Abstract
Long-term expression from helper virus-free Herpes Simplex Virus (HSV-1) vectors is required for many specific neural gene therapies and studies on neuronal physiology. We previously developed a promoter that supports long-term, neuron-specific expression by fusing the chicken ß-globin insulator (INS), followed by an upstream enhancer from the rat tyrosine hydroxylase (TH) promoter, to a neurofilament heavy gene (NFH) promoter. Here, we examined the capability of specific transcription factors to further improve long-term expression from this promoter. Following a HSV-1 virus infection, the virus genome is localized to promyelocytic leukemia protein (PML) nuclear bodies (NB). At these sites, specific cellular transcription factors interact with HSV-1 encoded transcription factors, and together regulate HSV-1 gene expression. Importantly, lysine-specific demethylase-1 (LSD1), CLOCK, and Co-Rest each activate HSV-1 gene expression. However, gene expression from HSV-1 vectors differs in a number of important aspects from the virus, including no HSV-1 genes are expressed. Nonetheless, these observations raise the possibility that specific transcription factors may improve long-term expression from specific promoters in HSV-1 vectors. Here, we show that overexpression of either LSD1 or CLOCK improves long-term expression from the INS-TH-NFH promoter, but overexpression of Co-Rest supports levels of long-term expression similar to those supported by a control vector. Further, overexpression of LSD1 is compatible with neuron-specific expression. Thus, overexpressing specific transcription factors can improve long-term expression from specific cellular promoters in HSV-1 vectors, and the chromatin structure of the vector has an important role in enabling expression.
Keywords: herpes simplex virus vector, long-term expression, transcription factor, chromatin modifying enzyme, enhancer, striatal neuron
1. Introduction
Direct gene transfer into neurons in the brain has supported studies on neuronal physiology and has significant potential for supporting gene therapies for specific neurological disorders. Herpes Simplex Virus (HSV-1) plasmid vectors (amplicons) have advantages that include high-efficiency gene transfer and a large capacity, supporting coexpression of multiple genes (Fraefel et al., 1996; Geller and Breakefield, 1988). Many applications of HSV-1 vectors to studying neuronal physiology or for specific gene therapies require long-term, neuron-specific expression. We developed a modified neurofilament promoter that supports greater than 90 % neuron-specific expression, and significant levels of long-term expression (Sun et al., 2004; Zhang et al., 2000; Zhang et al., 2005; Zhang et al., 2009). However, the levels of long-term expression from this promoter might be improved by overexpressing specific transcription factors or chromatin modifying enzymes.
A number of neuron-specific or viral promoters support only short-term expression from HSV-1 vectors, such as the neurofilament heavy gene (NFH) promoter ((Wang et al., 1999); reviewed in (Zhang et al., 2000)); nonetheless, a number of cellular promoters have been identified that support long-term expression. Neuron type-specific promoters that support significant levels of long-term expression from HSV-1 plasmid vectors include the tyrosine hydroxylase (TH) promoter, preproenkephalin (ENK) promoter, glutamic acid decarboxylase (GAD) promoter, phosphate-activated glutaminase (PAG) promoter, vesicular glutamate transporter-1 (VGLUT1) promoter, and specific fragments of the VGLUT1 promoter (Jin et al., 1996; Kaplitt et al., 1994; Rasmussen et al., 2007; Song et al., 1997; Wang et al., 1999; Zhang and Geller, 2010; Zhang et al., 2011). Interestingly, large fragments of the rat TH promoter (6.8 or 9 kb) support long-term, catecholaminergic-specific expression (Jin et al., 1996; Song et al., 1997; Wang et al., 1999), but a small fragment of this promoter (766 bp) supports only short-term expression (Wang et al., 1999), indicating that upstream sequences in the TH promoter support long-term expression. Thus, we constructed a chimeric promoter that supports long-term expression in forebrain neurons (Zhang et al., 2000) by fusing upstream sequences from the TH promoter (Brown et al., 1987) to the 5’ end of a NFH promoter (Schwartz et al., 1994). Further, we obtained higher levels of long-term expression by fusing the chicken ß-globin locus insulator (INS), the best characterized vertebrate INS, to the TH-NFH promoter (Zhang et al., 2000). The INS-TH-NFH promoter has supported both studies on visual learning (Zhang et al., 2005; Zhang et al., 2010) and gene therapy (Sun et al., 2004; Sun et al., 2005; Zhang et al., 2009).
Although relatively little is known about the organization of virus vector chromatin in the nucleus of neurons, much is known about the organization and regulation of HSV-1 virus chromatin in the nucleus of fibroblast cells, and this knowledge can inform studies to improve long-term expression from HSV-1 vectors. Following a virus infection, HSV-1 virus DNA, and other viral genomes, are localized in the nucleus to promyelocytic leukemia protein (PML) nuclear bodies (NB), or ND10 (Bernardi and Pandolfi, 2007; Everett and Maul, 1994; Everett and Murray, 2005; Everett et al., 2007; Lallemand-Breitenbach and de The, 2010; Sourvinos and Everett, 2002). In PML NB, specific chromatin proteins and transcription factors interact with HSV-1 encoded transcription factors, and together regulate HSV-1 gene expression. Of note, lysine-specific demethylase-1 (LSD1), CLOCK, and Co-Rest, particularly a deletion mutant, each activate HSV-1 virus gene expression in PML NB during a productive lytic infection of fibroblast cells, with some evidence for similar functions in neuronal cells (Gu et al., 2005; Gu and Roizman, 2007; Gu and Roizman, 2009a; Gu and Roizman, 2009b; Kalamvoki and Roizman, 2008; Kalamvoki and Roizman, 2010; Liang et al., 2009). However, gene expression from HSV-1 vectors is regulated differently than from the virus. Of note, localization of HSV-1 virus DNA to PML NB requires expression of specific HSV-1 genes (Tang et al., 2003) that are absent from helper virus-free HSV-1 vectors, as these vectors contain no HSV-1 genes. Overall, these observations raise the possibility that specific transcription factors may improve long-term expression from specific promoters in HSV-1 vectors.
Here, we tested three transcription factors that are known to activate HSV-1 virus gene expression for the capability to improve long-term expression from a cellular promoter in a HSV-1 vector. We constructed vectors that use a neuron-specific promoter, the INS-TH-NFH promoter, to express LSD1, CLOCK, or Co-Rest. After gene transfer into the striatum, for each vector, we compared the numbers of expressing cells in rats sacrificed at 4 days or 1 month after gene transfer. Overexpression of either LSD1 or CLOCK improved the stability of long-term expression, but overexpression of Co-Rest supported levels of long-term expression similar to those observed using a control vector that does express a transcription factor. Further, the vector that overexpresses LSD1 supports a level of neuron-specific expression similar to that previously observed using the modified neurofilament promoter. These results show that overexpressing specific transcription factors can improve long-term expression from cellular promoters in HSV-1 vectors, and suggest that the chromatin structure of the vector has an important role in supporting recombinant gene expression.
2. Results
2.1. Overexpression of LSD1 or CLOCK, but not Co-Rest, improves long-term expression from a modified neurofilament promoter
We constructed vectors that use the INS-TH-NFH promoter to express LSD1, or CLOCK, or Co-Rest, followed by an internal ribosome entry site (ires) and the Lac Z gene (Fig. 1). To detect these transcription factors, the flag tag was fused to LSD1, and the myc tag was fused to CLOCK or Co-Rest (Gu et al., 2005; Gu and Roizman, 2007; Gu and Roizman, 2009a; Gu and Roizman, 2009b; Kalamvoki and Roizman, 2008; Kalamvoki and Roizman, 2010; Liang et al., 2009). The resulting vectors were designated pINS-TH-NFHflag-lsd1/ires/lac, pINS-TH-NFHmyc-clock/ires/lac, or pINS-TH-NFHmyc-co-rest/ires/lac. The control vector has an analogous structure, but does not express a transcription factor, and instead expresses flag-TH, followed by an ires and the human aromatic amino acid decarboxylase (AADC) gene (pINS-TH-NFHflag-th/ires/aadc) (Sun et al., 2003).
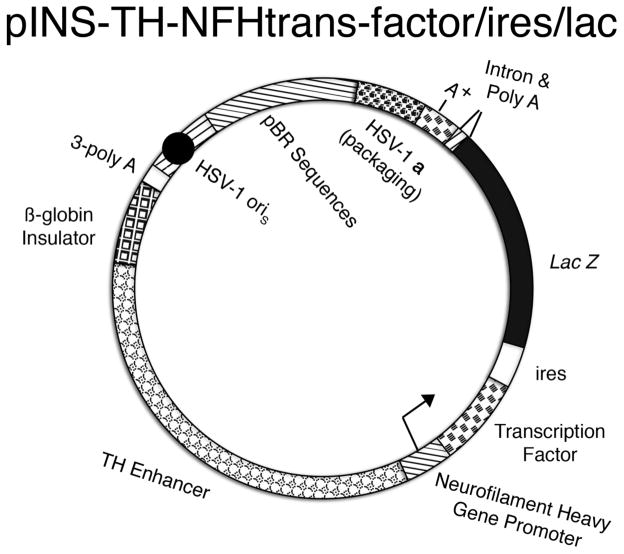
Fig. 1.
Schematic diagram of pINS-TH-NFHtrans-factor/ires/lac. The promoter contains the chicken ß-globin INS (block segment), an upstream enhancer from the rat TH promoter (alternating circle segment), and the mouse NFH promoter (diagonal line segment with arrow). The INS-TH-NFH promoter expresses a transcription factor (brick segment), an ires (clear segment), and the Lac Z gene (black segment), followed by the second intron from the mouse α-globin gene (triangle), and the SV40 polyadenylation signal (diagonal line and brick segments, respectively). A cassette of three polyadenylation sites (3-poly A, clear segment) was placed 5’ to the INS-TH-NFH promoter to reduce any effects on expression from the HSV-1 IE 4/5 promoter (diagonal line segment). The HSV-1 origin of DNA replication, small (oriS, black circle in diagonal line segment) and the HSV-1 a sequence, which contains the packaging site (clover segment), support the replication and packaging of the vector into HSV-1 particles, respectively. Sequences from pBR322 (horizontal line segment) enable propagation of the vector in E. coli.
These four vectors were packaged into HSV-1 particles using our helper virus-free packaging system. The resulting vector stocks were titered on Baby Hamster Kidney (BHK) fibroblast cells; at 24 hours after transduction of BHK cells, the numbers of infectious vector particles (IVP/ml) were determined. The stocks of vectors expressing specific transcription factors were diluted as indicated to achieve a concentration of 1 x 106 IVP/ml for each vector stock. The control vector was inadvertently diluted to a concentration of 2 x 106 IVP/ml; because, for each vector, we compared the numbers of expressing cells at 4 days and 1 month (see below), each vector has an internal control, and this small error in a vector concentration does not significantly affect the experimental design. We previously used a PCR assay to titer the number of vector genomes, and we found that vectors containing only the NFH promoter, the TH enhancer fragment fused to the NFH promoter, or the entire INS-TH-NFH promoter, were all efficiently packaged into HSV-1 particles with similar efficiencies, and these packaging efficiencies were similar to those observed using vectors containing other promoters (Gao et al., 2007; Zhang et al., 2000). Thus, we did not repeat the assay for vector genomes here.
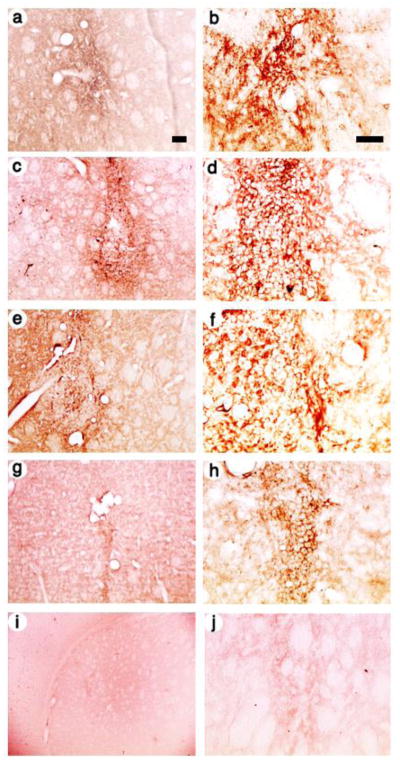
Each of these four vector stocks was microinjected into the striatum, the rats were sacrificed at either 4 days or 1 month after gene transfer, and expressing cells were visualized using an anti-flag or anti-c-myc antibody, a biotinylated secondary antibody, the avidin-biotinylated peroxidase complex (ABC) reagent, and the horse radish peroxidase (HRP) reaction. We chose a 4 day survival time to represent short-term expression and a 1 month survival time to represent long-term expression: A number of studies showed that the majority of the decrease in numbers of expressing cells occurs between 4 days and 2 weeks after gene transfer (Gao et al., 2007; Wang et al., 1999; Zhang et al., 2000); time courses showed the numbers of expressing cells is similar at 2 weeks and 1, 2, 4, or 6 months (Zhang et al., 2000); and similar numbers of expressing cells are present at 7 or 14 months (Sun et al., 2003; Sun et al., 2004; Sun et al., 2005). Thus, a survival time of 1 month is representative of long-term expression. In a rat sacrificed at 4 days after receiving the control vector, pINS-TH-NFHflag-th/ires/aadc, a low power view showed numerous flag-TH-immunoreactivity (IR) cells proximal to the needle track (Fig. 2A), and a high power view showed that many of these cells contain neuronal morphology, namely relatively large cell bodies at some distance from the needle track (Fig. 2B). Of note, the three vectors expressing specific transcription factors supported similar patterns of expressing cells as the control vector: Under low power, pINS-TH-NFHflag-lsd1/ires/lac supported numerous flag-LSD1-IR cells (Fig. 2C), pINS-TH-NFHmyc-clock/ires/lac supported numerous myc-CLOCK-IR cells (Fig. 2E), and pINS-TH-NFHmyc-co-rest/ires/lac supported numerous myc-Co-Rest-IR cells (Fig. 2G); these expressing cells were located proximal to the needle track. Further, high power views flag-LSD1-IR (Fig. 2D), myc-CLOCK-IR (Fig. 2F), or myc-Co-Rest-IR (Fig. 2H) revealed cells with neuronal morphology, specifically positive cells at some distance form the needle track with relatively large cell bodies, similar to pINS-TH-NFHflag-th/ires/aadc. Neuronal identity was confirmed by costaining for a neuronal marker, as detailed below. The intracellular location of the IR was different for each protein because flag-TH is found in the cytoplasm, whereas each of the three transcription factors is localized to different locations within the nucleus, and each protein is translated primarily in the cell body. The relatively clear ellipsoid-shaped regions in these photomicrographs is a characteristic of striatal morphology; similar regions are also observed in the Paxinos and Watson atlas using either an acetylcholinesterase assay or Cresyl violet staining (Paxinos and Watson, 1986). As a control, using sections from a rat that received pINS-TH-NFHflag-lsd1/ires/lac, omission of the primary antibody resulted in no flag-IR cells (Fig. 2I and J).
Fig. 2.
Low and high power views of recombinant-IR in striatal cells from rats sacrificed at 4 days after gene transfer with vectors that use the INS-TH-NFH promoter to express a specific transcription factor, or a control that expresses TH. Flag-TH (control), flag-LSD-1, c-myc-CLOCK, or c-myc-Co-Rest were detected using either an anti-flag or an anti-c-myc antibody that was visualized using a biotin-conjugated secondary antibody, the ABC reagent, and the HRP reaction. The photomicrographs show areas proximal to the needle tracks. (A and B) The control vector, pINS-TH-NFHflag-th/ires/aadc, TH was detected using flag-IR, (A) low power or (B) high power. (C and D) pINS-TH-NFHflag-lsd1/ires/lac, LSD1 was detected using flag-IR, (C) low power or (D) high power. (E and F) pINS-TH-NFHmyc-clock/ires/lac, CLOCK was detected using c-myc-IR, (E) low power or (F) high power. (G and H) pINS-TH-NFHmyc-clock/ires/lac, Co-REST was detected using c-myc-IR, (G) low power or (H) high power. Control: (I and J) pINS-TH-NFHflag-lsd1/ires/lac, no primary antibody, (I) low power or (J) high power. Scale bars: (A,C,E,G, and I) 100 μm; (B,D,F,H, and J) 50 μm.
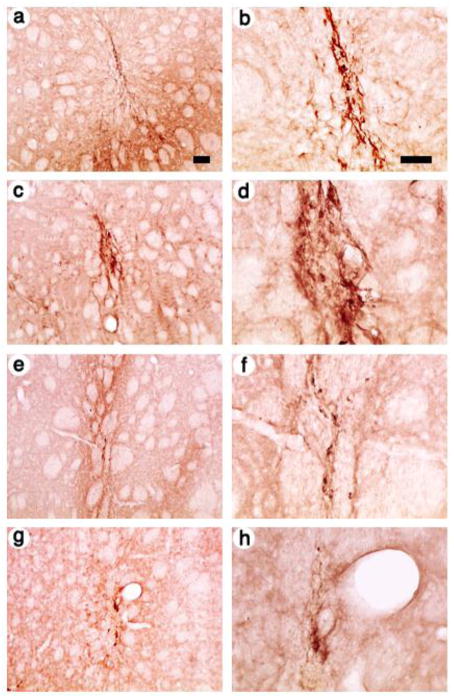
Next, we examined expression in rats sacrificed at 1 month after gene transfer. In a rat that received the control vector, pINS-TH-NFHflag-th/ires/aadc, a low power view showed flag-TH-IR cells proximal to the needle track (Fig. 3A), although apparently fewer cells than observed at 4 days, and a high power view showed that many of these cells displayed neuronal morphology (Fig. 3B). Of note, under low power, pINS-TH-NFHflag-lsd1/ires/lac supported numerous flag-LSD1-IR cells (Fig. 3C), and pINS-TH-NFHmyc-clock/ires/lac supported numerous myc-CLOCK-IR cells (Fig. 3E). Under high power, these cells displayed neuronal morphology (flag-LSD1-IR cells, Fig. 3D; myc-CLOCK-IR cells, Fig. 3F); the myc-CLOCK-IR was predominately in the nucleus. In contrast, pINS-TH-NFHmyc-co-rest/ires/lac supported myc-Co-Rest-IR cells (Fig. 3G), but there appeared to be fewer positive cells than at 4 days; these cells displayed neuronal morphology under high power (Fig. 3H).
Fig. 3.
Low and high power views of recombinant-IR in striatal cells from rats sacrificed at 1 month after gene transfer with vectors that express a specific transcription factor from the use the INS-TH-NFH promoter, or a control that expresses TH. The photomicrographs show areas proximal to the needle tracks. (A and B) The control vector, pINS-TH-NFHflag-th/ires/aadc, TH was detected using flag-IR, (A) low power or (B) high power. (C and D) pINS-TH-NFHflag-lsd1/ires/lac, LSD1 was detected using flag-IR, (C) low power or (D) high power. (E and F) pINS-TH-NFHmyc-clock/ires/lac, CLOCK was detected using c-myc-IR, (E) low power or (F) high power. (G and H) pINS-TH-NFHmyc-clock/ires/lac, Co-REST was detected using c-myc-IR, (G) low power or (H) high power. Scale bars: (A,C,E, and G) 100 μm; (B,D,F, and H) 50 μm.
The numbers of expressing cells was quantified by cell counts, and the level of long-term expression supported by each vector was quantified by comparing the numbers of cells at 4 days and 1 month. The results (Table 1) showed that the control, pINS-TH-NFHflag-th/ires/aadc, at 1 month after gene transfer, supported expression in 21 % of the number of cells as compared to 4 days. Of note, expression of either LSD1 or CLOCK improved the stability of long-term expression; pINS-TH-NFHflag-lsd1/ires/lac, at 1 month after gene transfer, supported expression in 34 % of the number of cells as compared to 4 days; and pINS-TH-NFHmyc-clock/ires/lac, at 1 month after gene transfer, supported expression in 30 % of the number of cells as compared to 4 days. In contrast, expression of Co-Rest did not improve the stability of long-term expression; pINS-TH-NFHmyc-co-rest/ires/lac, at 1 month after gene transfer, supported expression in 19 % of the number of cells as compared to 4 days; a stability of expression similar to that supported by the control, pINS-TH-NFHflag-th/ires/aadc. These differences were statistically significant: A one-way ANOVA showed a significant effect of vector (F3,12=10.7 p<0.002). Pairwise comparisons (Student-Neumann-Keuls test) confirmed that overexpression of either LSD1 or CLOCK supported higher levels of long-term expression than either overexpression of Co-REST or the control of no overexpression of a transcription factor (pINS-TH-NFHflag-lsd1/ires/lac vs. pINS-TH-NFHmyc-co-rest/ires/lac or pINS-TH-NFHflag-th/ires/aadc p<0.005; pINS-TH-NFHmyc-clock/ires/lac vs. pINS-TH-NFHmyc-co-rest/ires/lac or pINS-TH-NFHflag-th/ires/aadc p<0.02). Interestingly, overexpression of either LSD1 or CLOCK supported similar levels of long-term expression (p>0.05). Also, overexpression of Co-REST supported similar levels of long-term expression as the control (p>0.05). Thus, even though the INS-TH-NFH promoter supports significant levels of long-term expression at 1 month after gene transfer, this statistical analysis showed that overexpressing either LSD1 or CLOCK improves the stability of long-term expression.
TABLE 1.
The numbers of expressing cells, at 4 days or 1 month after gene transfer, supported by HSV-1 vectors that contain the INS-TH-NFH promoter and express LSD1, CLOCK, Co-Rest, or a control expressing TH.
| Numbers of cells | |||
|---|---|---|---|
| Vector | 4 days | 1 month | % long-term expressiona |
| pINS-TH-NFHflag-lsd1/ires/lac | 459±37 | 158±23 | 34±3 |
| pINS-TH-NFHmyc-clock/ires/lac | 411±37 | 123±15 | 30±1 |
| pINS-TH-NFHmyc-co-rest/ires/lac | 319±35 | 62±12 | 19±2 |
| pINS-TH-NFHflag-th/ires/aadc | 700±50 | 149±26 | 21±2 |
Four striata were analyzed for each condition. For each striata, the positive cells were counted in every 4th section, and these counts were multiplied by 4 to obtain the number of cells in each striata. The values shown are mean±s.e.m.
The number of expressing cells at 1 month divided by the number of expressing cells at 4 days, times 100. For each vector, the rank order of number of expressing cells in each striata, at each of the two time points, was used to calculate four ratios of % long-term expression, and the mean±s.e.m. is shown.
The numbers of positive cells were modest, ~100 to 200 at 1 months after gene transfer (Table 1). The experimental design used here differed in two critical aspects from our study that observed expression in ~11,400 striatal cells at 6 months (Sun et al., 2004); only one injection site, instead of three injection sites, was used, and the titers (IVP/ml) were ~10 to 20-fold lower (four vector stocks were used here, and specific stocks were diluted to yield a titer of 1 x 106 IVP/ml for each vector stock). The ~30- to 60-fold lower amount of vector injected into the striatum accounts for most of the difference in the numbers of positive cells between the current study (Table 1) and the ~11,400 positive cells observed in an earlier study (Sun et al., 2004).
2.2. pINS-TH-NFHflag-lsd1/ires/lac supports neuron-specific expression
In previous studies, we showed that the INS-TH-NFH promoter supports neuron-specific expression; we performed costaining for ß-gal-IR and a neuronal marker, NeuN (Sun et al., 2004; Zhang et al., 2006; Zhang et al., 2000; Zhang et al., 2005; Zhang et al., 2009). Here, we confirmed that in the presence of LSD1, one of the two transcription factors that improved long-term expression, the INS-TH-NFH promoter supports neuron-specific expression. Using sections from a rat that was sacrificed at 4 days after receiving pINS-TH-NFHflag-lsd1/ires/lac, we performed immunofluorescent costaining using antibodies against ß-gal, which is expressed from this vector, or NeuN. Photomicrographs revealed that most of the ß-gal-IR cells also contained NeuN-IR (Fig. 4A-C). As a control, omission of the primary antibodies resulted in no IR (Fig. 4D-F). Cell counts showed that pINS-TH-NFHflag-lsd1/ires/lac supported 95 % neuron-specific expression (102 ß-gal-IR cells were scored, and 97 of these cells also contained NeuN-IR); this level of neuron-specific expression is similar to that supported by the INS-TH-NFH promoter alone (Sun et al., 2004; Zhang et al., 2006; Zhang et al., 2000; Zhang et al., 2005; Zhang et al., 2009).
Fig. 4.
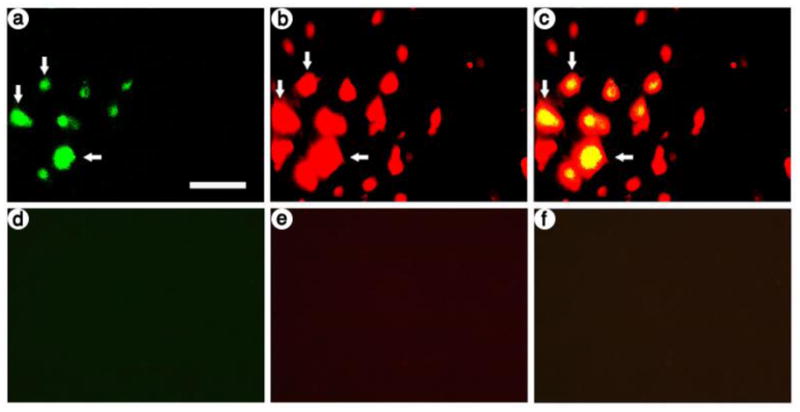
At 4 days after gene transfer with pINS-TH-NFHflag-lsd1/ires/lac, most ß-gal-IR striatal cells contain NeuN-IR. ß-gal-IR was detected with anti-E. coli ß-gal that was visualized using a fluorescein isothiocyanate-conjugated secondary antibody, and NeuN, a neuronal marker present in the nucleus, was detected with anti-NeuN that was visualized using a Texas red-conjugated secondary antibody. (A-C) pINS-TH-NFHflag-lsd1/ires/lac; (A) ß-gal-IR, (B) NeuN-IR, and (C) merged. Numerous ß-gal-IR cells were observed, and most of these cells contained NeuN-IR (arrows). (D-F) Omitting the primary antibodies from the assay resulted in background levels of fluorescence; (D) fluorescein-conjugated secondary antibody-IR, (E) Texas red-conjugated secondary antibody-IR, and (F) merge. Scale bar: 50 μm.
3. Discussion
In this study, we showed that overexpression of either of two transcription factors, LSD1 or CLOCK, improves the stability of long-term expression from a modified neurofilament promoter, but overexpression of Co-Rest does not affect the levels of long-term expression. Further, overexpression of LSD1 was compatible with neuron-specific expression from the modified neurofilament promoter. These results suggest that overexpressing specific transcription factors or chromatin modifying enzymes can improve long-term expression from specific cellular promoters in HSV-1 vectors.
Each of the three transcription factors tested here affects gene expression from the HSV-1 virus genome in PML NB. Overexpression of LSD1 activates expression from specific HSV-1 immediate early (IE) promoters by modifying the methylation levels of specific lys residues in specific histones, proximal to HSV-1 IE promoters (Liang et al., 2009). Further, inhibiting LSD1 activity, with specific monoamine oxidase inhibitors, blocks reactivation of latent HSV-1 virus in trigeminal ganglia neurons (Liang et al., 2009). Nonetheless, the histone deacetylase (HDAC1/2)-CoREST-REST complex represses the activity of specific promoters in the HSV-1 virus genome; this activity is modulated by specific HSV-1 proteins, including the transcription factor ICP0 and the protein kinases US3 and UL13 (Gu et al., 2005) (Andres et al., 1999; Chong et al., 1995; Gu and Roizman, 2009a; Gu and Roizman, 2009b). Of note, ICP0 relieves the repression of HSV-1 gene expression by the HDAC1/2-CoREST-REST complex by dissociating HDAC1 from the CoREST-REST complex. Further, overexpression of a truncated Co-Rest, lacking the ICP0 binding domain, also relieved the repression by the HDAC1/2-CoREST-REST complex by a similar mechanism, dissociating HDAC1 from the CoREST-REST complex (Gu and Roizman, 2007). Moreover, ICP0 interacts with Bmal1, a partner of the CLOCK histone acetyltransferase; interestingly, Bmal1 and CLOCK are present in PML NB, a wt HSV-1 infection stabilizes CLOCK, and overexpression of CLOCK can partially replace ICP0 (Kalamvoki and Roizman, 2010).
Nonetheless, gene expression from HSV-1 vectors is regulated differently than from the virus; in particular, localization of HSV-1 virus DNA to PML NB requires expression of specific HSV-1 genes (Tang et al., 2003) that are absent from this helper virus-free HSV-1 vector system; thus, specific components of PML NB have different effects on HSV-1 vector and HSV-1 virus gene expression. LSD1 and CLOCK each activate specific HSV-1 promoters in the virus and the modified neurofilament promoter in HSV-1 vectors. In contrast, the truncated Co-Rest activates specific HSV-1 promoters in the virus, but does not activate the modified neurofilament promoter in HSV-1 vectors, although Rest and Co-Rest both have important roles in regulating gene expression in neurons (Ballas and Mandel, 2005). Paradoxically, the HDAC1/2-CoREST-REST complex is regulated by the HSV-1 encoded protein kinases US3 and UL13, and mutation of each of these kinases improves expression from the modified neurofilament promoter in HSV-1 vectors (Liu et al., 2005; Liu et al., 2009; Yang et al., 2001). In summary, while the mechanisms that regulate HSV-1 virus gene expression in PML NB should be carefully considered in relation to HSV-1 vectors, the role of specific components of PML NB in regulating expression from specific cellular promoters in helper virus-free HSV-1 vectors can differ from regulating promoters in the virus, and should be directly studied.
The INS-TH-NFH promoter supports long-term expression in multiple types of neurons in multiple brain areas; but according to the Allen Brain Atlas, LSD1 is expressed at middle to high levels in all brain areas reported; whereas CLOCK is expressed at low levels in most brain areas, and is found at mid-range levels in a few specific areas. Of note, the INS-TH-NFH promoter supports expression in the striatum for 14 months (Sun et al., 2004), in the hippocampus for 1 month (Zhang et al., 2009), in postrhinal cortex for 1.75 months (Zhang et al., 2010), and in the substantia nigra for 1 month (Cao et al., 2008), the longest times tested. Moreover, the INS-TH-NFH promoter supports expression in both glutamatergic and GABAergic neurons in the hippocampus (Zhang et al., 2009); predominately glutamatergic and GABAergic neurons, but also cholinergic neurons, in postrhinal cortex (Zhang et al., 2005); GABAergic neurons in the striatum (Sun et al., 2004); and catecholaminergic neurons in the substantia nigra (Cao et al., 2008). Because the INS-TH-NFH promoter supports long-term expression in multiple types of neurons in multiple brain areas, additional transcription factors, besides LSD1 and CLOCK, may modulate expression from the INS-TH-NFH promoter, particularly in neuron types that lack LSD1 and/or CLOCK. Moreover, a number of neuron type-specific promoters support significant levels of long-term expression from HSV-1 plasmid vectors, including the TH, ENK, GAD, phosphate-activated glutaminase, and vesicular glutamate transporter-1 promoters (Jin et al., 1996; Kaplitt et al., 1994; Rasmussen et al., 2007; Song et al., 1997; Wang et al., 1999; Zhang and Geller, 2010; Zhang et al., 2011). It remains to be determined if either LSD1 or CLOCK can improve long-term expression from any of these neuron type-specific promoters. Identifying specific transcription factors and chromatin modifying enzymes that improve long-term expression from specific promoters may contribute to elucidating the mechanisms that govern long-term expression from HSV-1 vectors. Of note, overexpression of other transcription factors and chromatin modifying enzymes present in PML NB may also improve long-term expression. More generally, as most foreign DNA, from specific viruses or other delivery mechanisms, is localized to PML NB, the results reported here may be applicable to other delivery systems.
The INS-TH-NFH promoter has supported both gene therapies and basic neuroscience studies that require long-term expression. This promoter has supported correction of the rat model of Parkinson’s disease with expression for 7, 8, or 14 months (Sun et al., 2003; Sun et al., 2004; Sun et al., 2005). Further, the INS-TH-NFH promoter has enabled correction of spatial learning deficits in aged rats, by modifying hippocampal circuits (Zhang et al., 2009). Moreover, this promoter supported studies on visual object discrimination learning that identified a ~500 neuron circuit within rat postrhinal cortex that can encode some of the essential information for performance (Zhang et al., 2005; Zhang et al., 2010). HSV-1 vectors have a large capacity; coexpression of up to four genes by using two transcription units has been reported (Sun et al., 2004), and ~50 or 150 kb vectors have been isolated (Wade-Martins et al., 2003; Wang et al., 2000). Thus, it should be feasible to coexpress a transcription factor that improves long-term expression with genes that alter neuronal physiology. In summary, identifying specific transcription factors and chromatin modifying enzymes that can improve long-term expression from the INS-TH-NFH promoter, or particular neuron type-specific promoters, will benefit both gene therapy and basic neuroscience studies.
4. Materials and Methods
4.1. Materials
Genes were synthesized by GenScript. pcDNA1.1/Amp was from Invitrogen, pSP73 was from Promega, and pCMVth/ires/aadc was kindly provided by Dr. K. O’Malley. Restriction endonucleases were obtained from New England BioLabs. OPTI-MEM I, Dulbecco’s modified minimal essential medium (DMEM), fetal bovine serum (FBS), glutamine, and penicillin/streptomycin were from Invitrogen. G418 was obtained from RPI. 5-bromo-4-chloro-3-indoyl-ß-D-galactopyranoside (X-Gal) was obtained from Sigma. Mouse anti-flag and rabbit anti-E. coli ß-gal antibodies were obtained from Sigma, mouse anti-c-myc was obtained from Santa Cruz Biotechnology, and mouse anti-NeuN was obtained from Millpore. Alkaline phosphatase-conjugated horse anti-mouse immunoglobulin (Ig) G, biotinylated goat anti-mouse IgG, the ABC reagent, fluorescein isothiocyanate (FITC)-conjugated goat anti-rabbit IgG, and Texas red-conjugated horse anti-mouse IgG were from Vector Labs.
4.2. Vectors
pINS-TH-NFHtrans-factor/ires/lac is diagrammed in Fig. 1. The promoter in the transcription unit contains the chicken ß-globin insulator (INS; 1.2 kb), an enhancer from the rat tyrosine hydroxylase promoter (TH; −0.5 kb to −6.8 kb), and a mouse neurofilament heavy gene promoter (NFH; 0.6 kb) (Zhang et al., 2000). This promoter was used to express a specific transcription factor, an ires, and the Lac Z gene, followed by an intron and polyadenylation site. This vector contains a standard vector backbone: To reduce interactions between genetic elements in the vector backbone and the transcription unit, the HSV-1 oris fragment, which contains the HSV-1 IE 4/5 promoter, is followed by a cassette of three SV40 early region polyadenylation sites (Wang et al., 1999). Replication and packaging of the vector into HSV-1 particles is supported by an HSV-1 origin of DNA replication (oriS) and the HSV-1 a sequence, which contains the packaging site, respectively.
The flag-LSD1 coding sequence was as described (Lee et al., 2005), and was synthesized with an Asc I site at the 5’ end and a Xho I site at the 3’ end. The CLOCK coding sequence fused to the myc tag was as described (Kwon et al., 2006), and was synthesized with an Asc I site at the 5’ end and a Xho I site at the 3’ end. A truncated Co-Rest, containing amino acids 146–482 (aa 482 is the C-terminus), with the myc tag, was as described (Gu and Roizman, 2007), and was synthesized with an Asc I site at the 5’ end and a Xho I site at the 3’ end. Each of these three genes was inserted into pUC57 (pUC57fag-lsd1, pUC57myc-clock, pUC57myc-co-rest).
To simplify inserting these transcription factors into HSV-1 vectors, we first assembled a vector that contains the INS-TH-NFH promoter with preferred sites at the ends. The PkcΔ vector, pINS-TH-NFHpkcΔ\INS-TH-NFHlac (Wang et al., 2001; Zhang et al., 2005), was digested with Fse I to excise the PkcΔ transcription unit from the vector backbone and Lac Z transcription unit, and the vector was recyclized (pINS-TH-NFHlac-deletePkcΔ). This vector was partially digested with EcoR I, the linear fragment was isolated, two oligonucleotides were inserted to destroy an EcoR I site (sense 5’ AATTGGTCAAGCCTTGGCATACCATGGAGAC 3’, antisense 5’ 28AATTGTCTCCATGGTATGCCAAGGCTTGACC 3’), and candidates were screened to identify a vector that lacked an EcoR I site proximal to the HSV-1 a sequence (pINS-TH-NFHlac-deletePkcΔ - ΔRI). Similarly, pINS-TH-NFHlac-deletePkcΔ was partially digested with Hind III, the linear fragment was isolated, two oligonucleotides were inserted to destroy a Hind III site (sense 5’ AGCTGGCACAACGCTTGGACTACACTCCAGAC 3’, antisense 5’ AGCTGTCTGGAGTGTAGTCCAAGCGTTGTGCC 3’), and candidates were screened to identify a vector that lacked a Hind III site proximal to the cassette of three polyadenylation sites (pINS-TH-NFHlac-deletePkcΔ - ΔHIII). Next, a dendrite-targeted green fluorescent protein (dendrite-gfp) (Kameda et al., 2008) was synthesized with Hind III and Asc I sites at the 5’ end, and Bgl II, Pac I, and Eco RI sites at the 3’ end, and inserted into pUC57 (pUC57dendrite-gfp; this construct is for other studies yet to be published). Next, the INS-TH-NFH promoter fragment was excised from pINS-TH-NFHlac-deletePkcΔ - ΔHIII by digestion with Not I and Hind III, the vector backbone fragment was excised from pINS-TH-NFHlac-deletePkcΔ - ΔRI by digestion with Not I and EcoR I, the dendrite-gfp fragment was excised from pUC57 dendrite-gfp by digestion with Hind III and EcoR I, and these three fragments were ligated together to yield pINS-TH-NFHdendrite-gfp.
To further simplify inserting the transcription factors into HSV-1 vectors, we assembled a HSV-1 vector containing the ires/lac fragment. pcDNA1.1/Amp (Invitrogen) was digested with BamH I and Not I; a 4 kb fragment containing the Lac Z gene, intron, and polyadenylation site was isolated from pBR-8cutter-linker/TH-NFHlac (Wang et al., 2001) by a Not I complete and Hind III partial digestion; a 600 bp fragment containing an ires was isolated from pCMVth/ires/aadc (Moffat et al., 1997) by a BamH I complete and Hind III partial digestion; and these three fragments were ligated together to yield pcDNAires/lac. pcDNAires/lac was partially digested with EcoR I, the linear fragment was isolated, two oligonucleotides were inserted to destroy an EcoR I site and add a Bgl II site (sense 5’ AATTGGGACCCAGCGAGATCTTCTAGAGC 3’, antisense 5’ AATTGCTCTAGAAGATCTCGCTGGGTCCC 3’), and candidates were screened to identify a vector that lacked an EcoR I site at the 5’ end of the ires (pcDNAires/lac+BglII- ΔRI). pSP73 (Promega) was digested with Hind III and EcoR I; pcDNAires/lac+BglII- ΔRI was digested with Bgl II and EcoR I, and the 3.9 kb fragment containing the ires/lac segment was isolated; a rat synaptotagmin I cDNA was synthesized with a Hind III site at the 5’ end and Xho I and Bgl II sites at the 3’ end; and these three fragments were ligated together to yield pSP73syt/ires/lac (this construct is for other studies yet to be published). Next, the INS-TH-NFH promoter fragment was excised from pINS-TH-NFHlac-deletePkcΔ - ΔHIII by digestion with Not I and Hind III, the vector backbone fragment was excised from pINS-TH-NFHlac-deletePkcΔ - ΔRI by digestion with Not I and EcoR I, the syt/ires/lac fragment was isolated from pSP73syt/ires/lac by digestion with Hind III and EcoR I, and these three fragments were ligated together to yield pINS-TH-NFHsyt/ires/lac.
To assemble the three transcription factor vectors, the INS-TH-NFH promoter fragment was excised from pINS-TH-NFHdendrite-gfp by digestion with Not I and Asc I, the vector backbone-ires/lac fragment was excised from pINS-TH-NFHsyt/ires/lac by digestion with Not I and Xho I, each transcription factor was excised from pUC57fag-lsd1, pUC57myc-clock, pUC57myc-co-rest by digestion with Asc I and Xho I, and the appropriate three fragments were ligated together to yield pINS-TH-NFHflag-lsd1/ires/lac, pINS-TH-NFHmyc-clock/ires/lac, or pINS-TH-NFHmyc-co-rest/ires/lac
The control vector was pINS-TH-NFHflag-th/ires/aadc (Sun et al., 2003); this vector expresses flag-TH and AADC from the same vector backbone and promoter as the experimental vectors.
4.3. Cells
BHK21 cells (Yang et al., 2001) or 2–2 cells (Smith et al., 1992) were grown in DMEM supplemented with 4 mM glutamine, 10 % FBS, and penicillin/streptomycin. G418 (0.5 mg/ml) waqs present during the growth of 2–2 cells, and was removed before plating cells for vector packaging. Cells were grown at 37 °C in the presence of 5 % CO2 in humidified incubators.
4.4. Packaging vectors into HSV-1 particles
Vectors were packaged into HSV-1 particles using our helper virus-free packaging system (Fraefel et al., 1996), as modified to optimize the resulting titers (Sun et al., 1999). Purification and concentration of vector stocks was performed as described (Lim et al., 1996). The resulting vector stocks were titered by transducing BHK fibroblast cells and 1 day later quantifying the numbers expressing cells (IVP/ml) (Sun et al., 2004; Zhang et al., 2000). The numbers of infectious vector particles (IVP/ml) for pINS-TH-NFHflag-lsd1/ires/lac or pINS-TH-NFHflag-th/ires/aadc were determined by flag-IR, visualized using the alkaline phosphatase procedure, or the IVP/ml for pINS-TH-NFHmyc-clock/ires/lac or pINS-TH-NFHmyc-co-rest/ires/lac was determined by X-gal staining. BHK cells were used as the best available assay as these fibroblast cells form a monolayer; in contrast, most neuronal cell lines do not form a monolayer, and BHK cells support higher titers than PC12 cells (Yang et al., 2001; Zhang et al., 2000). The activity of the INS-TH-NFH promoter in BHK fibroblast cells represents ectopic activity that declines rapidly at longer times after gene transfer. Wild-type (wt) HSV-1 was not detected (<10 plaque forming units/ml) in any of these vector stocks.
4.5. Stereotactic injection of vector stocks into the striatum
All procedures with animals were approved by the W. Roxbury VA Hospital IACUC. Male Long Evans rats (175–200 gm) were used for these experiments. Vector stocks were delivered using stereotactic injections (2 sites per rat, 1 site per hemisphere, 3 μl/site) into the striatum (anterior-posterior (AP) +0.7, medial-lateral (ML) ±3.0, dorsal-ventral (DV) –5.5). AP is relative to bregma, ML is relative to the sagittal suture, and DV is relative to the bregma-lambda plane (Paxinos and Watson, 1986). A micropump (model 100, KD Scientific) was used to perform the injections; 3 μl of vector stock was injected over 8 minutes, and the needle was maintained in place for an additional 5 minutes and was then slowly withdrawn over approximately 5 minutes.
4.6. Immunohistochemistry
At 4 days or 1 month after gene transfer, the rats were perfused, and immunohistochemistry assays were performed, using previously described procedures (Zhang et al., 2000). Assays were peformed on 25 μm coronal brain sections that contained the striatum; sections were cut on a freezing microtome. To quantify the numbers of cells that contained a specific recombinant gene product, flag-LSD1-IR or flag-th-IR were detected using a mouse anti-flag antibody (1:400 dilution); myc-CLOCK-IR or myc-Co-Rest-IR were detected using a mouse anti-c-myc antibody (1:100 dilution); and each IR was visualized using a biotin-conjugated goat anti-mouse IgG (1:200 dilution), the ABC reagent, and the HRP reaction. To localize expression from pINS-TH-NFHflag-lsd1/ires/lac to neurons, sections were incubated with both rabbit anti-ß-gal and mouse anti-NeuN antibodies, and these antibodies were visualized using fluorescein isothiocyanate- or Texas red-conjugated secondary antibodies, as described (Zhang et al., 2000).
4.7. Cell counts
Coronal sections (25 μm) that contained areas proximal to an injection site were prepared. Recombinant gene expression was observed in ~35 sections per hemisphere, with the majority of the cells in ~20 sections per hemisphere. Every 4th section was analyzed for IR. Cells were counted under 60x magnification; each section was counted a minimum of two times, on different days; and the values differed by <10 % for each section.
To quantify neuron-specific expression, a video camera was used to capture photomicrographs, under 60x magnification. Each ß-gal-IR cell in a section was scored for containing, or lacking, NeuN-IR. Each section was counted a minimum of two times, and the values differed by <10 % for each section.
Statistical comparisons were performed using ANOVAs (SigmaStat).
4.8. Computer analyses
Expression of LSD1 or CLOCK in the mouse brain was analyzed using the Allen Brain Atlas (http://mouse.brain-map.org/welcome.do;jsessionid=B4CD215888034060325A29D70FA0EED1).
Highlights.
Promoters that support long-term expression are desirable
Overexpression of specific transcription factors can improve expression
Lysine-specific demethylase-1 or CLOCK improve long-term expression
Co-Rest does not improve long-term expression
Acknowledgments
We gratefully thank Dr. A. Davison for HSV-1 cosmid set C, Dr. K. O’Malley for the TH promoter and for pCMVth/ires/aadc, Dr. G. Felsenfeld for ß-globin insulator, and Dr. W. Schlaepfer for the NFH promoter. This work was supported by AG025894 (G.Z.) and NS045855, and NS057558 (A.I.G.).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Andres ME, Burger C, Peral-Rubio MJ, Battaglioli E, Anderson ME, Grimes J, Dallman J, Ballas N, Mandel G. CoREST: a functional corepressor required for regulation of neural-specific gene expression. Proc Natl Acad Sci USA. 1999;96:9873–8. doi: 10.1073/pnas.96.17.9873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ballas N, Mandel G. The many faces of REST oversee epigenetic programming of neuronal genes. Curr Opin Neurobiol. 2005;15:500–6. doi: 10.1016/j.conb.2005.08.015. [DOI] [PubMed] [Google Scholar]
- Bernardi R, Pandolfi PP. Structure, dynamics and functions of promyelocytic leukaemia nuclear bodies. Nat Rev Mol Cell Biol. 2007;8:1006–16. doi: 10.1038/nrm2277. [DOI] [PubMed] [Google Scholar]
- Brown ER, Coker GTD, O'Malley KL. Organization and evolution of the rat tyrosine hydroxylase gene. Biochemistry. 1987;26:5208–12. doi: 10.1021/bi00390a046. [DOI] [PubMed] [Google Scholar]
- Cao H, Zhang GR, Wang X, Kong L, Geller AI. Enhanced nigrostriatal neuron-specific, long-term expression by using neural-specific promoters in combination with targeted gene transfer by modified helper virus-free HSV-1 vector particles. BMC Neurosci. 2008;9:37. doi: 10.1186/1471-2202-9-37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chong JA, Tapia-Ramirez J, Kim S, Toledo-Aral JJ, Zheng Y, Boutros MC, Altshuller YM, Frohman MA, Kraner SD, Mandel G. REST: a mammalian silencer protein that restricts sodium channel gene expression to neurons. Cell. 1995;80:949–57. doi: 10.1016/0092-8674(95)90298-8. [DOI] [PubMed] [Google Scholar]
- Everett RD, Maul GG. HSV-1 IE protein Vmw110 causes redistribution of PML. Embo J. 1994;13:5062–9. doi: 10.1002/j.1460-2075.1994.tb06835.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Everett RD, Murray J. ND10 components relocate to sites associated with herpes simplex virus type 1 nucleoprotein complexes during virus infection. J Virol. 2005;79:5078–89. doi: 10.1128/JVI.79.8.5078-5089.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Everett RD, Murray J, Orr A, Preston CM. Herpes simplex virus type 1 genomes are associated with ND10 nuclear substructures in quiescently infected human fibroblasts. J Virol. 2007;81:10991–1004. doi: 10.1128/JVI.00705-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fraefel C, Song S, Lim F, Lang P, Yu L, Wang Y, Wild P, Geller AI. Helper virus-free transfer of herpes simplex virus type 1 plasmid vectors into neural cells. J Virol. 1996;70:7190–7. doi: 10.1128/jvi.70.10.7190-7197.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao Q, Sun M, Wang X, Geller AI. Isolation of an enhancer from the rat tyrosine hydroxylase promoter that supports long-term, neuronal-specific expression from a neurofilament promoter, in a helper virus-free HSV-1 vector system. Brain Res. 2007;1130:1–16. doi: 10.1016/j.brainres.2006.10.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geller AI, Breakefield XO. A defective HSV-1 vector expresses Escherichia coli beta-galactosidase in cultured peripheral neurons. Science. 1988;241:1667–9. doi: 10.1126/science.241.4873.1667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu H, Liang Y, Mandel G, Roizman B. Components of the REST/CoREST/histone deacetylase repressor complex are disrupted, modified, and translocated in HSV-1-infected cells. Proc Natl Acad Sci U S A. 2005;102:7571–6. doi: 10.1073/pnas.0502658102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu H, Roizman B. Herpes simplex virus-infected cell protein 0 blocks the silencing of viral DNA by dissociating histone deacetylases from the CoREST-REST complex. Proc Natl Acad Sci U S A. 2007;104:17134–9. doi: 10.1073/pnas.0707266104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu H, Roizman B. Engagement of the lysine-specific demethylase/HDAC1/CoREST/REST complex by herpes simplex virus 1. J Virol. 2009a;83:4376–85. doi: 10.1128/JVI.02515-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu H, Roizman B. The two functions of herpes simplex virus 1 ICP0, inhibition of silencing by the CoREST/REST/HDAC complex and degradation of PML, are executed in tandem. J Virol. 2009b;83:181–7. doi: 10.1128/JVI.01940-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin BK, Belloni M, Conti B, Federoff HJ, Starr R, Son JH, Baker H, Joh TH. Prolonged in vivo gene expression driven by a tyrosine hydroxylase promoter in a defective herpes simplex virus amplicon vector. Hum Gene Ther. 1996;7:2015–24. doi: 10.1089/hum.1996.7.16-2015. [DOI] [PubMed] [Google Scholar]
- Kalamvoki M, Roizman B. Nuclear retention of ICP0 in cells exposed to HDAC inhibitor or transfected with DNA before infection with herpes simplex virus 1. Proc Natl Acad Sci U S A. 2008;105:20488–93. doi: 10.1073/pnas.0810879105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalamvoki M, Roizman B. Circadian CLOCK histone acetyl transferase localizes at ND10 nuclear bodies and enables herpes simplex virus gene expression. Proc Natl Acad Sci U S A. 2010;107:17721–6. doi: 10.1073/pnas.1012991107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kameda H, Furuta T, Matsuda W, Ohira K, Nakamura K, Hioki H, Kaneko T. Targeting green fluorescent protein to dendritic membrane in central neurons. Neurosci Res. 2008;61:79–91. doi: 10.1016/j.neures.2008.01.014. [DOI] [PubMed] [Google Scholar]
- Kaplitt MG, Kwong AD, Kleopoulos SP, Mobbs CV, Rabkin SD, Pfaff DW. Preproenkephalin promoter yields region-specific and long-term expression in adult brain after direct in vivo gene transfer via a defective herpes simplex viral vector. Proc Natl Acad Sci USA. 1994;91:8979–83. doi: 10.1073/pnas.91.19.8979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwon H, Bai Q, Baek HJ, Felmet K, Burton EA, Goins WF, Cohen JB, Glorioso JC. Soluble V Domain of Nectin-1/HveC Enables Entry of Herpes Simplex Virus Type 1 (HSV-1) into HSV-Resistant Cells by Binding to Viral Glycoprotein D. J Virol. 2006;80:138–48. doi: 10.1128/JVI.80.1.138-148.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lallemand-Breitenbach V, de The H. PML nuclear bodies. Cold Spring Harb Perspect Biol. 2010;2:a000661. doi: 10.1101/cshperspect.a000661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee MG, Wynder C, Cooch N, Shiekhattar R. An essential role for CoREST in nucleosomal histone 3 lysine 4 demethylation. Nature. 2005;437:432–5. doi: 10.1038/nature04021. [DOI] [PubMed] [Google Scholar]
- Liang Y, Vogel JL, Narayanan A, Peng H, Kristie TM. Inhibition of the histone demethylase LSD1 blocks alpha-herpesvirus lytic replication and reactivation from latency. Nat Med. 2009;15:1312–7. doi: 10.1038/nm.2051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim F, Hartley D, Starr P, Lang P, Song S, Yu L, Wang Y, Geller AI. Generation of high-titer defective HSV-1 vectors using an IE 2 deletion mutant and quantitative study of expression in cultured cortical cells. Biotechniques. 1996;20:460–9. doi: 10.2144/19962003460. [DOI] [PubMed] [Google Scholar]
- Liu M, Tang J, Wang X, Yang T, Geller AI. Enhanced long-term expression from helper virus-free HSV-1 vectors packaged in the presence of deletions in genes that modulate the function of VP16, UL46 and UL47. J Neurosci Methods. 2005;145:1–9. doi: 10.1016/j.jneumeth.2004.09.030. [DOI] [PubMed] [Google Scholar]
- Liu M, Wang X, Geller AI. Helper virus-free HSV-1 vectors packaged in the presence of combinations of mutations in the protein kinase UL13 and the VP16 transcriptional complex enhance long-term expression. BMC Molecular Biology. 2009;10:58. doi: 10.1186/1471-2199-10-58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moffat M, Harmon S, Haycock J, O'Malley KL. L-Dopa and dopamine-producing gene cassettes for gene therapy approaches to Parkinson's disease. Exp Neurol. 1997;144:69–73. doi: 10.1006/exnr.1996.6390. [DOI] [PubMed] [Google Scholar]
- Paxinos G, Watson C. The rat brain in stereotaxic coordinates. Academic Press; Sidney: 1986. [DOI] [PubMed] [Google Scholar]
- Rasmussen M, Kong L, Zhang G, Liu M, Wang X, Szabo G, Curthoys NP, Geller AI. Glutamatergic or GABAergic neuron-specific, long-term expression in neocortical neurons from helper virus-free HSV-1 vectors containing the phosphate-activated glutaminase, vesicular glutamate transporter-1, or glutamic acid decarboxylase promoter. Brain Res. 2007;1144:19–32. doi: 10.1016/j.brainres.2007.01.125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwartz ML, Katagi C, Bruce J, Schlaepfer WW. Brain-specific enhancement of the mouse neurofilament heavy gene promoter in vitro. J Biol Chem. 1994;269:13444–50. [PubMed] [Google Scholar]
- Smith IL, Hardwicke MA, Sandri-Goldin RM. Evidence that the herpes simplex virus immediate early protein ICP27 acts post-transcriptionally during infection to regulate gene expression. Virology. 1992;186:74–86. doi: 10.1016/0042-6822(92)90062-t. [DOI] [PubMed] [Google Scholar]
- Song S, Wang Y, Bak SY, Lang P, Ullrey D, Neve RL, O'Malley KL, Geller AI. An HSV-1 vector containing the rat tyrosine hydroxylase promoter enhances both long-term and cell type-specific expression in the midbrain. J Neurochem. 1997;68:1792–803. doi: 10.1046/j.1471-4159.1997.68051792.x. [DOI] [PubMed] [Google Scholar]
- Sourvinos G, Everett RD. Visualization of parental HSV-1 genomes and replication compartments in association with ND10 in live infected cells. Embo J. 2002;21:4989–97. doi: 10.1093/emboj/cdf458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun M, Zhang GR, Yang T, Yu L, Geller AI. Improved titers for helper virus-free herpes simplex virus type 1 plasmid vectors by optimization of the packaging protocol and addition of noninfectious herpes simplex virus-related particles (previral DNA replication enveloped particles) to the packaging procedure. Hum Gene Ther. 1999;10:2005–11. doi: 10.1089/10430349950017365. [DOI] [PubMed] [Google Scholar]
- Sun M, Zhang G, Kong L, Holmes C, Wang X, Zhang W, Goldstein DS, Geller AI. Correction of a rat model of Parkinson’s disease by coexpression of tyrosine hydroxylase and aromatic amino acid decarboxylase from a helper virus-free herpes simplex virus type 1 vector. Hum Gene Ther. 2003;14:415–24. doi: 10.1089/104303403321467180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun M, Kong L, Wang X, Holmes C, Gao Q, Zhang W, Pfeilschifter J, Goldstein DS, Geller AI. Coexpression of Tyrosine Hydroxylase, GTP Cyclohydrolase I, Aromatic Amino Acid Decarboxylase, and Vesicular Monoamine Transporter-2 from a Helper Virus-Free HSV-1 Vector Supports High-Level, Long-Term Biochemical and Behavioral Correction of a Rat Model of Parkinson’s Disease. Hum Gene Ther. 2004;15:1177–1196. doi: 10.1089/hum.2004.15.1177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun M, Kong L, Wang X, Lu X, Gao Q, Geller AI. Comparison of protection of nigrostriatal neurons by expression of GDNF, BDNF, or both neurotrophic factors. Brain Res. 2005;1052:119–29. doi: 10.1016/j.brainres.2005.05.072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang Q, Li L, Ishov AM, Revol V, Epstein AL, Maul GG. Determination of minimum herpes simplex virus type 1 components necessary to localize transcriptionally active DNA to ND10. J Virol. 2003;77:5821–8. doi: 10.1128/JVI.77.10.5821-5828.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wade-Martins R, Saeki Y, Chiocca EA. Infectious delivery of a 135-kb LDLR genomic locus leads to regulated complementation of low-density lipoprotein receptor deficiency in human cells. Mol Ther. 2003;7:604–12. doi: 10.1016/s1525-0016(03)00060-1. [DOI] [PubMed] [Google Scholar]
- Wang X, Zhang G, Yang T, Zhang W, Geller AI. Fifty-one kilobase HSV-1 plasmid vector can be packaged using a helper virus-free system and supports expression in the rat brain. BioTechniques. 2000;27:102–6. doi: 10.2144/00281st05. [DOI] [PubMed] [Google Scholar]
- Wang X, Zhang G, Sun M, Geller AI. General strategy for constructing large HSV-1 plasmid vectors that co-express multiple genes. BioTechniques. 2001;31:204–12. doi: 10.2144/01311dd05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Yu L, Geller AI. Diverse stabilities of expression in the rat brain from different cellular promoters in a helper virus-free herpes simplex virus type 1 vector system. Hum Gene Ther. 1999;10:1763–71. doi: 10.1089/10430349950017446. [DOI] [PubMed] [Google Scholar]
- Yang T, Zhang G, Zhang W, Sun M, Wang X, Geller AI. Enhanced reporter gene expression in the rat brain from helper virus-free HSV-1 vectors packaged in the presence of specific mutated HSV-1 proteins that affect the virion. Molec Brain Res. 2001;90:1–16. doi: 10.1016/s0169-328x(01)00059-6. [DOI] [PubMed] [Google Scholar]
- Zhang C, Xuan Z, Otto S, Hover JR, McCorkle SR, Mandel G, Zhang MQ. A clustering property of highly-degenerate transcription factor binding sites in the mammalian genome. Nucleic Acids Res. 2006;34:2238–46. doi: 10.1093/nar/gkl248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang G, Wang X, Yang T, Sun M, Zhang W, Wang Y, Geller AI. A tyrosine hydroxylase--neurofilament chimeric promoter enhances long-term expression in rat forebrain neurons from helper virus-free HSV-1 vectors. Molec Brain Res. 2000;84:17–31. doi: 10.1016/s0169-328x(00)00197-2. [DOI] [PubMed] [Google Scholar]
- Zhang G, Wang X, Kong L, Lu X, Lee B, Liu M, Sun M, Franklin C, Cook RG, Geller AI. Genetic enhancement of visual learning by activation of protein kinase C pathways in small groups of rat cortical neurons. J Neurosci. 2005;25:8468–81. doi: 10.1523/JNEUROSCI.2271-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang G, Liu M, Cao H, Kong L, Wang X, Cook RG, Geller AI. Improved spatial learning in aged rats by genetic activation of protein kinase C in small groups of rat hippocampal neurons. Hippocampus. 2009;19:413–23. doi: 10.1002/hipo.20506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang G, Cao H, Kong L, O’Brien J, Baughns A, Jan M, Zhao H, Wang X, Lu X, Cook RG, Geller AI. Identified circuit in rat postrhinal cortex encodes essential information for performing specific visual shape discriminations. Proc Natl Acad Sci USA. 2010;107:14478–14483. doi: 10.1073/pnas.0912950107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang G, Geller AI. An HSV-1 vector containing the VGLUT1 promoter is expressed only in VGLUT1-, and not VGLUT2-, containing glutamatergic neurons. Brain Res. 2010;1331:12–19. doi: 10.1016/j.brainres.2010.03.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang G, Li X, Cao H, Zhao H, Geller AI. The vesicular glutamate transporter-1 upstream promoter and first intron each support glutamatergic-specific expression in rat postrhinal cortex. Brain Research. 2011;1377:1–12. doi: 10.1016/j.brainres.2010.12.040. [DOI] [PMC free article] [PubMed] [Google Scholar]