1658. [DOI] [PubMed] [Google Scholar]
- 4.Prat A, et al. Phenotypic and molecular characterization of the claudin-low intrinsic subtype of breast cancer. Breast Cancer Res. 2010;12:R68. doi: 10.1186/bcr2635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rakha EA, Reis-Filho JS, Ellis IO. Basal-like breast cancer: a critical review. J Clin Oncol. 2008;26:2568–2581. doi: 10.1200/JCO.2007.13.1748. [DOI] [PubMed] [Google Scholar]
- 6.Park SY, Gönen M, Kim HJ, Michor F, Polyak K. Cellular and genetic diversity in the progression of in situ human breast carcinomas to an invasive phenotype. J Clin Invest. 2010;120:636–644. doi: 10.1172/JCI40724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bonnet D, Dick JE. Human acute myeloid leukemia is organized as a hierarchy that originates from a primitive hematopoietic cell. Nat Med. 1997;3:730–737. doi: 10.1038/nm0797-730. [DOI] [PubMed] [Google Scholar]
- 8.Al-Hajj M, Wicha MS, Benito-Hernandez A, Morrison SJ, Clarke MF. Prospective identification of tumorigenic breast cancer cells. Proc Natl Acad Sci USA. 2003;100:3983–3988. doi: 10.1073/pnas.0530291100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Shackleton M, Quintana E, Fearon ER, Morrison SJ. Heterogeneity in cancer: Cancer stem cells versus clonal evolution. Cell. 2009;138:822–829. doi: 10.1016/j.cell.2009.08.017. [DOI] [PubMed] [Google Scholar]
- 10.Lim E, et al. kConFab Aberrant luminal progenitors as the candidate target population for basal tumor development in BRCA1 mutation carriers. Nat Med. 2009;15:907–913. doi: 10.1038/nm.2000. [DOI] [PubMed] [Google Scholar]
- 11.Hammond ME, et al. American Society of Clinical Oncology/College of American Pathologists guideline recommendations for immunohistochemical testing of estrogen and progesterone receptors in breast cancer. J Clin Oncol. 2010;28:2784–2795. doi: 10.1200/JCO.2009.25.6529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Horwitz KB, Dye WW, Harrell JC, Kabos P, Sartorius CA. Rare steroid receptor-negative basal-like tumorigenic cells in luminal subtype human breast cancer xenografts. Proc Natl Acad Sci USA. 2008;105:5774–5779. doi: 10.1073/pnas.0706216105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kabos P, et al. Cytokeratin 5 positive cells represent a steroid receptor negative and therapy resistant subpopulation in luminal breast cancers. Breast Cancer Res Treat. 2011;128:45–55. doi: 10.1007/s10549-010-1078-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Joensuu K, Heikkilä P, Andersson LC. Tumor dormancy: Elevated expression of stanniocalcins in late relapsing breast cancer. Cancer Lett. 2008;265:76–83. doi: 10.1016/j.canlet.2008.02.022. [DOI] [PubMed] [Google Scholar]
- 15.Horwitz KB, McGuire WL. Predicting response to endocrine therapy in human breast cancer: A hypothesis. Science. 1975;189:726–727. doi: 10.1126/science.168640. [DOI] [PubMed] [Google Scholar]
- 16.Nakshatri H, Badve S. FOXA1 in breast cancer. Expert Rev Mol Med. 2009;11:e8. doi: 10.1017/S1462399409001008. [DOI] [PubMed] [Google Scholar]
- 17.Stingl J, Raouf A, Emerman JT, Eaves CJ. Epithelial progenitors in the normal human mammary gland. J Mammary Gland Biol Neoplasia. 2005;10:49–59. doi: 10.1007/s10911-005-2540-7. [DOI] [PubMed] [Google Scholar]
- 18.Neve RM, et al. A collection of breast cancer cell lines for the study of functionally distinct cancer subtypes. Cancer Cell. 2006;10:515–527. doi: 10.1016/j.ccr.2006.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Raouf A, et al. Transcriptome analysis of the normal human mammary cell commitment and differentiation process. Cell Stem Cell. 2008;3:109–118. doi: 10.1016/j.stem.2008.05.018. [DOI] [PubMed] [Google Scholar]
- 20.Bouras T, et al. Notch signaling regulates mammary stem cell function and luminal cell-fate commitment. Cell Stem Cell. 2008;3:429–441. doi: 10.1016/j.stem.2008.08.001. [DOI] [PubMed] [Google Scholar]
- 21.Dontu G, et al. Role of Notch signaling in cell-fate determination of human mammary stem/progenitor cells. Breast Cancer Res. 2004;6:R605–R615. doi: 10.1186/bcr920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.McClelland RA, et al. Effects of short-term antiestrogen treatment of primary breast cancer on estrogen receptor mRNA and protein expression and on estrogen-regulated genes. Breast Cancer Res Treat. 1996;41:31–41. doi: 10.1007/BF01807034. [DOI] [PubMed] [Google Scholar]
- 23.Ross DT, Perou CM. A comparison of gene expression signatures from breast tumors and breast tissue derived cell lines. Dis Markers. 2001;17:99–109. doi: 10.1155/2001/850531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Creighton CJ, et al. Residual breast cancers after conventional therapy display mesenchymal as well as tumor-initiating features. Proc Natl Acad Sci USA. 2009;106:13820–13825. doi: 10.1073/pnas.0905718106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mani SA, et al. The epithelial-mesenchymal transition generates cells with properties of stem cells. Cell. 2008;133:704–715. doi: 10.1016/j.cell.2008.03.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Proia TA, et al. Genetic predisposition directs breast cancer phenotype by dictating progenitor cell fate. Cell Stem Cell. 2011;8:149–163. doi: 10.1016/j.stem.2010.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Leong KG, et al. Jagged1-mediated Notch activation induces epithelial-to-mesenchymal transition through Slug-induced repression of E-cadherin. J Exp Med. 2007;204:2935–2948. doi: 10.1084/jem.20071082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lee CW, et al. A functional Notch-survivin gene signature in basal breast cancer. Breast Cancer Res. 2008;10:R97. doi: 10.1186/bcr2200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Dong Y, Li A, Wang J, Weber JD, Michel LS. Synthetic lethality through combined Notch-epidermal growth factor receptor pathway inhibition in basal-like breast cancer. Cancer Res. 2010;70:5465–5474. doi: 10.1158/0008-5472.CAN-10-0173. [DOI] [PubMed] [Google Scholar]
- 30.Rizzo P, et al. Cross-talk between notch and the estrogen receptor in breast cancer suggests novel therapeutic approaches. Cancer Res. 2008;68:5226–5235. doi: 10.1158/0008-5472.CAN-07-5744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Molyneux G, et al. BRCA1 basal-like breast cancers originate from luminal epithelial progenitors and not from basal stem cells. Cell Stem Cell. 2010;7:403–417. doi: 10.1016/j.stem.2010.07.010. [DOI] [PubMed] [Google Scholar]
- 32.Bayliss J, Hilger A, Vishnu P, Diehl K, El-Ashry D. Reversal of the estrogen receptor negative phenotype in breast cancer and restoration of antiestrogen response. Clin Cancer Res. 2007;13:7029–7036. doi: 10.1158/1078-0432.CCR-07-0587. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.