Abstract
Cutaneous malignant melanoma is considered one of the most deadly human cancers, based on both its penchant for metastatic spread and its typical resistance to currently available therapy. Long known to harbor oncogenic NRAS mutations, melanomas were more recently reported to be frequent bearers of activating mutations in BRAF, one of the effectors situated downstream of wild-type NRAS. NRAS and BRAF mutations are rarely found in the same melanoma, suggesting that they may possess important overlapping oncogenic activities. Here, we compare and contrast the oncogenic roles of the three major NRas downstream effectors, Raf, phosphatidylinositol 3-kinase (PI3K) and Ral guanine exchange factor (RalGEF), using genetically engineered Arf-deficient immortalized mouse melanocytes as a model system. Although no single downstream pathway could recapitulate all of the consequences of oncogenic NRas expression, our data indicate a prominent role for BRaf and PI3K in melanocyte senescence and invasiveness, respectively. More surprisingly, we discovered that constitutive RalGEF activation had a major impact on several malignant phenotypes, particularly anchorage-independent growth, indicating that this often overlooked pathway should be more carefully evaluated as a possible therapeutic target.
Keywords: anchorage-independent growth, BRaf, melanoma, NRas, PI3K, RalGEF
RAS gene products (HRas, KRas and NRas) are 21 kd G-proteins that serve as molecular switches converting cell-surface kinase activation events to nuclear events, thus influencing cell behavior. The major downstream effectors of Ras are the Rafs (ARaf, BRaf and CRaf), phosphatidylinositol 3-kinase (PI3K) and the Ral guanine exchange factors (RalGEFs) (Figure 1a) (Downward, 2003). Before the beginning of the new millennium, activating mutations in NRAS constituted the most common oncogenic gain-of-function genomic event documented in cutaneous malignant melanoma, with up to a 25% incidence (Gray-Schopfer et al., 2007). NRAS mutations, most commonly at Q61K, result in constitutive activation of NRas and its downstream effectors. More recently, activating mutations were discovered in BRAF ranging from 44 to 70% of melanomas and nevi, predominantly consisting of V600E (Brose et al., 2002; Davies et al., 2002; Pollock and Meltzer, 2002; Pollock et al., 2003; Bennett, 2008). These constitutive activation BRAF mutations were mutually exclusive of mutant NRAS, highlighting the importance of the Ras-Raf-MEK-ERK, or MAPK, pathway. With respect to the PI3K pathway, although alterations in PI3K itself are rare in melanoma, mutations in its negative regulator PTEN are found in up to 40%of melanoma cell lines and ~15% of primary melanoma (Wu et al., 2003; Dahl and Guldberg, 2007; Bennett, 2008). PTEN antagonizes PI3K and negatively regulates AKT activity, which can affect migration and apoptosis as well as proliferation. PTEN mutations are typically found to be mutually exclusive of NRAS mutations, indicative of the importance of the NRas downstream effector pathways in melanomagenesis (Tsao et al., 1998; Goel et al., 2006; Gray-Schopfer et al., 2007). The third major effector, RalGEF, activates the small GTPases RalA and RalB when instructed by activated RAS (Feig, 2003). Activated RalGEFs can transform human and rodent cells (Urano et al., 1996; Hamad et al., 2002; Rangarajan et al., 2004; Lim et al., 2005), but activating mutations have yet to be detected in human melanoma (Omholt and Hansson, 2007). Loss of Ral function in genetically engineered mice and in cultured cells through RNAi-mediated knockdown results in reduction in the malignant phenotype (Chien and White, 2003; Oxford et al., 2005; Chien et al., 2006; Lim et al., 2006; Rosse et al., 2006; de Gorter et al., 2008; C Counter, personal communication). Moreover, the RalGEF-Ral pathway has been shown to promote oncogenic transformation in several KRas- and HRas-driven cancer cell lines and tumor tissues (Lim et al., 2005, 2006; Smith et al., 2007). Here, we examine in immortalized mouse melanocytes the consequences of the activation of these three pathways, Raf, PI3K and Ral, alone and in combination, as compared with the full transforming phenotype observed with the expression of oncogenic NRasQ61K.
Figure 1.
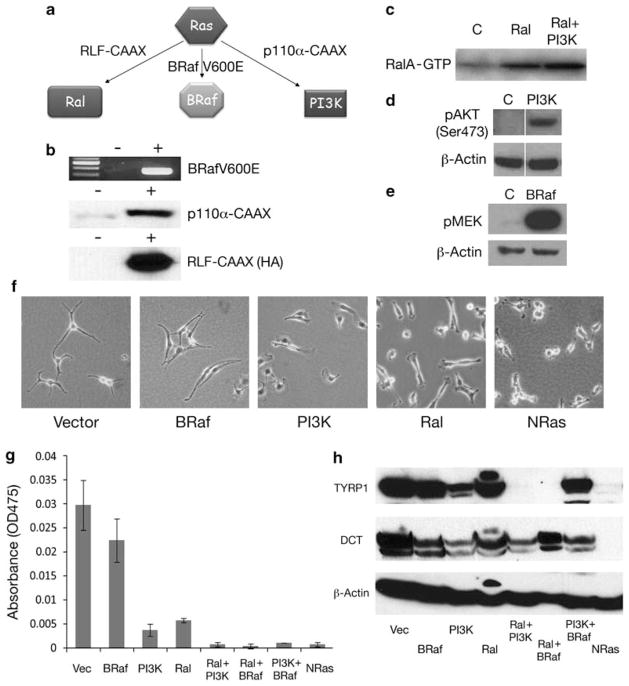
Expression and effects of NRas and its downstream effectors on melanocyte morphology and pigmentation. (a) Highly simplified schematic of NRas downstream pathway members RalGEF, BRaf and PI3K, showing the activating mutants used in this study. (b) Panels show evidence for expression (+) of BrafV600E RNA (RT–PCR), and p110α-CAAX and RLF-CAAX (HA tagged) protein (western blot). Arf-deficient mouse melanocytes were used in all studies to avoid the senescence-prone phenotype (Ha et al., 2007). (c) RalGEF activity in vector control and RLF-CAAX-transfected melanocytes, assayed by measuring the quantity of RalA-GTP bound to RALBP-1 in a pull-down assay (Yin et al., 2007). (d) PI3K pathway activation in p110α-CAAX melanocytes assayed by visualizing phospho-AKP (Ser473) and total AKT levels by western blotting. (e) Activation of BRaf pathway in BrafV600E melanocytes assayed by visualizing phospho-MEK levels by western blotting. (f) Morphological alterations resulting from constitutive activation of: control (vector), BRaf (expressing BrafV600E), PI3K (expressing p110α-CAAX), RalGEF (expressing RLF-CAAX) or NRas (expressing NRasQ61K). Bright field images are shown (20×). NRas, PI3K and BRaf mouse melanocytes were generated using LZRS retroviruses, as described earlier (Chudnovsky et al., 2005; Ha et al., 2007). Ral and RalBN28 melanocytes were generated using pBABE retroviruses (Yin et al., 2007). For pBABE infections, EP293 RLF-CAAX cells were grown overnight in DMEM with 10% FBS, L-glutamine (no puromycin selection). A measure of 8 μg/ml polybreene was added to the media and used to infect Arf-deficient melanocytes for 8 h with pBABE retroviruses. Melanocytes were grown in CMGM overnight and the infection was repeated the next day. Then, 24 h after the second pBABE infection, melanocytes were thereafter grown in CMGM plus 1.5 μg/ml puromycin. Vec, control vector. Note that melanocyte cell lines were taken through ~15 passages after transfection before conducting experiments, under selection where appropriate. (g) Measurement of melanin content in genetically engineered melanocytes showing the pathway activated (see Figure 1 for expressed mutant proteins). Cells were lysed in 1N NaOH at 37 °C overnight and the melanin content was measured by optical density at 475nm (Virador et al., 1999). Error bars represent standard deviation (s.d.). (h) The protein expression levels of key mouse pigmentation genes, tyrosinase-related protein 1 (TYRP1) and DCT were determined by western blotting using the rabbit antibodies PEP7 and PEP8H, respectively (Recio et al., 2002). β-actin was used as a loading control. Western blots were performed using standard procedures.
To dissect the role of Ras downstream signaling pathways, we used early passage cultures of mouse melanocytes deficient in the tumor suppressor p19Arf (p14ARF in humans) to avoid the rapid senescence that characterizes cultured primary wild-type mouse melanocytes (Ha et al., 2007). ARF is a frequent target of deletion, inactivating mutation or methylation suppression in human melanoma (Curtin et al., 2005; Chin et al., 2006; Freedberg et al., 2008); therefore, ARF deficiency represents a relevant genetic context for studying the consequences of activated RAS and downstream factors such as Ral, Raf and PI3K in melanomagenesis.
Retroviral vectors encoding mutationally activated forms of NRas (NRasQ61K), RalGEF (RLF-CAAX), BRaf (V600E) and PI3K (p110α-CAAX) were expressed in the Arf-deficient melanocytes (Figure 1b; Chudnovsky et al., 2005; Yin et al., 2007). Cultures were grown at 37 °C in RPMI 1640 medium with 10% FBS, 200 nM 12-O-tetradecanoyl phorbol 13-acetate, and 200 pM cholera toxin, referred to as complete melanocyte growth medium (CMGM) (Ha et al., 2007). The expression and activity of RLF-CAAX, p110α-CAAX and BRafV600E were confirmed by measuring GTP-RalA activity (Figure 1c; Yin et al., 2007), phospho-AKT (Ser473) (Figure 1d) and phospho-MEK (Figure 1e), respectively. As previously reported for Ink4a/Arf-deficient cells, mutant NRasQ61K expression in Arf-deficient mouse melanocytes resulted in a highly transformed, refractile appearance (Figure 1f; Ha et al., 2007). The morphological features of immortalized melanocytes transduced with the different downstream effector expression vectors varied greatly: the cells expressing BRafV600E looking most like vector controls, whereas those expressing RLF-CAAX looked most like the NRasQ61K-expressing cells (Figure 1f). In contrast, melanocytes expressing p110α-CAAX exhibited a more flattened morphology. Examination of these images, as well as the cell’s conditioned media, also suggested that pigmentation varied between these cell lines.
We therefore determined the effect of NRasQ61K and downstream pathway activation on differentiation by measuring the melanin content and expression of pigmentary genes. Briefly, cells were lysed in NaOH and the melanin content measured by optical density at 475nm (Virador et al., 1999). Activation of NRas and/or two different pathways downstream of NRas, PI3K and to a lesser extent Ral, resulted in loss of pigmentation in the melanocytes (Figure 1g). In contrast, BRafV600E-expressing immortalized melanocytes were fully pigmented. To probe these pathways further, we generated Arf-deficient melanocytes that had two pathways constitutively activated (expressing BRafV600E plus p110α-CAAX, BRafV600E plus RLF-CAAX, or p110α-CAAX plus RLF-CAAX). Any of the two-pathway combinations resulted in complete loss of melanin production (Figure 1g). Next, the levels of the key pigmentary gene products tyrosine-related protein 1 (TYRP1) and dopachrome tautomerase (DCT) were determined using western blotting. Expression of NRasQ61K had a profound effect on TYRP1 and DCT expression, as did activation of the PI3K pathway, albeit to a lesser extent (Figure 1h). The individual BRaf and Ral pathways appeared to have little effect on the expression of these pigment genes, in general agreement with the observed effects on melanin levels. However, the combinations of p110α-CAAX plus RLF-CAAX, and p110α-CAAX plus BRafV600E both had a dramatic effect on pigment gene expression, especially TYRP1 (Figure 1h). Moreover, PI3K pathway activation seemed to be most responsible for suppression of DCT expression, alone or in various combinations (Figure 1h).
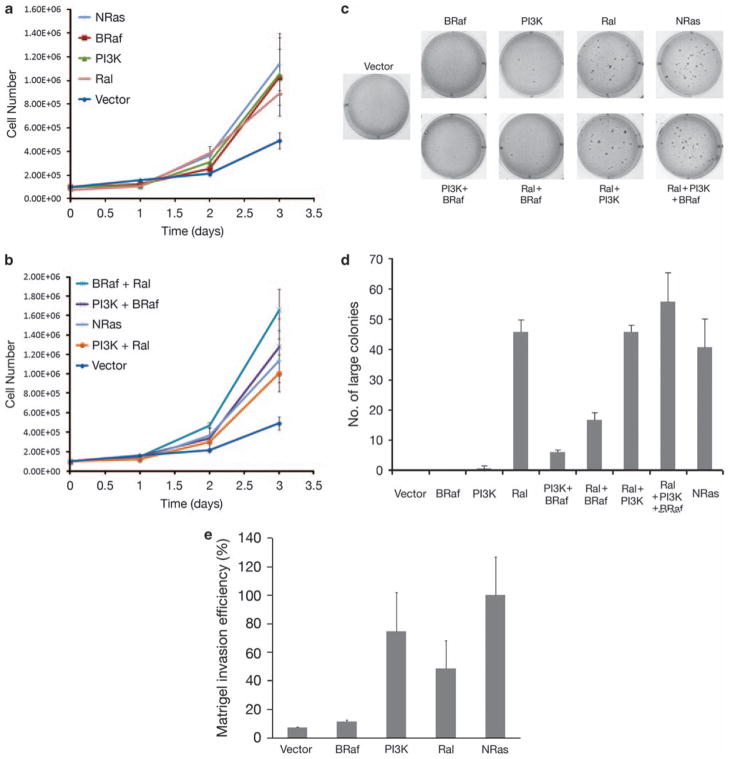
We next quantified the anchorage-dependent proliferation of these melanocytes using a hemocytometer to count trypsin-suspended cells. Transfection of vectors expressing NRasQ61K or the effectors, BRafV600E, p110α-CAAX and RLF-CAAX, individually or in combination, gave the melanocytes a clear growth advantage (Figures 2a and b). We then determined the effects of these activated pathways on growth in soft agar. The ability of NRasQ61K and effectors to induce the growth of anchorage-independent colonies was tested by growing the cells in soft agar for 2 weeks, as described earlier (Ha et al., 2007). It is interesting to note that melanocytes expressing RLF-CAAX, individually or in combination with p110α-CAAX, grew very efficiently in soft agar (Figure 2c) and formed large colonies similar to NRasQ61K-expressing melanocytes. In contrast, BRafV600E and p110α-CAAX (individually or together) formed fewer soft agar colonies, which were uniformly small. Quantification of all large colonies (Figure 2d) clearly showed that constitutive RalGEF activation was critical in conferring anchorage-independent growth in immortalized melanocytic mouse cells. RLF-CAAX-expressing melanocytes were generated by retroviral infection of Arf-deficient cells on two separate occasions, and on soft agar analysis the same results were obtained.
Figure 2.
Consequences of expression of activated NRas and downstream effectors on anchorage-dependent growth, anchorage-independent growth and invasiveness. (a and b) The growth rates in 2D culture of melanocytes genetically modified to bear constitutively activated NRas or its individual effectors. Proliferation was quantified in triplicate using a hemocytometer to count trypsin-suspended cells, which were replated at 3×104 cells/ml, 2 ml/dish. Error bars represent s.d. (c) The anchorage-independent growth ability of melanocytes expressing activated NRas, its individual effectors, or combinations of these targets, as assayed by growth of the cells in 3% semi-solid agar medium as previously described (Ha et al., 2007). Briefly, 5×104 cells/well were seeded in triplicate in six-well plates in growth medium containing 3% agar. After 2 weeks of inoculation, colonies that developed in soft agar were counted. (d) Quantification of the soft agar colonies are shown in (c). Melanocyte cell lines were taken through about 15 passages after transfection before conducting experiments. (e) Invasive potential of the melanocytes was assayed by matrigel invasion assay using previously published methods (Albini et al., 1987). Unlike control and BRafV600E-expressing melanocytes, PI3K and RalGEF activation in melanocytes conferred invasiveness.
The invasive potential of NRasQ61K and its effectors was determined using matrigel invasion assays (Albini et al., 1987). Briefly, the basement membrane matrix was reconstituted using the matrix onto a filter in a Boyden chamber, and the ability of various melanocytes to penetrate through the coated filter was determined. Melanocytes expressing p110α-CAAX were highly invasive in matrigel, similar to NRasQ61K-expressing melanocytes, as were RLF-CAAX-expressing melanocytes (Figure 2e and Supplementary Figure 1). Fewer control and BRafV600E-expressing melanocytes invaded through matrigel. Our data suggest that similar to NRasQ61K-expressing melanocytes, activation of both PI3K and RalGEF confer invasiveness to melanocytic cells (Figure 2e).
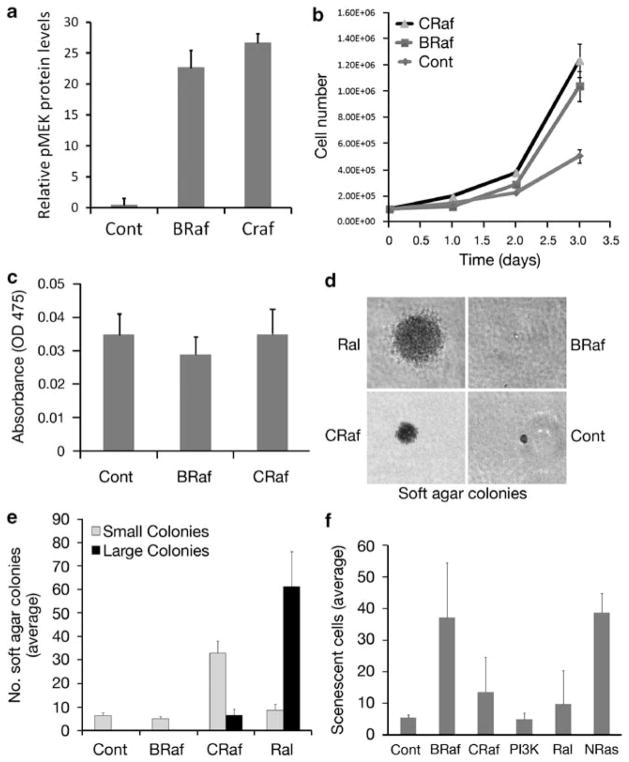
Although RLF-CAAX expression strongly induced soft agar growth on its own, in contrast BRafV600E failed to overtly transform melanocytes; moreover, BRafV600E appeared to actually have a dominant negative effect on the anchorage-independent growth effects elicited by RLF-CAAX expression (Figures 2c and d). As it was recently shown that oncogenic NRas does not signal through BRaf, but rather through CRaf (Dumaz et al., 2006), we compared the effects of BRaf and CRaf activation on melanocyte growth, transformation, differentiation and senescence (Figure 3). Melanocytes were transfected with an activated CRaf construct (pEF-CAAX-Raf-1) (Heidecker et al., 1990), and BRaf and CRaf activity was confirmed by measuring levels of phospho-MEK (Figure 3a). Both anchorage-dependent growth and melanin content of CAAX-Raf-1-expressing cells were comparable to that of BrafV600E-expressing cells (Figures 3b and c). However, CAAX-Raf-1-expressing melanocytes formed more small and large soft agar colonies compared with BrafV600E-expressing melanocytes (Figures 3d and e). We note that RLF-CAAX-expressing melanocytes still formed more large colonies compared with CAAX-Raf-1, confirming a role for RalGEF in melanocyte transformation.
Figure 3.
Comparative effects of activated CRaf and BRaf on melanocyte growth, pigmentation, transformation and senescence. (a) The activity of CRaf in melanocytes transfected with the pEF-CAAX-Raf-1 vector, using Lipofectamine 2000 (Invitrogen) transfection reagent as per the manufacturer’s protocol, was confirmed by measuring and quantifying phospho-MEK levels. (b) Anchorage-dependent growth was quantified in triplicate by counting trypsin-suspended cells using an automated cell counter Countess (Invitrogen); cells were replated in six-well plates, 1×105 cells/well. Error bars represent s.d. (c) Melanin content was assayed by lysing the cells in NaOH and measuring the optical density at 475 nm. (d and e) The anchorage-independent growth ability of melanocytes expressing activated BRaf, CRaf and Ral was assayed by growing melanocytes in 3% semi-solid agar as previously described (Ha et al., 2007). Briefly, 2×104 cells/well were seeded in triplicate in six-well plates in growth medium containing 3% agar. After 2 weeks of inoculation, colonies that developed in soft agar were counted. Individual soft agar colonies are shown in (d) and quantitation is shown in (e). Error bars represent s.d. (f) The ability of melanocytes to induce senescence was quantified using senescence-associated β-galactosidase (SA-β-gal) staining and senescent morphology, using previously published methods (Michaloglou et al., 2005; Ha et al., 2007). Error bars represent s.d.
We further evaluated the role of NRasQ61K and downstream pathway members to induce senescence in melanocytes. The ability of melanocytes to induce senescence was quantified using senescence-associated-β-galactosidase (SA-β-gal) staining and senescent morphology, as described earlier (Michaloglou et al., 2005; Ha et al., 2007). Senescence induced by activation of either BRaf or, to a lesser extent, by CRaf was comparable to that induced by NRasQ61K (Figure 3f). Cellular senescence was relatively low in control melanocytes, as well as in those expressing activated PI3K and Ral (Figure 3f and Supplementary Figure 2).
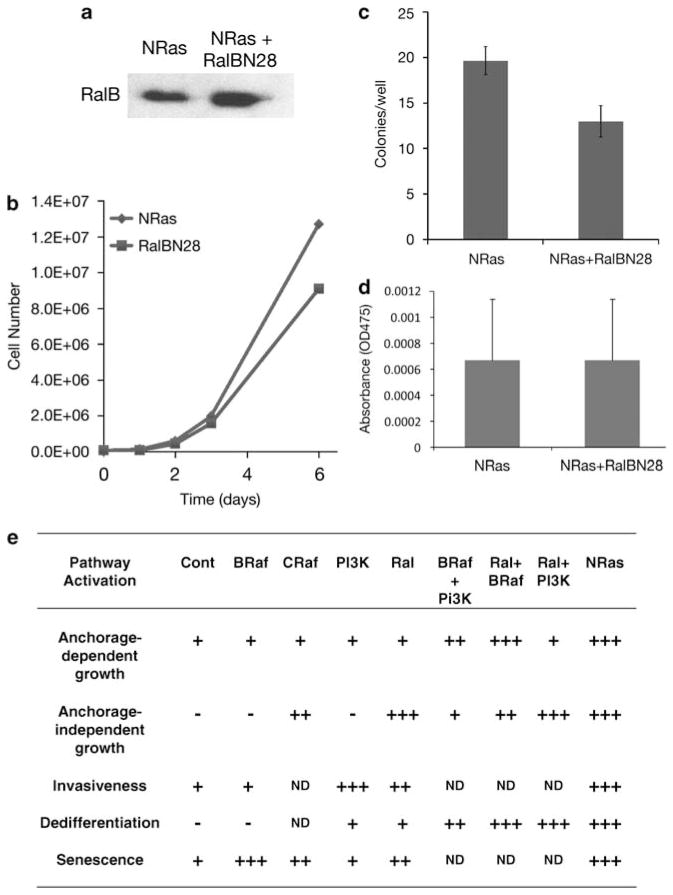
We next confirmed that transformation based on Ral signaling was dependent on high levels of RalGEF activity. We explored the effect of suppressing RalGEF activity in NRasQ61K-transformed melanocytes by transfection with a dominant-negative Ral (RalBN28) that tightly binds and inhibits RalGEF (Figure 4a; Lim et al., 2005; Yin et al., 2007). RalBN28 inhibition of RalGEF in the NRasQ61K-expressing cells had a noticeable but limited effect on the anchorage-dependent growth rate (Figure 4b). In contrast, a significant effect (P<0.05) of RalGEF inhibition was observed on the ability of NRasQ61K-transformed melanocytes to grow under anchorage-independent conditions (Figure 4c), confirming that the transformed phenotype of NRasQ61K-expressing cells is, at least in part, regulated through RalGEF-Ral pathways. The consequences of expressing the dominant-negative Ral were specific, as RalBN28 expression did not rescue pigmentation in NRasQ61K-transformed melanocytes (Figure 4d), suggesting that anchorage-independent growth is more sensitive to reduced RalGEF activity.
Figure 4.
Consequences of expression of an Ral-dominant negative protein (RalBN38) on growth, and summary of all phenotypes associated with constitutive pathway activation. (a) Western blot showing a higher level of RalB owing to expression of the dominant-negative RalBN38, which binds to and inhibits RalGEF (Yin et al., 2007). (b) RalBN38 reduced the growth rate of the NRasQ61K-expressing melanocytes in 2D culture. (c) RalBN38 reduced the anchorage-independent growth of the NRasQ61K-expressing melanocytes in soft agar (P<0.05; error bars show s.d.). Briefly, 2×104 cells/well were seeded in triplicate in six-well plates in growth medium containing 3% agar. After 2 weeks of inoculation, colonies that developed in soft agar were counted. (d) RalBN38 had no significant effect on pigmentation in NRasQ61K-expressing melanocytes. (e) Summary of phenotypes associated with constitutive activation of NRas and its downstream effectors. The dedifferentiation phenotype included loss of melanin and decrease in expression of DCT and TYRP1. The BRaf, CRaf, PI3K and Ral pathways were constitutively activated by BRafV600E, CAAX-RIF-1, p110a-CAAX and RLF-CAAX, respectively. NRas pathways were activated by expression of NRasQ61K. All melanocyte lines were passaged ~15 times before analysis. (−) No phenotype; (+) mild phenotype; (+ +) moderate phenotype; (+ + +) strong phenotype.
To summarize our findings (Figure 1e), we here show that activation of each of the three major downstream pathways of NRas has overt phenotypic effects on immortalized mouse melanocytes, many distinct. It should be noted that cross talk can occur between the downstream NRas pathway members; for example, Ral can be activated by members of the PI3K pathway in melanomas (Figure 1c; Gray-Schopfer et al., 2007; Bodemann and White, 2008). Not unexpectedly, expression of activated BRaf and PI3K enhanced senescence and invasiveness, respectively, to levels approaching NRasQ61K. Importantly, we found that constitutively activated RalGEF stimulated melanocyte invasiveness and anchorage-independent growth almost as impressively as did NRasQ61K. The suppressive effects of expression of a dominant negative Ral corroborated the role of the RalGEF-Ral pathway in anchorage-independent growth. We also confirmed that melanocytes expressing constitutively activated mutants of all three NRas downstream factors (RLF-CAAX plus p110α-CAAX plus BRafV600E) achieved soft agar colony-forming efficiency that was fully comparable to those expressing NRasQ61K alone (Figures 2c and d).
The strong association of activated RalGEF with transformed behavior was not anticipated; RalGEF mutations have, to our knowledge, not been detected in melanoma (Omholt and Hansson, 2007). Here, we show that constitutive activation of the RalGEF-RAL pathway through expression of RLF-CAAX has a profound effect on the morphology, invasiveness, and anchorage-dependent and -independent growth of immortalized mouse melanocytes. Counter and colleagues have detected activation of RalA in human melanoma cell lines, besides discovering that shRNA-mediated knockdown of RalA and RalB had a strong inhibitory effect on the growth of human melanoma cells in immuno-compromised mice (personal communication). Hence, the RalGEF pathway is now implicated in several phenotypes associated with transformation of mouse and human melanocytes.
Given the recent potent melanomagenesis reported in vivo in mice that experience simultaneous activation of BRaf and inactivation of PTEN in melanocytes (Dankort et al., 2009), it was surprising that the phenotypic consequences of combined activation of PI3K and BRaf in our immortalized mouse melanocyte model was not strongly additive; we observed only modest combined effects on pigmentation and 2D growth in culture, and almost no effect on soft agar growth. One explanation is that interaction of melanocytes with stromal elements is required for the robustness of melanoma development in vivo. Alternatively, full inactivation of PTEN may have additional consequences that go beyond PI3K activation through p110α-CAAX expression in melanoma; for example, the protein phosphatase activity of PTEN may have an important role (Stiles, 2009). Additional experiments will be required to better understand this observation.
Another surprise was that BRafV600E alone was relatively ineffective at transforming Arf-deficient mouse melanocytes, and not at all the equivalent of the activity of NRasQ61K. This could be due to the presence of functional Ink4a, as BRAFV600E has been reported to transform mouse melanocytes lacking both functional Ink4a and Arf (Wellbrock et al., 2004). The oncogenic potential of BRafV600E could be undermined by its ability to induce cellular senescence in Arf-deficient mouse melanocytes. In fact, unlike activated PI3K and RalGEF, we found that activation of BRaf and, to a lesser extent, CRaf stimulated an obvious senescent phenotype in Arf-deficient melanocytes. The relative ineffectiveness of BRafV600E could also be related to reports that oncogenic NRas actually targets CRaf, not BRaf (Dumaz et al., 2006). Accordingly, we found that melanocytes expressing CAAX-Raf-1 exhibited a more transformed phenotype relative to those expressing BrafV600E.
The discovery of a significant role for the RalGEF pathway in melanomagenesis may have important clinical implications. It is noted that RalA and RalB were recently shown to be geranylgeranylated and targets of geranylgeranyltransferase I inhibitors, which hamper anchorage-dependent and -independent growth (Falsetti et al., 2007), phenotypes stimulated by RLF-CAAX expression in our study. The RalGEF arm of the NRas pathway is relatively understudied in melanoma. Our data, especially when considered in concert with those of Counter and colleagues (personal communication), suggest that the RalGEF pathway represents a promising new target as we as a community attempt to identify more efficacious anti-melanoma treatments.
Supplementary Material
Acknowledgments
We thank Dr Christopher Counter (Duke University) for useful discussions, and for communicating data before publication. We acknowledge Dr Paul Khavari (Stanford University) for gifting the NRas, PI3K and BRaf retroviral vectors, and Drs Frederique Zindy and Charles Sherr (St Jude Children’s Research Hospital) for the Arf-deficient mouse skins from which the immortalized melanocytes were generated. The PEP7 and PEP8H antibodies were a gift from Dr Vince Hearing (NCI). The pEF-CAAX-Raf-1 vector was a gift from Dr Silvio J Gutkind (NIH/NIDCR). This work was supported in part by the Intramural Research Program of the NCI, National Institutes of Health, in part by NCI Contract N01-CO-12400, and in part by Wellcome Trust Program Grant 078327 (to EVS).
Footnotes
Conflict of interest
The authors declare no conflict of interest.
Supplementary Information accompanies the paper on the Oncogene website (http://www.nature.com/onc)
References
- Albini A, Iwamoto Y, Kleinman HK, Martin GR, Aaronson SA, Kozlowski JM, et al. A rapid in vitro assay for quantitating the invasive potential of tumor cells. Cancer Res. 1987;47:3239–3245. [PubMed] [Google Scholar]
- Bennett DC. How to make a melanoma: what do we know of the primary clonal events? Pig Cell Melan Res. 2008;21:27–38. doi: 10.1111/j.1755-148X.2007.00433.x. [DOI] [PubMed] [Google Scholar]
- Bodemann BO, White MA. Ral GTPases and cancer: linchpin support of the tumorigenic platform. Nat Rev. 2008;8:133–140. doi: 10.1038/nrc2296. [DOI] [PubMed] [Google Scholar]
- Brose MS, Volpe P, Feldman M, Kumar M, Rishi I, Gerrero R, et al. BRAF and RAS mutations in human lung cancer and melanoma. Cancer Res. 2002;62:6997–7000. [PubMed] [Google Scholar]
- Chien Y, Kim S, Bumeister R, Loo YM, Kwon SW, Johnson CL, et al. RalB GTPase-mediated activation of the IkappaB family kinase TBK1 couples innate immune signaling to tumor cell survival. Cell. 2006;127:157–170. doi: 10.1016/j.cell.2006.08.034. [DOI] [PubMed] [Google Scholar]
- Chien Y, White MA. RAL GTPases are linchpin modulators of human tumour-cell proliferation and survival. EMBO Rep. 2003;4:800–806. doi: 10.1038/sj.embor.embor899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chin L, Garraway LA, Fisher DE. Malignant melanoma: genetics and therapeutics in the genomic era. Genes Dev. 2006;20:2149–2182. doi: 10.1101/gad.1437206. [DOI] [PubMed] [Google Scholar]
- Chudnovsky Y, Adams AE, Robbins PB, Lin Q, Khavari PA. Use of human tissue to assess the oncogenic activity of melanoma-associated mutations. Nat Genet. 2005;37:745–749. doi: 10.1038/ng1586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Curtin JA, Fridlyand J, Kageshita T, Patel HN, Busam KJ, Kutzner H, et al. Distinct sets of genetic alterations in melanoma. New Engl J Med. 2005;353:2135–2147. doi: 10.1056/NEJMoa050092. [DOI] [PubMed] [Google Scholar]
- Dahl C, Guldberg P. The genome and epigenome of malignant melanoma. APMIS. 2007;115:1161–1176. doi: 10.1111/j.1600-0463.2007.apm_855.xml.x. [DOI] [PubMed] [Google Scholar]
- Dankort D, Curley DP, Cartlidge RA, Nelson B, Karnezis AN, Damsky WE, Jr, et al. Braf(V600E) cooperates with Pten loss to induce metastatic melanoma. Nat Genet. 2009;41:544–552. doi: 10.1038/ng.356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davies H, Bignell GR, Cox C, Stephens P, Edkins S, Clegg S, et al. Mutations of the BRAF gene in human cancer. Nature. 2002;417:949–954. doi: 10.1038/nature00766. [DOI] [PubMed] [Google Scholar]
- de Gorter DJ, Reijmers RM, Beuling EA, Naber HP, Kuil A, Kersten MJ, et al. The small GTPase Ral mediates SDF-1-induced migration of B cells and multiple myeloma cells. Blood. 2008;111:3364–3372. doi: 10.1182/blood-2007-08-106583. [DOI] [PubMed] [Google Scholar]
- Downward J. Targeting RAS signalling pathways in cancer therapy. Nat Rev. 2003;3:11–22. doi: 10.1038/nrc969. [DOI] [PubMed] [Google Scholar]
- Dumaz N, Hayward R, Martin J, Ogilvie L, Hedley D, Curtin JA, et al. In melanoma, RAS mutations are accompanied by switching signaling from BRAF to CRAF and disrupted cyclic AMP signaling. Cancer Res. 2006;66:9483–9491. doi: 10.1158/0008-5472.CAN-05-4227. [DOI] [PubMed] [Google Scholar]
- Falsetti SC, Wang DA, Peng H, Carrico D, Cox AD, Der CJ, et al. Geranylgeranyltransferase I inhibitors target RalB to inhibit anchorage-dependent growth and induce apoptosis and RalA to inhibit anchorage-independent growth. Mol Cell Biol. 2007;27:8003–8014. doi: 10.1128/MCB.00057-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feig LA. Ral-GTPases: approaching their 15 minutes of fame. Trends Cell Biol. 2003;13:419–425. doi: 10.1016/s0962-8924(03)00152-1. [DOI] [PubMed] [Google Scholar]
- Freedberg DE, Rigas SH, Russak J, Gai W, Kaplow M, Osman I, et al. Frequent p16-independent inactivation of p14ARF in human melanoma. J Natl Cancer Inst. 2008;100:784–795. doi: 10.1093/jnci/djn157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goel VK, Lazar AJ, Warneke CL, Redston MS, Haluska FG. Examination of mutations in BRAF, NRAS, and PTEN in primary cutaneous melanoma. J Invest Derm. 2006;126:154–160. doi: 10.1038/sj.jid.5700026. [DOI] [PubMed] [Google Scholar]
- Gray-Schopfer V, Wellbrock C, Marais R. Melanoma biology and new targeted therapy. Nature. 2007;445:851–857. doi: 10.1038/nature05661. [DOI] [PubMed] [Google Scholar]
- Ha L, Ichikawa T, Anver M, Dickins R, Lowe S, Sharpless NE, et al. ARF functions as a melanoma tumor suppressor by inducing p53-independent senescence. Proc Natl Acad Sci USA. 2007;104:10968–10973. doi: 10.1073/pnas.0611638104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamad NM, Elconin JH, Karnoub AE, Bai W, Rich JN, Abraham RT, et al. Distinct requirements for Ras oncogenesis in human versus mouse cells. Genes Dev. 2002;16:2045–2057. doi: 10.1101/gad.993902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heidecker G, Huleihel M, Cleveland JL, Kolch W, Beck TW, Lloyd P, et al. Mutational activation of c-raf-1 and definition of the minimal transforming sequence. Mol Cell Biol. 1990;10:2503–2512. doi: 10.1128/mcb.10.6.2503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim KH, Baines AT, Fiordalisi JJ, Shipitsin M, Feig LA, Cox AD, et al. Activation of RalA is critical for Ras-induced tumorigenesis of human cells. Cancer Cell. 2005;7:533–545. doi: 10.1016/j.ccr.2005.04.030. [DOI] [PubMed] [Google Scholar]
- Lim KH, O’Hayer K, Adam SJ, Kendall SD, Campbell PM, Der CJ, et al. Divergent roles for RalA and RalB in malignant growth of human pancreatic carcinoma cells. Curr Biol. 2006;16:2385–2394. doi: 10.1016/j.cub.2006.10.023. [DOI] [PubMed] [Google Scholar]
- Michaloglou C, Vredeveld LC, Soengas MS, Denoyelle C, Kuilman T, van der Horst CM, et al. BRAFE600-associated senescence-like cell cycle arrest of human naevi. Nature. 2005;436:720–724. doi: 10.1038/nature03890. [DOI] [PubMed] [Google Scholar]
- Omholt K, Hansson J. No evidence of RALGDS mutations in cutaneous melanoma. Melan Res. 2007;17:410–412. doi: 10.1097/CMR.0b013e3282ef4178. [DOI] [PubMed] [Google Scholar]
- Oxford G, Owens CR, Titus BJ, Foreman TL, Herlevsen MC, Smith SC, et al. RalA and RalB: antagonistic relatives in cancer cell migration. Cancer Res. 2005;65:7111–7120. doi: 10.1158/0008-5472.CAN-04-1957. [DOI] [PubMed] [Google Scholar]
- Pollock PM, Harper UL, Hansen KS, Yudt LM, Stark M, Robbins CM, et al. High frequency of BRAF mutations in nevi. Nat Genet. 2003;33:19–20. doi: 10.1038/ng1054. [DOI] [PubMed] [Google Scholar]
- Pollock PM, Meltzer PS. A genome-based strategy uncovers frequent BRAF mutations in melanoma. Cancer Cell. 2002;2:5–7. doi: 10.1016/s1535-6108(02)00089-2. [DOI] [PubMed] [Google Scholar]
- Rangarajan A, Hong SJ, Gifford A, Weinberg RA. Species-and cell type-specific requirements for cellular transformation. Cancer Cell. 2004;6:171–183. doi: 10.1016/j.ccr.2004.07.009. [DOI] [PubMed] [Google Scholar]
- Recio JA, Noonan FP, Takayama H, Anver MR, Duray P, Rush WL, et al. Ink4a/arf deficiency promotes ultraviolet radiation-induced melanomagenesis. Cancer Res. 2002;62:6724–6730. [PubMed] [Google Scholar]
- Rosse C, Hatzoglou A, Parrini MC, White MA, Chavrier P, Camonis J. RalB mobilizes the exocyst to drive cell migration. Mol Cell Biol. 2006;26:727–734. doi: 10.1128/MCB.26.2.727-734.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith SC, Oxford G, Baras AS, Owens C, Havaleshko D, Brautigan DL, et al. Expression of ral GTPases, their effectors, and activators in human bladder cancer. Clin Cancer Res. 2007;13:3803–3813. doi: 10.1158/1078-0432.CCR-06-2419. [DOI] [PubMed] [Google Scholar]
- Stiles BL. Phosphatase and tensin homologue deleted on chromosome 10: extending its PTENtacles. Internatl J Biochem Cell Biol. 2009;41:757–761. doi: 10.1016/j.biocel.2008.09.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsao H, Zhang X, Benoit E, Haluska FG. Identification of PTEN/MMAC1 alterations in uncultured melanomas and melanoma cell lines. Oncogene. 1998;16:3397–3402. doi: 10.1038/sj.onc.1201881. [DOI] [PubMed] [Google Scholar]
- Urano T, Emkey R, Feig LA. Ral-GTPases mediate a distinct downstream signaling pathway from Ras that facilitates cellular transformation. EMBO J. 1996;15:810–816. [PMC free article] [PubMed] [Google Scholar]
- Virador VM, Kobayashi N, Matsunaga J, Hearing VJ. A standardized protocol for assessing regulators of pigmentation. Anal Biochem. 1999;270:207–219. doi: 10.1006/abio.1999.4090. [DOI] [PubMed] [Google Scholar]
- Wellbrock C, Ogilvie L, Hedley D, Karasarides M, Martin J, Niculescu-Duvaz D, et al. V599EB-RAF is an oncogene in melanocytes. Cancer Res. 2004;64:2338–2342. doi: 10.1158/0008-5472.can-03-3433. [DOI] [PubMed] [Google Scholar]
- Wu H, Goel V, Haluska FG. PTEN signaling pathways in melanoma. Oncogene. 2003;22:3113–3122. doi: 10.1038/sj.onc.1206451. [DOI] [PubMed] [Google Scholar]
- Yin J, Pollock C, Tracy K, Chock M, Martin P, Oberst M, et al. Activation of the RalGEF/Ral pathway promotes prostate cancer metastasis to bone. Mol Cell Biol. 2007;27:7538–7550. doi: 10.1128/MCB.00955-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.