Abstract
The gene and protein structure of the mouse UBF (mUBF), a transcription factor for mouse ribosomal RNA gene, have been determined by cDNA and genomic clones. The unique mUBF gene consists of 21 exons spanning over 13 kb. Two mRNAs coding for mUBF1 and mUBF2 having 765 a.a. and 728 a.a., respectively, are produced by an alternative splicing of exon 8. It specifies 37 amino acids constituting a part of the regions homologous to high mobility group proteins (HMG box 2). A human UBF (hUBF) cDNA obtained by polymerase chain reaction also indicates the presence of two kinds of mRNAs, the shorter form lacking the same region as mUBF2. Comparison of the cDNAs from hUBF and mUBF revealed an unusual conservation of nucleotide sequence in the 3'-terminal non-coding region. We examined the relative amounts of expression of mUBF1 and mUBF2. The eight tissues studied contained both molecular species, although mUBF2 was the predominant form of UBF. The mRNA of mUBF1 was expressed one half of the mUBF2 in quiescent mouse fibroblasts but reached the same amount in growing state.
Full text
PDF




Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Aviv H., Leder P. Purification of biologically active globin messenger RNA by chromatography on oligothymidylic acid-cellulose. Proc Natl Acad Sci U S A. 1972 Jun;69(6):1408–1412. doi: 10.1073/pnas.69.6.1408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bachvarov D., Moss T. The RNA polymerase I transcription factor xUBF contains 5 tandemly repeated HMG homology boxes. Nucleic Acids Res. 1991 May 11;19(9):2331–2335. doi: 10.1093/nar/19.9.2331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bateman E., Paule M. R. Events during eucaryotic rRNA transcription initiation and elongation: conversion from the closed to the open promoter complex requires nucleotide substrates. Mol Cell Biol. 1988 May;8(5):1940–1946. doi: 10.1128/mcb.8.5.1940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bateman E., Paule M. R. Regulation of eukaryotic ribosomal RNA transcription by RNA polymerase modification. Cell. 1986 Nov 7;47(3):445–450. doi: 10.1016/0092-8674(86)90601-x. [DOI] [PubMed] [Google Scholar]
- Bell S. P., Jantzen H. M., Tjian R. Assembly of alternative multiprotein complexes directs rRNA promoter selectivity. Genes Dev. 1990 Jun;4(6):943–954. doi: 10.1101/gad.4.6.943. [DOI] [PubMed] [Google Scholar]
- Bell S. P., Learned R. M., Jantzen H. M., Tjian R. Functional cooperativity between transcription factors UBF1 and SL1 mediates human ribosomal RNA synthesis. Science. 1988 Sep 2;241(4870):1192–1197. doi: 10.1126/science.3413483. [DOI] [PubMed] [Google Scholar]
- Bell S. P., Pikaard C. S., Reeder R. H., Tjian R. Molecular mechanisms governing species-specific transcription of ribosomal RNA. Cell. 1989 Nov 3;59(3):489–497. doi: 10.1016/0092-8674(89)90032-9. [DOI] [PubMed] [Google Scholar]
- Cassidy B., Haglund R., Rothblum L. I. Regions upstream from the core promoter of the rat ribosomal gene are required for the formation of a stable transcription initiation complex by RNA polymerase I in vitro. Biochim Biophys Acta. 1987 Jul 14;909(2):133–144. doi: 10.1016/0167-4781(87)90035-2. [DOI] [PubMed] [Google Scholar]
- Cavanaugh A. H., Thompson E. A., Jr Hormonal regulation of transcription of rDNA: glucocorticoid effects upon initiation and elongation in vitro. Nucleic Acids Res. 1985 May 10;13(9):3357–3369. doi: 10.1093/nar/13.9.3357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clos J., Buttgereit D., Grummt I. A purified transcription factor (TIF-IB) binds to essential sequences of the mouse rDNA promoter. Proc Natl Acad Sci U S A. 1986 Feb;83(3):604–608. doi: 10.1073/pnas.83.3.604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gregor P. D., Sawadogo M., Roeder R. G. The adenovirus major late transcription factor USF is a member of the helix-loop-helix group of regulatory proteins and binds to DNA as a dimer. Genes Dev. 1990 Oct;4(10):1730–1740. doi: 10.1101/gad.4.10.1730. [DOI] [PubMed] [Google Scholar]
- Grummt I. Specific transcription of mouse ribosomal DNA in a cell-free system that mimics control in vivo. Proc Natl Acad Sci U S A. 1981 Feb;78(2):727–731. doi: 10.1073/pnas.78.2.727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gubler U., Hoffman B. J. A simple and very efficient method for generating cDNA libraries. Gene. 1983 Nov;25(2-3):263–269. doi: 10.1016/0378-1119(83)90230-5. [DOI] [PubMed] [Google Scholar]
- Haltiner M. M., Smale S. T., Tjian R. Two distinct promoter elements in the human rRNA gene identified by linker scanning mutagenesis. Mol Cell Biol. 1986 Jan;6(1):227–235. doi: 10.1128/mcb.6.1.227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henikoff S., Kelly J. D., Cohen E. H. Transcription terminates in yeast distal to a control sequence. Cell. 1983 Jun;33(2):607–614. doi: 10.1016/0092-8674(83)90441-5. [DOI] [PubMed] [Google Scholar]
- Jantzen H. M., Admon A., Bell S. P., Tjian R. Nucleolar transcription factor hUBF contains a DNA-binding motif with homology to HMG proteins. Nature. 1990 Apr 26;344(6269):830–836. doi: 10.1038/344830a0. [DOI] [PubMed] [Google Scholar]
- Kato H., Nagamine M., Kominami R., Muramatsu M. Formation of the transcription initiation complex on mammalian rDNA. Mol Cell Biol. 1986 Oct;6(10):3418–3427. doi: 10.1128/mcb.6.10.3418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kishimoto T., Nagamine M., Sasaki T., Takakusa N., Miwa T., Kominami R., Muramatsu M. Presence of a limited number of essential nucleotides in the promoter region of mouse ribosomal RNA gene. Nucleic Acids Res. 1985 May 24;13(10):3515–3532. doi: 10.1093/nar/13.10.3515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kornfeld K., Saint R. B., Beachy P. A., Harte P. J., Peattie D. A., Hogness D. S. Structure and expression of a family of Ultrabithorax mRNAs generated by alternative splicing and polyadenylation in Drosophila. Genes Dev. 1989 Feb;3(2):243–258. doi: 10.1101/gad.3.2.243. [DOI] [PubMed] [Google Scholar]
- Learned R. M., Cordes S., Tjian R. Purification and characterization of a transcription factor that confers promoter specificity to human RNA polymerase I. Mol Cell Biol. 1985 Jun;5(6):1358–1369. doi: 10.1128/mcb.5.6.1358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Learned R. M., Learned T. K., Haltiner M. M., Tjian R. T. Human rRNA transcription is modulated by the coordinate binding of two factors to an upstream control element. Cell. 1986 Jun 20;45(6):847–857. doi: 10.1016/0092-8674(86)90559-3. [DOI] [PubMed] [Google Scholar]
- Miller K. G., Tower J., Sollner-Webb B. A complex control region of the mouse rRNA gene directs accurate initiation by RNA polymerase I. Mol Cell Biol. 1985 Mar;5(3):554–562. doi: 10.1128/mcb.5.3.554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mishima Y., Financsek I., Kominami R., Muramatsu M. Fractionation and reconstitution of factors required for accurate transcription of mammalian ribosomal RNA genes: identification of a species-dependent initiation factor. Nucleic Acids Res. 1982 Nov 11;10(21):6659–6670. doi: 10.1093/nar/10.21.6659. [DOI] [PMC free article] [PubMed] [Google Scholar]
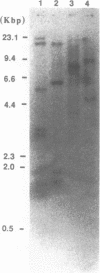
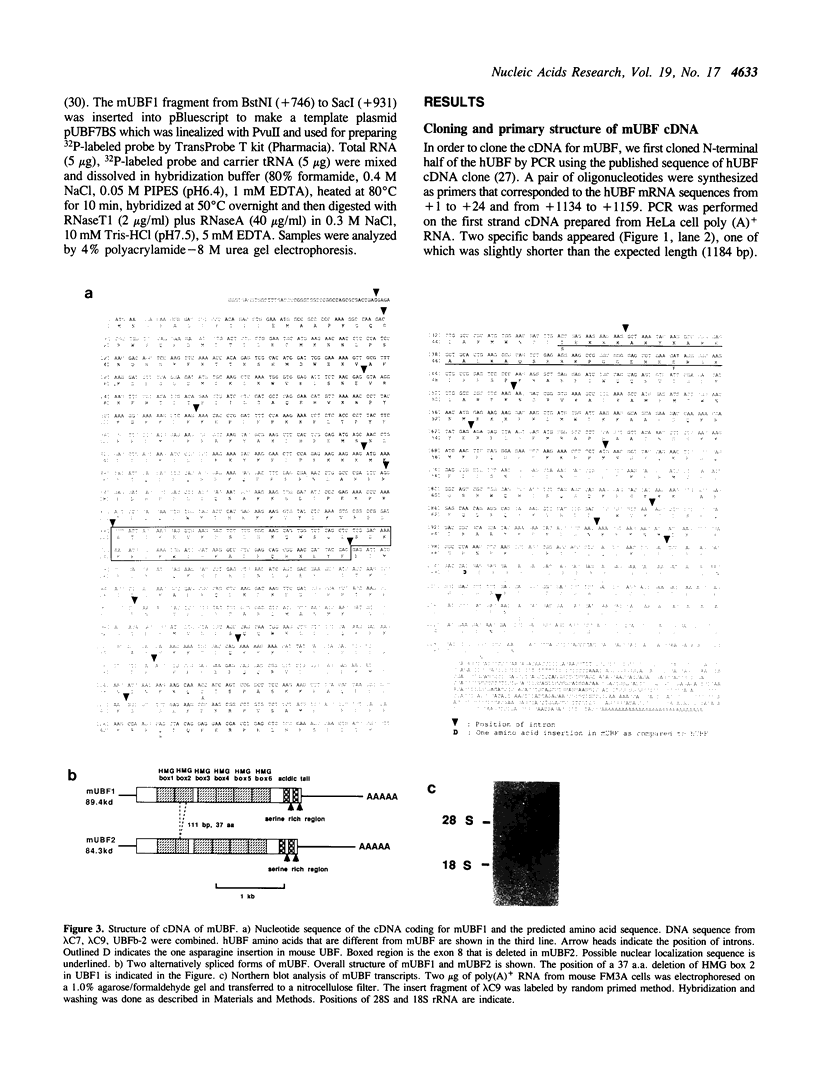
- Mishima Y., Yamamoto O., Kominami R., Muramatsu M. In vitro transcription of a cloned mouse ribosomal RNA gene. Nucleic Acids Res. 1981 Dec 21;9(24):6773–6785. doi: 10.1093/nar/9.24.6773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagamine M., Kishimoto T., Aono J., Kato H., Kominami R., Muramatsu M. Sequestration analysis for RNA polymerase I transcription factors with various deletion and point mutations reveals different functional regions of the mouse rRNA gene promoter. Mol Cell Biol. 1987 Apr;7(4):1486–1495. doi: 10.1128/mcb.7.4.1486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakabeppu Y., Nathans D. A naturally occurring truncated form of FosB that inhibits Fos/Jun transcriptional activity. Cell. 1991 Feb 22;64(4):751–759. doi: 10.1016/0092-8674(91)90504-r. [DOI] [PubMed] [Google Scholar]
- O'Mahony D. J., Rothblum L. I. Identification of two forms of the RNA polymerase I transcription factor UBF. Proc Natl Acad Sci U S A. 1991 Apr 15;88(8):3180–3184. doi: 10.1073/pnas.88.8.3180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pikaard C. S., McStay B., Schultz M. C., Bell S. P., Reeder R. H. The Xenopus ribosomal gene enhancers bind an essential polymerase I transcription factor, xUBF. Genes Dev. 1989 Nov;3(11):1779–1788. doi: 10.1101/gad.3.11.1779. [DOI] [PubMed] [Google Scholar]
- Pikaard C. S., Smith S. D., Reeder R. H., Rothblum L. rUBF, an RNA polymerase I transcription factor from rats, produces DNase I footprints identical to those produced by xUBF, its homolog from frogs. Mol Cell Biol. 1990 Jul;10(7):3810–3812. doi: 10.1128/mcb.10.7.3810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santoro C., Mermod N., Andrews P. C., Tjian R. A family of human CCAAT-box-binding proteins active in transcription and DNA replication: cloning and expression of multiple cDNAs. Nature. 1988 Jul 21;334(6179):218–224. doi: 10.1038/334218a0. [DOI] [PubMed] [Google Scholar]
- Schnapp A., Clos J., Hädelt W., Schreck R., Cvekl A., Grummt I. Isolation and functional characterization of TIF-IB, a factor that confers promoter specificity to mouse RNA polymerase I. Nucleic Acids Res. 1990 Mar 25;18(6):1385–1393. doi: 10.1093/nar/18.6.1385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schnapp A., Pfleiderer C., Rosenbauer H., Grummt I. A growth-dependent transcription initiation factor (TIF-IA) interacting with RNA polymerase I regulates mouse ribosomal RNA synthesis. EMBO J. 1990 Sep;9(9):2857–2863. doi: 10.1002/j.1460-2075.1990.tb07475.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shirakawa H., Tsuda K., Yoshida M. Primary structure of non-histone chromosomal protein HMG2 revealed by the nucleotide sequence. Biochemistry. 1990 May 8;29(18):4419–4423. doi: 10.1021/bi00470a022. [DOI] [PubMed] [Google Scholar]
- Smale S. T., Tjian R. Transcription of herpes simplex virus tk sequences under the control of wild-type and mutant human RNA polymerase I promoters. Mol Cell Biol. 1985 Feb;5(2):352–362. doi: 10.1128/mcb.5.2.352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith S. D., Oriahi E., Lowe D., Yang-Yen H. F., O'Mahony D., Rose K., Chen K., Rothblum L. I. Characterization of factors that direct transcription of rat ribosomal DNA. Mol Cell Biol. 1990 Jun;10(6):3105–3116. doi: 10.1128/mcb.10.6.3105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka N., Kato H., Ishikawa Y., Hisatake K., Tashiro K., Kominami R., Muramatsu M. Sequence-specific binding of a transcription factor TFID to the promoter region of mouse ribosomal RNA gene. J Biol Chem. 1990 Aug 15;265(23):13836–13842. [PubMed] [Google Scholar]
- Tower J., Culotta V. C., Sollner-Webb B. Factors and nucleotide sequences that direct ribosomal DNA transcription and their relationship to the stable transcription complex. Mol Cell Biol. 1986 Oct;6(10):3451–3462. doi: 10.1128/mcb.6.10.3451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsuda K., Kikuchi M., Mori K., Waga S., Yoshida M. Primary structure of non-histone protein HMG1 revealed by the nucleotide sequence. Biochemistry. 1988 Aug 9;27(16):6159–6163. doi: 10.1021/bi00416a050. [DOI] [PubMed] [Google Scholar]
- Yamamoto K. K., Gonzalez G. A., Menzel P., Rivier J., Montminy M. R. Characterization of a bipartite activator domain in transcription factor CREB. Cell. 1990 Feb 23;60(4):611–617. doi: 10.1016/0092-8674(90)90664-z. [DOI] [PubMed] [Google Scholar]
- Yamamoto O., Takakusa N., Mishima Y., Kominami R., Muramatsu M. Determination of the promoter region of mouse ribosomal RNA gene by an in vitro transcription system. Proc Natl Acad Sci U S A. 1984 Jan;81(2):299–303. doi: 10.1073/pnas.81.2.299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaret K. S., Sherman F. DNA sequence required for efficient transcription termination in yeast. Cell. 1982 Mar;28(3):563–573. doi: 10.1016/0092-8674(82)90211-2. [DOI] [PubMed] [Google Scholar]
- Zheng X. M., Black D., Chambon P., Egly J. M. Sequencing and expression of complementary DNA for the general transcription factor BTF3. Nature. 1990 Apr 5;344(6266):556–559. doi: 10.1038/344556a0. [DOI] [PubMed] [Google Scholar]