Abstract
Hendra virus (HeV) continues to cause morbidity and mortality in both humans and horses with a number of sporadic outbreaks. HeV has two structural membrane glycoproteins that mediate the infection of host cells: the attachment (G) and the fusion (F) glycoproteins that are essential for receptor binding and virion-host cell membrane fusion, respectively. N-linked glycosylation of viral envelope proteins are critical post-translation modifications that have been implicated in roles of structural integrity, virus replication and evasion of the host immune response. Deciphering the glycan composition and structure on these glycoproteins may assist in the development of glycan-targeted therapeutic intervention strategies. We examined the site occupancy and glycan composition of recombinant soluble G (sG) glycoproteins expressed in two different mammalian cell systems, transient human embryonic kidney 293 (HEK293) cells and vaccinia virus (VV)-HeLa cells, using a suite of biochemical and biophysical tools: electrophoresis, lectin binding and tandem mass spectrometry. The N-linked glycans of both VV and HEK293-derived sG glycoproteins carried predominantly mono- and disialylated complex-type N-glycans and a smaller population of high mannose-type glycans. All seven consensus sequences for N-linked glycosylation were definitively found to be occupied in the VV-derived protein, whereas only four sites were found and characterized in the HEK293-derived protein. We also report, for the first time, the existence of O-linked glycosylation sites in both proteins. The striking characteristic of both proteins was glycan heterogeneity in both N- and O-linked sites. The structural features of G protein glycosylation were also determined by X-ray crystallography and interactions with the ephrin-B2 receptor are discussed.
Keywords: glycopeptides, Hendra virus (HeV), mass spectrometry, N- and O-linked glycosylation
Introduction
N-linked glycosylation of proteins occurs on the amino acid sequon NXS/T, whereas O-glycosylation is on serine and/or threonine residues. Biosynthesis of both N- and O-glycans involves the attachment of sugar monosaccharides in a sequential enzymatic process occurring in the endoplasmic reticulum and Golgi apparatus. The process involves attachment, trimming and modification of the glycan precursor to form the final branched glycan structures that are classified as high-mannose complex or hybrid glycans. These are both species- and tissue-specific. Glycosylated proteins often exhibit both macroheterogeneity (variable occupancy of glycosylation sites) and microheterogeneity (variable degree of type, trimming and elongation of the glycan attached to one glycosylation site) adding to their complexity. Glycosylation plays an important part in a number of biological roles, including cell–cell communication and interaction, development, morphogenesis, embryogenesis, immunity, protein folding, transport, blood protein modification, mucosal development and differentiation (Lis and Sharon 1993; Varki 1993; Weerapana and Imperiali 2006; Gao and Mehta 2007). One of the most intriguing areas of the glycan–protein interaction is in the union between glycan and virus. Many pathogenic microorganisms have long glycan structures on their surface, with viral carbohydrates being shown to play a crucial role in active transmission into host cells as well as providing a mechanism for host immune system evasion (Vigerust and Shepherd 2007; Li et al. 2008).
The Paramyxoviridae family includes a number of highly contagious human and animal pathogens such as measles virus, mumps virus, Newcastle disease virus, Sendai virus, human respiratory syncytial virus, Hendra virus (HeV) and Nipah virus (NiV). HeV and NiV are the members of the Henipavirus genus, a new class of virus in the Paramyxoviridae family (Chua et al. 2000; Wang et al. 2000). HeV has been isolated only in the Australian states of Queensland and New South Wales (Epstein et al. 2006). HeV is a zoonotic biosafety level 4 pathogen that continues to cause morbidity and mortality in both humans and horses with a number of sporadic outbreaks. The last outbreak resulting in a human fatality was recorded in 2009.
HeV possesses two structural membrane proteins, the attachment glycoprotein (G) and the fusion glycoprotein (F) that together mediate virus entry and infection of host cells and facilitate the receptor binding and virion-host cell membrane fusion processes, respectively. The henipavirus G glycoprotein mediates binding to the host cell via ephrin-B2 or -B3 receptors (Bonaparte et al. 2005; Carter et al. 2005; Eaton et al. 2005; Mungall et al. 2006; Bowden et al. 2008), which in turn triggers the F glycoprotein-mediated, pH-independent, membrane fusion between the virus and its host cell (Lamb et al. 2006; Bossart and Broder 2009). Prediction software maps seven potential N-linked glycosylation sites for the HeV G protein. In recent work, the crystal structure of the unliganded six-bladed β-propeller domain of HeV G was compared with the previously reported structure of the HeV G protein in complex with its cellular receptor, ephrin-B2 (Bowden et al. 2011). This study also reported the N-linked glycosylation of this domain.
There are currently a number of expression systems used to produce human or humanized recombinant proteins all with advantages and disadvantages relating to expression, yield and display of the correct post-translational modifications (Patel et al. 1992; Brooks 2004). In this study, we analyzed two forms of recombinant soluble G (sG) protein of HeV. The first sG was expressed in HeLa cells, using a recombinant vaccinia virus (VV) system, and a second sG was produced in stably transfected human embryonic kidney 293 (HEK293) cells using the phCMV-1 vector. The respective glycoproteins are referred to as VV-sG and HEK293-sG. We compared the two expression systems, not only to determine the glycan composition of each sG protein, but also to examine how each expression system would affect the resultant glycosylation profiles. Structural implications of the HeV attachment glycoprotein in its interaction with the ephrin-B2 receptor are also discussed.
In order to investigate the glycan composition and site occupancy, we expressed the HeV sG protein in two mammalian cell expression systems and used one- and two-dimensional gel electrophoresis (1-DE and 2-DE), and lectin-binding assays to determine the carbohydrates present in each system. Furthermore, mass spectrometry was employed for the investigation of the heterogeneity and complexity of both N- and O-linked glycans. We report here the population of N-glycans observed and the first observation of O-glycosylation of these viral glycoproteins.
Results
Electrophoretic mobilities and lectin affinity analysis
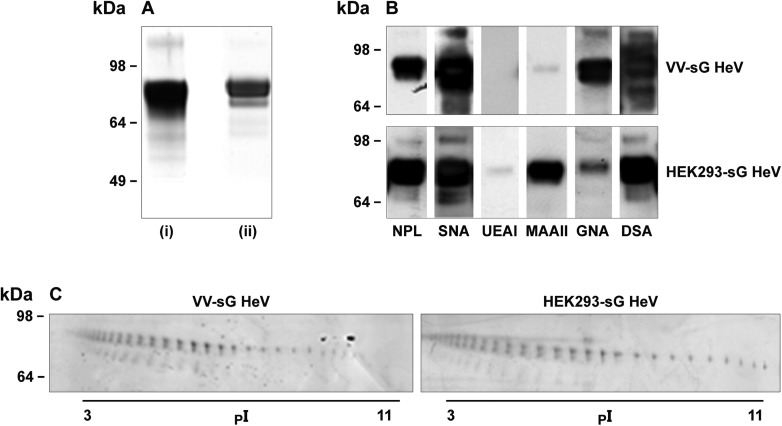
The 1-DE separation of VV-sG and HEK293-sG was performed under reducing conditions (Figure 1A). Both proteins had molecular mass of ∼70–80 kDa, which correlated well with the experimentally determined molecular weight of a previous study (Bossart et al. 2005).
Fig. 1.
Electrophoretic mobility, heterogeneity and lectin affinities of sG glycoproteins. (A) VV-sG HeV (i) and HEK293-sG HeV (ii) separated by SDS–PAGE on 4–12% gels and (B) transferred to PVDF membranes for detection with biotinylated lectins: NPL, SNA, UEAI, MAAII, GNA and DSA. Lectin affinities are shown for each of the sG glycoproteins. (C) VV-sG or HEK293-sG (15 μg) was separated by 2-DE using 7 cm (pH 3–11) non-linear immobilized pH gradient (IPG) strips and run on 4–20% SDS–PAGE gels. The protein spots were visualized by silver staining.
The carbohydrates present on the glycoproteins were initially detected using a series of lectins that specifically bind to different glycan components (Figure 1B). To further characterize the type of oligosaccharides present, the viral proteins were also subjected to various glycosidase treatments prior to separation and the results are summarized in Table I. Datura stramonium agglutinin (DSA) showed very strong binding to both proteins, indicating that β1,4-N-acetylglucosamine (GlcNAc) is an abundant component of the glycan structure and treatment with peptide-N-glycosidase F (PNGase-F), an enzyme that cleaves between the innermost GlcNAc and the Asn residue resulted in a complete loss of reactivity. Treatment with endoglycosidase H (Endo H), which cleaves off asparagine-linked mannose-rich oligosaccharides between two GlcNAc residues proximal to the asparagine, and neuraminidase, which cleaves the glycosidic linkages of neuraminic acid/sialic acid, resulted in no change to DSA binding, thus indicating that a large proportion of the glycan population was of the complex and/or high mannose type. Galanthus nivalis agglutinin (GNA) and Narcissus pseudonarcissus lectin (NPL) binding to both proteins were observed confirming the presence of an α1,3-mannose component. Maackia amurensis lectin II (MAAII) reacted strongly only with HEK293-sG, suggesting the presence of sialic acid in an α2,3-configuration on this protein (Geisler and Jarvis 2011). Sambucus nigra agglutinin (SNA) binds preferentially to sialic acid attached to terminal galactose in an α2,6- and to a lesser degree in an α2,3-configuration and this lectin reacted strongly with both proteins. Ulex europaeus agglutinin I (UEAI), a lectin that shows specificity toward α-fucose (Fuc), only displayed reactivity toward the HEK293-sG glycoprotein.
Table I.
Observed reactivity of biotinylated lectins toward VV-sG and HEK293-sG proteins
| Lectin | Major specificity | VV-sG |
HEK293-sG |
||||||
|---|---|---|---|---|---|---|---|---|---|
| Untreated | PNGase-F | Endo H | NA | Untreated | PNGase-F | Endo H | NA | ||
| DSA | β1,4 GlcNAc | +++ | − | +++ | +++ | +++ | − | +++ | +++ |
| GNA | α1,3 mannose | +++ | − | + | +++ | ++ | − | + | ++ |
| MAAII | α2,3 NeuAc | + | − | − | − | +++ | − | − | − |
| SNA | α2,6 NeuAc | +++ | − | − | +++ | − | − | ||
| UEAI | α-Fuc | − | − | − | + | − | + | ||
| NPL | α1,6 mannose | +++ | − | +++ | − | ||||
The viral glycoproteins were transferred to PVDF membrane for visualization following electrophoresis. Reactivity was assessed against untreated sG proteins and after PNGase-F treatment, Endo H treatment or neuraminidase (NA) treatment. DSA, Datura stramonium agglutinin; GNA, Galanthus nivalis agglutinin; MAAII, Maackia amurensis agglutinin II; NPL, Narcissus pseudonarcissus lectin; SNA Sambucus nigra agglutinin; UEAI, Ulex europaeus agglutinin I.
The 2-DE separation of both VV-sG and HEK293-sG glycoproteins (Figure 1C) revealed a train of spots with slightly descending molecular mass and pI values ranging from 3 to 11. It was initially speculated that the more acidic isoforms of this train of spots (pI 3–5) would contain glycans with a higher content of sialic acid. In addition to the increased molecular masses, the glycans may represent more diverse tri- and tetra-antennary N-linked oligosaccharides. Therefore, the more basic isoforms (pI 5–9) would contain low levels of or no sialic acid, and as they exhibited lower molecular masses, a larger proportion of glycans with simple branching would be present. A doublet of spots is observed that can be explained as two distinct isoforms by Edman sequencing (Supplementary data, Figure S1). The larger isoform arises from the expected cleavage site of the immunoglobulin κ-chain leader sequence, and the smaller isoform from a truncation of an additional 11 amino acids. A third train of spots at lower molecular weight can be seen suggesting further truncation, but this minor species could not be isolated in sufficient quantity for sequence analysis.
Identification of glycopeptides and determination of glycan composition
In silico proteolytic digestion of the viral glycoproteins was performed to determine the mass of proteolytic products required for characterization of all potential glycopeptides. Trypsin generated fragments varied in size from 9 to 25 amino acids (a size amenable to tandem mass spectrometric analysis; MS/MS) with the exception of a single peptide of 57 amino acids in length. The identification of this potential glycopeptide (containing the NXT/S motif at 302N) required the use of chymotrypsin to reduce the peptide size to a mass amenable to MS/MS analysis and sequencing. The theoretical peptides that would be produced from the tryptic and chymotryptic digests are listed in Table II.
Table II.
Proteolytic products of sG constructs containing potential sites of N- and O-linked glycosylation
| # | Mass (mono)a | Start | End | Glycopeptide | RT (min)b | Enzyme | Glycan type |
|---|---|---|---|---|---|---|---|
| HT1 | 1052.50 | 63 | 71 | (R) STTM*QNYTR (T) | 27.2 | T | N |
| HT2 | 1852.88 | 151 | 165 | (K) IHEC(Cam)NISC(Cam)PNPLPFR (E) | 40.1 | T | N |
| HT4 | 2024.88 | 369 | 384 | (R) TEFQYNDSNC(Cam)PIIHC(Cam)K (Y) | 36.8 | T | N |
| HT5 | 2263.19 | 412 | 431 | (K) YNLSLGGDIILQFIEIADNR (L) | ND | T | N |
| HT6 | 1698.83 | 477 | 491 | (R) NNSVISRPGQSQC(Cam)PR (F) | 26.8 | T | N |
| HT7 | 2767.40 | 513 | 537 | (R) LNWVSAGVYLNSNQTAENPVFAVFK (D) | 61.4 | T | N |
| HT8 | 1002.53 | 72 | 80 | (R) TTDNQALIK (E) | 26.6 | T | O |
| HT9 | 1286.68 | 81 | 91 | (K) ESLQSVQQQIK (A) | 33.8 | T | O |
| HT10 | 1341.75 | 92 | 104 | (K) ALTDK^IGTEIGPK (V) | 34.7 | T | O |
| HT11 | 813.46 | 97 | 104 | (K) IGTEIGPK (V) | 27.7 | T | O |
| HT12 | 2199.24 | 105 | 126 | (K) VSLIDTSSTITIPANIGLLGSK (I) | 56.7 | T | O |
| HT13 | 1634.77 | 127 | 141 | (K) ISQSTSSINENVNDK (C) | 28.2 | T | O |
| HT14 | 787.43 | 401 | 406 | (K) SHYILR (S) | 24.9 | T | O |
| HC1 | 1955.92 | 289 | 306 | (Y) TLC(Cam)AVSHVGDPILNSTSW (T) | 53.5 | C | N |
Potential N-linked tryptic glycopeptides are referred to as HT1–HT7 and O-linked glycopeptides as HT8 to HT14. HC1 is a chymotryptic glycopeptide. HT3 was over 40 amino acids in length and as unsuitable for MS/MS analysis has not been included in the table. K^ refers to a missed cleavage peptide. M* refers to the decomposition of carboxymethylated methionine, a known modification observed after treatment with iodoacetamide and exposure to acid (Jones et al. 1994). The retention times (RTs) were observed after PNGase-F treatment of VV-sG HeV.
aMonoisotopic mass of each peptide.
bRetention time determined for deglycosylated peptides after PNGase-F treatment; ND, not determined.
Enzymatic treatment with PNGase-F was performed to remove all N-linked glycan structures and identify the deglycosylated peptides. The retention time for each observed deglycosylated peptide is listed in Table II and was used for prediction of the relative retention times of the glycosylated forms. Deamidation of the Asn residue at the glycan attachment site is seen following PNGase-F treatment and was used as a preliminary indicator of potential glycosylation at each site.
Two complementary liquid chromatography (LC)-MS/MS workflows were employed on a linear ion trap mass spectrometer (QTRAP) in order to capture glycopeptide identities. Using an information-dependent acquisition (IDA) experiment, both peptides and glycopeptides were analyzed in a single LC-MS/MS run. Precursor ion scanning experiments were also employed to specifically target the glycopeptides present in the digests by triggering MS/MS upon the observation of the carbohydrate-specific ions, such as the oxonium ion of m/z 366 [hexose (Hex)-N-acetylhexosamine (HexNAc)]. In parallel, an IDA workflow on a quadrupole-time-of-flight (Q-TOF) instrument was also employed. The product ion spectra of all glycopeptides showed a very characteristic pattern. There were intense oligosaccharide-derived peaks of m/z 204 (HexNAc), 366 (Hex-HexNAc), 186 (HexNAc-H2O) and 168 (HexNAc-2H2O), and if present, 163 (Hex), 292 (NeuAc) and 274 (NeuAc-H2O). The presence of these diagnostic ions allowed glycopeptide precursor ions to be easily distinguished from unmodified peptide precursors. Overall, glycan composition and heterogeneity at specific N-linked sites in both VV-sG and HEK293-sG glycoproteins were determined and results are presented in Table III and Supplementary data, Tables SIA and B and SIIA and B.
Table III.
Characterization of N-linked glycopeptides of VV-sG and HEK293-sG HeV
| Peptide (glycan) | VV-sG | HEK293-sG |
|---|---|---|
| HT1 (68N), STTM*QNYTR | (GlcNAc)2(Man)3 + (HexNAc)1 | ND |
| HT2 (155N), IHECNISCPNPLPFR | (GlcNAc)2(Man)3 + (Hex)2 | (GlcNAc)2(Man)3 + (Hex)2 |
| (GlcNAc)2(Man)3 + (Hex)3 | (GlcNAc)2(Man)3 + (HexNAc)3(Hex)2 | |
| (GlcNAc)2(Man)3 + (Hex)4 | (GlcNAc)2(Man)3 + (HexNAc)2(Fuc)1 | |
| (GlcNAc)2(Man)3 + (Hex)5 | (GlcNAc)2(Man)3 + (HexNAc)2(Hex)1(Fuc)1 | |
| (GlcNAc)2(Man)3 + (HexNAc)2(Hex)2 | (GlcNAc)2(Man)3 + (HexNAc)2(Hex)2(Fuc)1 | |
| (GlcNAc)2(Man)3 + (HexNAc)3(Hex)2 | (GlcNAc)2(Man)3 + (HexNAc)3(Hex)1(Fuc)1 | |
| (GlcNAc)2(Man)3 + (HexNAc)2(Hex)2(Fuc)1 | (GlcNAc)2(Man)3 + (HexNAc)3(Hex)2(NeuAc)1 | |
| (GlcNAc)2(Man)3 + (HexNAc)2(Hex)2(NeuAc)1 | (GlcNAc)2(Man)3 + (HexNAc)2(Hex)2(Fuc)1(NeuAc)1 | |
| (GlcNAc)2(Man)3 + (HexNAc)3(Hex)2(NeuAc)1 | (GlcNAc)2(Man)3 + (HexNAc)2(Hex)2(Fuc)1(NeuAc)2 | |
| (GlcNAc)2(Man)3 + (HexNAc)3(Hex)2(NeuAc)2 | (GlcNAc)2(Man)3 + (HexNAc)3(Hex)1(Fuc)1(NeuAc)1 | |
| (GlcNAc)2(Man)3 + (HexNAc)2(Hex)2(Fuc)1(NeuAc)1 | (GlcNAc)2(Man)3 + (HexNAc)3(Hex)2(Fuc)1 | |
| (GlcNAc)2(Man)3 + (HexNAc)2(Hex)2(Fuc)1(NeuAc)2 | (GlcNAc)2(Man)3 + (HexNAc)3(Hex)3(Fuc)1 | |
| (GlcNAc)2(Man)3 + (HexNAc)3(Hex)3(Fuc)1(NeuAc)1 | ||
| (GlcNAc)2(Man)3 + (HexNAc)3(Hex)3(Fuc)1(NeuAc)2 | ||
| (GlcNAc)2(Man)3 + (HexNAc)4(Hex)4(Fuc)1(NeuAc)3 | ||
| HT4 (374N), TEFQYNDSNCPIIHCK | GlcNAc)2(Man)3 + (HexNAc)2(Hex)2 | (GlcNAc)2(Man)3 + (HexNAc)1(Hex)1(Fuc)1 |
| GlcNAc)2(Man)3 + (HexNAc)2(Hex)2(Fuc)1 | (GlcNAc)2(Man)3 + (HexNAc)2(Hex)2(Fuc)1 | |
| GlcNAc)2(Man)3 + (HexNAc)2(Hex)2(Fuc)1(NeuAc)1 | (GlcNAc)2(Man)3 + (HexNAc)2(Hex)2(Fuc)1(NeuAc)1 | |
| GlcNAc)2(Man)3 + (HexNAc)2(Hex)2(Fuc)1(NeuAc)2 | (GlcNAc)2(Man)3 + (HexNAc)2(Hex)2(Fuc)1(NeuAc)2 | |
| (GlcNAc)2(Man)3 + (HexNAc)3(Hex)1(Fuc)1 | ||
| (GlcNAc)2(Man)3 + (HexNAc)3(Hex)1(Fuc)1(NeuAc)1 | ||
| (GlcNAc)2(Man)3 + (HexNAc)3(Hex)2(Fuc)1 | ||
| (GlcNAc)2(Man)3 + (HexNAc)3(Hex)2(Fuc)1(NeuAc)1 | ||
| (GlcNAc)2(Man)3 + (HexNAc)3(Hex)2(NeuAc)2 | ||
| (GlcNAc)2(Man)3 + (HexNAc)3(Hex)3(Fuc)1 | ||
| (GlcNAc)2(Man)3 + (HexNAc)3(Hex)3(Fuc)1(NeuAc)1 | ||
| (GlcNAc)2(Man)3 + (HexNAc)3(Hex)3(Fuc)1(NeuAc)2 | ||
| (GlcNAc)2(Man)3 + (HexNAc)4(Hex)4(Fuc)1(NeuAc)2 | ||
| HT5 (413N), YNLSLGGDIILQFIEIADNR | (GlcNAc)2(Man)3 + (Hex)4 | (GlcNAc)2(Man)3 + (HexNAc)2(Hex)2(Fuc)1 |
| (GlcNAc)2(Man)3 + (Hex)5 | (GlcNAc)2(Man)3 + (HexNAc)2(Hex)2(NeuAc)1 | |
| (GlcNAc)2(Man)3 + (HexNAc)2(Hex)2 | (GlcNAc)2(Man)3 + (HexNAc)3(Hex)3 | |
| (GlcNAc)2(Man)3 + (HexNAc)1 | (GlcNAc)2(Man)3 + (HexNAc)3(Hex)3(Fuc)1 | |
| (GlcNAc)2(Man)3 + (HexNAc)1(Hex)1(NeuAc)1 | (GlcNAc)2(Man)3 + (HexNAc)3(Hex)4(Fuc)1(NeuAc)1 | |
| (GlcNAc)2(Man)3 + (HexNAc)2(Hex)2(Fuc)1 | ||
| (GlcNAc)2(Man)3 + (HexNAc)2(Hex)2(NeuAc)1 | ||
| (GlcNAc)2(Man)3 + (HexNAc)2(Hex)2(NeuAc)2 | ||
| HT6 (477N), NNSVISRPGQSQCPR | (GlcNAc)2(Man)3 + (Hex)2 | (GlcNAc)2(Man)3 + (HexNAc)1(Hex)1(Fuc)1 |
| (GlcNAc)2(Man)3 + (Hex)3 | (GlcNAc)2(Man)3 + (HexNAc)1(Hex)1(Fuc)1(NeuAc)1 | |
| (GlcNAc)2(Man)3 + (Hex)4 | (GlcNAc)2(Man)3 + (HexNAc)1(Hex)2 | |
| (GlcNAc)2(Man)3 + (HexNAc)2(Hex)2 | (GlcNAc)2(Man)3 + (HexNAc)1(Hex)2(Fuc)1 | |
| (GlcNAc)2(Man)3 + (HexNAc)1(Hex)1 | (GlcNAc)2(Man)3 + (HexNAc)1(Hex)2(NeuAc)1 | |
| (GlcNAc)2(Man)3 + (HexNAc)1(Hex)2 | (GlcNAc)2(Man)3 + (HexNAc)1(Hex)2(Fuc)1(NeuAc)1 | |
| (GlcNAc)2(Man)3 + (HexNAc)1(Hex)1(NeuAc)1 | (GlcNAc)2(Man)3 + (HexNAc)1(Hex)3 | |
| (GlcNAc)2(Man)3 + (HexNAc)2(Hex)2(Fuc)1 | (GlcNAc)2(Man)3 + (HexNAc)2(Hex)1(Fuc)1 | |
| (GlcNAc)2(Man)3 + (HexNAc)2(Hex)2(NeuAc)1 | (GlcNAc)2(Man)3 + (HexNAc)2(Hex)2 | |
| (GlcNAc)2(Man)3 + (HexNAc)2(Hex)4(NeuAc)1 | (GlcNAc)2(Man)3 + (HexNAc)2(Hex)2(Fuc)1 | |
| (GlcNAc)2(Man)3 + (HexNAc)2(Hex)2(Fuc)1(NeuAc)1 | (GlcNAc)2(Man)3 + (HexNAc)2(Hex)2(Fuc)1(NeuAc)1 | |
| (GlcNAc)2(Man)3 + (HexNAc)3(Hex)1(Fuc)1 | ||
| HT7 (525N), LNWVSAGVYLNSNQTAENPVFAVFK | (GlcNAc)2(Man)3 + (HexNAc)2(Hex)2(Fuc)1 | ND |
| (GlcNAc)2(Man)3 + (HexNAc)2(Hex)2(NeuAc)1 | ||
| (GlcNAc)2(Man)3 + (HexNAc)2(Hex)2(Fuc)1(NeuAc)2 | ||
| HC1 (302N), TLCAVSHVGDPILNSTSW | (GlcNAc)2(Man)3 + (Hex)2 | ND |
| (GlcNAc)2(Man)3 + (HexNAc)2(Hex)2 | ||
| (GlcNAc)2(Man)3 + (HexNAc)2(Hex)2(Fuc)1 | ||
| (GlcNAc)2(Man)3 + (HexNAc)2(Hex)2(Fuc)1(NeuAc)1 |
The N-glycan structures identified using parallel workflows on a linear ion trap (QTRAP) and a Q-TOF mass spectrometer are shown. The retention time, m/z and mass for each glycopeptide are given in Supplementary data, Tables SI and SII. ND, not detected. M* refers to the decomposition of carboxymethylated methionine.
All seven of the predicted glycan sites were determined to be occupied for VV-sG, whereas only four of the seven sites were found to be occupied for HEK293-sG, with the two larger hydrophobic peptides undetectable. The first peptide (HT1), containing the 68NYT motif, proved difficult to analyze because of the presence of an unusual modification on the Met residue (Jones et al. 1994). A single glycosylated form of this peptide was observed and is listed in Table III. The deglycosylated form was detected without modification (other than deamidation of Asn) after PNGase-F treatment. In addition to the N-linked glycopeptides, a suite of O-linked glycopeptides were detected with greater diversity observed for the HEK293-sG glycoprotein (Table IV; Supplementary data, Tables SIC and SIIC).
Table IV.
Characterization of O-linked glycopeptides of VV-sG and HEK293-sG HeV
| Peptide | Glycopeptide | VV-sG | HEK293-sG |
|---|---|---|---|
| HT8 | TTDNQALIK | (HexNAc)1(Hex)1 | (HexNAc)1(Hex)1(Fuc)1 |
| (HexNAc)1(Hex)1(NeuAc)1 | (HexNAc)1(Hex)1(NeuAc)2 | ||
| (HexNAc)1(Hex)1(NeuAc)2 | (HexNAc)2(Hex)2(NeuAc)2 | ||
| HT9 | ESLQSVQQQIK | ND | (HexNAc)1(Hex)1 |
| (HexNAc)1(Hex)1(NeuAc)2 | |||
| (HexNAc)2(Hex)2(NeuAc)1 | |||
| (HexNAc)2(Hex)2(NeuAc)2 | |||
| HT10 | ALTDK^IGTEIGPK | (HexNAc)1(Hex)1(NeuAc)1 | (HexNAc)1(Hex)1(NeuAc)2 |
| (HexNAc)2(Hex)2(NeuAc)2 | |||
| HT11 | IGTEIGPK | ND | (HexNAc)1(Hex)1(NeuAc)2 |
| (HexNAc)2(Hex)2(NeuAc)2 | |||
| HT12 | VSLIDTSSTITIPANIGLLGSK | (HexNAc)1(Hex)1 | (Hex)1 |
| (HexNAc)1(Hex)1(NeuAc)1 | (HexNAc)1(Hex)1 | ||
| (HexNAc)1(Hex)1(NeuAc)2 | (HexNAc)1(Hex)2 | ||
| (HexNAc)1(NeuAc)1 | |||
| (HexNAc)1(Hex)1(NeuAc)1 | |||
| (HexNAc)1(Hex)1(NeuAc)2 | |||
| (HexNAc)1(Hex)2(NeuAc)1 | |||
| (HexNAc)2(Hex)2 | |||
| (HexNAc)2(Hex)1(Fuc)1(NeuAc)1 | |||
| (HexNAc)2(Hex)2(NeuAc)1 | |||
| (HexNAc)2(Hex)2(NeuAc)2 | |||
| HT13 | ISQSTSSINENVNDK | ND | (HexNAc)3(Hex)3 |
| (HexNAc)4(Hex)4 | |||
| (HexNAc)2(Hex)2(NeuAc)1 | |||
| HT14 | SHYILR | ND | (HexNAc)4(Hex)4(Fuc)1 |
The O-glycan structures identified using parallel workflows on a linear ion trap (QTRAP) and a Q-TOF mass spectrometer are shown. The retention time, m/z and mass for each glycopeptide are given in Supplementary data, Tables SI and SII. ND, not detected. K^ refers to a missed cleavage peptide.
Glycan heterogeneity of VV-sG
The glycopeptides derived from VV-sG were assigned based on the examination of product ion spectra and the determination of the relative order of retention times of each of the deglycosylated peptides after PNGase-F treatment. Table III and Supplementary data, Table SI list all the VV-sG glycopeptides identified, highlighting the heterogeneity in the glycan structures. In VV-sG, the modifications were identified as predominantly asialo-, mono- and/or disialylated complex-type N-glycans, which occurred with or without Fuc. A population of high-mannose glycans was also identified. These varied from five to eight mannose residues. The libraries of MS/MS spectra acquired were searched against the UniProt database (http://www.uniprot.org) to which the recombinant protein sequences had been added. Using the ProteinPilot program, non-glycosylated peptides and peptides containing modifications, such as deamidation, oxidation and ubiquitination, were detected. In addition, this software package was able to detect several examples of simple O-glycosylated peptides. The majority of glycopeptides, however, were manually assigned.
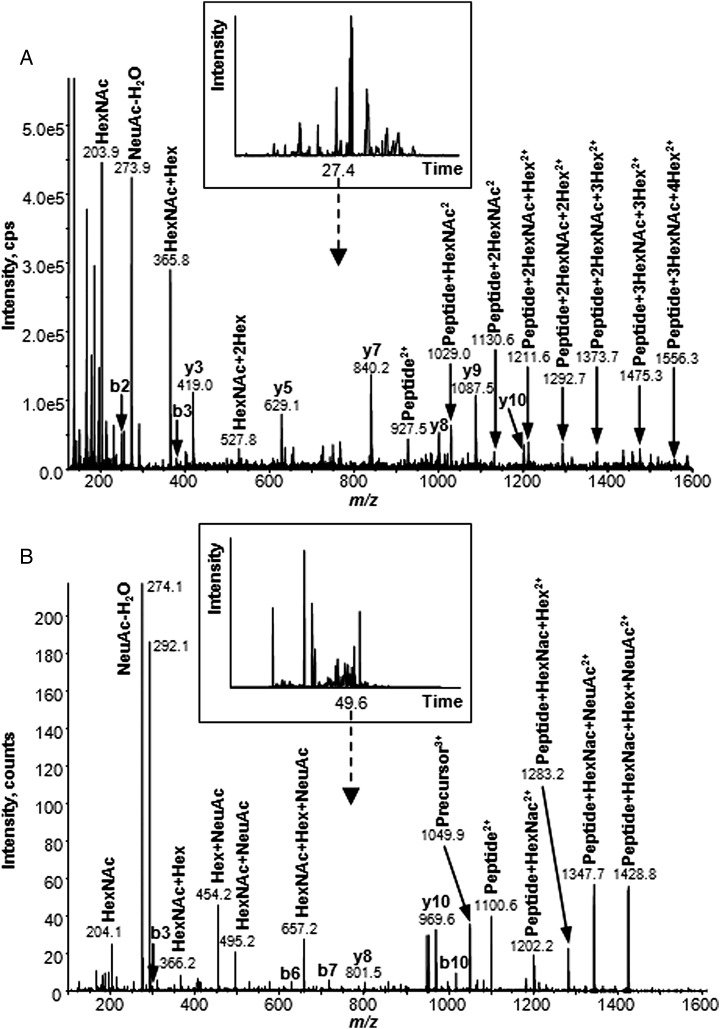
A representative MS/MS spectrum acquired on the QTRAP instrument of the precursor ion m/z 1305.213+ at 27.4 min observed for VV-sG is shown in Figure 2A. Abundant ions derived from carbohydrates, such as m/z 203.9, 273.9, 292.0, 365.8 and 527.8, are seen in the lower m/z region. The mass of the unglycosylated peptide was determined to be 1853.0 Da based on the observed doubly protonated ion at m/z 927.5. A series of peptide sequence ions was observed and confirmed the peptide sequence as HT2. For example, b2, b3 and b4 were observed at m/z 251.1, 380.2 and 540.2, respectively, and y1, y3, y5, y7, y8, y9 and y10 were seen at m/z 175.1, 419.0, 629.1, 840.2, 1000.5, 1087.5 and 1200.6, respectively. The presence of these ions definitively identifies the site of glycosylation as Asn5 (155N) in IHEC(Cam) NISC(Cam)PNPLPFR. A m/z difference of 101.5 between the fragment ions at m/z 927.52+ [peptide + 2H]2+ and m/z 1029.0 [peptide + HexNAc + 2H]2+ correlates with the addition of the first HexNAc residue to the peptide core. The pattern of ions observed in the MS/MS spectrum shows subsequent addition of carbohydrate groups to this peptide core. Based on the precursor m/z value of 1305.2, the molecular mass of the glycopeptide was calculated to be 3912.6 Da. The molecular mass of the carbohydrate moiety was calculated to be 2059.6 Da, which corresponds with a carbohydrate composition of (HexNAc)4(Hex)5(Fuc)1(NeuAc)1. The experimentally determined mass for this putative glycopeptide differs from the theoretical mass of 3912.6 by only 0.03 Da. By following the additions of carbohydrate groups to the peptide core, the predicted glycan composition was confirmed. Interestingly, we are the first to report the existence of a number of O-linked glycosylation sites on the VV-sG glycoproteins (Table IV).
Fig. 2.
Glycopeptide identification and compositional analysis by LC-MS/MS. (A) Product ion spectra acquired on a linear ion trap mass spectrometer of the triply charged precursor ion at m/z 1305.21 eluting at 27.4 min from a tryptic digest of the VV-sG protein. Several b- and y-ions confirming the peptide as IHEC(Cam)NISC(Cam)PNPLPFR (HT2) are labeled. Carbohydrate diagnostic oxonium ions at m/z 204, 274, 366 and 528 were observed. The fragmentation observed from the spectra m/z 150–1600 revealed an N-glycan with the composition (HexNAc)4(Hex)5(NeuAc)1(Fuc)1. Note that not all y- and b-ions are labeled in this figure but are described in the text. (B) Product ion spectra acquired on a Q-TOF mass spectrometer of the triply charged precursor ion at m/z 1049.85 eluting at 49.6 min from a tryptic digest of the HEK293-sG protein. In this example, the peptide sequence was determined to be VSLIDTSSTITIPANIGLLGSK (HT12) and was O-glycosylated with a glycan of composition (HexNAc)1(Hex)1(NeuAc)2.
Analysis of the tryptic peptide products clearly demonstrated that six sites on the VV-sG glycoprotein were occupied. However, one sequon in this protein could not be assessed using trypsin as the resulting peptide was 57 amino acids in length with a molecular mass >6 kDa. In order to produce a peptide more suitable for MS/MS detection and characterization, a chymotryptic digest was performed. The expected peptide (encompassing the uncharacterized sequon NSTS) resulting from the chymotrypsin digest would be 18 amino acids in length and thus of a size amenable to MS/MS analysis. The peptide TLCAVSHVGDPILNSTSW (HC1) was identified and shown to be glycosylated. After treatment with PNGase-F, the deglycosylated form of this peptide was detected. Endo H digestion followed by chymotrypsin digestion would be expected to result in proteolytic fragments consisting of a single GlcNAc residue attached to each modified asparagine residue. Peptide HC1 was detected with a HexNAc attached to Asn14 (302N) confirming the occupancy of this site.
Glycan heterogeneity of HEK293-sG
The glycopeptides derived from HEK293-sG were assigned as described for the VV-sG glycoprotein. The N-linked glycans identified in HEK293-sG are shown in Table III and were greater in both number and variability (29 different glycan structures) than that observed for VV-sG (18 glycan structures). Glycans identified in the HEK293-sG were predominantly asialo-, mono- and/or disialylated complex-type, which occurred with or without Fuc. A single example of a trisialylated complex-type glycan was observed at 155N. Only a single high-mannose glycan (comprised of five mannose residues) was identified. Tryptic digestion of the HEK293-sG glycoprotein revealed glycan occupancy at four of the seven predicted sites. The N-terminal peptide containing the sequon NYTR was not detected either before or after deglycosylation.
As demonstrated for the VV-sG glycoprotein, O-linked glycopeptides were also observed for the HEK293-sG (Table IV). However, seven different peptides primarily located in the N-terminal region of HEK293-sG were observed compared with only three for VV-sG. Figure 2B demonstrates the assignment of an O-glycan that was observed in the LC-MS/MS analysis of HEK293-sG undertaken on the Q-TOF instrument. In this example, a number of ions derived from carbohydrates, such as m/z 204.1, 274.1, 366.2, 454.2 and 657.2 in the lower m/z region, were observed. The experimental mass of the unglycosylated peptide was observed at 1100.6 Da as a doubly protonated ion that corresponded to the mass of the tryptic peptide VSLIDTSSTITIPANIGLLGSK (HT12). Confirmation of the peptide was achieved by the observation of a series of b- and y-ions. The peptide had an attached disialylated glycan (HexNAc)1(Hex)1(NeuAc)2. In this relatively simple glycan structure, the branching could be determined with the sialic acid groups attached to both the HexNAc residue and the Hex. The same glycopeptide was also detected in VV-derived sG (Table IV).
Overall, analysis by LC-MS/MS resulted in sequence coverage between 70 and 85% for trypsin and 40 and 68% for chymotrypsin reflecting the differences in the size of peptides produced and increased ionization efficiency for tryptic products.
Role of glycosylation in sG binding to ephrin-B2
The G protein globular head domain expressed in the baculovirus/insect-cell system and the ephrin-B2 ectodomain expressed in HEK293 cells were mixed to form a complex and crystallized. An X-ray crystallographic structure of this complex was determined at 2.7 Å resolution (PDB code: 3UWF) and figures were generated with the program PyMOL, using this structure. The overall structure and conformation of the G protein globular head domain both alone and in complex with ephrin-B2 is quite similar to those reported by Bowden et al. (2008, 2010) and Xu et al. (2008). In this instance, we focused on the description of the glycan chains only in the crystal structure with detailed overall description planned for another publication.
N-linked glycans were modeled at all five predicted sites on the G protein head domain (302N, 374N, 413N, 477N and 525N; Figure 3). Glycan attached at 525N appeared the largest (refer to HT7 in VV-sG; Table III) and was located the closest to the receptor-binding region (Figure 3A and B). In the crystal structure of a sG/ephrin-B2 complex, we observed that glycan 525N forms close contacts with ephrin-B2 with several charged residues in ephrin-B2, including 107D and 109K, likely involved in this interaction (Supplementary data, Table SIII). The total buried surface area in this glycan/ephrin contact interface is 62 Å2.
Fig. 3.

Three-dimensional model of the HeV attachment glycoprotein and receptor complex. Three-dimensional models of a globular head domain of HeV sG (A) and in complex with ephrin-B2 (B). The glycans were modeled according to the electron density map of the crystal structure of sG/ephrin-B2 complex at 2.7 Å resolution. The glycan branches and some of the core structures were not modeled due to the weak signal that resulted from the flexibility of the glycan chains. N-linked glycosylation is shown at all five predicted sites on the head domain of the G protein. Glycan 525N forms close contacts with ephrin-B2.
Because of the close location and the flexibility of the glycan chain, the longer and more-branched glycan chain in mammalian cell-expressed sG are more likely to be in contact with the residues on a receptor. Indeed, there are several charged residues, such as D59, K64, D107 and K109, located on the same side of ephrin-B2 as the glycan 525N. We thus proposed that the glycan chain on HeV sG protein may be involved in receptor attachment.
To examine this possibility, we have performed a preliminary experiment comparing the receptor-binding affinity of sG constructs with different glycosylation levels obtained in different expression systems. For this purpose, we compared the insect cell-expressed sG head domain construct (low glycosylation level and lower molecular weight) with that expressed in the HEK293 cells, using an endogenous tryptophan fluorescence-quenching assay (Figure 4). Indeed, insect cell-expressed sG required a higher concentration of ephrin-B2 for maximum quenching (saturation of HeV G protein's receptor-binding site) suggesting that the type and level of glycosylation on the sG protein affects its affinity with ephrin-B2 receptors. In other words, the glycan chain on HeV, and possibly NiV G proteins, is likely to be involved in the viral-receptor attachment process.
Fig. 4.
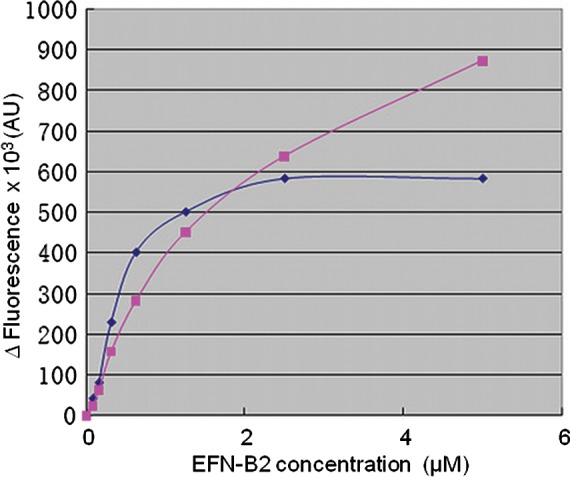
Binding of different forms of the HeV-sG protein to ephrin-B2. The determination of interactions between different constructs of the sG (1 µM) and ephrin-B2 (EFN-B2) was performed using a tryptophan fluorescence quench assay on a SPEX FluoroMax-2 spectrofluorimeter (25°C, 10 mm path-length cuvette, excitation and emission wavelengths of 295 and 350 nm, respectively). Titrations were performed by a stepwise addition of small volumes of concentrated (0.5 mM) ephrin-B2 solutions to a 1 µM solution of sG constructs at pH 7.2. The difference between the fluorescence units of the complex and the sum of individual components was used to plot the results (filled square), insect cell-expressed sG head domain construct (low glycosylation level) and (filled diamond) HEK293 cell-expressed sG head domain construct.
Discussion
VV-sG and HEK293-sG glycoproteins were analyzed using a combination of glycosidase digestion and lectin binding followed by comprehensive MS/MS analysis. The heterogeneity of glycosylation of the VV-sG and HEK293-sG glycoproteins was apparent upon 2-DE separation with spot trains observed across the entire pI range (Figure 1C). This phenomenon could be attributed to chemical modification (carbamylation) of the proteins, variable sialic acid content and/or generation of protein hetero-oligomers. From the MS analysis, we conclude that the proteins were not adversely affected by carbamylation. It is very likely that the spot distribution was due to the charge distribution arising from the presence of a large number of sialic acid residues, most likely, in conjunction with protein oligomerization. Indeed, the presence of monomeric and oligomeric forms of recombinantly expressed hemagglutinin–neuraminidase glycoproteins of paramyxovirus PIV5 (Yuan et al. 2008) and the G glycoproteins of HeV and NiV (Bossart et al. 2005; Bowden et al. 2008, 2010) has been reported previously.
In our preliminary mass analyses, we utilized both IDA and precursor ion scanning on an LC/MS triple quadrupole instrument to identify glycan composition and site heterogeneity for the recombinant sG glycoproteins. A major advantage of precursor ion scanning is that carbohydrate-specific ions can be selectively detected at high sensitivity during the chromatographic separation of complex digest mixtures. Glycopeptides may be identified at the low picomole level by the appearance of specific oxonium ions. Subsequent analyses were performed using IDA-triggered MS/MS on a hybrid Q-TOF instrument. By using two independent mass spectrometry methods, we were able to assess overall glycan composition as well as compositional heterogeneity at specific N-linked glycan sequons of the G glycoproteins produced in two different expression systems.
Overall, the VV-sG and HEK293-sG predominantly carried two glycan types: complex and high mannose. Taking into account the monosaccharide components, it was predicted that complex-type glycans ranged between bi-, di- and tri-antennary structures; however, without linkage data, the specific branching patterns cannot be confirmed. Mono- and disialylated capping of complex-type N-glycans with or without Fuc were present in both VV-sG and HEK293-sG glycoproteins. The population of high-mannose glycans ranged from five to eight mannose residues per glycan structure and these residues were more predominant in the Vaccinia expression system (nine structures observed) than in the HEK293 expression system (single structure observed). In a recent study (Bowden et al. 2010), matrix-assisted laser desorption/ionization time-of-flight (MALDI-TOF) and electrospray-MS analysis of N-linked glycans from HEK293 cells revealed the presence of complex-type glycans as the predominant form, with a subset of high-mannose glycans also present correlating well with the results presented in our study.
The HeV sG contains seven potential sites of N-glycosylation. Figure 3 shows a three-dimensional structure model of the HeV G protein head domain with complex N-glycan structures attached to the predicted sites. In this study, we identified 38 different glycan structures attached to N-glycosylation sites. Interestingly, of the O-glycans observed, the majority were simple structures attached to Thr/Ser via N-acetylgalactosamine (Peter-Katalinic 2005) in the N-terminal region of the protein (15 known glycan structures observed), but several (seven) high-hexose structures were observed on a single peptide (405SHYILR) in the C-terminal region (Supplementary data, Table SIIA). However, these structures could not be explained by comparison with any known O-linked glycans (Peter-Katalinic 2005; Lommel and Strahl 2009; Nakamura et al. 2010; Zaia 2010) and were deemed biosynthetically impossible. The complete structural characterization of these high-hexose glycans will be the subject of future investigations.
It is clear from our data that the overall structure and conformation of the G protein globular head domain, both alone and in complex with ephrin-B2, was quite similar to those reported by Bowden et al. (2008, 2010). Our focus in the present study was to detail the glycosylation patterns of HeV sG and to put these data in context with the protein structure; a detailed comparison of the present structures to those described earlier will be described elsewhere. Here, we have not only observed carbohydrate moieties at all five predicted N-linked glycosylation sites in the head domain of the G protein (Figure 3A) but have also provided further physiological details on the carbohydrate structures. Furthermore, the ephrin-B2 utilized in the complex structure (Figure 3B) was also expressed in HEK293, suggesting a more relevant glycosylation pattern and, indeed, an additional N-linked glycosylation modification was revealed. Interestingly, in the crystal structure of the sG protein head domain and the ephrin-B2 complex, glycan 525N appears to form close contacts with ephrin-B2 with several charged residues on the receptor likely involved in the interaction. These observations together with a tryptophan-quenching assay suggest that this carbohydrate moiety, possibly in conjunction with the conformation-dependent binding domain (Bishop et al. 2007), participates in a receptor attachment. This may affect virus host-cell entry.
The most striking difference observed between the glycoproteins produced in the two different expression systems was in the suite of O-linked glycans present. Not only do we report for the first time the presence of O-linked glycosylation for HeV, but we report three glycopeptides with three different glycan structures in VV-sG and seven glycopeptides with up to 15 different glycan structures in HEK293-sG. It is interesting to note that O-glycosylation prediction algorithms available did not indicate this type of glycosylation with significant probability. The question of how these glycoprotein structures might affect biological activity remains to be solved, but it is clear that the production of glycoproteins is largely dependent on the expression system used. It is known that both the VV-sG and HEK293-sG are expressed as oligomers, can bind ephrin-B2 and -B3 receptors and induce protective antibody responses when used as immunogens (Mungall et al. 2006; McEachern et al. 2008; D Middleton and CC Broder, personal communication). Variation in the glycosylation profile may impact these functions.
With the process of glycosylation being a non-template driven occurrence, glycoproteins typically exist as an assortment of glycoforms. Since N-glycans may influence circulation half-life, tissue distribution and biological activity, it is important to consider that each glycoform has its own pharmacokinetic, pharmacodynamic and efficacy profile. In the production of recombinant proteins for research and possible human or animal administration, it is important to note that any deviations from the desired oligosaccharide composition resulting from the use of a particular expression system could result in undesired immunogenicity (Jacobs and Callewaert 2009; Kim et al. 2009; Liu et al. 2011). In this study, we have characterized and compared the glycosylation profiles of the HeV attachment glycoprotein in two commonly used expression systems.
Material and methods
Soluble recombinant glycoprotein production
Constructs of HeV sG with the transmembrane cytoplasmic-tail deletion and S-tag incorporated were made by polymerase chain reaction amplification. A series of cloning optimizations produced a sG-encoding cassette that was subcloned into pMCO2 to generate pKB16 which was used to produce recombinant VV vKB16 (sGS-tag) as detailed previously (Bossart et al. 2005). For expression of the VV-sG HeV glycoprotein, 6 × 850 cm2 roller bottles of HeLa cells were washed three times with phosphate-buffered saline (PBS) and then infected with vKB16 (multiplicity of infection of 3) for 2 h in Opti-MEM serum-free media (Gibco, Invitrogen Australia, Victoria), following that 120 mL of serum-free media was added and the roller bottle cultures allowed to incubate at 37°C for an additional 36 h. The sG containing supernatant was harvested, centrifuged for 20 min at 860 × g and filtered with a 0.22-μm polyethersulfone (PES, low protein-binding) filter unit. Triton-X 100 was added to a final concentration of 0.5%, and the sG was purified by S-Agarose affinity chromatography. For the production of cell-derived sG, the sG open reading frame from pKB16 was cloned into phCMV-1 (Gelantis, San Diego, CA). The plasmid (phCMV-1-HeV-sG) was transfected into HEK293 cells (ATCC, Manassas, VA) using the Fugene reagent following the manufacturer's instruction. At 48 h post-transfection, the culture medium was replaced with Dulbecco's modified Eagle medium-10 supplemented with 500 µg/mL of geneticin for the selection of sG-expressing cells. Standard limiting dilution procedures were carried out to isolate a single colony yielding a high level of expression as determined by supernatant screening from individual clones. For protein production, the isolated clone was cultured in 293 SFM II serum-free medium (Gibco) as a suspension culture. After 4 days, the culture supernatant was harvested and centrifuged for 20 min at 860 × g, recovered and centrifuged again for 30 min at 15,300 × g, then filtered through a 0.22-µm PES membrane filter unit. Typically 1.6 L of sG containing supernatant was prepared. The sG was purified by S-Agarose affinity chromatography (Novagen, San Diego, CA) on an XK26 column (GE Healthcare, Carlsbad, CA) with a bed volume of 16 mL of S-Agarose. The sG containing supernatant was applied at 2 mL/min, then washed with six column volumes of PBS. The bound sG was eluted with 16 mL of 0.2 M citric acid (pH 2). The eluate was neutralized with Tris–HCl (pH 8), then concentrated and dialyzed against PBS. The sG was then fractionated into its oligomeric forms using a preparative S200 Superdex gel-filtration column in PBS. Individual 1 mL fractions were analyzed by non-denaturing polyacrylamide gel electrophoresis (PAGE) and pooled according to species’ molecular weight, then quantitated, sterile-filtered and stored at −80°C. Recombinant sG preparation typically consisted of ∼20% tetramer and dimer mix, 75% dimer and 5% monomer. The dimer fractions of VV-sG and HEK293-sG were used in this study.
For structure determination studies, the recombinant sG protein-coding sequence (globular head domain, residues 171–604; refer to Table II) was subcloned into the pAcGP67 vector and expressed in baculovirus expression system (BD Biosciences, Franklin Lakes, NJ), and ephrin-B2-coding region (residue 27–167; gi|4758250|ref|NP_004084.1|) was cloned into the pCDNA3.1 vector and expressed in HEK293 cells (Invitrogen). Both proteins were purified as described previously (Xu et al. 2008).
Glycosidase treatment
Analysis of purified recombinant proteins was performed before and after treatment with glycosidases. Approximately 30 μg of protein samples were denatured in 0.05% sodium dodecyl sulfate (SDS) and 15 mM dithiothreitol (DTT) for 5 min at 100°C and incubated with 1 mU of neuraminidase (Arthrobacter ureafaciens), 5 mU of Endo H (recombinant from gene from Streptomyces plicatus), 3 mU of recombinant N-glycosidase F (recombinant from gene from Flavobacterium meningosepticum) and 1 mU of O-glycosidase (bovine serum albumin-free from Diplococcus pneumoniae) for 16 h at 37°C in optimal buffer and pH conditions specified by enzymes’ manufacturer (Roche Applied Science, Mannheim, Germany). Fetuin, a glycoprotein known to contain N- and O-linked glycans and sialic acid (Roche Applied Science), was used to determine enzyme specificity and effectiveness.
One-dimensional electrophoresis
Pre-cast NuPAGE 4–12% bis–tris polyacrylamide gels (Invitrogen) were used for the electrophoretic separation of protein samples under reducing conditions. Samples were mixed with 4× lithium dodecyl sulfate (LDS) sample buffer (Invitrogen) and 0.5 M DTT in a 65:25:10 volume ratio, and then denatured by incubation at 100°C for 5 min. Samples were then run on gels at 120–150 V in a 2-(N-morpholino) ethane sulphonic acid running buffer in a Mini-cell apparatus (Invitrogen). SeeBlue Plus2 Pre-Stained Standards (Invitrogen) were loaded with each gel to allow for the size estimation of target proteins. Protein bands were visualized with Coomassie blue or silver staining.
Two-dimensional electrophoresis
All recombinant viral protein samples (15 μg) were prepared for 2-DE analysis by first boiling in 100 mM DTT and 0.3% SDS for 10 min to denature the proteins. Samples were allowed to cool for 30 min at room temperature, then 130 μL of isoelectric focusing (IEF) rehydration solution containing 8 M urea, 130 mM DTT, 4% CHAPS and 2.0% v/v immobilized pH gradient (IPG) buffer (pH 3–11; GE Healthcare) was added. Proteins prepared in this manner were used to rehydrate 7 cm (pH 3–11) IPG strips (GE Healthcare Bio-Sciences, NSW, Australia) for 12 h, at 50 V, in an IPGphorIII IEF apparatus (GE Healthcare). Iso-electric focusing was then performed using a step-voltage program comprising 1 h at 500 V, 1 h at 1000 V followed by 2 h at 8000 V for a total focusing time of 15 kVh. After focusing, strips were directly equilibrated in 50 mM Tris–HCl, 2% w/v SDS, 6 M urea, 35% v/v glycerol and 10 mg/mL DTT for 15 min followed by 15 min incubation in 50 mM Tris–HCl, 2% w/v SDS, 6 M urea, 35% v/v glycerol and 25 mg/mL iodoacetamide. For second-dimension separations, IPG strips were sealed with 0.5% agarose onto pre-cast 4–20% Tris–glycine SDS–PAGE zoom mini-gels (Invitrogen). Electrophoresis was conducted at 125 V over 2 h. Proteins were detected by silver staining and gel images were acquired at 600 dpi using an Image-Master (GE Healthcare) desktop scanner and Labscan software, version 3.00 (GE Healthcare).
Lectin detection
Following electrophoresis, separated proteins were electro-transferred onto BioTrace™ polyvinylidene fluoride (PVDF) transfer membranes (Pall) for 1 h at 180 mA then blocked overnight in 1% w/v gelatin in Tris-buffered saline/Tween (TBST) buffer (20 mM Tris–HCl, 150 mM NaCl and 0.5% Tween-20, pH 7.4, containing Ca2+, Mg2+ and Mn2+ at 1 mM each). Membranes were washed three times with TBST followed by 1 h incubation with biotinylated lectins. DSA, GNA, MAAII, NPL, SNA and UEAI (Vector Labs, Burlingame, CA) used at 1:40,000 dilution. After lectin incubation, the blots were washed three times in TBST for 10 min and then incubated for 1 h with streptavidin–horseradish peroxidase (HRP) conjugate (1:20,000; Dako, NSW, Australia) in Tris-buffered saline (TBS) followed by three 10 min washes, two in TBST and one in TBS. The lectin-bound strepavidin–HRP was then incubated with enhanced chemiluminescence substrate (GE Healthcare Bio-Sciences) and visualized by exposure on an X-ray film.
Reduction, alkylation and enzymatic digestion
Enzymatic digestion was undertaken using 25 μg of aliquots of each sG protein dissolved in 50 mM NH4HCO3. Disulfide bonds were reduced by addition of an equal volume of 10 mM DTT in 50 mM NH4HCO3 at 55°C for 1 h. Reduced samples were then alkylated with an equal volume of 50 mM iodoacetamide in 50 mM NH4HCO3 and were added and incubated in the dark at room temperature for 20 min. Reduced and alkylated samples were then digested with either trypsin (proteomics grade; Sigma, St Louis, MO) or chymotrypsin (sequencing grade; Roche Applied Science), using a 1:50 enzyme to sample ratio in 50 mM NH4HCO3 at 37°C over 16 h. The digestion was terminated by acidification with formic acid to 0.2% v/v final composition.
N-terminal amino acid sequencing
PVDF immobilized proteins were subjected to amino acid sequence analysis in an automated sequenator (Procise 492, Applied Biosystems, Foster City, CA).
Mass spectrometry
Linear ion trap (triple quadrupole) MS
For glycosylation site determination and glycan composition of sG, reduced and alkylated tryptic peptides were analyzed on a 4000 QTRAP mass spectrometer (Applied Biosystems) equipped with a TurboV ionization source operated in a positive ion mode. Samples were chromatographically separated on a Dionex Ultimate 3000 high-performance liquid chromatography (HPLC) (Dionex, Sunnyvale, CA) using a Phenomenex C18 (2.1 mm × 25 cm) column (Torrance, CA) with a linear gradient of 0–40% solvent B over 40 min with a flow rate of 250 μL/min. The mobile phases consisted of solvent A (0.1% formic acid) and solvent B (0.1% formic acid/90% acetonitrile/10% water). The eluent from the HPLC was directly coupled to the mass spectrometer. Data were acquired and processed using Analyst V1.5 software™. IDA analyses were performed using an enhanced MS scan over the mass range 350–1500 as the survey scan and triggered the acquisition of tandem mass spectra. The top two ions of charge state +2 or +3 that exceeded a defined threshold value (100,000 counts) were selected and first subjected to an enhanced resolution scan prior to acquiring an enhanced product scan over the mass range 125–1600. Precursor ion scanning experiments with a mass range from 400 to 1600 were also used to focus specifically on glycopeptides containing either the two carbohydrate oxonium ions of m/z 204 (HexNAc) or m/z 366 (Hex-HexNAc). For both IDA and precursor ion scanning experiments, the scan speed was set at 1000 Da/s and peptides were fragmented in the collision cell with nitrogen gas using rolling collision energy dependent on the size and charge of the precursor ion.
Q-TOF mass spectrometry
Samples were chromatographically separated on an Agilent 1100 Capillary HPLC system (Palo Alto, CA) using a Vydac MS C18 300 Å, column (150 mm × 2 mm) with a particle size of 5 μm (Grace Davison, Deerfield) using a linear gradient of 2–42% solvent B over 60 min at a flow rate of 4 μL/min. The mobile phases consisted of solvent A (0.1% formic acid) and solvent B (0.1% formic acid/90% acetonitrile/10% water). A Q-Star Elite QqTOF mass spectrometer (Applied Biosystems) was used in a standard MS/MS data-dependent acquisition mode with a nano-electrospray ionization source. Survey MS spectra were collected (m/z 400–1500) for 1 s followed by three MS/MS measurements on the most intense parent ions (20 counts/s threshold, 2+ to 5+ charge state and m/z 100–1500 mass range for MS/MS), using the manufacturer's “Smart Exit”. Parent ions previously targeted were excluded from repetitive MS/MS acquisition for 60 s (mass tolerance of 0.250 Da).
Analysis of mass spectra and database searching
ProteinPilot™ 2.0.1 software (Applied Biosystems) with the Paragon algorithm was used for the identification of proteins. Tandem mass spectrometry data were searched against in silico tryptic digests of the Swissprot (version 20081216) database. All search parameters were defined as iodoacetamide modified with cysteine alkylation, with either trypsin or chymotrypsin as the digestion enzyme. Modifications were set to the “generic workup” and “biological” modification sets provided with this software package, which consisted of 126 possible modifications, e.g. acetylation, methylation and phosphorylation. The generic workup modifications set contains 51 potential modifications that may occur as a result of sample handling, e.g. oxidation, dehydration and deamidation. Peptides with one missed cleavage were included in the analysis. Individual ion scores at P < 0.05 or better were indicative of identity. Manual interrogation of the glycopeptide MS/MS data was required as neither ProteinPilot™ nor Mascot software packages were able to elucidate the identification of the complex glycan side chains. Glycosylation prediction algorithms (e.g. GlycoMod) and other proteomics tools (Peptide Mass) were accessed from the ExPASy Proteomics Server and ExPASy beta Bioinformatics Resource Portal (http://au.expasy.org).
X-ray crystallography
The complex of the recombinant soluble globular head domain G protein and the ephrin-B2 ectodomain was generated by mixing HeV G and ephrin-B2 in a 1:2 molar ratio and passing them through a gel-filtration column. Crystals of this complex grew in vapor diffusion sitting drops with a reservoir of 27.5% PEG2000MME and 0.09% MG7. The X-ray diffraction data set of this crystal was used for structure determination (K Xu and D Nikolov, personal communication, manuscript submitted). The structure was determined at 2.7 Å resolution with space group P1. There are four molecules of complex in each asymmetric unit of the crystal. The structure was refined with program package Phenix (Terwilliger et al. 2008) to 23.3% Rfree. There were only 0.3% of residues in a disallowed region (Supplementary data, Table SIII). Figures were created in the program PyMOL (http://www.pymol.org), using the PDB code 3UWF with the X-ray crystallography structure. The glycans were modeled according to the electron density map. The glycan branches and some of the core structures were not modeled due to the weak signal resulting from the flexibility of the glycan chains.
Tryptophan fluorescence quench assay
For the determination of interactions between different constructs of the sG and ephrin-B2, a tryptophan fluorescence quench assay was performed on a SPEX FluoroMax-2 spectrofluorimeter (Horiba Jobin Yvon, Edison, NJ) at a constant 25°C, using a 10-mm-path-length cuvette, with excitation and emission wavelengths of 295 and 350 nm, respectively. Titrations were performed by a stepwise addition of small volumes of concentrated (0.5 mM) ephrin-B2 solutions to a cuvette containing 2 mL of a 1 µM solution of sG in 4-(2-Hydroxyethyl)piperazine-1-ethanesulfonic acid buffer (pH 7.2) and allowing the mixture to equilibrate for 5 min. The volume of added ephrin-B2 never exceeded 5% of the total volume. The difference between the fluorescence units of the complex and the sum of individual components was used to plot the results.
Supplementary data
Supplementary data for this article is available online at http://glycob.oxfordjournals.org/.
Funding
H.J.S. was funded by a CSIRO Livestock Industries PhD scholarship with additional supervision and guidance from Prof. Tony Bacic, School of Botany, The University of Melbourne. This work was supported in part by National Institutes of Health (AI054715 and AI077995) to C.C.B.
Conflict of interest
None declared.
Abbreviations
2-DE, two-dimensional electrophoresis; DSA, Datura stramonium agglutinin; DTT, dithiothreitol; Endo H, endoglycosidase H; Fuc, fucose; GlcNAc, N-acetylglucosamine; GNA, Galanthus nivalis agglutinin; HEK, human embryonic kidney; HeV, Hendra virus; Hex, hexose; HexNAc, N-acetyl hexosamine; HPLC, high-performance liquid chromatography; HRP, horseradish peroxidase; IDA, information-dependent acquisition; IEF, isoelectric focusing; IPG, immobilized pH gradient; LC-MS, liquid chromatography mass spectrometry; LDS, lithium dodecyl sulfate; MAAII, Maackia amurensis agglutinin II; MALDI-TOF, matrix-assisted laser desorption/ionization time-of-flight; Man, mannose; MS, mass spectrometry; MS/MS, tandem mass spectrometry; NeuAc, N-acetylneuraminic acid; NiV, Nipah virus; NPL, Narcissus pseudonarcissus lectin; PAGE, polyacrylamide gel electrophoresis; PBS, phosphate-buffered saline; PES, polyethersulfone; PNGase-F, peptide-N-glycosidase F; Q-TOF, quadrupole-time of flight; SDS, sodium dodecyl sulfate; sG, soluble glycoprotein; SNA, Sambucus nigra agglutinin; TBST, Tris-buffered saline/Tween; UEAI, Ulex europaeus agglutinin I; VV, vaccinia virus.
Supplementary Material
Acknowledgements
The authors are thankful to Mr Gary Beddome (CSIRO) for involvement in N-terminal sequencing. We would also like to thank the Molecular and Cellular Proteomics Facility at the University of Queensland, Institute for Molecular Bioscience for access to the mass spectrometers used in this study.
References
- Bishop KA, Stantchev TS, Hickey AC, Khetawat D, Bossart KN, Krasnoperov V, Gill P, Feng YR, Wang L, Eaton BT, et al. Identification of Hendra virus G glycoprotein residues that are critical for receptor binding. J Virol. 2007;81:5893–5901. doi: 10.1128/JVI.02022-06. doi:10.1128/JVI.02022-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonaparte MI, Dimitrov AS, Bossart KN, Crameri G, Mungall BA, Bishop KA, Choudhry V, Dimitrov DS, Wang LF, Eaton BT, et al. Ephrin-B2 ligand is a functional receptor for Hendra virus and Nipah virus. Proc Natl Acad Sci USA. 2005;102:10652–10657. doi: 10.1073/pnas.0504887102. doi:10.1073/pnas.0504887102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bossart KN, Broder CC. Paramyxovirus entry. In: Pöhlmann S, Simmons G, editors. Viral Entry into Host Cells. Austin (TX): Landes Bioscience; 2009. [Google Scholar]
- Bossart KN, Crameri G, Dimitrov AS, Mungall BA, Feng YR, Patch JR, Choudhary A, Wang LF, Eaton BT, Broder CC. Receptor binding, fusion inhibition, and induction of cross-reactive neutralizing antibodies by a soluble G glycoprotein of Hendra virus. J Virol. 2005;79:6690–6702. doi: 10.1128/JVI.79.11.6690-6702.2005. doi:10.1128/JVI.79.11.6690-6702.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowden TA, Aricescu AR, Gilbert RJ, Grimes JM, Jones EY, Stuart DI. Structural basis of Nipah and Hendra virus attachment to their cell-surface receptor ephrin-B2. Nat Struct Mol Biol. 2008;15:567–572. doi: 10.1038/nsmb.1435. doi:10.1038/nsmb.1435. [DOI] [PubMed] [Google Scholar]
- Bowden TA, Crispin M, Harvey DJ, Jones EY, Stuart DI. Dimeric architecture of the Hendra virus attachment glycoprotein: Evidence for a conserved mode of assembly. J Virol. 2010;84:6208–6217. doi: 10.1128/JVI.00317-10. doi:10.1128/JVI.00317-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowden TA, Jones EY, Stuart DI. Cells under siege: Viral glycoprotein interactions at the cell surface. J Struct Biol. 2011;175:120–126. doi: 10.1016/j.jsb.2011.03.016. doi:10.1016/j.jsb.2011.03.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brooks SA. Appropriate glycosylation of recombinant proteins for human use: Implications of choice of expression system. Mol Biotechnol. 2004;28:241–255. doi: 10.1385/MB:28:3:241. doi:10.1385/MB:28:3:241. [DOI] [PubMed] [Google Scholar]
- Carter JR, Pager CT, Fowler SD, Dutch RE. Role of N-linked glycosylation of the Hendra virus fusion protein. J Virol. 2005;79:7922–7925. doi: 10.1128/JVI.79.12.7922-7925.2005. doi:10.1128/JVI.79.12.7922-7925.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chua KB, Bellini WJ, Rota PA, Harcourt BH, Tamin A, Lam SK, Ksiazek TG, Rollin PE, Zaki SR, Shieh W, et al. Nipah virus: A recently emergent deadly paramyxovirus. Science. 2000;288:1432–1435. doi: 10.1126/science.288.5470.1432. doi:10.1126/science.288.5470.1432. [DOI] [PubMed] [Google Scholar]
- Eaton BT, Broder CC, Wang LF. Hendra and Nipah viruses: Pathogenesis and therapeutics. Curr Mol Med. 2005;5:805–816. doi: 10.2174/156652405774962308. doi:10.2174/156652405774962308. [DOI] [PubMed] [Google Scholar]
- Epstein JH, Field HE, Luby S, Pulliam JR, Daszak P. Nipah virus: Impact, origins, and causes of emergence. Curr Infect Dis Rep. 2006;8:59–65. doi: 10.1007/s11908-006-0036-2. doi:10.1007/s11908-006-0036-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao Y, Mehta K. N-linked glycosylation of CD38 is required for its structure stabilization but not for membrane localization. Mol Cell Biochem. 2007;295:1–7. doi: 10.1007/s11010-006-9265-9. doi:10.1007/s11010-006-9265-9. [DOI] [PubMed] [Google Scholar]
- Geisler C, Jarvis DL. Effective glycoanalysis with Maackia amurensis lectins requires a clear understanding of their binding specificities. Glycobiology. 2011;21:988–993. doi: 10.1093/glycob/cwr080. doi:10.1093/glycob/cwr080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacobs PP, Callewaert N. N-glycosylation engineering of biopharmaceutical expression systems. Curr Mol Med. 2009;9:774–800. doi: 10.2174/156652409789105552. doi:10.2174/156652409789105552. [DOI] [PubMed] [Google Scholar]
- Jones MD, Merewether LA, Clogston CL, Lu HS. Peptide map analysis of recombinant human granulocyte colony stimulating factor: Elimination of methionine modification and nonspecific cleavages. Anal Biochem. 1994;216:135–146. doi: 10.1006/abio.1994.1017. doi:10.1006/abio.1994.1017. [DOI] [PubMed] [Google Scholar]
- Kim PJ, Lee DY, Jeong H. Centralized modularity of N-linked glycosylation pathways in mammalian cells. PLoS One. 2009;4:e7317. doi: 10.1371/journal.pone.0007317. doi:10.1371/journal.pone.0007317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamb RA, Paterson RG, Jardetzky TS. Paramyxovirus membrane fusion: Lessons from the F and HN atomic structures. Virology. 2006;344:30–37. doi: 10.1016/j.virol.2005.09.007. doi:10.1016/j.virol.2005.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Cleveland B, Klots I, Travis B, Richardson BA, Anderson D, Montefiori D, Polacino P, Hu SL. Removal of a single N-linked glycan in human immunodeficiency virus type 1 gp120 results in an enhanced ability to induce neutralizing antibody responses. J Virol. 2008;82:638–651. doi: 10.1128/JVI.01691-07. doi:10.1128/JVI.01691-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lis H, Sharon N. Protein glycosylation. Structural and functional aspects. Eur J Biochem. 1993;218:1–27. doi: 10.1111/j.1432-1033.1993.tb18347.x. doi:10.1111/j.1432-1033.1993.tb18347.x. [DOI] [PubMed] [Google Scholar]
- Liu C, Dong S, Xu XJ, Yin Y, Shriver Z, Capila I, Myette J, Venkataraman G. Assessment of the quality and structural integrity of a complex glycoprotein mixture following extraction from the formulated biopharmaceutical drug product. J Pharm Biomed Anal. 2011;54:27–36. doi: 10.1016/j.jpba.2010.07.044. doi:10.1016/j.jpba.2010.07.044. [DOI] [PubMed] [Google Scholar]
- Lommel M, Strahl S. Protein O-mannosylation: Conserved from bacteria to humans. Glycobiology. 2009;19:816–828. doi: 10.1093/glycob/cwp066. doi:10.1093/glycob/cwp066. [DOI] [PubMed] [Google Scholar]
- McEachern JA, Bingham J, Crameri G, Green DJ, Hancock TJ, Middleton D, Feng YR, Broder CC, Wang LF, Bossart KN. A recombinant subunit vaccine formulation protects against lethal Nipah virus challenge in cats. Vaccine. 2008;26:3842–3852. doi: 10.1016/j.vaccine.2008.05.016. doi:10.1016/j.vaccine.2008.05.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mungall BA, Middleton D, Crameri G, Bingham J, Halpin K, Russell G, Green D, McEachern J, Pritchard LI, Eaton BT, et al. Feline model of acute Nipah virus infection and protection with a soluble glycoprotein-based subunit vaccine. J Virol. 2006;80:12293–12302. doi: 10.1128/JVI.01619-06. doi:10.1128/JVI.01619-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura N, Lyalin D, Panin VM. Protein O-mannosylation in animal development and physiology: From human disorders to Drosophila phenotypes. Semin Cell Dev Biol. 2010;21:622–630. doi: 10.1016/j.semcdb.2010.03.010. doi:10.1016/j.semcdb.2010.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel AH, Subak-Sharpe JH, Stow ND. The N-terminal 22 amino acids encoded by the gene specifying the major secreted protein of vaccinia virus, strain Lister, can function as a signal sequence to direct the export of a foreign protein. Virus Res. 1992;26:197–212. doi: 10.1016/0168-1702(92)90013-y. doi:10.1016/0168-1702(92)90013-Y. [DOI] [PubMed] [Google Scholar]
- Peter-Katalinic J. Methods in enzymology: O-glycosylation of proteins. Methods Enzymol. 2005;405:139–171. doi: 10.1016/S0076-6879(05)05007-X. doi:10.1016/S0076-6879(05)05007-X. [DOI] [PubMed] [Google Scholar]
- Terwilliger TC, Grosse-Kunstleve RW, Afonine PV, Moriarty NW, Zwart PH, Hung LW, Read RJ, Adams PD. Iterative model building, structure refinement and density modification with the PHENIX AutoBuild wizard. Acta Crystallogr D Biol Crystallogr. 2008;64:61–69. doi: 10.1107/S090744490705024X. doi:10.1107/S090744490705024X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varki A. Biological roles of oligosaccharides: All of the theories are correct. Glycobiology. 1993;3:97–130. doi: 10.1093/glycob/3.2.97. doi:10.1093/glycob/3.2.97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vigerust DJ, Shepherd VL. Virus glycosylation: Role in virulence and immune interactions. Trends Microbiol. 2007;15:211–218. doi: 10.1016/j.tim.2007.03.003. doi:10.1016/j.tim.2007.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang LF, Yu M, Hansson E, Pritchard LI, Shiell B, Michalski WP, Eaton BT. The exceptionally large genome of Hendra virus: Support for creation of a new genus within the family Paramyxoviridae. J Virol. 2000;74:9972–9979. doi: 10.1128/jvi.74.21.9972-9979.2000. doi:10.1128/JVI.74.21.9972-9979.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weerapana E, Imperiali B. Asparagine-linked protein glycosylation: From eukaryotic to prokaryotic systems. Glycobiology. 2006;16:91R–101R. doi: 10.1093/glycob/cwj099. doi:10.1093/glycob/cwj099. [DOI] [PubMed] [Google Scholar]
- Xu K, Rajashankar KR, Chan YP, Himanen JP, Broder CC, Nikolov DB. Host cell recognition by the henipaviruses: Crystal structures of the Nipah G attachment glycoprotein and its complex with ephrin-B3. Proc Natl Acad Sci USA. 2008;105:9953–9958. doi: 10.1073/pnas.0804797105. doi:10.1073/pnas.0804797105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan P, Leser GP, Demeler B, Lamb RA, Jardetzky TS. Domain architecture and oligomerization properties of the paramyxovirus PIV 5 hemagglutinin-neuraminidase (HN) protein. Virology. 2008;378:282–291. doi: 10.1016/j.virol.2008.05.023. doi:10.1016/j.virol.2008.05.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaia J. Mass spectrometry and glycomics. OMICS. 2010;14:401–418. doi: 10.1089/omi.2009.0146. doi:10.1089/omi.2009.0146. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.