Abstract
Among many methods used to investigate proteins/protein interactions, chemical cross-linking combined with mass spectrometry remains a vital experimental approach. Mapping peptides modified by cross-linker provides clues about proteins’ interacting domains. One complication is that such modification may result from intra- but not intermolecular interactions. Therefore, for overall data interpretation, a combination of results from various platforms is necessary. It is postulated that the secretory isoform of gelsolin regulates several biological processes through interactions with proteins such as actin, fibronectin, vitamin D binding protein and unidentified receptors on the surface of eukaryotic; it also has been shown to self-assemble eventually leading to the formation of homo-multimers. As such, it is an excellent model for this study. We used four cross-linkers with arm length ranging from 7.7Å to 21.7Å and MALDI-TOF/TOF mass spectrometry as the analytical platform. Results of this study show that MALDI based mass spectrometry generates high quality data to show lysine residues modified by cross-linkers and combined with existing data based on crystallography (Protein Data Bank, PDB) can be used to discriminate between inter- and intra-molecular linking.
Keywords: gelsolin, chemical cross-linking, MALDI-TOF/TOF, protein-protein interaction, mass spectrometry
Introduction
A variety of methods are used to study 3-dimensional structures of proteins, their self-assembly into homo-polymeric complexes, interactions with other proteins and non-proteinaceous molecules. Methods used for such studies include: X-ray crystallography, fluorescenseresonance energy transfer (FRET), chemical cross-linking, mass spectrometry [1] also with ion mobility technique[2], circular dichroism (CD), atomic force microscopy [3], as well as a combination of these methods [4], each being complementary with specific strengths and limitations[5; 6; 7; 8; 9; 10; 11; 12]. For example, FRET can be used if the proximity of two fluorescent groups is between 10 to 100Å; however, it requires labeling of proteins[13]. X-ray crystallography requires growing of crystals and can be affected by disorder in unit cell or distortion in crystallization.
Chemical cross-linking has been utilized since the 1940s as a tool to study protein complexes, structural features and stability as well as for practical applications such as formation of toxoids[14; 15]. Recently, chemical cross-linkers regained interest. Cross-linking reactions combined with mass spectrometry can be used in rapid detection of interacting domains of proteins[16]. Further development of cleavable and non-cleavable cross-linkers of various lengths that target specific chemical groups, advancement in high-throughput crystallography[17], and mass spectrometry of proteins and peptides allowed a much broader application of cross-linking to address questions related to protein-protein interaction, changes in three-dimensional structures etc.[18].
Matrix-assisted laser desorption ionization (MALDI) is a soft ionization technique widely used in proteomic studies [19]. Moreover, MALDI-TOF (time of flight) mass spectrometry is broadly applicable to investigate many post-translational modifications and protein/peptide quantitation, including isobaric Tags for Relative and Absolute Quantitation (iTRAQ), formylation of serine (Ser) and threonine (Thr) residues [20], protein modifications induced by polyacrylamide gel [21] and various aspects of signal transduction [22]. Although development of electrospray ionization based mass spectrometry unquestionably advanced the application of this type of ionization to study post-translational modification, MALDI-TOF/TOF with collision induced dissociation (CID) remains one of the major platforms to study protein and protein/peptide drug modification in biopharmaceutical research [23]. Furthermore, new cross-linkers such as the α-cyano-4-hydroxycinnamic acid (CHCA)-tagged cross-linker JMV 3378 have been developed to selectively enhance MALDI-TOF signal and compare spectra using either CHCA or the α-cyano-4-hydroxycinnamic methyl ester matrix to discriminate between modified and non-modified peptides [24].
Gelsolin (GSN), an 86-kD protein, is the most widely expressed member of actin severing proteins. Isoform 1 is a secretory (plasma, pGSN) protein and is longer by 51 amino acids at the N-terminal end than isoform 2 (cytoplasmic, cGSN). The increasing body of experimental evidence shows that pGSN is a more versatile molecule than previously thought; more regulatory functions based on interactions with other proteins have been described. Indeed, it has been postulated that pGSN interacts with lysophosphatidic acid (LPA) [25], fibronectin [26], Vitamin D-Binding Protein [27] and adenosine triphosphate ATP [28] acquiring new functional properties. Although these interactions have been shown at the functional level [29], there are no reports about structural changes and conformational requirements that may occur upon such interactions. Although Lind and Janmey using Scatchard plot analysis showed that pGSN interacts with fibronectin[26], our initial experiments suggested that chemical cross-linking in vitro favored pGSN self-assembly into homo-polymers and/or intramolecular interactions rather than interactions with fibronectin. This observation prompted more a general question about interpretation of interactions based on modifications of Lys residues as measured by mass spectrometry. Therefore, we initiated systematic studies of pGSN’s, a model protein, self-interactions using crosslinking technology and MALDI-TOF/TOF mass spectrometry in combination with X-ray based crystallography structural features already available through open source databases.
Material and Methods
Reagents
pGSN from human plasma, CHCA, trifluoroacetic acid (TFA), sodium phosphate, calcium chloride, ammonium bicarbonate and iodoacetamide were purchased from Sigma Aldrich (St. Louis, MO, USA). Cross-linkers, Bis(sulfosuccinimidyl)glutarate-d0 (BS2G-d0), Bis(sulfosuccinimidyl)suberate-d0 (BS3G-d0), Bis-N-succinimidyl(PEG)5 (BS(PEG)5) and 3,3'-Dithiobis(sulfosuccinimidylpropionate) (DTSSP) were purchased from Thermo Scientific (San Jose, CA, USA). Glycine was purchased from Biorad (Hercules, CA, USA). Sequencing grade modified trypsin and Dithiothreitol (DTT) were from Promega (Madison, WI, USA). Acetonitrile (ACN), methanol and acetic acid were purchased from Fisher Scientific (Pittsburg, PA, USA). 1D-gel reagents, i.e. sample buffer, NuPAGE 4–12% gels and running buffer were purchased from Invitrogen Corp. (Carlsbad, CA, USA).
Protein crosslinking
Gelsolin (5µg) was diluted with a 1:1 ratio in a 20mM sodium phosphate by adding 1 volume of 50mM calcium chloride solution for 10 volumes of reacting solution (5mM of CaCl2 final). The sample was then incubated for 10 min at room temperature to allow the protein-protein interaction. Then we added 1 volume of 20mM of cross-linker for 10 volume of sample (2mM of cross-linker final). The cross-linking reaction was performed for 30min at room temperature. After incubation, the reaction was quenched by addition of 1 volume of 40mM glycine per 10 volume of solution (4mM of glycine final).
Protein separation and in-gel digestion
Protein samples (5µg) were first dried and re-suspended in 20µL of non-reducing SDS-PAGE sample buffer. Samples were separated on NuPAGE 4–12% gradient gels. After the separation, the gels were then fixed for 30 min at room temperature in a 50% methanol and 10% acetic acid solution and stained for 2 h with Commassie Brilliant Blue. Visible bands of proteins were cut using a razor blade and destained by two washes in 50% ACN followed by 30 min incubation in 20mM ammonium bicarbonate buffer. Proteins were then reduced by incubation of gel cubes in 10mM DTT for 45min at 60°C and 45 min in 50mM iodoacetamide at room temperature in the dark. Trypsin digestion was performed for 16h at 37°C. Peptide extraction was performed in two steps, first, 20 min incubation in 50 ACN with agitation and second 20 min incubation in 100% ACN. Extracted peptides were dried using SpeedVac®. Each sample was then re-suspended in 10 µl of 0.1% TFA and frozen at −80°C until further use.
Mass spectrometry analyses
The samples of digested proteins were spotted on the MALDI plate, 1 µl for each sample. On top of the dried samples, we added 1 µl of 5mg/ml of CHCA solubilized in 50% ACN and 0.1% TFA.
Spotted samples were submitted for data acquisition on a 4800 MALDI-TOF/TOF mass spectrometer (ABsciex, Foster City, CA). MS spectra were acquired from 800 to 6000 m/z, for a total of 1000 laser shots by an Nd-YAG laser operating at 355 nm and 200 Hz. Laser intensity remained fixed for all the analyses. MS/MS analyses were performed using 1 kV collision energy with air as CID gas. Metastable ions were suppressed for a total of 1000 laser shots. Spectra analysis was performed manually.
Results and Discussion
Our studies using western blot analyses showed an ability of pGSN to spontaneously form homo-dimers and homo-trimers, which we detected in samples of human plasma[30]. Quantitative comparison based on immunoreactivity shows that homo-dimer is more prominent than homo-trimer. However, if reactivity of antibodies is conformation dependent and epitopes partially accessible, it is likely that such analytical tests will be misleading. Importantly, pGSN isolated from its environment of plasma or cerebrospinal fluid shows a higher degree of multimerization[30]. Mass spectrometry analysis of tryptic digest of these forms confirmed the presence of pGSN and absence of other proteins showing that these forms sustain heating in the presence of SDS and are not part of hetero-complex.1-dimensional SDS-PAGE electrophoresis (1-DE) analysis shown in Fig. 1a indicates that under experimental conditions applied in this study, spontaneous multimerization leading to formation of heat and SDS resistant forms is less prominent (Fig. 1a lane G). However, chemical cross-linking makes such multimeric forms permanent. We also observed bands of high molecular weight that barely entered the gel and which may represent an even higher order of multimerization. Taking this together, we showed that pGSN could spontaneously form multi-mers. Importantly, this process occurs concurrently during in vitro studies of interactions of pGSN in the presence offibronectin (data not shown). Therefore, detection of peptides with cross-linker modification(s) without knowing which interactions are inter- and intra-molecular may lead to false interpretation of experimental data. Information presented in this study will aid in interpretation of subsequent experiments of pGSN interaction with other proteins and/or non-proteinaceous molecules. More broadly, this study evaluates the utility of chemical-cross-linking to map residues involved in protein-protein interactions for which we used pGSN as a model protein and how mass spectrometry analysis can discriminate between intra- and inter-molecular interactions.
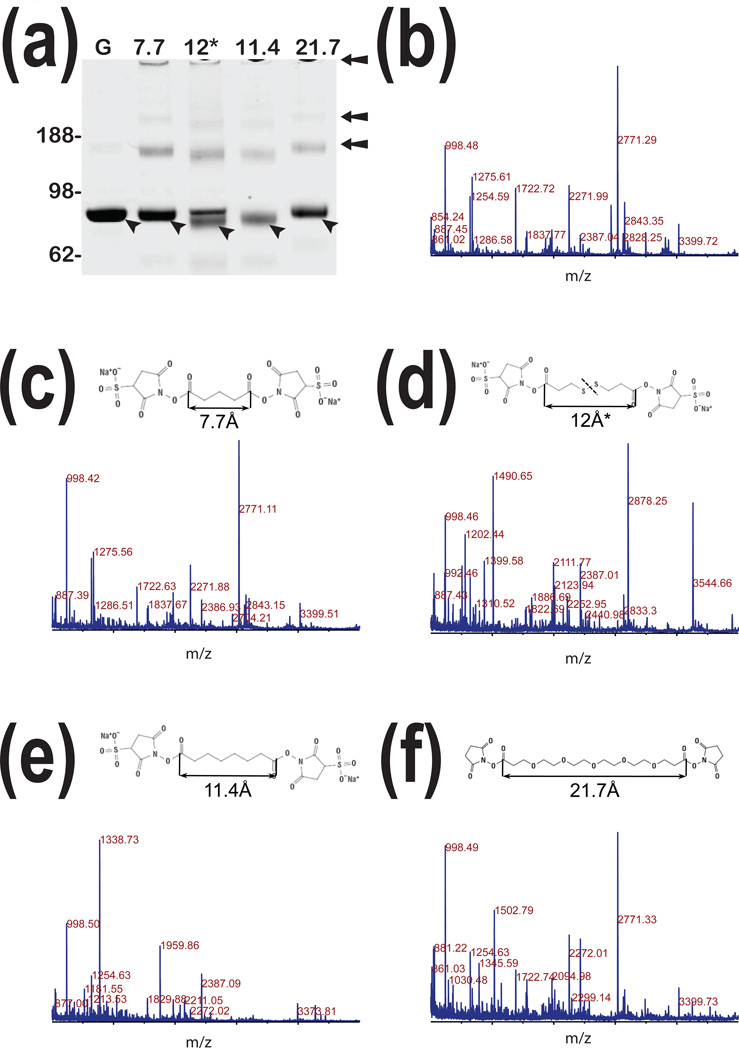
Figure 1. Chemical cross-linking of human pGSN.
Panel a: 1-DE analysis of native human pGSN purified from human plasma (G). Lanes represent 1-DE analysis of the same preparation of pGSN after chemical cross-linking with four linkers. Chemical structures and space lengths of respective linkers are presented in panelsc – f. Panel b:Representative MALDI-TOF spectrum of pGSN tryptic digests. Panels c – f represent MALDI-TOF spectra of tryptic digests of pGSN after cross-linking with respective linkers showing significant changes in MALDI-TOF spectra resulting from cross-linking.
From many linkers which are commercially available we used four: a short one of 7.7Å; two with medium length: 11.4Å and 12.0Å non cleavable and cleavable respectively; and a long one of 21.7Å. Our reasoning for using such selection was to facilitate cross-linking of residues located at various distances in 3-dimensional space which we gathered from crystallography studies posted in Protein Data Bank (PDB, http://www.pdb.org/pdb/home/home.do).
Chemical cross-linkers
As expected MALDI-TOF analysis of tryptic digests shown in Fig. 1 c – f presents different spectra that are compared to those obtained from non-cross-linked pGSN (Fig. 1b). Nevertheless, the cross-linking with a short arm (Fig. 1c) and a long arm (Fig. 1f) does not induce strong changes compared to the non-cross-linked pGSN, and intense peaks such as 1254.59, 1722.72, 2271.99, 2771.29, and 3399.72m/z representing fragments of pGSN are common between these three spectra. However, the 12.0Å and the 11.4Å cross-linkers, although similar in size, differ from each other and induce substantially different profiles with very few exceptions such as 998.50 m/z fragment ion. This result was unexpected because linkers with such comparable length and chemistry should facilitate linkage of the same lysine residues. In addition, cross-linking with 11.4Å and 12.0Å linkers show additional forms of pGSN that have higher mobility in 1-DE than original native pGSN (Fig.1a lane 12* and lane 11.4). As further presented, we conclude that due to internal cross-linking leading to more compact conformation yielding monomers acquire higher electrophoretic mobility. Interestingly, a 12.0Å linker did not support complete intra-molecular cross-linking of monomer like 11.4Å cross-linker resulting in the presence of two bands in 1-DE analysis. The same effect was observed for pGSN homo-dimers and homo-trimers reflected by various electrophoretic mobility of bands within 170 kDa region and above. We further postulate that such stepwise process is driven by change inequilibrium between monomer, homo-dimer and homo-trimer. Combining these results, we conclude that chemical cross-linking of pGSN facilitates intra-molecular and inter-molecular reactions. We further conclude that such effect is less dependent on the length of linkers and will mostly depend on availability of ε amines of lysineresidues.
Inter- and intra-molecular cross-linking
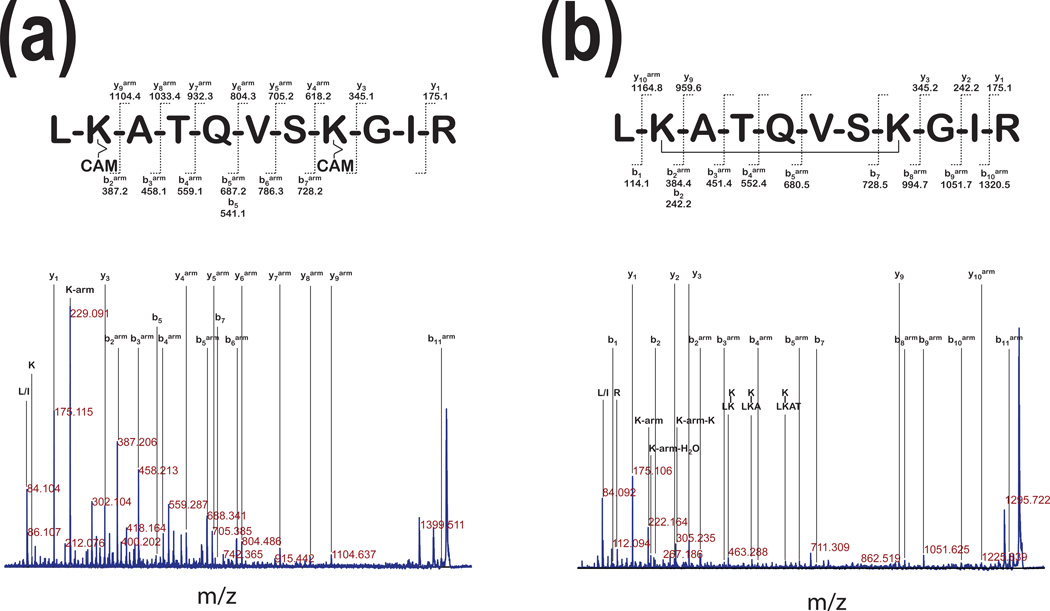
Fig. 2 shows an example of two MS/MS spectra of peptide H2N- (211)LKATQVSKGIRD(223)-COOH after pGSN cross-linking. This peptide was derived from forms representing monomers of pGSN (lower bands on Fig. 1a, lanes 12* and 11.4). Fig. 2a shows fragment ion assignment of this peptide when cleavable 12.0Å linker and reducing condition was used. This spectrum provides very good coverage for y- and b- series ions with one arm of cleavable linker modification labeled with “arm” superscript in this figure. The same experiment performed using non-cleavable 11.4Å linker (Fig. 2b) provides less coverage in y-ions series. Mass difference between y10arm and y3arm ions and b1 and b8arm ions strongly indicates intra-molecular linking of two lysine residues. These results indicate that whether 12.0Å cleavable or 11.4Å non-cleavable linker was used, modification of lysine residues at positions 213 and 219 represent intra-molecular linking. Intra-molecular linking also occurred in different regions of pGSN.
Figure 2. Comparison of cross-linking with cleavable or non-cleavable linkers.
Panel a: The MS/MS analysis of precursor ions with 1490.7 m/z (~86 kDa, Fig. 1a) of pGSN after linking with a cleavable 12Å cross-linker shows fragment ion assignment for peptideH2N-LKATQVSKGIR-COOH with modifications resulting from the spacer arm cleaved by reduction and a carbamidomethylation (CAM) of the free -SH. In this spectrum, a 229.09 m/z fragment corresponds to the immonium ion of the lysine residue linked to the arm with the CAM. Panel b: The MS/MS analysis of precursor ions with 1338.7 m/z from the digest of the major band of pGSN cross-linked with a 11.4 Å linker, shows fragment ion assignment for the same peptide as in panel (a). This peptide contains non-cleavable 11.4Å linker. This MS/MS spectrum shows ions specific for the following modifications: 222.16 m/z, the immonium ion of lysine with the arm; 242.20 m/z, the immonium ion of lysine linked to the arm and water; 305.24 m/z the arm linked on both sides to immonium ions of lysine. The lysine residue modifications were labeled with a superscript (arm).
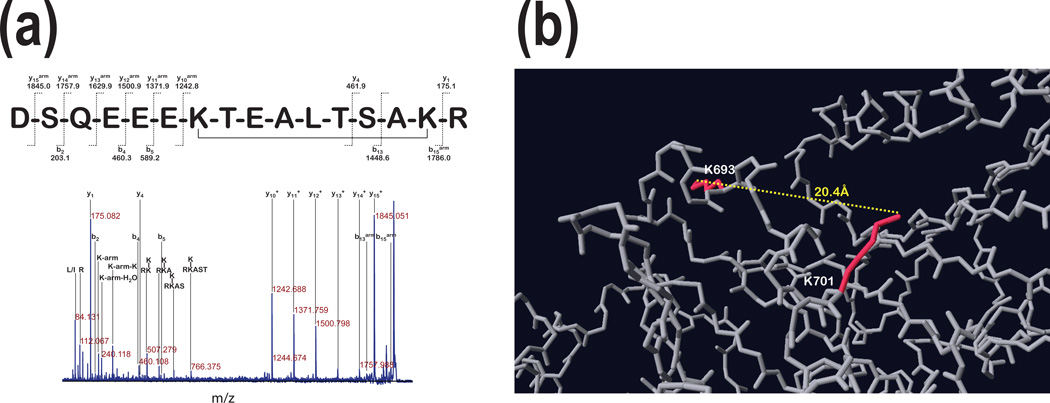
Fig. 3 shows MS/MS spectrum of 16 a.a. long H2N- (687)DSQEEEKTEALTSAKR(702)-COOH peptide modified by 11.4Å cross-linker. This linkage occurred between Lys (693) and Lys (701) as depicted in Fig. 3b. Sequence of this modified peptide was based on assignment of large fragment with mass corresponding to a gap between y1 and y10 ions plus linker (1242.8 m/z). Only two additional fragments y4 (461.9 m/z)and b13 (1448.6 m/z) but not y4arm and b13arm were detected suggesting that fragmentation at Thr-Ser and Ser-Ala peptide bonds was associated with loss of linker arm. In fact, the link between lysine residues and linker arm is an amide, which can be lost upon CID fragmentation, leaving a free lysine side chain without modification. This was not the case when H2N-(212)LKATQVSKGIRD(223)- COOH was fragmented. Also, loss of linker’s arm during fragmentation was not always complete as observedin the presence of b5 and b5arm (Fig. 2a) as well as b2 and b2arm (Fig. 2b). We propose that a peptide’s a.a. sequence is influenced whether there is loss of linker’s arm or not, which would need additional studies to confirm. Nevertheless, we conclude that this spectrum represents intra-molecular linking.
Figure 3. Internal cross-linking distant lysine residues using non-cleavable linker.
Panel a: The MS/MS analysis of the precursor ion with 1959.89 m/z (~86kDa in Fig. 1 a) linked with 11.4Å linker presents fragment ion assignment for peptide H2N-DSQEEEKTEALTSAKR-COOH with the cross-linker molecule linking both lysine residues present in this peptide. The ions containing the mass of the arm were labeled with subscript (arm). Panel b shows that the linked lysine side chains are in opposite directions and the distance between the ε-amines of both lysine residues is 20.4Å.
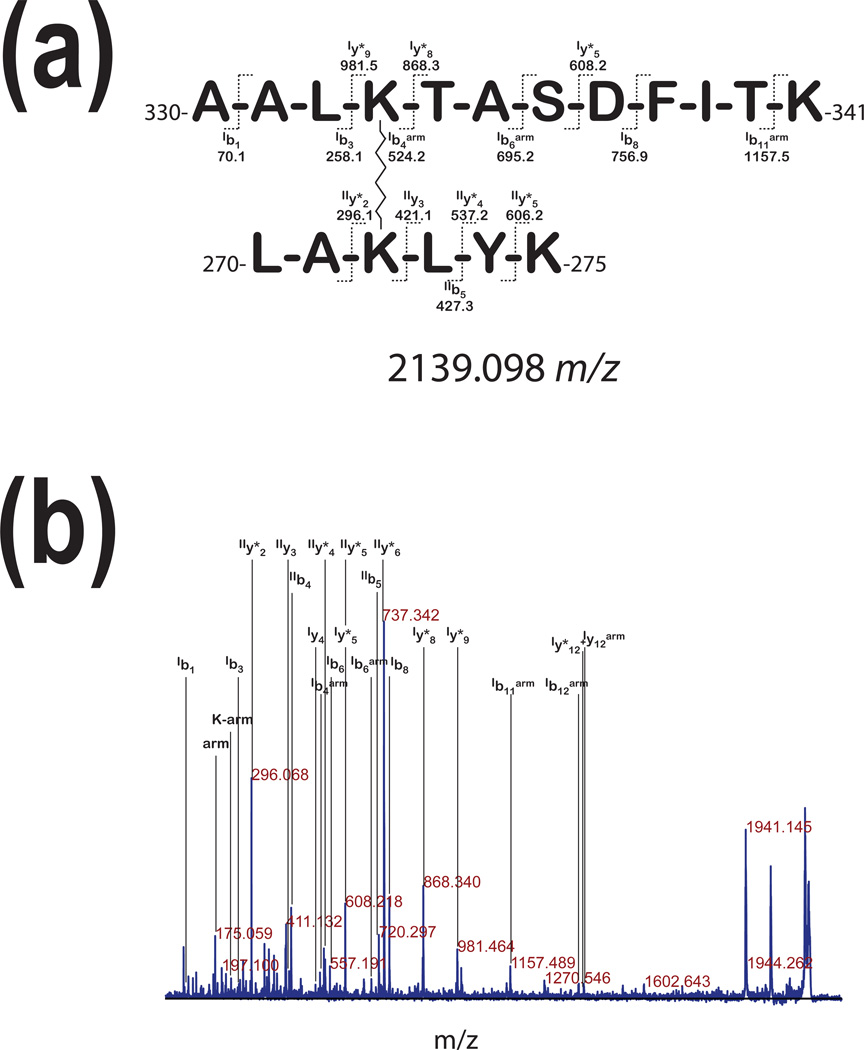
Fig. 4a shows two peptides linked with non-cleavable 11.4Å linker based on MS/MS spectrum presented in Fig. 4b. In this case, we conclude that this is the link between two pGSN molecules because this fragment was found in a tryptic digest of a band that corresponded to electrophoretic migration of homo-dimer and was not found in tryptic digest of bands migrating as non-cross-linked (control) monomer of pGSN (Fig.1, lane 11.4). Fragmentation of these two cross-linked peptides yielded ions corresponding to Lys-arm and arm of the linker itself (Fig. 4b).
Figure 4. Inter-peptide cross-linking.
The MS/MS analysis of precursor ion with 1959.88 m/z, derived from the tryptic digest of the upper band of pGSN (~170kDa in Fig. 1 a) after cross-linking with the 11.4Å linker. This spectrum presents fragment ion assignment for two peptides, first H2N-(357)AALKTASDFITK(368)-COOH (the ions for this peptide are labeled with subscriptI) and second H2N-(297)LAKLYK(302)-COOH (the ions for this peptide are labeled with subscriptII). The ions labeled with (arm) superscript the fragments containing the masses with linker modifications.
Distance between lysine residues and the length of cross-linker
The next question we wanted to address in this study was the relationship between the length of cross-linker and distance between lysine residues to be successfully linked and whether the choice of the specific length of cross-linker can be used to preferentially link selected lysine residues. If the 3-dimensional structure of protein or its fragments is rigid, then we expect that a specific length of linker is required. On the other hand, if lysine residues are much further apart than the length of linker and are still being linked, we may interpret this finding as a result of substantial flexibility of protein’s 3-dimensional structure. Indeed, during the course of this study, we have found that based on PDB model (Fig. 3b) ε-amines of two lysine residues, which are 20.4Å apart and oriented to different directions in 3-dimensional space, were cross-linked by 11.4Å linker. This indicates that in this region pGSN polypeptide chain has substantial flexibility in an aqueous environment. This observation may have profound effects on interpretation of results of future experiments investigating conformational changes associated with biological functions as well as interaction of pGSN with other proteins.
Modification of precursor ions of peptides derived from chemically cross-linked proteins
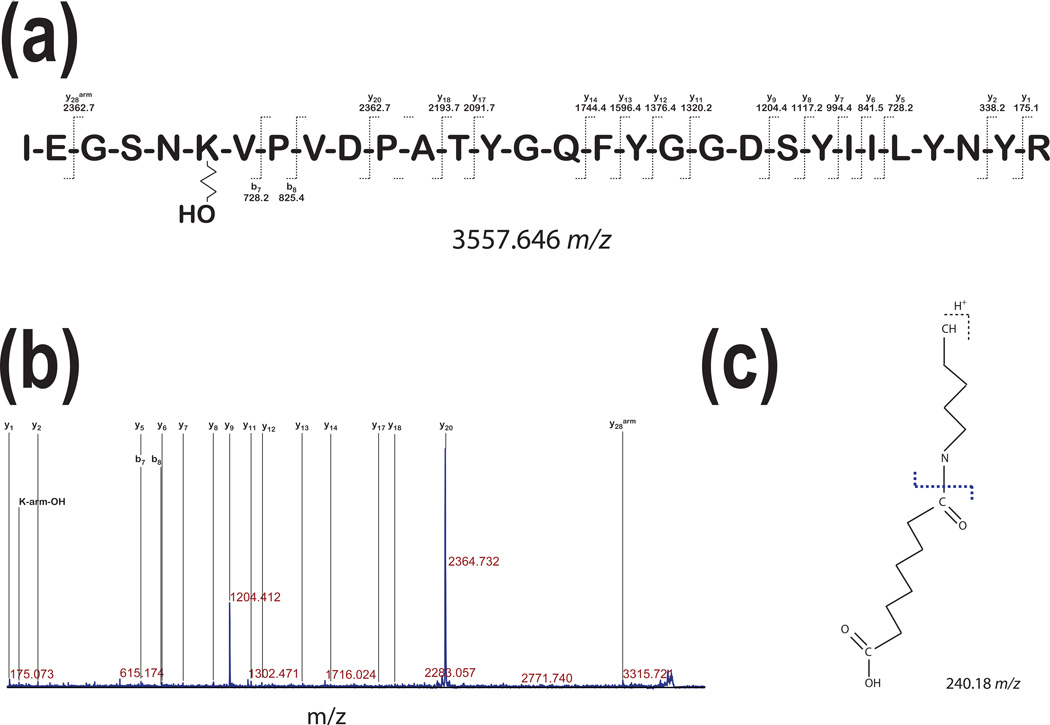
One of the challenges in the interpretation of spectra with peptide modifications is correct assignment of a precursor ion. In our subsequent experiment, we have found that 30 a.a. long peptide H2N-(425)IEGSNKVPVDPATYGQFYGGDSYIILYNYR(454)- COOH had a non cleavable linker attached with additional hydroxyl group increasing m/z of the precursor ion by 17 to 3557.646 (Fig. 5a). In Fig. 5b we show fragmentation of this peptide. Lysine at position 6 has noncleavable 11.4Å linker attached with a terminal −OH group. Interestingly, b7 and b8 ions do not show any modification indicating that fragmentation occurred at the nitrogen – carbon bond between ε amine group of lysine and the linker (Fig. 5c). Fragmentation spectrum presented in Fig. 5b shows relatively poor CID fragmentation of this peptide yielding mostly y-series ions and only two b-series ions. Two most intense peaks represent y9 and y20 ions. High efficiency ingenerating those ions can be explained by previous reports showing that peptide bonds C-terminal of aspartic acid are prone to CID induced fragmentation. This peptide contains only two proline residues (6.7%). As much as proline residues affect CID fragmentation in proline rich peptides [31], in this case, Pro-Val bond is fragmented (b8 ion) at low, yet detectable levels. We have also found immonium ion of Lysine-linker – OH with m/z of 240.18 (Fig. 5c) present in the spectrum that aided in the interpretation and final assignment of the sequence.
Figure 5. Modification of peptides without cross-linking.
The fragmentation spectrum of the precursor ion with 3557.60 m/z was derived from the tryptic digest of the major band of pGSN (~86kDa in Fig. 1 a) incubated with a 11.4Å spacer arm and showed fragment ion assignment for peptide H2N-IEGSNKVPVDPATYGQFYGGDSYIILYNYR-COOH. It shows a modification of the lysine residue in which the cross-linker ends with H2O indicating that this linker was unable to cross-link two lysine residues. We labeled with (arm) superscript the fragments containing the masses with linker modifications.
Summary of chemical cross-linking of pGSN
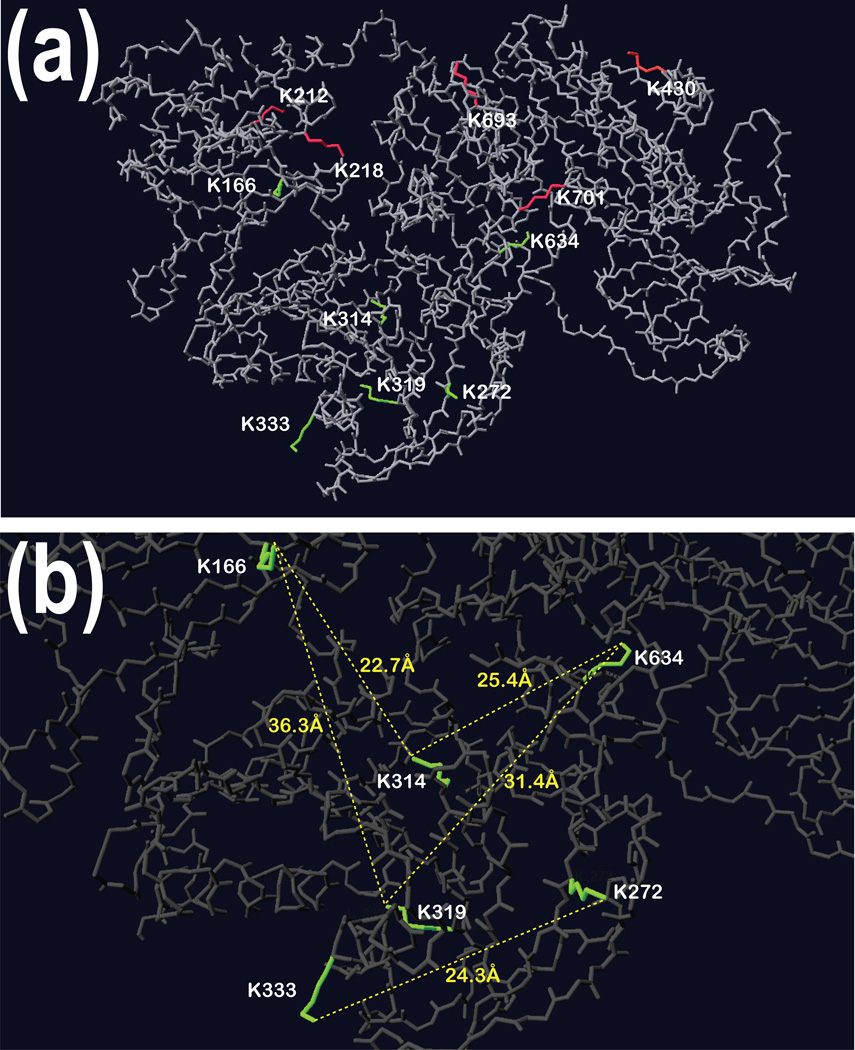
Summary of our findings are presented in Fig. 6. Panels (a) and (b) in this figure show 3-dimensional structure of GSN obtained from PDB. In panel (a) we marked side chains of all lysine residues that were modified. Importantly, residues colored green are those involved in inter-molecular linking while those colored in red are involved in intra molecular linking. In panel (b), we show distances in Å between the ε-amine groups of the lysine residues we found linked by the 11.4Å cross-linker. Although we showed that an 11.4Å cross-linker can reach two lysine residues 20.4Å apart, the distances shown in Fig. 6b are longer than 25Å and the presence of those cross-linked peptides in the band which correspond to electrophoretic migration of homo-dimer of pGSN suggests an inter-molecular cross-linking. In some instances our approach still has limitations. For example, based on presented results, we cannot distinguish whether lysine 166 and lysine 634 were specifically linked to lysine 314 or lysine 319.
Figure 6. An overlay of identified modified lysine residues on 3-dimensional structure of GSN.
Panels show the analysis of the modified lysines on the 3D structure of gelsolin. Panel a: Red labeled Lys residues linked together within the same peptide, as well as the dead-end modifications (Lys 420) are located on the surface of gelsolin. Green labeled Lys residues involved in the cross-linking of the dimer of gelsolin are mostly found in cavities of the protein. Pane b: The distances between the ε amine of Lys residues involved in the dimerization reveal a distance that is longer for the cross-linker to reach both amines.
As described above, the cGSN form that was used for crystallographic studies is shorter by 23 a.a. on the N-terminal end than pGSN.
Conclusions
Several conclusions can be drawn based on experimental data presented in this study. First, when interpreting results of in vitro cross-linking interacting proteins, a possibility of self cross-linking of any of interacting proteins should be considered. Dimerization or self-assembly to multi-monomer structures is a commonly observed effect in in vitro experiments that can lead to undesired effects. Second, critical residues for forming hetero-complexes can be blocked and new and not biologically relevant interacting domains can be created. Also in vitro experimental conditions might favor homo- over hetero- interactions with another protein in other’s or our systems. Third, flexibility of some regions within the protein polypeptide chain, as we have shown here, may lead to linking of distant residues where linker length is half the distance thus such assumption cannot be made. X-ray crystallography data did not indicate such flexibility in the region where such intra-molecular cross-linking occurred. Fourth, our strategy used cleavable and non-cleavable cross-linkers; the non-cleavable one provided confirmation of intra-molecular cross-linking, while cleavable cross-linker did not. Fifth, because of the possibility of a loss during the gas phase of modifications, resulting from chemical cross-linking, a thorough manual interrogation of MS spectra for precursor ions needs to be performed. The manual interpretation of MS/MS spectra allowed us to gather more specific information, e.g., the dissociation of the arm/peptide during CID. Such information provides easier ways to parse pairs of cross-linked peptides and in fine this would help to complete software for the analysis of modified peptides. Sixth, despite experimental limitations including low intensity of fragment ions, chemical cross-linking using MALDI-TOF/TOF mass spectrometry remains a vital method to study protein interactions.
Acknowledgments
We would like to thank Ms. Jayme Wiederin and Ms. Robin Taylor at the University of Nebraska Medical Center for help in preparation of this manuscript. We also thank Melinda Wojtkiewicz from the Mass Core Facility at the University of Nebraska Medical Center for providing assistance with mass spectrometry analyses. This work was partially supported by the National Institutes of Health grants 5P01DA026146, 5R01DA030962, 2P30MH062261, 5P20RR016469 and Nebraska Research Initiative.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Literature
- 1.Ramisetty SR, Washburn MP. Unraveling the dynamics of protein interactions with quantitative mass spectrometry. Crit Rev Biochem Mol Biol. 2011;46:216–228. doi: 10.3109/10409238.2011.567244. [DOI] [PubMed] [Google Scholar]
- 2.Schaferling M, Nagl S. Forster resonance energy transfer methods for quantification of protein-protein interactions on microarrays. Methods Mol Biol. 2011;723:303–320. doi: 10.1007/978-1-61779-043-0_19. [DOI] [PubMed] [Google Scholar]
- 3.Alessandrini A, Facci P. Unraveling lipid/protein interaction in model lipid bilayers by Atomic Force Microscopy. J Mol Recognit. 2011;24:387–396. doi: 10.1002/jmr.1083. [DOI] [PubMed] [Google Scholar]
- 4.Blaum BS, Deakin JA, Johansson CM, Herbert AP, Barlow PN, Lyon M, Uhrin D. Lysine and arginine side chains in glycosaminoglycan-protein complexes investigated by NMR, cross-linking, and mass spectrometry: a case study of the factor H-heparin interaction. J Am Chem Soc. 2010;132:6374–6381. doi: 10.1021/ja1000517. [DOI] [PubMed] [Google Scholar]
- 5.Drenth J. Principles of protein x-ray crystallography. New York: Springer Verlag; 1999. [Google Scholar]
- 6.Morelli X, Bourgeas R, Roche P. Chemical and structural lessons from recent successes in protein-protein interaction inhibition (2P2I) Curr Opin Chem Biol. 2011;15:475–481. doi: 10.1016/j.cbpa.2011.05.024. [DOI] [PubMed] [Google Scholar]
- 7.Kim C, Ye F, Ginsberg MH. Regulation of Integrin Activation. Annu Rev Cell Dev Biol. 2011:321–345. doi: 10.1146/annurev-cellbio-100109-104104. [DOI] [PubMed] [Google Scholar]
- 8.Vidal M, Cusick ME, Barabasi AL. Interactome networks and human disease. Cell. 2011;144:986–998. doi: 10.1016/j.cell.2011.02.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Andersson CR, Gustafsson MG, Strombergsson H. Quantitative Chemogenomics: Machine-Learning Models of Protein-Ligand Interaction. Curr Top Med Chem. 2011;11:1978–1993. doi: 10.2174/156802611796391249. [DOI] [PubMed] [Google Scholar]
- 10.Volkmer R, Kretzschmar I, Tapia V. Mapping receptor-ligand interactions with synthetic peptide arrays: Exploring the structure and function of membrane receptors. Eur J Cell Biol. 2011 doi: 10.1016/j.ejcb.2011.03.004. [Epub ahead of print.] [DOI] [PubMed] [Google Scholar]
- 11.Tuncbag N, Gursoy A, Keskin O. Prediction of protein-protein interactions: unifying evolution and structure at protein interfaces. Phys Biol. 2011;8:035006. doi: 10.1088/1478-3975/8/3/035006. [DOI] [PubMed] [Google Scholar]
- 12.Aubry S, Aussedat B, Delaroche D, Jiao CY, Bolbach G, Lavielle S, Chassaing G, Sagan S, Burlina F. MALDI-TOF mass spectrometry: a powerful tool to study the internalization of cell-penetrating peptides. Biochim Biophys Acta. 2010;1798:2182–2189. doi: 10.1016/j.bbamem.2009.11.011. [DOI] [PubMed] [Google Scholar]
- 13.Selvin PR. The renaissance of fluorescence resonance energy transfer. Nat Struct Biol. 2000;7:730–734. doi: 10.1038/78948. [DOI] [PubMed] [Google Scholar]
- 14.Means GE, Feeney RE. Chemical modifications of proteins: history and applications. Bioconjug Chem. 1990;1:2–12. doi: 10.1021/bc00001a001. [DOI] [PubMed] [Google Scholar]
- 15.Wong SS, Wong LJ. Chemical crosslinking and the stabilization of proteins and enzymes. Enzyme Microb Technol. 1992;14:866–874. doi: 10.1016/0141-0229(92)90049-t. [DOI] [PubMed] [Google Scholar]
- 16.Sinz A, Kalkhof S, Ihling C. Mapping protein interfaces by a trifunctional cross-linker combined with MALDI-TOF and ESI-FTICR mass spectrometry. J Am Soc Mass Spectrom. 2005;16:1921–1931. doi: 10.1016/j.jasms.2005.07.020. [DOI] [PubMed] [Google Scholar]
- 17.Rosano C, Stec-Martyna E, Lappano R, Maggiolini M. Structure-based approach for the discovery of novel selective estrogen receptor modulators. Curr Med Chem. 2011;18:1188–1194. doi: 10.2174/092986711795029645. [DOI] [PubMed] [Google Scholar]
- 18.Tang X, Bruce JE. A new cross-linking strategy: protein interaction reporter (PIR) technology for protein-protein interaction studies. Mol Biosyst. 2010;6:939–947. doi: 10.1039/b920876c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sedo O, Sedlacek I, Zdrahal Z. Sample preparation methods for MALDI-MS profiling of bacteria. Mass Spectrom Rev. 2011;30:417–434. doi: 10.1002/mas.20287. [DOI] [PubMed] [Google Scholar]
- 20.Hamdan M, Bordini E, Galvani M, Righetti PG. Protein alkylation by acrylamide, its N-substituted derivatives and cross-linkers and its relevance to proteomics: a matrix assisted laser desorption/ionization-time of flight-mass spectrometry study. Electrophoresis. 2001;22:1633–1644. doi: 10.1002/1522-2683(200105)22:9<1633::AID-ELPS1633>3.0.CO;2-C. [DOI] [PubMed] [Google Scholar]
- 21.Hamdan M, Galvani M, Righetti PG. Monitoring 2-D gel-induced modifications of proteins by MALDI-TOF mass spectrometry. Mass Spectrom Rev. 2001;20:121–141. doi: 10.1002/mas.10000. [DOI] [PubMed] [Google Scholar]
- 22.Sheffield LG, Gavinski JJ. Proteomics methods for probing molecular mechanisms in signal transduction. J Anim Sci. 2003;81 Suppl 3:48–57. doi: 10.2527/2003.81suppl_348x. [DOI] [PubMed] [Google Scholar]
- 23.Kafka AP, Kleffmann T, Rades T, McDowell A. The application of MALDI TOF MS in biopharmaceutical research. Int J Pharm. 2010;417:70–82. doi: 10.1016/j.ijpharm.2010.12.010. [DOI] [PubMed] [Google Scholar]
- 24.Paramelle D, Cantel S, Enjalbal C, Amblard M, Forest E, Heymann M, Geourjon C, Martinez J, Subra G. A new generation of cross-linkers for selective detection by MALDI MS. Proteomics. 2009;9:5384–5388. doi: 10.1002/pmic.200900562. [DOI] [PubMed] [Google Scholar]
- 25.Bucki R, Byfield FJ, Kulakowska A, McCormick ME, Drozdowski W, Namiot Z, Hartung T, Janmey PA. Extracellular gelsolin binds lipoteichoic acid and modulates cellular response to proinflammatory bacterial wall components. J Immunol. 2008;181:4936–4944. doi: 10.4049/jimmunol.181.7.4936. [DOI] [PubMed] [Google Scholar]
- 26.Lind SE, Janmey PA. Human plasma gelsolin binds to fibronectin. J Biol Chem. 1984;259:13262–13266. [PubMed] [Google Scholar]
- 27.Lind SE, Smith DB, Janmey PA, Stossel TP. Role of plasma gelsolin and the vitamin D-binding protein in clearing actin from the circulation. J Clin Invest. 1986;78:736–742. doi: 10.1172/JCI112634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kambe H, Ito H, Kimura Y, Okochi T, Yamamoto H, Hashimoto T, Tagawa K. Human plasma gelsolin reversibly binds Mg-ATP in Ca(2+)-sensitive manner. J Biochem. 1992;111:722–725. doi: 10.1093/oxfordjournals.jbchem.a123825. [DOI] [PubMed] [Google Scholar]
- 29.Bohgaki M, Matsumoto M, Atsumi T, Kondo T, Yasuda S, Horita T, Nakayama KI, Okumura F, Hatakeyama S, Koike T. Plasma gelsolin facilitates interaction between beta(2) glycoprotein I and alpha5beta1 integrin. J Cell Mol Med. 2009;15:141–151. doi: 10.1111/j.1582-4934.2009.00940.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Haverland N, Pottiez G, Wiederin J, Ciborowski P. Immunoreactivity of anti-gelsolin antibodies: implications for biomarker validation. J Transl Med. 2010;8:137. doi: 10.1186/1479-5876-8-137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Leymarie N, Berg EA, McComb ME, O'Connor PB, Grogan J, Oppenheim FG, Costello CE. Tandem mass spectrometry for structural characterization of proline-rich proteins: application to salivary PRP-3. Anal Chem. 2002;74:4124–4132. doi: 10.1021/ac0255835. [DOI] [PubMed] [Google Scholar]