Abstract
Pairwise comparison of long stretches of genomic DNA sequence can identify regions conserved across species, which often indicate functional significance. However, the novel insights frequently must be windowed from a flood of information; for instance, running an alignment program on two 50-kilobase sequences might yield over a hundred pages of alignments. Direct inspection of such a volume of printed output is infeasible, or at best highly undesirable, and computer tools are needed to summarize the information, to assist in its analysis, and to report the findings. This paper describes two such software tools. One tool prepares publication-quality pictorial representations of alignments, while another facilitates interactive browsing of pairwise alignment data. Their effectiveness is illustrated by comparing the beta-like globin gene clusters between humans and rabbits. A second example compares the chloroplast genomes of tobacco and liverwort.
Full text
PDF
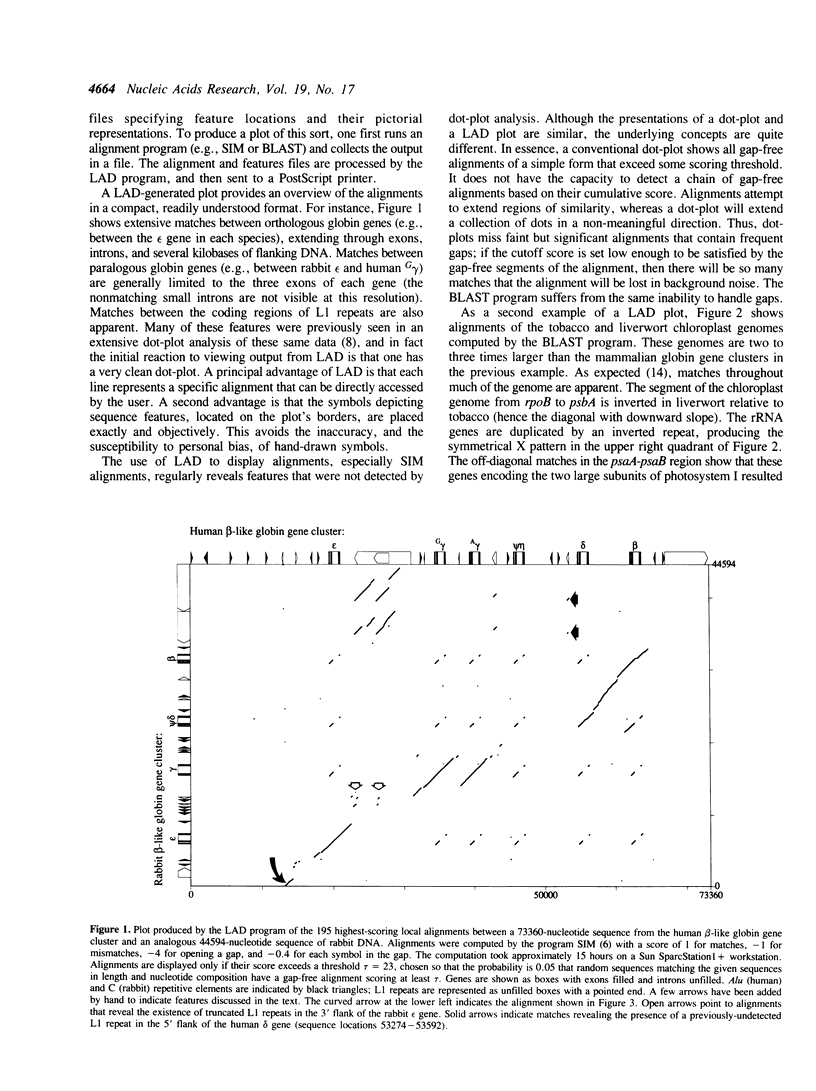
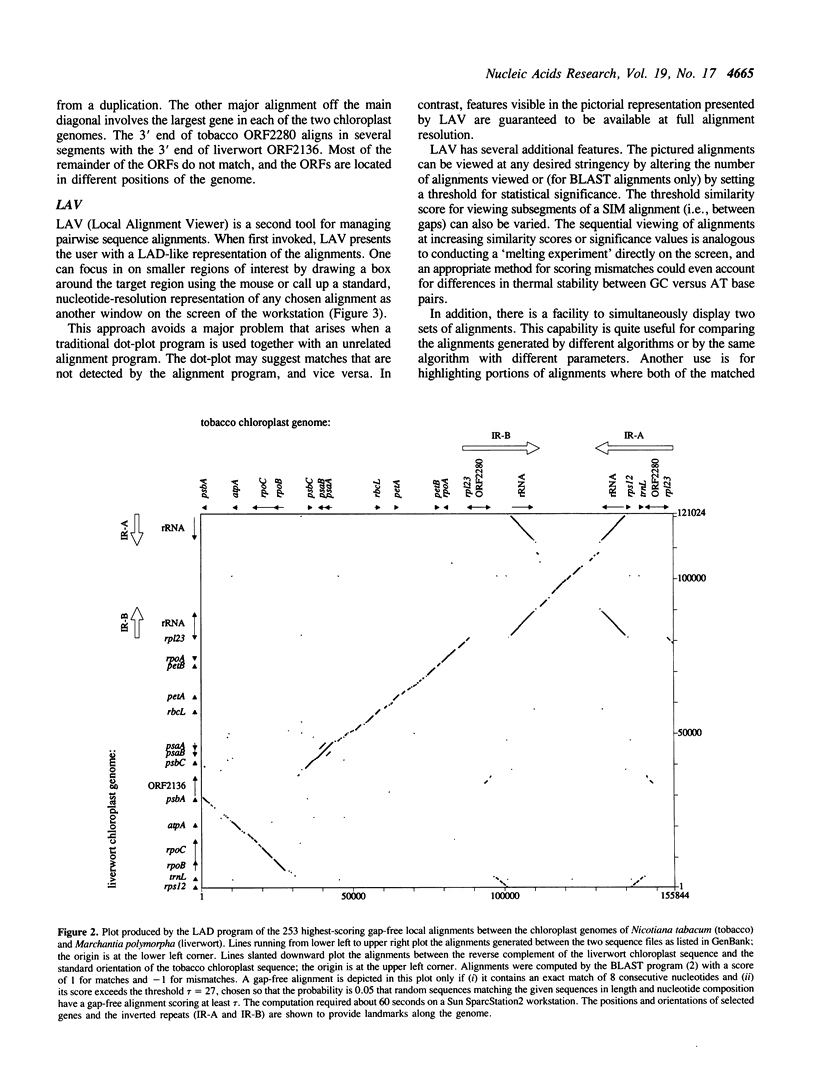
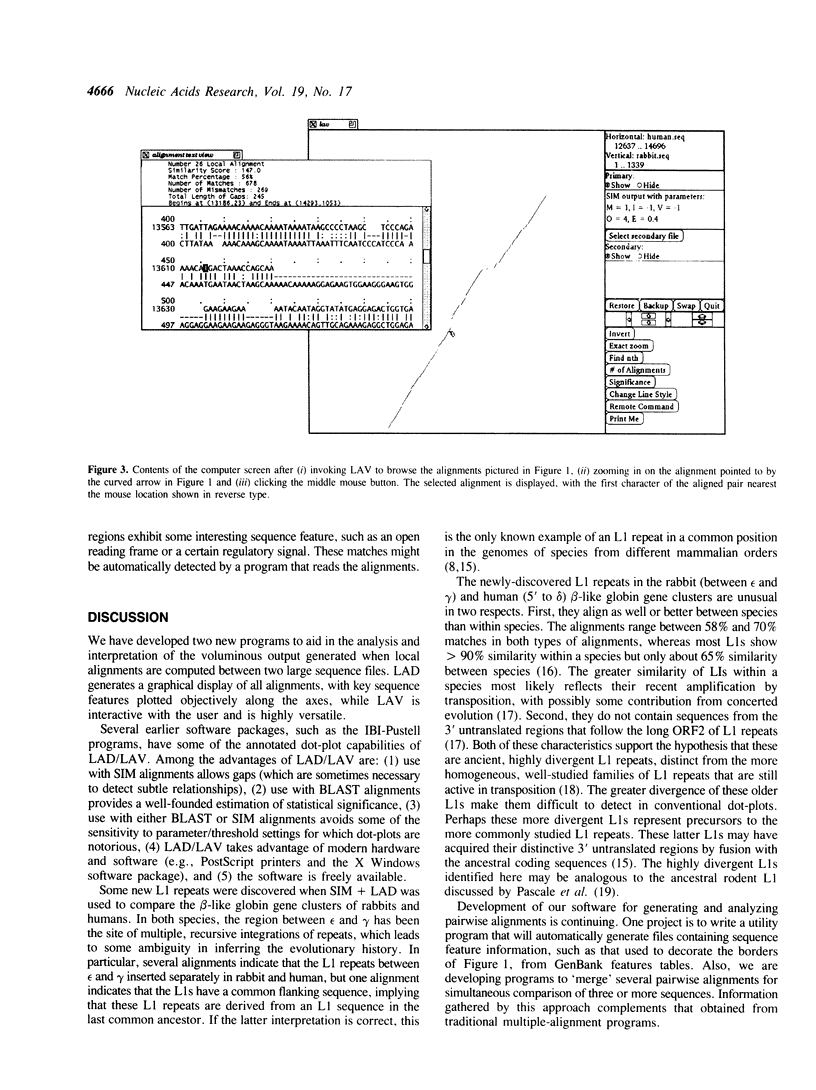

Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Altschul S. F., Gish W., Miller W., Myers E. W., Lipman D. J. Basic local alignment search tool. J Mol Biol. 1990 Oct 5;215(3):403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- Collins F. S., Weissman S. M. The molecular genetics of human hemoglobin. Prog Nucleic Acid Res Mol Biol. 1984;31:315–462. doi: 10.1016/s0079-6603(08)60382-7. [DOI] [PubMed] [Google Scholar]
- Demers G. W., Matunis M. J., Hardison R. C. The L1 family of long interspersed repetitive DNA in rabbits: sequence, copy number, conserved open reading frames, and similarity to keratin. J Mol Evol. 1989 Jul;29(1):3–19. doi: 10.1007/BF02106177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang X. Q., Hardison R. C., Miller W. A space-efficient algorithm for local similarities. Comput Appl Biosci. 1990 Oct;6(4):373–381. doi: 10.1093/bioinformatics/6.4.373. [DOI] [PubMed] [Google Scholar]
- Karlin S., Altschul S. F. Methods for assessing the statistical significance of molecular sequence features by using general scoring schemes. Proc Natl Acad Sci U S A. 1990 Mar;87(6):2264–2268. doi: 10.1073/pnas.87.6.2264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kazazian H. H., Jr, Wong C., Youssoufian H., Scott A. F., Phillips D. G., Antonarakis S. E. Haemophilia A resulting from de novo insertion of L1 sequences represents a novel mechanism for mutation in man. Nature. 1988 Mar 10;332(6160):164–166. doi: 10.1038/332164a0. [DOI] [PubMed] [Google Scholar]
- Maizel J. V., Jr, Lenk R. P. Enhanced graphic matrix analysis of nucleic acid and protein sequences. Proc Natl Acad Sci U S A. 1981 Dec;78(12):7665–7669. doi: 10.1073/pnas.78.12.7665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Margot J. B., Demers G. W., Hardison R. C. Complete nucleotide sequence of the rabbit beta-like globin gene cluster. Analysis of intergenic sequences and comparison with the human beta-like globin gene cluster. J Mol Biol. 1989 Jan 5;205(1):15–40. doi: 10.1016/0022-2836(89)90362-8. [DOI] [PubMed] [Google Scholar]
- Pascale E., Valle E., Furano A. V. Amplification of an ancestral mammalian L1 family of long interspersed repeated DNA occurred just before the murine radiation. Proc Natl Acad Sci U S A. 1990 Dec;87(23):9481–9485. doi: 10.1073/pnas.87.23.9481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pearson W. R., Lipman D. J. Improved tools for biological sequence comparison. Proc Natl Acad Sci U S A. 1988 Apr;85(8):2444–2448. doi: 10.1073/pnas.85.8.2444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pearson W. R. Rapid and sensitive sequence comparison with FASTP and FASTA. Methods Enzymol. 1990;183:63–98. doi: 10.1016/0076-6879(90)83007-v. [DOI] [PubMed] [Google Scholar]
- Pustell J., Kafatos F. C. A high speed, high capacity homology matrix: zooming through SV40 and polyoma. Nucleic Acids Res. 1982 Aug 11;10(15):4765–4782. doi: 10.1093/nar/10.15.4765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott A. F., Schmeckpeper B. J., Abdelrazik M., Comey C. T., O'Hara B., Rossiter J. P., Cooley T., Heath P., Smith K. D., Margolet L. Origin of the human L1 elements: proposed progenitor genes deduced from a consensus DNA sequence. Genomics. 1987 Oct;1(2):113–125. doi: 10.1016/0888-7543(87)90003-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shinozaki K., Ohme M., Tanaka M., Wakasugi T., Hayashida N., Matsubayashi T., Zaita N., Chunwongse J., Obokata J., Yamaguchi-Shinozaki K. The complete nucleotide sequence of the tobacco chloroplast genome: its gene organization and expression. EMBO J. 1986 Sep;5(9):2043–2049. doi: 10.1002/j.1460-2075.1986.tb04464.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith T. F., Waterman M. S. Identification of common molecular subsequences. J Mol Biol. 1981 Mar 25;147(1):195–197. doi: 10.1016/0022-2836(81)90087-5. [DOI] [PubMed] [Google Scholar]
- Weiner A. M., Deininger P. L., Efstratiadis A. Nonviral retroposons: genes, pseudogenes, and transposable elements generated by the reverse flow of genetic information. Annu Rev Biochem. 1986;55:631–661. doi: 10.1146/annurev.bi.55.070186.003215. [DOI] [PubMed] [Google Scholar]



