Abstract
Almost all eukaryotic mRNAs possess 3′ ends with a polyadenylate (poly(A)) tail. This poly(A) tail is not encoded in the genome but is added by the process of polyadenylation. Polyadenylation is a two-step process, and this process is accomplished by multisubunit protein factors. Here, we comprehensively compare the protein machinery responsible for polyadenylation of mRNAs across many evolutionary divergent species, and we have found these protein factors to be remarkably conserved in nature. These data suggest that polyadenylation of mRNAs is an ancient process.
1. Introduction
Almost all eukaryotic mRNAs have a poly(A) tail at their 3′ ends, with the most notable exception being histone mRNAs. The process by which mRNAs acquire a poly(A) tail is termed polyadenylation. Polyadenylation is a tightly coupled, two-step process that first endonucleolytically cleaves the pre-mRNA and subsequently adds an unencoded poly(A) tail (reviewed in [1–7]). Poly(A) tails serve the mRNA in many ways, aiding in mRNA translation, facilitating transport from the nucleus to the cytoplasm, and promoting stability [8–12]. The addition of the poly(A) tail is a highly coordinated event, requiring cooperation from both cis-acting RNA sequence elements and trans-acting protein factors to complete the process [13, 14]. Alternative or regulated polyadenylation likely requires further cooperation and integration of efforts.
Two sequence elements in mammals serve as the core polyadenylation elements: the AAUAAA or a variant, and a U/GU-rich element located downstream 10–30 nts of the actual site of polyadenylation (Figure 1, [15, 16] and references therein). The cleavage site, where the poly(A) tail is added, is located in between these two sequence elements and is often a CA dinucleotide, but it has some variability ([15] and references therein). The AAUAAA element serves as a binding site for the CPSF (cleavage and polyadenylation specificity factor) complex, a complex of four subunits, while the U/GU-rich element binds the CstF (cleavage stimulation factor) complex, a trimeric complex of proteins (Figure 1). Yeast polyadenylation signals have a slightly different composition but bind similar protein complexes with slightly different orientation.
Figure 1.

Human polyadenylation factors. Human basal polyadenylation factors are composed of many multisubunit complexes: CPSF, CstF, CFIm, and CFIIm. There are also many other auxiliary factors that contribute to polyadenylation; representative factors are listed at the top right.
The protein factors that make up the basal polyadenylation machinery in mammalian cells were purified, isolated, and cloned by many laboratories in the 1990s (including [17–23]). Additional proteins that influence or regulate polyadenylation have also been identified over the past decade or more (including [24–27]). Many of the basal polyadenylation factors from mammalian cells, and some additional factors, have been shown to have orthologues or homologs in other organisms. A report has compared the mammalian polyadenylation machinery with that of the protozoan Entamoeba histolytica [27]; however, no comprehensive study has been undertaken to compare and contrast the polyadenylation machinery from a number of different species. Here, we have compared basal polyadenylation factors from human to species ranging from mouse to plants and archaea and have found most of them to be remarkably conserved. These findings are consistent with the universal eukaryotic nature of mRNAs having a poly(A) tail.
2. Materials and Methods
2.1. Homologous Human Polyadenylation Factors
The human polyadenylation factors were compared to 14 different species that are shown in Table 1. Using the NCBI protein-protein BLAST (blastp, version 2.2.25), we compared the human polyadenylation factor protein sequences to homologous sequences present in the other species through the nonredundant database (nr). The highest ranked protein with a bit score of 50 or greater was chosen as the homolog. These proteins were compared to the human factor in question by the number of amino acids present in the homolog relative to the human factor, as well as by amino acid alignment of the same or similar amino acids.
Table 1.
Species included in the phylogenetic comparison. Common and scientific names are included. The common name will be used in the comparison presented here.
| Common Name | Scientific Name |
|---|---|
| Mouse | Mus musculus |
| Chicken | Gallus gallus |
| Fly | Drosophila melanogaster |
| Mosquito | Anopheles gambiae |
| Purple sea urchin | Strongylocentrotus purpuratus |
| Trypanosome | Trypanosoma brucei |
| Trypanosoma cruzi | |
| Nematode | Caenorhabditis elegans |
| Rice | Oryza sativa |
| Thale cress | Arabidopsis thaliana |
| Wine grape | Vitis vinifera |
| Fission yeast | Schizosaccharomyces pombe |
| Budding yeast | Saccharomyces cerevisiae |
| Archaea | Haloferax volcanii |
2.2. Domain Comparison
The NCBI conserved domain database was used to find the domains in each of the human polyadenylation factor proteins as well as known published human domains. The presence of these domains was determined in each of its corresponding homologs. The domains were aligned using the same parameters of comparison as the whole protein comparison.
3. Results and Discussion
By comparing basal polyadenylation factors from a phylogenetic perspective, we can gain insight into functional and mechanistic differences that may exist in different species. We have compared and contrasted polyadenylation factors from a number of different species for their overall homology and percent identity relative to human, as well as for their similarity in specific protein domains. The species we analyzed from mouse to archaea are shown in Table 1. Tables 2 and 3 show the specific locus name for a given polyadenylation factor for each species. In some instances, the locus name may not reveal much. CPSF 1, 2, 3, and 4 are also known as CPSF 160, 100, 73, and 30, respectively. CSTF 1, 2, and 3 are known as CstF 55, 64, and 77, respectively; CPSF 6 is also known as CFIm68; PAPOLA is poly(A) polymerase.
Table 2.
Homologs of human polyadenylation factors in vertebrates, insects, sea urchin, and trypanosomes. Protein sequences of basal polyadenylation factors from human were compared to other species found in Table 1 to find the homologous factors.
| Human | Mouse | Chicken | Fly | Mosquito | Purple sea urchin | Trypanosomes (T. cruzi) | Trypanosome (T. brucei) | |
|---|---|---|---|---|---|---|---|---|
| CPSF complex | CPSF1 | CPSF1 | LOC770075 | CPSF160 isoform A | AGAP011340-PA | LOC584773 | Tc00.1047053506871.140 | Tb11.01.6170 |
| CPSF160 isoform B | ||||||||
| CPSF2 | CPSF2 | CPSF2 | CPSF100 isoform A | AGAP002474-PA | LOC582050 | Tc00.1047053504109.110 | Tb11.03.0910 | |
| CPSF100 isoform B | ||||||||
| CPSF3 | CPSF3 | CPSF3 | CPSF73 | AGAP001224-PA | LOC591455 | Tc00.1047053511003.221 | Tb927.4.1340 | |
| CPSF4 isoform 1 | CPSF4 | CPSF4 | CLP | AGAP005735-PA | LOC765046 | Tc00.1047053511555.40 | Tb11.01.4600 | |
| CPSF4 isoform 2 | ||||||||
| FIP1L1 isoform 1 | FIP1L1 | FIP1L1 | FIP1 | AGAP001514-PA | LOC580164 | Tc00.1047053507601.80 | Tb927.5.4320 | |
| FIP1L1 isoform 2 | ||||||||
| FIP1L1 isoform 3 | ||||||||
|
| ||||||||
| CstF complex | CSTF1 isoform 1 | CSTF1 | CSTF1 | CST-50 isoform A | AGAP002776-PA | LOC582854 | Tc00.1047053511365.10 | Tb10.61.0570 |
| CSTF1 isoform 2 | CST-50 isoform B | |||||||
| CSTF1 isoform 3 | ||||||||
| CSTF2 | CSTF2 | CSTF2 | CSTF-64 | AGAP010918-PA | LOC759858 | Tc00.1047053506795.10 | Tb927.7.3730 | |
| CSTF2T | CSTF2T | |||||||
| CSTF3 isoform 1 | CSTF3 | CSTF3 | SU(F) | AGAP003019-PA | LOC582899 | |||
| CSTF3 isoform 2 | LOC591939 | |||||||
| CSTF3 isoform 3 | ||||||||
|
| ||||||||
| CF1m | CPSF6 | CPSF6 | CPSF6 | CG7185 | AGAP005062-PA | LOC577326 | ||
| CSPF7 | CPSF7 | CPSF7 | ||||||
| NUDT21 | NUDT21 | AMFR | CG3689 isoform B | AGAP007242-PA | LOC579716 | Tc00.1047053509509.40 | Tb927.7.1620 | |
| CG3689 isoform C | Tc00.1047053508207.220 | |||||||
|
| ||||||||
| CFIIm | CLP1 | CLP1 | CLP1 | CBC | AGAP007701-PA | LOC763581 | Tc00.1047053507027.59 | Tb927.6.3690 |
| Tc00.1047053506941.229 | ||||||||
| PCF11 | PCF11 | PCF11 | PCF11 | AGAP001271-PA | LOC582414 | |||
|
| ||||||||
| Other factors | PAPOLA | PAPOLA | PAPOLA | hrg isoform A | LOC575500 | Tc00.1047053506795.50 | Tb927.7.3780 | |
| hrg isoform B | ||||||||
| hrg isoform C | ||||||||
| PAPOLB | PAPOLB | |||||||
| PAPOLG | ||||||||
| SYMPK | SYMPK | SYM | AGAP002618-PA | SYMPK | ||||
|
| ||||||||
| PABP | PABPC1 | PABPC1 | PABPC1 | PABP | AGAP011092-PA | PABP | Tc00.1047053506885.70 | Tb09.211.2150 |
| PABPC3 | PABPC6 | |||||||
| PABPC4 | PABPC4 | PABPC4 | Tc00.1047053506885.70 | Tb09.211.2150 | ||||
| PABPN1 | PABPN1 | PABPN1 | PABP2 | AGAP005117-PA | LOC594592 | Tc00.1047053511741.40 | Tb09.211.4120 | |
|
| ||||||||
| Homologs of yeast polyadenylation factors | WDR33 | WDR33 | WDR33 | CG1109 | AGAP001362-PA | LOC574793 | TC00.1047053511491.140 | Tb927.6.1830 |
| RBBP6 | RBBP6 | RBBP6 | SNAMA | AGAP011217-PA | LOC584197 | |||
| PPP1CA | PPP1CA | PPP1CC | PP1alpha-96A | AGAP011166-PA | LOC586142 | Tc00.1047053508815.110 | Tb11.01.0450 | |
| PPP1CB | PPP1CB | PPP1CB | PP1alpha-96A | AGAP003114-PA | LOC752338 | Tc00.1047053508815.110 | Tb11.01.0450 | |
Table 3.
Homologs of human polyadenylation factors in nematode, plants, yeast, and archaea. Protein sequences of basal polyadenylation factors from human were compared to other species found in Table 1 to find the homologous factors.
| Human | Nematode | Rice | Thale cress | Wine grape | Fission yeast | Budding yeast | Archaea | |
|---|---|---|---|---|---|---|---|---|
| CPSF complex | CPSF1 | CPSF-1 | Os04g0252200 | CPSF160 | LOC100256706 | CFT1 | CFT1p | |
| CPSF2 | CPSF-2 | Os09g0569400 | CPSF100 | LOC100267865 | CFT2 | CFT2p | EPF1 | |
| CPSF3 | CPSF-3 | Os03g0852900 | CPSF73-I | LOC100261042 | YSH1 | YSH1 | EPF2 | |
| CPSF73-II | ||||||||
| CPSF4 isoform 1 | CPSF-4 | Os06g0677700 | CPSF30 | LOC100253258 | YTH1 | YTH1 | ||
| CPSF4 isoform 2 | ||||||||
| FIP1L1 isoform 1 | F32D1.9 | Os01g0377500 | FIP1[V] | LOC100251960 | SPAC22G7.10 | Fip1p | ||
| FIP1L1 isoform 2 | ||||||||
| FIP1L1 isoform 3 | ||||||||
|
| ||||||||
| CstF complex | CSTF1 isoform 1 | CPF-1 | Os03g0754900 | AT5G60940 | LOC100267233 | |||
| CSTF1 isoform 2 | ||||||||
| CSTF1 isoform 3 | ||||||||
| CSTF2 | CPF-2 | Os11g0176100 | CSTF64 | LOC100256296 | CTF1 | RNA15 | ||
| CSTF2T | ||||||||
| CSTF3 isoform 1 | SUF-1 | Os12g0571900 | CSTF77 | LOC100262033 | RNA14 | RNA14 | ||
| CSTF3 isoform 2 | ||||||||
| CSTF3 isoform 3 | ||||||||
|
| ||||||||
| CF1m | CPSF6 | D1046.1 | Os09g0476100 | AT5G55670 | LOC100268141 | |||
| CSPF7 | AT1G13190 | |||||||
| NUDT21 | F43G9.5 | Os04g0683100 | AT4G25550 | LOC100261950 isoform 1 | ||||
| CFIIM-25 | LOC100261950 isoform 2 | |||||||
|
| ||||||||
| CFIIm | CLP1 | F59A2.4 | Os02g0217500 | CLPS5 | LOC100242380 | SPAC22H10.05c | Clp1p | |
| PCF11 | R144.2 | Os09g0566100 | PCFS4 | LOC100251089 | SPAC4G9.04c | PCF11 | ||
|
| ||||||||
| Other factors | PAPOLA | Pap-1 | Os06g0319600 | PAPS1 | LOC100252483 | Pla1 | Pap1 | |
| PAPOLB | Os06g0558700 | PAPS2 | LOC100263460 | |||||
| PAPOLG | ||||||||
| SYMPK | F25G6.2 | Os07g0693900 | ESP4 | LOC100266091 | PTA1 | PTA1 | ||
|
| ||||||||
| PABP | PABPC1 | PAB-1 | Os08g0314800 | PAB2 | LOC100262903 | PABP | PAB1 | |
| PABPC3 | PABP5 | LOC100255846 | ||||||
| PABPC4 | PAB5 | LOC100255846 | ||||||
| PABPN1 | PABP-2 | Os06g0219600 | AT5G10350 | LOC100242522 | PAB2 | SGN1 | ||
|
| ||||||||
| Homologs of yeast polyadenylation factors | WDR33 | R06A4.9 | Os04g0599800 | FY | LOC100263567 | PFS2 | PFS2 | |
| RBBP6 | TAG-214 | Os10g0431000 | AT5G47430 | LOC100252571 | SPBP8B7.15c | MPE1 | ||
| PPP1CA | GSP-2 | OS03g0268000 | TOPP7 | LOC100256994 | DIS2 | GLC7 | ||
| PPP1CB | GSP-1 | Os06g0164100 | TOPP4 | LOC100258649 | DIS2 | GLC7 | ||
Human polyadenylation factor homologs were found for most of the species with the major exception of archaea and yeast (Tables 2 and 3). Archaea only had homologs in the CPSF complex. A polymer “A” tail is not found in H. volcanii [28]. In some archaea, a random copolymer tail is added by the exosome or PnPase [29]. Therefore, most of the human polyadenylation factors evolved after archaea.
Both yeast species did not contain homologs for the entire CFIm complex and CSTF1 (Table 3). This emphasizes a major difference in yeast and human polyadenylation (reviewed in [1, 13]). CFIm is involved in early steps of polyadenylation and recruits other polyadenylation factors [14, 30, 31]. This is achieved by NUDT21 binding to a UGUA sequence [32]. The Hrp1p complex in yeast likely plays a similar role as CFIm. Hrp1p binds to the polyadenylation enhancer element [33] and interacts with RNA14 and RNA15 [34]. RNA14 and RNA15 are homologs of the CSTF2 and CSTF3 human proteins. Therefore, Hrp1p may abrogate the need for CSTF1 and CFIm complex in yeast.
The malaria mosquito (Anopheles gambiae) did not contain any poly(A) polymerase homologs (Table 2). This is most likely due to missing gene annotation because the yellow fever mosquito (Aedes aegypti) and southern house mosquito (Culex quinquefasciatus) contain a poly(A) polymerase homolog.
Humans have gene variant forms of CSTF2, PABPC, and PAPOLA that are tissue-specific. CSTF2T (CstF-64 tau) is expressed in the testis and brain and is found in meiotic and postmeiotic germ cells where CSTF2 is inactivated [35]. This variant was only found in the human and mouse species. Cytoplasmic PABP has two cell-specific isoforms, PABPC3 and PABPC4. PABPC3 is found in the testis and has a lower binding affinity to RNA [36], and PABPC4 is inducible in T cells [37]. Both of these proteins are found in mouse and the eudicot plants. PABPC4 is also found in chicken, trypanosomes, and eudicot plants. Poly(A) polymerase has a testis-specific gene variant form, PAPOLB [38]. Homologs are also found in mouse and plants. PAPOLG homolog was only found in mouse. The human gene variant homologs of PABPC and PAPOLA found in plants emphasize the difference in plant and human polyadenylation (reviewed in [39]). Thale cress contains at least eight isoforms of PABP and four isoforms of PAP [40, 41]. Homologs for most tissue-specific human polyadenylation factors are more recently evolved since homologs are only found in mouse.
Humans have several isoforms of the polyadenylation factors FIPI1L, CSTF1, and CSTF3 (Tables 2 and 3). Multiple isoforms of these factors were not found in any of the other species. The NUDT21 complex contained the most evolutionary conserved multiple isoforms with isoforms only in Drosophila, T. cruzi, and eudicots. Drosophila has the most species-specific isoforms for human factors CPSF1, CPSF2, CSTF1, NUDT21, and PAPOLA, but there is generally only one isoform of these factors in the other species. Therefore, isoforms of some polyadenylation factors are not evolutionary conserved and often their function is species specific.
We concluded from this comparison that human basal polyadenylation factors are quite well conserved evolutionarily with the exceptions of archaea and some yeast factors, tissue-specific gene variants, and protein isoforms.
We next further analyzed the identified homologs of the human polyadenylation factor protein sequences to see how stringently the factors were conserved by two different means: conservation of protein length and conservation of the amino acids in the alignment with the same or a similar amino acid (Table 4). These analyses were performed using the NCBI databases and BLAST alignment tools.
Table 4.
Phylogenetic comparison of human basal polyadenylation factors. Human basal polyadenylation factors were compared to homologous factors in other species by two criteria: percent length is the change in the number of amino acids as compared to the specific human polyadenylation factor. Positive identity is the percentage of amino acids that align to the human polyadenylation factor that are the same or similar to amino acids.
| Species | Homolog | % length | % positive Identity | |
|---|---|---|---|---|
| CPSF1 | Mouse | CPSF1 | 100 | 98 |
| Chicken | LOC770075 | 5 | 91 | |
| Fly | CPSF160 iso. A | 101 | 63 | |
| CPSF160 iso. B | 98 | 61 | ||
| Mosquito | AGAP011340-PA | 99 | 65 | |
| Purple sea urchin | LOC584773 | 85 | 70 | |
| Trypanosome (T. cruzi) | Tc00.1047053506871.140 | 100 | 41 | |
| Trypanosome (T. brucei) | Tb11.01.6170 | 100 | 41 | |
| Nematode | Cpsf-1 | 101 | 52 | |
| Rice | Os04g0252200 | 30 | 57 | |
| Thale cress | CPSF160 | 100 | 50 | |
| Wine grape | LOC100256706 | 100 | 49 | |
| Fission yeast | CTF1 | 100 | 46 | |
| Budding yeast | CTF1 | 94 | 44 | |
|
| ||||
| CPSF2 | Mouse | CPSF2 | 100 | 99 |
| Chicken | CPSF2 | 100 | 97 | |
| Fly | CPSF100 iso. A | 97 | 69 | |
| CPSF100 iso. B | 85 | 68 | ||
| Mosquito | AGAP002474-PA | 95 | 71 | |
| Purple sea urchin | LOC582050 | 99 | 75 | |
| Trypanosome (T. cruzi) | Tc00.1047053504109.110 | 103 | 42 | |
| Trypanosome (T. brucei) | Tb11.03.0910 | 105 | 42 | |
| Nematode | CPSF-2 | 108 | 60 | |
| Rice | Os09g0569400 | 94 | 56 | |
| Thale cress | CPSF100 | 95 | 57 | |
| Wine grape | LOC100267865 | 95 | 62 | |
| Fission yeast | CFT2 | 102 | 49 | |
| Budding yeast | CFT2 | 110 | 46 | |
| Archaea (H. volcanii) | EPF1 | 82 | 40 | |
|
| ||||
| CPSF3 | Mouse | CPSF3 | 100 | 99 |
| Chicken | CSPF3 | 101 | 97 | |
| Fly | CPSF73 | 100 | 79 | |
| Mosquito | AGAP001224-PA | 85 | 88 | |
| Purple sea urchin | LOC591455 | 24 | 89 | |
| Trypanosome (T. cruzi) | Tc00.1047053511003.221 | 63 | 78 | |
| Trypanosome (T. brucei) | Tb927.4.1340 | 113 | 73 | |
| Nematode | CPSF-3 | 103 | 75 | |
| Rice | Os03g0852900 | 102 | 72 | |
| Thale cress | CPSF73-I | 101 | 72 | |
| CPSF73-II | 90 | 72 | ||
| Wine grape | LOC100261042 | 101 | 72 | |
| Fission yeast | YSH1 | 113 | 67 | |
| Budding yeast | YSH1 | 114 | 60 | |
| Archaea | EPF2 | 60 | 45 | |
|
| ||||
| CPSF4 | Mouse | CPSF4 | 82 | 75 |
| Chicken | CPSF4 | 90 | 88 | |
| Fly | Clp | 110 | 64 | |
| Mosquito | AGAP005735-PA | 290 | 47 | |
| Purple sea urchin | LOC765046 | 109 | 66 | |
| Trypanosome (T. cruzi) | Tc00.1047053511555.40 | 101 | 48 | |
| Trypanosome (T. brucei) | Tb11.01.4600 | 103 | 48 | |
| Nematode | CPSF-4 | 112 | 62 | |
| Rice | Os06g0677700 | 273 | 64 | |
| Thale cress | CPSF30 | 102 | 52 | |
| Wine grape | LOC100253258 | 275 | 67 | |
| Fission yeast | YTH1 | 63 | 72 | |
| Budding yeast | Yth1p | 78 | 64 | |
|
| ||||
| FIP1L1 | Mouse | FIP1L | 98 | 92 |
| Chicken | FIP1L | 130 | 88 | |
| Fly | FIP1 | 118 | 58 | |
| Mosquito | AGAP001514-PA | 96 | 63 | |
| Purple sea urchin | LOC580164 | 142 | 60 | |
| Trypanosome (T. cruzi) | Tc00.1047053507601.80 | 48 | 60 | |
| Trypanosome (T. brucei) | Tb927.5.4320 | 47 | 65 | |
| Nematode | F32D1.9 | 86 | 79 | |
| Rice | Os01g0377500 | 73 | 58 | |
| Thale cress | FIP1[V] | 203 | 68 | |
| Wine grape | LOC100251960 | 251 | 89 | |
| Fission yeast | SPAC22G7.10 | 58 | 82 | |
| Budding yeast | Fip1 | 55 | 52 | |
|
| ||||
| CstF1 | Mouse | Cstf1 | 100 | 99 |
| Chicken | Cstf1 | 125 | 99 | |
| Fly | CstF-50 isoform A | 98 | 87 | |
| CstF-50 isoform B | 74 | 65 | ||
| Mosquito | AGAP002776-PA | 93 | 72 | |
| Purple sea urchin | LOC582854 | 95 | 74 | |
| Trypanosome (T. cruzi) | Tc00.1047053511365.10 | 121 | 42 | |
| Trypanosome (T. brucei) | Tb10.61.0570 | 120 | 43 | |
| Nematode | cpf-1 | 100 | 69 | |
| Rice | Os03g0754900 | 109 | 58 | |
| Thale cress | AT5G60940 | 100 | 57 | |
| Wine grape | LOC100267233 | 113 | 57 | |
|
| ||||
| CstF2 | Mouse | CSTF2 | 101 | 96 |
| Chicken | CSTF2 | 82 | 70 | |
| Fly | CstF-64 | 73 | 82 | |
| Mosquito | AGAP010918-PA | 68 | 81 | |
| Purple sea urchin | LOC759858 | 118 | 75 | |
| Trypanosome (T. cruzi) | Tc00.1047053506795.10 | 59 | 62 | |
| Trypanosome (T. brucei) | Tb927.7.3730 | 59 | 63 | |
| Nematode | cpf-2 | 62 | 73 | |
| Rice | OSs11g0176100 | 88 | 55 | |
| Thale cress | CSFF64 | 80 | 47 | |
| Wine grape | LOC100256296 | 94 | 49 | |
| Fission yeast | CFT1 | 63 | 73 | |
| Budding yeast | RNA15 | 51 | 75 | |
|
| ||||
| CstF2T | Mouse | CSTF2t | 103 | 93 |
|
| ||||
| CstF3 | Mouse | Cstf3 | 100 | 99 |
| Chicken | Cstf3 | 100 | 99 | |
| Fly | su(f) | 102 | 74 | |
| Mosquito | AGAP003019-PA | 710 | 75 | |
| Purple sea urchin | LOC591939 | 78 | 87 | |
| LOC582899 | 90 | 74 | ||
| Nematode | Suf-1 | 103 | 68 | |
| Rice | Os12g0571900 | 709 | 71 | |
| Thale cress | CSTF77 | 713 | 71 | |
| Wine grape | LOC100262033 | 747 | 69 | |
| Fission yeast | RNA14 | 102 | 52 | |
| Budding yeast | RNA14 | 94 | 49 | |
|
| ||||
| CPSF6 | Mouse | CPSF6 | 100 | 99 |
| Chicken | CPSF6 | 100 | 98 | |
| Fly | CG7185 | 118 | 94 | |
| Mosquito | AGAP005062-PA | 117 | 64 | |
| Purple sea urchin | LOC577326 | 163 | 62 | |
| Nematode | D1046.1 | 89 | 43 | |
| Rice | Os09g0475100 | 110 | 60 | |
| Thale cress | AT5G55670 | 106 | 50 | |
| Wine grape | LOC100268141 | 116 | 51 | |
|
| ||||
| CPSF7 | Mouse | CPSF7 | 100 | 99 |
| Chicken | CPSF7 | 98 | 92 | |
| Thale cress | AT1G13190 | 122 | 46 | |
|
| ||||
| NUDT21 | Mouse | NUDT21 | 100 | 99 |
| Chicken | AMFR | 336 | 99 | |
| Fly | CG3689 isoform B | 89 | 83 | |
| CG3689 isoform C | 104 | 85 | ||
| Mosquito | AGAP007242-PA | 102 | 86 | |
| Purple sea urchin | LOC579716 | 100 | 96 | |
| Trypanosome (T. cruzi) | Tc00.1047053509509.40 | 129 | 51 | |
| Tc00.1047053508207.220 | 129 | 51 | ||
| Trypanosome (T. brucei) | Tb927.7.1620 | 132 | 49 | |
| Nematode | F43G9.5 | 100 | 84 | |
| Rice | Os04g0683100 | 114 | 73 | |
| Thale cress | AT4G25550 | 88 | 73 | |
| CFIM-25 | 98 | 67 | ||
| Wine grape | LOC100261950 isoform 1 | 88 | 73 | |
| LOC100261950 isoform 2 | 92 | 70 | ||
|
| ||||
| Clp1 | Mouse | Clp1 | 100 | 99 |
| Chicken | Clp1 | 100 | 98 | |
| Fly | cbc | 99 | 75 | |
| Mosquito | AGAP007701-PA | 117 | 65 | |
| Purple sea urchin | LOC763581 | 85 | 70 | |
| Trypanosome (T. cruzi) | Tc00.1047053507027.59 | 97 | 47 | |
| Tc00.1047053506941.229 | 97 | 47 | ||
| Trypanosome (T. brucei) | Tb927.6.3690 | 100 | 43 | |
| Nematode | F59A2.4 | 101 | 68 | |
| Rice | Os02g0217500 | 120 | 58 | |
| Thale cress | CLPS5 | 118 | 46 | |
| CLPS3 | 123 | 60 | ||
| Wine grape | LOC100242380 | 118 | 60 | |
| Fission yeast | SPAC22H10.05c | 108 | 54 | |
| Budding yeast | Clp | 104 | 47 | |
|
| ||||
| PCF11 | Mouse | PCF11 | 100 | 97 |
| Chicken | PCF11 | 97 | 77 | |
| Fly | PCF11 | 126 | 59 | |
| Mosquito | AGAP001271-PA | 120 | 56 | |
| Purple sea urchin | LOC582414 | 170 | 64 | |
| Nematode | R144.2 | 53 | 52 | |
| Rice | Os09g0566100 | 69 | 58 | |
| Thale cress | PCFS4 | 52 | 54 | |
| Wine grape | LOC100251089 | 70 | 55 | |
| Fission yeast | SPAC4G9.04c | 41 | 65 | |
| Budding yeast | PCF11 | 40 | 56 | |
|
| ||||
| WDR33 | Mouse | WDR33 | 100 | 96 |
| Chicken | WDR33 | 98 | 88 | |
| Fly | CG1109 | 60 | 80 | |
| Mosquito | AGAP001362-PA | 271 | 74 | |
| Purple sea urchin | LOC574793 | 86 | 82 | |
| Trypanosome (T. cruzi) | Tc00.1047053511491.140 | 33 | 53 | |
| Trypanosome (T. brucei) | Tb927.6.1830 | 33 | 52 | |
| Nematode | R06A4.9 | 61 | 57 | |
| Rice | Os04g0599800 | 155 | 47 | |
| Thale cress | FY | 198 | 65 | |
| Wine grape | LOC100263567 | 237 | 70 | |
| Fission yeast | PFS2 | 38 | 64 | |
| Budding yeast | PFS2 | 35 | 58 | |
|
| ||||
| RBBP6 | Mouse | RBBP6 | 100 | 93 |
| Chicken | RBBP6 | 101 | 82 | |
| Fly | SNAMA | 69 | 59 | |
| Mosquito | AGAP011217-PA | 69 | 60 | |
| Purple sea urchin | LOC584197 | 36 | 63 | |
| Nematode | TAG-214 | 63 | 51 | |
| Rice | Os10g0431000 | 26 | 48 | |
| Thale cress | AT5G47430 | 50 | 47 | |
| Wine grape | LOC100252571 | 101 | 62 | |
| Fission yeast | SPBP8B7.15c | 27 | 51 | |
| Budding yeast | MPE1 | 25 | 49 | |
|
| ||||
| PPP1CA | Mouse | PPP1CA | 100 | 100 |
| Chicken | PPP1CC | 98 | 94 | |
| Fly | PP1alpha-96A | 99 | 92 | |
| Mosquito | AGAP011166-PA | 96 | 90 | |
| Purple sea urchin | LOC586142 | 100 | 94 | |
| Trypanosome (T. cruzi) | Tc00.1047053508815.110 | 92 | 89 | |
| Trypanosome (T. brucei) | Tb11.01.0450 | 92 | 90 | |
| Nematode | GSP-2 | 100 | 95 | |
| Rice | OS03g0268000 | 95 | 89 | |
| Thale cress | TOPP7 | 94 | 84 | |
| Wine grape | LOC100256994 | 94 | 86 | |
| Fission yeast | DIS2 | 99 | 94 | |
| Budding yeast | GLC7 | 95 | 94 | |
|
| ||||
| PPP1CB | Mouse | PPP1CB | 100 | 100 |
| Chicken | PPP1CB | 100 | 100 | |
| Fly | PP1Alpha-96A | 100 | 93 | |
| Mosquito | AGAP003114-PA | 97 | 93 | |
| Purple sea urchin | LOC752338 | 99 | 97 | |
| Trypanosome (T. cruzi) | Tc00.1047053508815.110 | 91 | 88 | |
| Trypanosome (T. brucei) | Tb11.01.0450 | 91 | 89 | |
| Nematode | GSP-1 | 100 | 97 | |
| Rice | Os06g0164100 | 98 | 92 | |
| Thale cress | TOPP4 | 98 | 90 | |
| Wine grape | LOC100258649 | 104 | 89 | |
| Fission yeast | DIS2 | 100 | 93 | |
| Budding yeast | GLC7 | 94 | 91 | |
Protein length can change through evolution by many mechanisms, including insertions, deletions, and transposable elements. The general belief is that protein length increases through evolution [42]. While there tends to be a protein lengthening from E. coli to yeast, nematode, and humans, species of fungi, animals, and plants tend to have a conservation of protein length [43]. The majority of the polyadenylation factor homologs remained within 20% of the same size as the corresponding human polyadenylation factor (Figure 2). CSTF2, FIP1L1, and PABPN1 shortened as the species became evolutionary more diverse and the yeast homologs are ~50% of the size of their human counterparts. The PCF11 protein length was relatively conserved evolutionary down to purple sea urchin but nematode, plants, and yeast homologs are only half the size of the human protein.
Figure 2.

Protein length changes in polyadenylation factors. The changes in length of polyadenylation factors between homologs were compared to the human homolog of each specific polyadenylation factor. See the top left for color code of species. If the homolog was greater than 2 times the length of the human protein, the value is given above the bar.
There are specific species that do not follow the evolutionary trends. In insects, purple sea urchin, and plants, the protein lengths of the homologs tend to increase in size dramatically when protein length is not conserved. CSTF3 homologs in plants and mosquito are seven times larger than the human protein. While more uncommon, there are some truncated proteins within these species. For example, the CPSF1 homolog in rice and the CPSF3 homolog in purple sea urchin are ~25% of the human protein length (Figure 2).
The protein length of the chicken homologs of CPSF1 and NUDT21 provides evidence for some errors in the species gene annotation. The chicken CPSF1 homolog is only 5% of the length of human CPSF1 (Figure 2) and is not large enough to be a functional human homolog. Zebra finch (Taeniopygia guttata) and wild turkey (Meleagris gallopavo) have CPSF1 homologs that were about 75% the size of the human protein (data not shown). Therefore, it is likely that the chicken CPSF1 gene annotation is incorrect. The chicken NUDT21 homolog is three times larger than the human homolog. The zebra finch (Taeniopygia guttata) NUDT21 homolog is 110% the size of the human protein length. The chicken autocrine motility factor receptor (AMFR) is annotated incorrectly and contains two genes: the human NUDT21 and AMFR human homologs.
We concluded that while most of the polyadenylation machinery was similar in protein length as compared to the corresponding human proteins, there were some significant differences in either direction in insects, purple sea urchin, and plants. Also, some homologs did show a lengthening trend in proteins through evolution from yeast to human.
Another way to determine the conservation of polyadenylation factors is to determine how the amino acid sequence has changed through evolution. The protein sequence that aligned to the human polyadenylation factor identity was compared to determine how many amino acids were the same or similar. We performed this analysis by aligning the two protein sequences in NCBI and recording the percent positive. As to be expected, most of the factors decreased in similarity as the comparison was performed from mouse to yeast and plants. Most of the factors retained at least 40% of the human amino acid sequence (Figure 3). PPP1CA and PPP1CB, which are homologous factors of the yeast polyadenylation factor GLC7, were surprisingly the most conserved among all the factors with at least 90% positive identity.
Figure 3.

Conservation of protein sequences in polyadenylation factors. The protein sequence for each factor or complex of the human basal polyadenylation machinery was compared to the homologous factors in each species to determine how much of the protein sequence is changed.
To further look into the phylogenetic comparison, protein domains present in the human basal polyadenylation factors were compared to the domains present in the homologous factors in other species using the same methods as we used in analyzing the whole protein. This analysis with published human domains can help verify homologs and determine if the polyadenylation factors retain their same function(s) throughout evolution. The same protein domains were found in many, but not all, of the homologous factors.
CPSF1 (CPSF-160) has four domains found in human (Figure 4). The CPSF A domain was found in all the homologous factors. The CPSF A domain is a region that may be involved in RNA/DNA binding but its function is unknown. The beta-propeller domains were found in all the homologs except the truncated rice homolog. The beta-propeller domain contains five propeller repeats and is required for RNA binding in the yeast homolog [44]. Two RNP type binding motifs are present in CPSF1 and may be involved in RNP binding [45]. These motifs were evolutionary conserved down to trypanosome. None of the domains amino acid sequences were more conserved than the entire CPSF1 (Figure 5).
Figure 4.

Domain homology of CPSF. Schematics of proteins are shown approximately to scale. The domains were identified by the NCBI conserved domain database or known published domains. CPSF1 contains domains involved in RNA binding: two RNP binding motifs, CPSF_A and beta-propeller domains. CPSF3 contains a YSH1 domain that contains the lactamase, beta-caspase, and RNA-metabolizing metallo-beta-lactamase (RRMBL) domains. CPSF73-100_C is the conserved C-terminal domain of CPSF3. CPSF2 contains the lactamase, beta-caspase, and RRMBL domains. CPSF4 contains a protein-protein interaction YTH1 domain that contains five zinc finger domains. FIP1L1 contains an acidic, conserved, proline-rich, RD repeats and arginine-rich domains involved in protein-protein interactions.
Figure 5.
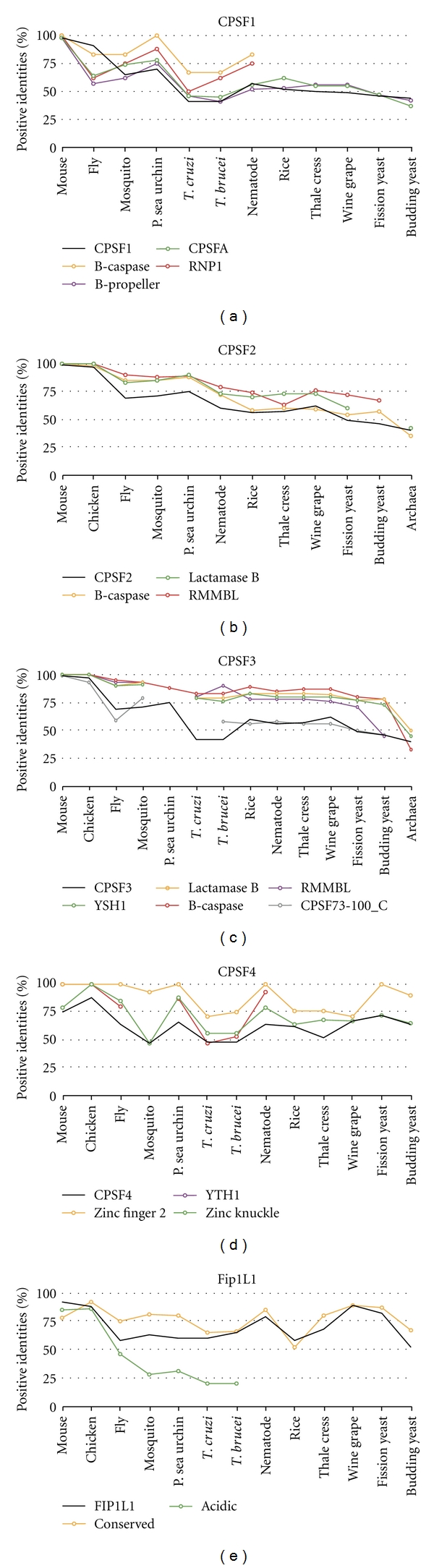
Conservation of protein sequence between the protein domains of the CPSF subunits. The amino acid sequence of human CPSF subunits and domains were compared to the homologous factor protein and domains in other species.
CPSF3 (CPSF-77) has five highly conserved domains (Figure 4). The YSH1 domain is the yeast homolog of CPSF3 which contains the entire metallo-beta-lactamase domain. Many metallo-beta lactamases are zinc-dependent nucleases [46], and CSPF3 is the predicted pre-mRNA 3′ end processing nuclease [47, 48]. The lactamase B domain contains four out of the five canonical metallo-beta-lactamase sequence motifs. RNA-metabolizing metallo-beta-lactamase (RMMBL) domain contains the fifth motif. B-caspase is a cassette inserted between the fourth and fifth beta-lactamase motifs. The B-caspase and lactamase domains form an interface around the active site [48]. The CPSF73-100_C domain is the conserved C-terminal region of CPSF3. These domains were found in all species examined except the purple sea urchin, Trypanosome (T. cruzi), and archaea. These species had missing domains due to the fact that the homologs were truncated. Except for CPSF73-100_C, all of the domains amino acid sequences were more conserved than the entire protein in all species excluding archaea (Figure 5). Therefore, the domains within the CPSF3 protein, except for the sea urchin homolog, may be conserved to maintain the endonuclease function.
CPSF2 (CPSF-100) is similar to CPSF3 and both proteins share all but one domain (Figure 4). CPSF2 is an inactive nuclease with an inability to bind two zinc molecules [48] and its function is unknown. Trypanosomes are missing the entire metallo-beta lactamase domain. Sequence conservation of these domains is only slightly higher compared to the entire protein (Figure 5).
The CPSF4 (CPSF-30) protein has YTH1, zinc knuckle, and five zinc finger domains (Figure 4). The YTH1 domain is the yeast homolog of CPSF4 and encompasses all five zinc fingers. This domain was found in all species analyzed. The zinc knuckle CCHC motif aids in binding to polyU RNA [49]. This domain was absent in plants and yeast homologs. Two zinc knuckles are present in trypanosomes and Drosophila. Zinc fingers are involved in protein and RNA interactions [50]. All five zinc finger CCCH motifs were found in most of the species examined with four motifs present in fission yeast and three in plants and mosquito homologs. The second zinc finger domain is most conserved in yeast and is lethal when deleted [50]. This conservation was also maintained with at least 90% positive identities in all the species, except trypanosomes and plants which maintain at least 70% positive identity (Figure 5). Yeast homologs have all five zinc finger CCCH motifs; however, excluding the second zinc finger domains, none of the zinc finger domains maintained more than 65% positive identities to human. The zinc knuckle domain (when present) and multiple zinc finger motifs are highly conserved and may maintain the ability of CPSF4 homologs to bind to RNA.
FIP1L1 has four domains involved in protein-protein interactions, and these domains are present in most species (Figure 4). The acidic domain binds to PAP [51, 52]. An acidic domain was found in all species except rice. The conserved region is found in all the species and interacts with CPSF4 [52]. The pro-rich domain function is unknown but was found to be evolutionary conserved to nematode. The C-terminal portion of FIP1L1 is made up of RD repeats and an arginine-rich region; it binds to CPSF1 and to U-rich RNA [52]. These two domains were found in all species except trypanosomes, plants, and yeast. None of the domains amino acid sequences were conserved more than the entire protein (Figure 5). However, the presence of these domains suggests that the FIP1L1 homologs retain their binding ability to PAP and the CPSF complex, while the interaction of FIP1L1 directly with RNA may be lost in trypanosomes, plants, and yeast.
CSTF1 (CstF-50) has two domains, WD40 and a dimerization domain (Figure 6). The WD40 domain has seven beta-transducin repeats, and deletion of this domain in CSTF1 reduces binding to CSTF3 [53]. This domain was found in all species analyzed. The conservation of amino acids of the domain was similar to the entire protein (Figure 7), but this is most likely due to the domain comprising 75% of the entire protein. The dimerization domain is involved in homodimerization of CSTF1 [53, 54]; this domain can also bind to the CTD of RNA polymerase II (RNA pol II) [55]. The dimerization domain was present in all species except for trypanosomes and plants. Therefore, all the CSTF1 homologs may bind to the CSTF2 homologs or a similar protein. Plants and trypanosome CSTF1 homologs may not self-dimerize or associate with RNA pol II.
Figure 6.

Domain homology of the CstF complex. Schematics of proteins are shown approximately to scale. The domains were identified by the NCBI conserved domain database or known published domains. CSTF1 contains dimerization and WD40 domains involved in protein-protein interactions. CSTF2 has five domains: RRM, hinge, proline/glycine rich, MEARA/G, and CTD. The RRM is involved in CSTF2 RNA binding. The hinge and CTD domains are involved in protein-protein interactions. CSTF3 has three protein interacting domains: HAT-N, HAT-C, and proline-rich domains.
Figure 7.

Conservation of protein sequence between the protein domains of the CSTF subunits. The amino acid sequence of human CSTF subunits and domains were compared to the homologous factor protein and domains in other species.
CSTF2 (CstF-64) has five domains: an RNA recognition motif (RRM), hinge, MEARA/G, pro-rich, and CTD domains (Figure 6). The RRM is involved in sequence-specific RNA recognition [53, 56–58]. Within this domain are two RNP binding motifs. All the species examined contained the RRM domain and RNP motifs. Trypanosomes have only the second RNP motif. The RRM domain is conserved more than the entire protein in all species examined except nematode, trypanosomes, and yeast (Figure 7). The hinge domain is involved in protein-protein interactions with CSTF3 and SYMPK [53]. This domain is also involved in nuclear localization [59]. This domain is present in all species examined except trypanosomes, and the domain amino acid sequence is conserved more than the protein in all species except insects and yeast (Figure 7). The CTD domain is a three-helix bundle and involves protein-protein interactions with CSTF2 and PCF11 in the yeast homologs [60]. The CTD domain is found in all species except trypanosomes. Before the CTD domain is a proline/glycine-rich domain (pro-rich) and a 12 repeat MEARA/G domain. The functions of these domains are unknown and they only are present in mouse and chicken homologs. Therefore, CSTF2 homologs may maintain the same functions except for the trypanosome homologs.
CSTF3 (CstF-77) has three domains: HAT-N, HAT-C, and pro-rich domains (Figure 6). The HAT (half-A-TPR) domain is a variant of the tetratricopeptide repeat (TPR) domain. CSTF3 contains 12 HAT motifs [61]. HAT-N contains motifs 1–5 and HAT-C contains motifs 6–11. The function of the HAT-N domain is unknown. The HAT-C domain is involved in many protein-protein interactions. This includes self-dimerization and interaction with the second beta-propeller motif of CPSF1 [61, 62]. Both HAT-N and HAT-C motifs are found in all species examined. The pro-rich domain interacts with the WD40 region in CSTF1 and the hinge region in CSTF2 [53]. This domain was found to be evolutionary conserved down to purple sea urchin but was not found in plants and yeast (Figure 7). Therefore, most of the CSTF3 homologs may perform the same functions as the human counterparts. Plant and yeast CSTF3 homologs do not have the pro-rich domain and may not associate with CSTF1 and CSTF2 homologs.
The CFIm complex domains are very well conserved. CPSF6 (CFIm68) and CPSF7 (CFIm59) are very similar proteins and share their three domains: RRM, proline-rich, and RS domains (Figure 8). These domains were present in all CPSF6 and CPSF7 homologs. The RRM domain was the only domain where the amino acid sequence was more conserved than the entire protein (Figure 9). The RRM domain of CPSF6 does not bind to RNA but is required to bind to NUDT21 [63]. The proline-rich domain may be a weak nuclear localization signal [63]. The RS domain is a dipeptide repeat region of RS, RE, or RD and associates with spliceosomal SR proteins [63, 64]. NUDT21 (CFIm25) has two domains: loop-helix and Nudix domains. These two domains form a complex to bind UGUA RNA sequence elements and eliminate the typical Nudix hydrolase activity [32]. These domains were found in all species except trypanosomes which do not have the loop binding domain. Therefore, the CFIm homologs may form a complex and perform similar functions as the human counterparts.
Figure 8.

Domain homology of CFIm and CFIIm. Schematics of proteins are shown approximately to scale. The domains were identified by the NCBI conserved domain database or known published domains. CPSF6 contains an RRM, a proline-rich, and RS domains involved in protein-protein interaction. NUDT21 has two domains: a loop-helix domain and a Nudix domain that binds RNA. CLP1 has N-terminal, central, and C-terminal domains. PCF11 has a CTD interacting domain (CID), a Clp binding domain, and two zinc fingers.
Figure 9.

Conservation of protein sequence between the protein domains of the CFIm and CFIIm subunits. The amino acid sequence of human CFIm and CFIIm protein subunits and domains were compared to the homologous factor proteins and domains in other species.
CLP1 contains three domains that are not more conserved than the entire protein (Figure 8). The N-terminal and central domains are found in all homologs examined. The C-terminal domain is only conserved evolutionary until insects. The central domain contains the Walker motif which binds ATP/GTP [65]. Clp1 is a kinase involved in tRNA splicing [66]. Therefore, the CLP1 homologs may have the same kinase activity. PCF11 has three domains, CTD interacting domain (CID), CLP1 binding domain (CLP BD), and two zinc fingers. These domains were slightly more conserved than the entire protein (Figure 9). The CID domain is found in all homologs. At least one zinc domain was found in all species except nematode. Clp binding domain was found evolutionary conserved down to sea urchin and yeast. Budding yeast has additional unique features of a Q20 and RNA14/15 binding domain. PCF11 homologs maintain the CTD and some protein-protein interactions.
The nuclear and cytoplasmic PABP proteins contain well-conserved RRM domains that bind to the poly(A) tail (Figure 10). PABPN1 has one RRM domain that is found in all the homologs. The RNP motifs are found in all species except thale cress. PAPBC1 has four RRM domains but not all of them are required for RNA binding [67]. These domain and RNP binding motifs were found in all species examined. The nematode homolog only contains three RRM domains. PABPC1 also contains a PABPC domain, which includes a MLLE motif and is involved in protein-protein interactions [68, 69]. The PABPC domain was found in all homologs examined. The RRM and PABPC domains are more conserved than the entire protein in all species except for in trypanosomes (Figure 11). Therefore, the PABP homologs may retain the same functions as the human proteins with protein-protein interactions and binding to poly(A) sequences.
Figure 10.

Domain homology of SYMP, PAPOLA, PABPN1, and PABPC4. Schematics of proteins are shown approximately to scale. The domains were identified by the NCBI conserved domain database or known published domains. Symplekin has SYMP-N, SYMP-C, and CstF binding domains. PAPOLA contains many domains including N-terminal, catalytic central, nuclear localization signal (NLS), serine/threonine-rich, and C-terminal domains. The nuclear and cytoplasmic PABP proteins contain RRM domains. PABPC1 has a protein-protein interacting PABC domain.
Figure 11.

Conservation of protein sequence between the protein domains of SYMP, PAPOLA, PABPN1, and PABPC4. The amino acid sequence of human polyadenylation factor protein and domains were compared to the homologous factor protein and domains in other species.
SYMPK has three domains: SYMP-N, SYMP-C, and CstF binding domain, none of which are well conserved (Figure 10). SYMP-N contains HEAT repeats that are involved in protein-protein interactions including Ssu72 [70]. SYMP-N is found in all homologs except for wine grape and budding yeast. The CstF binding domain binds to the hinge region of CSTF2 [71]. This domain was not found in mosquito, eudicots, or budding yeast. SYMP-C contains the domain involved in tight junctions [72]. This domain was found in all species examined except for yeast. Only the SYMP-C domain is more conserved than the entire protein (Figure 11). Therefore, the function of these homologs, especially in budding yeast, may be through different means.
PAPOLA homologs contain most of the domains except for the C-terminal domain (Figure 10). The domains present are the N-terminal, catalytic, central, NLS, Ser/Thr-rich, and C-terminal domains. None of the domains have an amino acid sequence which is more conserved than the entire protein (Figure 11). The N-terminal domain contains the catalytic domain which is the nucleotidyltransferase [73]. The N-terminal as well as the central domain was conserved in all species. The entire C-terminal domain was only conserved in vertebrates. The Ser/Thr-rich regions are found in all homologs but the amino acid sequence is not conserved per se. This region is involved in protein-protein interactions [74] and can be phosphorylated to affect poly(A) polymerase activity [75]. Therefore, all the homologs may maintain the same polymerase activity as the human PAPOLA.
Taken together, protein domains present in the basal polyadenylation factors were for the most part very well conserved between species and therefore most likely maintain similar functions as the human polyadenylation factors.
4. Conclusions
Comparison of the protein machinery involved in mRNA 3′ end formation and how this machinery is conserved in a number of representative species reveals that positive selection has been imposed on retaining the salient functional features of most of the factors. Since humans diverged from yeast and plants approximately 1 billion years ago (990 million years ago for Drosophila and nematode, 31 million years ago for chicken, and 91 million years ago for mouse), it is apparent that polyadenylation of mRNAs is an ancient process indeed.
Acknowledgments
The authors wish to thank Bin Tian for advice on development of the project and for critical reading of the papers. They also thank NIH grant award RHG005129A for support to C. S. Lutz.
References
- 1.Zhao J, Hyman L, Moore C. Formation of mRNA 3’ ends in eukaryotes: mechanism, regulation, and interrelationships with other steps in mRNA synthesis. Microbiology and Molecular Biology Reviews. 1999;63(2):405–445. doi: 10.1128/mmbr.63.2.405-445.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Edmonds M. A history of poly A sequences: from formation to factors to function. Progress in Nucleic Acid Research and Molecular Biology. 2002;71:285–389. doi: 10.1016/s0079-6603(02)71046-5. [DOI] [PubMed] [Google Scholar]
- 3.Lutz CS. Alternative polyadenylation: a twist on mRNA 3′ end formation. ACS Chemical Biology. 2008;3(10):609–617. doi: 10.1021/cb800138w. [DOI] [PubMed] [Google Scholar]
- 4.Lutz CS, Moreira A. Alternative mRNA polyadenylation in eukaryotes: an effective regulator of gene expression. WIREs RNA. 2011;2(1):23–31. doi: 10.1002/wrna.47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Millevoi S, Vagner S. Molecular mechanisms of eukaryotic pre-mRNA 3’ end processing regulation. Nucleic Acids Research. 2009;38(9):2757–2774. doi: 10.1093/nar/gkp1176. Article ID gkp1176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Proudfoot NJ. Ending the message: poly(A) signals then and now. Genes & Development. 2011;25(14):1770–1782. doi: 10.1101/gad.17268411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chan S, Choi EA, Shi Y. Pre-mRNA 3’-end processing complex assembly and function. Wiley Interdisciplinary Reviews RNA. 2011;2(3):321–335. doi: 10.1002/wrna.54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lewis JD, Gunderson SI, Mattaj IW. The influence of 5’ and 3’ end structures on pre-mRNA metabolism. Journal of Cell Science. 1995;108(19):13–19. doi: 10.1242/jcs.1995.supplement_19.2. [DOI] [PubMed] [Google Scholar]
- 9.Jacobson A, Peltz SW. Interrelationships of the pathways of mRNA decay and translation in eukaryotic cells. Annual Review of Biochemistry. 1996;65:693–739. doi: 10.1146/annurev.bi.65.070196.003401. [DOI] [PubMed] [Google Scholar]
- 10.Sachs AB, Sarnow P, Hentze MW. Starting at the beginning, middle, and end: translation initiation in eukaryotes. Cell. 1997;89(6):831–838. doi: 10.1016/s0092-8674(00)80268-8. [DOI] [PubMed] [Google Scholar]
- 11.Wickens M, Anderson P, Jackson RJ. Life and death in the cytoplasm: messages from the 3’ end. Current Opinion in Genetics and Development. 1997;7(2):220–232. doi: 10.1016/s0959-437x(97)80132-3. [DOI] [PubMed] [Google Scholar]
- 12.Zhang X, Virtanen A, Kleiman FE. To polyadenylate or to deadenylate: that is the question. Cell Cycle. 2010;9(22):4437–4449. doi: 10.4161/cc.9.22.13887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mandel CR, Bai Y, Tong L. Protein factors in pre-mRNA 3′-end processing. Cellular and Molecular Life Sciences. 2008;65(7-8):1099–1122. doi: 10.1007/s00018-007-7474-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yang Q, Doublie S. Structural biology of poly(A) site definition. Wiley Interdisciplinary Reviews RNA. 2011;2(5):732–747. doi: 10.1002/wrna.88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tian B, Hu J, Zhang H, Lutz CS. A large-scale analysis of mRNA polyadenylation of human and mouse genes. Nucleic Acids Research. 2005;33(1):201–212. doi: 10.1093/nar/gki158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nunes NM, Li W, Tian B, Furger A. A functional human Poly(A) site requires only a potent DSE and an A-rich upstream sequence. The EMBO Journal. 2010;29(9):1523–1536. doi: 10.1038/emboj.2010.42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bienroth S, Christofori G, Lang KM, Wahle E, Keller W. Components involved in 3′ processing of precursors to polyadenylated messenger RNA. Molecular Biology Reports. 1990;14(2-3):p. 197. doi: 10.1007/BF00360471. [DOI] [PubMed] [Google Scholar]
- 18.Bienroth S, Wahle E, Suter-Crazzolara C, Keller W. Purification of the cleavage and polyadenylation factor involved in the 3’-processing of messenger RNA precursors. Journal of Biological Chemistry. 1991;266(29):19768–19776. [PubMed] [Google Scholar]
- 19.Keller W, Bienroth S, Lang KM, Christofori G. Cleavage and polyadenylation factor CPF specifically interacts with the pre-mRNA 3’ processing signal AAUAAA. The EMBO Journal. 1991;10(13):4241–4249. doi: 10.1002/j.1460-2075.1991.tb05002.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Takagaki Y, Ryner LC, Manley JL. Four factors are required for 3’-end cleavage of pre-mRNAs. Genes & Development. 1989;3(11):1711–1724. doi: 10.1101/gad.3.11.1711. [DOI] [PubMed] [Google Scholar]
- 21.Takagaki Y, Manley JL, MacDonald CC, Wilusz J, Shenk T. A multisubunit factor, CstF, is required for polyadenylation of mammalian pre-mRNAs. Genes & Development. 1990;4(12 A):2112–2120. doi: 10.1101/gad.4.12a.2112. [DOI] [PubMed] [Google Scholar]
- 22.Wilusz J, Shenk T, Takagaki Y, Manley JL. A multicomponent complex is required for the AAUAAA-dependent cross-linking of a 64-kilodalton protein to polyadenylation substrates. Molecular and Cellular Biology. 1990;10(3):1244–1248. doi: 10.1128/mcb.10.3.1244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gilmartin GM, Nevins JR. An ordered pathway of assembly of components required for polyadenylation site recognition and processing. Genes & Development. 1989;3(12 B):2180–2190. doi: 10.1101/gad.3.12b.2180. [DOI] [PubMed] [Google Scholar]
- 24.Wallace AM, Denison TL, Attaya EN, MacDonald CC. Developmental distribution of the polyadenylation protein CstF-64 and the variant tauCstF-64 in mouse and rat testis. Biology of Reproduction. 2004;70(4):1080–1087. doi: 10.1095/biolreprod.103.022947. [DOI] [PubMed] [Google Scholar]
- 25.Shi Y, Di Giammartino DC, Taylor D, et al. Molecular architecture of the human pre-mRNA 3′ processing complex. Molecular Cell. 2009;33(3):365–376. doi: 10.1016/j.molcel.2008.12.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nagaike T, Logan C, Hotta I, Rozenblatt-Rosen O, Meyerson M, Manley J. Transcriptional activators enhance polyadenylation of mRNA precursors. Molecular Cell. 2011;41(4):409–418. doi: 10.1016/j.molcel.2011.01.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lopez-Camarillo C, Orozco E, Marchat LA. Entamoeba histolytica: comparative genomics of the pre-mRNA 3′ end processing machinery. Experimental Parasitology. 2005;110(3):184–190. doi: 10.1016/j.exppara.2005.02.024. [DOI] [PubMed] [Google Scholar]
- 28.Portnoy V, Evguenieva-Hackenberg E, Klein F, et al. RNA polyadenylation in Archaea: not observed in Haloferax while the exosome polynucleotidylates RNA in Sulfolobus. EMBO Reports. 2005;6(12):1188–1193. doi: 10.1038/sj.embor.7400571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Slomovic S, Portnoy V, Yehudai-Resheff S, Bronshtein E, Schuster G. Polynucleotide phosphorylase and the archaeal exosome as poly(A)-polymerases. Biochimica et Biophysica Acta. 2008;1779(4):247–255. doi: 10.1016/j.bbagrm.2007.12.004. [DOI] [PubMed] [Google Scholar]
- 30.Brown KM, Gilmartin GM. A mechanism for the regulation of pre-mRNA 3′ processing by human cleavage factor Im. Molecular Cell. 2003;12(6):1467–1476. doi: 10.1016/s1097-2765(03)00453-2. [DOI] [PubMed] [Google Scholar]
- 31.Ruegsegger U, Blank D, Keller W. Human pre-mRNA cleavage factor Im Is related to spliceosomal SR proteins and can be reconstituted in vitro from recombinant subunits. Molecular Cell. 1998;1(2):243–253. doi: 10.1016/s1097-2765(00)80025-8. [DOI] [PubMed] [Google Scholar]
- 32.Yang Q, Gilmartin GM, Doublié S. Structural basis of UGUA recognition by the Nudix protein CFIm25 and implications for a regulatory role in mRNA 3′ processing. Proceedings of the National Academy of Sciences of the United States of America. 2010;107(22):10062–10067. doi: 10.1073/pnas.1000848107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pérez-Cãadillas JM. Grabbing the message: structural basis of mRNA 3′UTR recognition by Hrp1. The EMBO Journal. 2006;25(13):3167–3178. doi: 10.1038/sj.emboj.7601190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kessler MM, Henry MF, Shen E, et al. Hrp1, a sequence-specific RNA-binding protein that shuttles between the nucleus and the cytoplasm, is required for mRNA 3’-end formation in yeast. Genes & Development. 1997;11(19):2545–2556. doi: 10.1101/gad.11.19.2545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wallace AM, Dass B, Ravnik SE, et al. Two distinct forms of the 64,000 Mr protein of the cleavage stimulation factor are expressed in mouse male germ cells. Proceedings of the National Academy of Sciences of the United States of America. 1999;96(12):6763–6768. doi: 10.1073/pnas.96.12.6763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Feral C, Guellaen G, Pawlak A. Human testis expresses a specific poly(A)-binding protein. Nucleic Acids Research. 2001;29(9):1872–1883. doi: 10.1093/nar/29.9.1872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Okochi K, Suzuki T, Inoue JI, Matsuda S, Yamamoto T. Interaction of anti-proliferative protein Tob with poly(A)-binding protein and inducible poly(A)-binding protein: implication of Tob in translational control. Genes to Cells. 2005;10(2):151–163. doi: 10.1111/j.1365-2443.2005.00826.x. [DOI] [PubMed] [Google Scholar]
- 38.Lee YJ, Lee Y, Chung JH. An intronless gene encoding a poly(A) polymerase is specifically expressed in testis. FEBS Letters. 2000;487(2):287–292. doi: 10.1016/s0014-5793(00)02367-x. [DOI] [PubMed] [Google Scholar]
- 39.Hunt AG. Messenger RNA 3′ end formation in plants. Current Topics in Microbiology and Immunology. 2008;326:151–177. doi: 10.1007/978-3-540-76776-3_9. [DOI] [PubMed] [Google Scholar]
- 40.Chekanova JA, Belostotsky DA. Evidence that poly(A) binding protein has an evolutionarily conserved function in facilitating mRNA biogenesis and export. RNA. 2003;9(12):1476–1490. doi: 10.1261/rna.5128903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Addepalli B, Meeks LR, Forbes KP, Hunt AG. Novel alternative splicing of mRNAs encoding poly(A) polymerases in Arabidopsis. Biochimica et Biophysica Acta. 2004;1679(2):117–128. doi: 10.1016/j.bbaexp.2004.06.001. [DOI] [PubMed] [Google Scholar]
- 42.Trifonov EN, Berezovsky IN. Evolutionary aspects of protein structure and folding. Current Opinion in Structural Biology. 2003;13(1):110–114. doi: 10.1016/s0959-440x(03)00005-8. [DOI] [PubMed] [Google Scholar]
- 43.Wang D, Hsieh M, Li WH. A general tendency for conservation of protein length across eukaryotic kingdoms. Molecular Biology and Evolution. 2005;22(1):142–147. doi: 10.1093/molbev/msh263. [DOI] [PubMed] [Google Scholar]
- 44.Dichtl B, Blank D, Sadowski M, Hübner W, Weiser S, Keller W. Yhh1p/Cft1p directly links poly(A) site recognition and RNA polymerase II transcription termination. The EMBO Journal. 2002;21(15):4125–4135. doi: 10.1093/emboj/cdf390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Murthy KGK, Manley JL. The 160-kD subunit of human cleavage-polyadenylation specificity factor coordinates pre-mRNA 3’-end formation. Genes & Development. 1995;9(21):2672–2683. doi: 10.1101/gad.9.21.2672. [DOI] [PubMed] [Google Scholar]
- 46.Dominski Z. Nucleases of the metallo-β-lactamase family and their role in DNA and RNA metabolism. Critical Reviews in Biochemistry and Molecular Biology. 2007;42(2):67–93. doi: 10.1080/10409230701279118. [DOI] [PubMed] [Google Scholar]
- 47.Ryan K, Calvo O, Manley JL. Evidence that polyadenylation factor CPSF-73 is the mRNA 3′ processing endonuclease. RNA. 2004;10(4):565–573. doi: 10.1261/rna.5214404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Mandel CR, Kaneko S, Zhang H, et al. Polyadenylation factor CPSF-73 is the pre-mRNA 3′-end-processing endonuclease. Nature. 2006;444(7121):953–956. doi: 10.1038/nature05363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Barabino SML, Hubner W, Jenny A, Minvielle-Sebastia L, Keller W. The 30-kd subunit of mammalian cleavage and polyadenylation specificity factor and its yeast homolog are rna-binding zinc finger proteins. Genes & Development. 1997;11(13):1703–1716. doi: 10.1101/gad.11.13.1703. [DOI] [PubMed] [Google Scholar]
- 50.Barabino SM, Ohnacker M, Keller W. Distinct roles of two Yth1p domains in 3’-end cleavage and polyadenylation of yeast pre-mRNAs. The EMBO Journal. 2000;19(14):3778–3787. doi: 10.1093/emboj/19.14.3778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Helmling S, Zhelkovsky A, Moore CL. Fip1 regulates the activity of poly(A) polymerase through multiple interactions. Molecular and Cellular Biology. 2001;21(6):2026–2037. doi: 10.1128/MCB.21.6.2026-2037.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kaufmann I, Martin G, Friedlein A, Langen H, Keller W. Human Fip1 is a subunit of CPSF that binds to U-rich RNA elements and stimulates poly(A) polymerase. The EMBO Journal. 2004;23(3):616–626. doi: 10.1038/sj.emboj.7600070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Takagakit Y, Manley JL. Complex protein interactions within the human polyadenylation machinery identify a novel component. Molecular and Cellular Biology. 2000;20(5):1515–1525. doi: 10.1128/mcb.20.5.1515-1525.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Moreno-Morcillo M, Minvielle-Sebastia L, Mackereth C, Fribourg S. Hexameric architecture of CstF supported by CstF-50 homodimerization domain structure. RNA. 2011;17(3):412–418. doi: 10.1261/rna.2481011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.McCracken S, Fong N, Rosonina E, et al. 5’-Capping enzymes are targeted to pre-mRNA by binding to the phosphorylated carboxy-terminal domain of RNA polymerase II. Genes & Development. 1997;11(24):3306–3318. doi: 10.1101/gad.11.24.3306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Takagaki Y, Manley JL. RNA recognition by the human polyadenylation factor CstF. Molecular and Cellular Biology. 1997;17(7):3907–3914. doi: 10.1128/mcb.17.7.3907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Perez Canadillas JM, Varani G. Recognition of GU-rich polyadenylation regulatory elements by human CstF-64 protein. The EMBO Journal. 2003;22(11):2821–2830. doi: 10.1093/emboj/cdg259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Pancevac C, Goldstone DC, Ramos A, Taylor IA. Structure of the Rna15 RRM-RNA complex reveals the molecular basis of GU specificity in transcriptional 3’-end processing factors. Nucleic Acids Research. 2010;38(9):3119–3132. doi: 10.1093/nar/gkq002. Article ID gkq002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Hockert JA, Yeh HJ, MacDonald CC. The hinge domain of the cleavage stimulation factor protein CstF-64 is essential for CstF-77 interaction, nuclear localization, and polyadenylation. Journal of Biological Chemistry. 2010;285(1):695–704. doi: 10.1074/jbc.M109.061705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Qu X, Perez-Canadillas JM, Agrawal S, et al. The C-terminal domains of vertebrate CstF-64 and its yeast orthologue Rna15 form a new structure critical for mRNA 3′-end processing. Journal of Biological Chemistry. 2007;282(3):2101–2115. doi: 10.1074/jbc.M609981200. [DOI] [PubMed] [Google Scholar]
- 61.Legrand P, Pinaud N, Minvielle-Sebastia L, Fribourg S. The structure of the CstF-77 homodimer provides insights into CstF assembly. Nucleic Acids Research. 2007;35(13):4515–4522. doi: 10.1093/nar/gkm458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Bai Y, Auperin TC, Chou CY, Chang GG, Manley JL, Tong L. Crystal Structure of Murine CstF-77: dimeric Association and Implications for Polyadenylation of mRNA Precursors. Molecular Cell. 2007;25(6):863–875. doi: 10.1016/j.molcel.2007.01.034. [DOI] [PubMed] [Google Scholar]
- 63.Dettwiler S, Aringhieri C, Cardinale S, Keller W, Barabino SML. Distinct sequence motifs within the 68-kDa subunit of cleavage factor Im mediate RNA binding, protein-protein interactions, and subcellular localization. Journal of Biological Chemistry. 2004;279(34):35788–35797. doi: 10.1074/jbc.M403927200. [DOI] [PubMed] [Google Scholar]
- 64.Millevoi S, Loulergue C, Dettwiler S, et al. An interaction between U2AF 65 and CF Im links the splicing and 3′ end processing machineries. The EMBO Journal. 2006;25(20):4854–4864. doi: 10.1038/sj.emboj.7601331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Walker JE, Saraste M, Runswick MJ, Gay NJ. Distantly related sequences in the alpha- and beta-subunits of ATP synthase, myosin, kinases and other ATP-requiring enzymes and a common nucleotide binding fold. The EMBO Journal. 1982;1(8):945–951. doi: 10.1002/j.1460-2075.1982.tb01276.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Weitzer S, Martinez J. The human RNA kinase hClp1 is active on 3′ transfer RNA exons and short interfering RNAs. Nature. 2007;447(7141):222–226. doi: 10.1038/nature05777. [DOI] [PubMed] [Google Scholar]
- 67.Deo RC, Bonanno JB, Sonenberg N, Burley SK. Recognition of polyadenylate RNA by the poly(A)-binding protein. Cell. 1999;98(6):835–845. doi: 10.1016/s0092-8674(00)81517-2. [DOI] [PubMed] [Google Scholar]
- 68.Kozlov G, Trempe JF, Khaleghpour K, Kahvejian A, Ekiel I, Gehring K. Structure and function of the C-terminal PABC domain of human poly(A)-binding protein. Proceedings of the National Academy of Sciences of the United States of America. 2001;98(8):4409–4413. doi: 10.1073/pnas.071024998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Kozlov G, Menade M, Rosenauer A, Nguyen L, Gehring K. Molecular determinants of PAM2 recognition by the MLLE domain of poly(A)-binding protein. Journal of Molecular Biology. 2010;397(2):397–407. doi: 10.1016/j.jmb.2010.01.032. [DOI] [PubMed] [Google Scholar]
- 70.Xiang K, Nagaike T, Xiang S, et al. Crystal structure of the human symplekin-Ssu72-CTD phosphopeptide complex. Nature. 2010;467(7316):729–733. doi: 10.1038/nature09391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Ruepp MD, Schweingruber C, Kleinschmidt N, Schumperli D. Interactions of CstF-64, CstF-77, and symplekin: implications on localisation and function. Molecular Biology of the Cell. 2011;22(1):91–104. doi: 10.1091/mbc.E10-06-0543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Keon BH, Schafer S, Kuhn C, Grund C, Franke WW. Symplekin, a novel type of tight junction plaque protein. Journal of Cell Biology. 1996;134(4):1003–1018. doi: 10.1083/jcb.134.4.1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Martin G, Keller W. Mutational analysis of mammalian poly(A) polymerase identifies a region for primer binding and a catalytic domain, homologous to the family X polymerases, and to other nucleotidyltransferases. The EMBO Journal. 1996;15(10):2593–2603. [PMC free article] [PubMed] [Google Scholar]
- 74.Vagner S, Vagner C, Mattaj IW. The carboxyl terminus of vertebrate poly(A) polymerase interacts with U2AF 65 to couple 3’-end processing and splicing. Genes & Development. 2000;14(4):403–413. [PMC free article] [PubMed] [Google Scholar]
- 75.Colgan DF, Murthy KGK, Prives C, Manley JL. Cell-cycle related regulation of poly(A) polymerase by phosphorylation. Nature. 1996;384(6606):282–285. doi: 10.1038/384282a0. [DOI] [PubMed] [Google Scholar]


