Abstract
DNA methylation is the most common form of DNA modification in prokaryotic and eukaryotic genomes. We have applied the method of single-molecule, real-time (SMRT®) DNA sequencing that is capable of direct detection of modified bases at single-nucleotide resolution to characterize the specificity of several bacterial DNA methyltransferases (MTases). In addition to previously described SMRT sequencing of N6-methyladenine and 5-methylcytosine, we show that N4-methylcytosine also has a specific kinetic signature and is therefore identifiable using this approach. We demonstrate for all three prokaryotic methylation types that SMRT sequencing confirms the identity and position of the methylated base in cases where the MTase specificity was previously established by other methods. We then applied the method to determine the sequence context and methylated base identity for three MTases with unknown specificities. In addition, we also find evidence of unanticipated MTase promiscuity with some enzymes apparently also modifying sequences that are related, but not identical, to the cognate site.
INTRODUCTION
Methylation of DNA bases, catalyzed by DNA methyltransferases (MTases), is the most abundant form of post-replicative DNA modification found in the genomes of prokaryotic and higher eukaryotic organisms. Three functional classes of MTases have been identified in bacteria and archaea. Two of these transfer a methyl group from S-adenosyl-l-methionine (SAM) to the exocyclic amino groups of adenine and cytosine bases in duplex DNA, yielding N6-methyladenine (m6A) and N4-methylcytosine (m4C), respectively (1). A third, and mechanistically distinct, class transfers the methyl group of SAM to C5 of cytosine to produce 5-methylcytosine (m5C) (2,3).
Most bacterial and archaeal MTases are associated with sequence-specific restriction-modification (RM) systems that protect the prokaryotic cell from invasion by DNA bacteriophages, and examples have been identified that recognize several hundred distinct DNA sequences (4). However, some well-characterized prokaryotic MTases do not appear to be associated with a cognate restriction endonuclease (REase) and some of these ‘orphan’ MTases perform different cellular functions. For example, the product of the Escherichia coli deoxyadenosine methyltransferase (dam) gene, M.EcoKDam, which modifies adenine residues in the sequence 5′-GATC-3′, is involved in both chromosomal replication initiation and in the maintenance of genomic integrity [reviewed earlier (5)].
Most eukaryotic MTases are of the m5C class and are related to their prokaryotic equivalents through a common reaction mechanism that is reflected in the conservation of tertiary structural elements within the enzyme active sites (6). Mammalian m5C-MTase activity is predominantly targeted to CpG dinucleotides and three different enzymes (DNMT1, DNMT3A and 3B) are known (7). Such DNA methylation is a key component of the epigenetic control of gene expression [reviewed earlier (8)]. CpG methylation is also a key player in genomic imprinting and in female X-inactivation (9,10). An additional modified base, 5-hydroxymethylcytosine (5-hmC), was presumed to be a product of DNA damage (11), but has recently been found to be a normal component of mammalian DNA (12,13) and appears to be generated by oxidation of m5C in a reaction catalyzed by the ten eleven translocation (Tet)-family of enzymes (13).
A number of diagnostic tools have been developed to detect DNA methylation, to establish what type of modification is associated with a particular MTase and to determine the DNA sequence context in which such methylation occurs. Nucleosides containing m6A, m4C and m5C can be resolved from their unmodified equivalents by chromatographic methods, thereby facilitating their detection in total DNA hydrolysates (14,15). Alternatively, polyclonal antisera that specifically recognize m6A or m4C have been used for immunological detection of such modifications in order to ascribe function to multiple putative MTase genes in Helicobacter pylori (16).
Bioinformatic analysis of a large number of genes that encode well-characterized prokaryotic MTases have identified groups of conserved sequence motifs that are diagnostic for DNA MTases and permit the accurate prediction of m5C-MTases, but the amino m6A- and m4C-MTases cannot be unequivocally distinguished (1,2). In the case of methylation activity that is part of a Type II RM system, the sequence specificity of the MTase is expected to be the same as the cleavage specificity of the associated REase. However, very little experimental evidence has been generated to support this, and biochemical characterization of the target specificity of most MTases remains to be gathered. Furthermore, the exact sites of modification are often uncertain in cases where the recognition sequence contains multiple potential target bases (i.e. A or C) or where separate MTases act on the two strands independently (17).
The experimental approaches currently available to fully characterize prokaryotic and eukaryotic MTases are labor-intensive, requiring radioactive labeling with [3H] S-adenosylmethionine and mapping and sequencing of individual sites (17,18). One method based on Sanger sequencing can also detect methylated bases, but this is not a high-throughput method (19). Methods currently in use to discriminate between m5C and unmodified cytosines at CpG sequences, such as differential sensitivity to cleavage by REases (20) and methylation-specific polymerase chain reaction (PCR) of bisulfite-modified DNA (21,22) do not permit genome-wide analysis. The state-of-the-art method for m5C methylome studies is MethylC-Seq (23,24), but it is indirect and also requires extensive experimentation. In particular, the first requisite is the complete DNA sequence of the DNA to be analyzed. Only then can methylome analysis be undertaken. Some additional new technologies have been described recently that have potential applications in methylome analysis such as methods to map genomic 5-hmC by enzyme-catalyzed glucosylation (25,26) and a family of REases that specifically recognize m5CpG and m5CpWpG sequences (27). The latter enzymes are of interest because members of this MspJI family of REases excise a 32–33 bp fragment that includes the methylated bases in a central position and the products lend themselves to high-throughput sequencing approaches. However, they are partially constrained by the sequence specificity of the REases (27).
A method has been described previously to directly detect methylated DNA bases during single-molecule, real-time (SMRT) DNA sequencing (28). This method takes advantage of kinetic data pertaining to the rate of incorporation of each dNTP in the form of two parameters—the pulse width (PW) and the interpulse duration (IPD). Significant changes in these kinetic parameters were observed during SMRT sequencing when the DNA polymerase encounters m6A, m5C or 5-hmC on the template strand. These distinct kinetic signatures allow for the identification of the type and position of the base modification in the DNA template.
Here, we extend the SMRT sequencing method to combine complete DNA sequence determination and methylated base analysis to characterize MTase specificities in a single operation (28,29). We analyzed a set of 16 DNA substrates that were methylated in vivo by a range of single prokaryotic MTases expressed in an E. coli strain that lacks additional MTase genes. The samples included MTases introducing m6A, m4C or m5C modifications, either from MTases whose substrate specificity was previously known, or from some whose specificity was unknown. The results allowed us to determine the absolute sequence specificity of the MTase, as well as the precise location of the methylated base.
MATERIALS AND METHODS
Materials
All restriction endonucleases, Phusion-HF DNA polymerase, Antarctic Phosphatase, T4-DNA ligase, SAM, MTase genes and E. coli cells were from New England Biolabs Inc. (Ipswich, MA, USA). Synthetic oligonucleotides were purchased from Integrated DNA Technologies (Coralville, IA, USA).
Escherichia coli strains
C2523 (NEB-Express): fhuA2 [lon] ompT gal sulA11 R(mcr-73::miniTn10–TetS)2 [dcm] R(zgb-210::Tn10–TetS) endA1 Δ(mcrC-mrr)114::IS10
C2925 (NEB dam-/dcm-): ara-14 leuB6 fhuA31 lacY1 tsx78 glnV44 galK2 galT22 mcrA dcm-6 hisG4 rfbD1 R(zgb210::Tn10) TetS endA1 rspL136 (StrR) dam13::Tn9 (CamR) xylA-5 mtl-1 thi-1 mcrB1 hsdR2
ER2796 (=DB24): fhuA2 Δ (lacZ)r1 glnV44 trp-31 dcm-6 his-1 zed-501::Tn10 argG6 rpsL104 dam-16::Kan xyl-7 mtl-2 metR1 mcr-62 Δ (mcrB-hsd-mrr)114
Methyltransferase cloning
In total, 16 known and putative MTase genes (Table 1) were amplified from bacterial genomic or recombinant plasmid DNA sources with Phusion-HF DNA polymerase using gene-specific oligonucleotide primers (Supplementary Table S1). The 5′-end oligonucleotides incorporated a PstI site (SbfI site in the case of the gene encoding M.SacI which contains an internal PstI site), followed by the sequence 5′-TTAAGG-3′ (to terminate translation of the lac α-peptide reading frame of the pRRS plasmid vector and reinitiate translation of the cloned MTase genes), followed by an eight nucleotide spacer sequence 5′-TTAATCAT-3′ and sequences complementary to the 5′-end of the relevant MTase coding sequence. 3′-end oligonucleotides were complementary to the 3′-end of the MTase coding sequences, including translation termination codons and either BamHI or BglII restriction sites. Because the predicted sequences of the pRRS constructs containing the M.EsaBC2I and M.SacI genes do not contain any restriction sites that are diagnostic of the activity of these MTases, additional restriction site sequences (for SalI and SacI, respectively) were included in the 3′-oligonucleotides, positioned between the termination codon and BamHI site in each case. PCR amplicons were restricted with PstI (or SbfI) and BamHI (or BglII) and ligated to PstI–BamHI restricted pRRS plasmid DNA (Genbank accession number JN569339) that had been dephosphorylated using Antarctic Phosphatase. All methylase genes were under the control of the same E. coli promoter present in the pRRS vector.
Table 1.
MTases subjected to specificity characterization by SMRT DNA sequencing
| No. | Methyltransferase name | Methylation context | Plasmid size (bp) | No. of sites |
|---|---|---|---|---|
| 1 | M.CviQI | GT(m6A)C | 3566 | 3 |
| 2 | M.RsaI | GTA(m4C) | 3986 | 2 |
| 3 | M.EcoKDam | G(m6A)TC | 3592 | 23 |
| 4 | M.EsaLHCI | GAT(m4C) | 3590 | 19 |
| 5 | M.Sau3AI | GAT(m5C) | 3995 | 19 |
| 6 | M.AluI | AG(m5C)T | 4331 | 19 |
| 7 | M.EsaBC1I | AG(m4C)T | 3797 | 18 |
| 8 | M.BstNI | C(m4C)WGG | 4142 | 5 |
| 9 | M.EcoKDcm | C(m5C)WGG | 4175 | 7 |
| 10 | M.EsaBC2I | T(m4C)GA | 3566 | 2 |
| 11 | M.NspI | R(m5C)ATGY | 3950 | 3 |
| 12 | M.HpaII | C(m5C)GG | 3833 | 15 |
| 13 | M.SacI | GAG(m5C)TC | 3936 | 1 |
| 14 | M.Tsp509I | AATT (m6A; position unknown) | 3863 | 7 |
| 15 | M.AatII | GACGTC (m6A/m4C; position unknown) | 3750 | 2 |
| 16 | M.BceJI | Unknown | 4760 | Unknown |
Isolation of plasmid DNA and determination of methylation status
Ligation products were used to transform NEB-Express E. coli (or NEB dam-/dcm- E. coli in the case of the M.Sau3AI ligation) and recombinant plasmid DNAs were isolated from ampicillin-resistant transformants and the presence of inserts of the expected size was confirmed by restriction analysis. Plasmids were then used to transform ER2796, also called DB24, a strain that lacks all known E. coli MTase genes (16). Plasmid DNAs were reisolated from ER2796 cells and their methylation status was assessed by restriction with PstI plus the relevant cognate restriction endonuclease in the cases of constructs containing M.AatII, M.AluI, M.BstNI, M.CviQI, M.HpaII, M.NspI, M.RsaI, M.SacI, M.Sau3AI and M.Tsp509I. Constructs containing genes encoding M.EcoKDam, M.EcoKDcm, M.EsaBC1I, M.EsaBC2I and M.EsaLHCI were assessed by restriction with PstI plus MboI, PspGI, AluI, SalI and MboI endonucleases, respectively. Unmethylated control substrates for each construct were produced by PCR amplification (using Phusion-HF polymerase) of the complete plasmids—using oligonucleotide primers that anneal to opposite strands of the vector DNA at a position 18 nucleotides 5′ of the vector SbfI/PstI site. Control substrates were restricted with the same enzymes as the methylated plasmids but without PstI (except in the case of M.SacI due to the internal PstI site within the gene).
Sample preparation and SMRT sequencing
An aliquot of ∼25 ng of plasmid DNA was whole-genome amplified (WGA) using the REPLI-g Midi Kit (Qiagen, Valencia, CA, USA). Amplification factors were typically approximately 1600× (∼40 ug output), translating to <0.06% of residual methylated DNA in the control samples. Further trimming of 5% of data from the top and bottom of the IPD distributions (see data analysis section) removed any remaining spurious signal from modified DNA. WGA and native plasmid DNA was sheared to an average size of 300 bp via adaptive focused acoustics (Covaris, Woburn, MA, USA). SMRTbell template sequencing libraries were prepared as previously described (30). Briefly, sheared DNA was end repaired, A-tailed and hairpin adapters with a single T-overhang were ligated. Incompletely formed SMRTbell templates were degraded with a combination of Exonuclease III (New England Biolabs; Ipswich, MA, USA) and Exonuclease VII (USB; Cleveland, OH, USA). Primer was annealed and samples were sequenced on the PacBio RS as previously described (31,32).
Data processing
Reads were processed and mapped to the respective reference sequences for each plasmid using the Basic Local Alignment with Successive Refinement (BLASR) mapper (http://www.pacbiodevnet.com/smrtanalysis/software/blasr) and the Pacific Biosciences SMRT Analysis pipeline (http://www.pacbiodevnet.com/smrtanalysis/software/smrtpipe) using the standard mapping protocol. Interpulse durations (IPDs) were measured as previously described (28) for all pulses aligned to each position in the reference sequence. For each position, the distribution of IPDs was compared between the native and WGA samples separately for the forward and reverse strands. The comparison was made using baseline-corrected IPD ratios. Baseline correction was applied by dividing the IPD mean for each position by the mean of mean IPDs over all positions in the plasmid, excluding those positions where the known methyltransferase recognition motif was detected and a window of 6 bases in each direction around such positions. This correction was applied to both the native sample IPD mean and the WGA control mean before obtaining the ratio of the two. In addition, 5% of outlier values were trimmed from both sides of the IPD distribution at each position before taking the ratio. IPD ratio plots were visualized using Circos (33).
The uncertainty in the IPD ratio was quantified by calculating the standard error of the mean of the IPD ratio using the delta method:
 |
where µ1 and µ2 are the average IPD values of the native and control, s1 and s2 are their standard deviations and n is the lower sequencing coverage of the two samples. The relationship between this standard error and the sequencing fold coverage is shown in Supplementary Figure S1 for three different methyltransferases (M.EcoKDam, M.EsaLHCI, M.Sau3AI).
RESULTS
SMRT DNA sequencing of plasmids containing MTase-mediated methylation
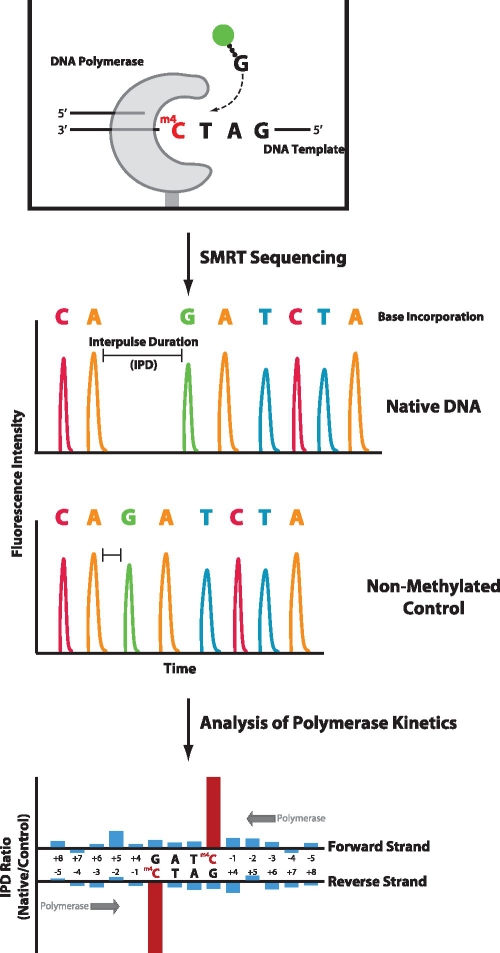
Plasmids containing cloned DNA-methyltransferase genes were expressed in an E. coli strain that otherwise lacked endogenous methyltransferase activities. The expressed methyltransferase acts to modify a specific recognition sequence by adding a methyl group to individual bases forming m6A, m4C or m5C. The methyltransferase will modify both the genomic and plasmid DNA. Plasmid DNAs containing the modifications were isolated and converted into SMRTbell templates to facilitate sequencing (30). Plasmids were also subjected to whole-genome amplification (WGA) to create a control DNA template lacking any DNA base modifications. Native and control samples were subjected to SMRT DNA sequencing (Figure 1). SMRT DNA Sequencing involves monitoring an individual DNA polymerase while it is replicating the input DNA, employing phospholinked nucleotides with different fluorophores for each of the four bases to produce fluorescence pulses at each incorporation step (29,32). As the DNA polymerase is sensitive to even subtle perturbations in the DNA template, it is possible to detect the presence of a methylated base by analyzing the polymerase kinetics as it moves across the DNA (28). This is manifested by the polymerase slowing down in a predictable manner upon encountering a base modification relative to the speed of copying the same non-modified control sequence. Thus, the time between fluorescent pulses (interpulse duration or IPD) is longer for a DNA template that contains a methylated base than for one that does not (28). One simple means of visualizing the differences in polymerase kinetics between a native and control sample is to graph the ratio of IPDs (methylated/control) for each position along the DNA template. The extent and magnitude of these kinetic signals is dependent on the nature of the base modification and the local sequence context. Because the polymerase is in contact with the modification for several bases before and after occupying the polymerase active site, the kinetic signal can encompass multiple nucleotide incorporations surrounding the methylated position (28).
Figure 1.
Principle of characterizing MTase specificities by SMRT DNA sequencing. In SMRT sequencing, single molecules of an engineered phi29-based polymerase are monitored in real-time, using fluorescently-labeled phospholinked nucleotides, as they synthesize a complementary strand from the DNA template strand that contains methylated bases. The timing of fluorescence pulses corresponding to nucleotide incorporations is analyzed and compared with a control template lacking methylated bases. The kinetics of DNA synthesis is affected by the presence of a methylated base in the template, e.g. by increasing the time prior to nucleotide binding across the methylated base, resulting in an increased IPD. The ratio of IPDs between native and control samples for each template position yield kinetic signatures for identifying the presence of methylated bases in the DNA template, thus defining MTase specificities.
We confirmed the activities of the different MTases for all samples by restriction digestion (Figure 2). In all cases, the DNAs were protected completely from restriction digestion by the specific REase that corresponds to the particular MTase expressed on the plasmid, indicating complete methylation at the cognate sites (Figure 2). The validity of using WGA for generating the control data set for reference sequence and kinetics was confirmed by analyzing a portion of the plasmid common to all samples, comparing the kinetics using as the control data either native, unamplified DNA from the plasmid lacking an MTase gene, or whole-genome amplified (WGA) DNA derived from the MTase plasmid (Supplementary Figure S2).
Figure 2.
Confirmation of activity of cloned methyltransferase genes using methylation protection assays. Plasmids containing genes encoding the MTases (identified in the text above the gel images) were isolated from E. coli ER2796, a strain which lacks endogenous methyltransferase activities. Methylated plasmid DNAs (right lane in each case) were linearized using PstI then challenged with restriction endonucleases (REases) that are expected to be blocked by the action of the relevant cloned MTase. CviQI, RsaI, Sau3AI, Tsp509I, AatII, AluI, BstNI, NspI, HpaII and SacI samples were assayed using the equivalent cognate REases. EcoKDam and EsaLHCI samples were assayed using REase MboI, EsaBC1I using REase AluI, EcoKDcm with REase PspGI and EsaBC2I with REase SalI. Control PCR product DNAs (left lane in each case) were restricted with the same enzymes as the methylated plasmids but without PstI. The SacI unmodified (PCR product) DNA was also cleaved with PstI as this construct contains an additional PstI site within the coding sequence of the SacI MTase gene. M = 1 kb DNA-Ladder Marker.
Characterization of MTase specificities
We carried out SMRT sequencing on DNA templates with methylation marks from 16 different bacterial MTases, including enzymes with both known and unknown specificities (Table 1). Plasmid sequences were obtained through applying standard analysis tools (www.pacb.com/devnet), yielding high fold coverage over the entire DNA template from a single sequencing run (minimum of 300-fold; Supplementary Table S2), and are available at http://pacificbiosciences.com/devnet/files/how-tos/dna-methyltransferase/1.0/index.html. Consensus sequences of each of the plasmids can be found in Supplementary File S1. Results from the kinetic analysis are shown by example in Figure 3 for the two MTases, M.CviQI and M.RsaI, which modify the sequence context 5′-GTAC-3′ to impart m6A or m4C, respectively. For each MTase, the left panel shows a global analysis of the IPD ratio data for both strands of the entire plasmid. The right panels show detailed bar graphs of IPD ratios for two representative template positions. The large peaks of increased IPD ratio are present at sites of DNA modification and correspond to the presumed recognition sequences for each bacterial DNA MTase, based on the known specificity of the cognate restriction enzymes. As expected, the SMRT sequencing data from a plasmid lacking an MTase gene was devoid of any methylation-specific kinetic signals (Supplementary Figure S3).
Figure 3.
MTase specificities determined from SMRT sequencing. The sequence context 5′-GTAC-3′ is methylated by (A) M.CviQI (m6A) and (B) M.RsaI (m4C). The left panel shows IPD ratio data for both strands over the entire plasmid, with the inner and outer circles representing the reverse and forward DNA template strands, respectively. Template positions are indicated by the numbered track middle circle, with blue markers denoting occurrences of MTase target sequence contexts. The right panels show IPD ratios for two representative template positions containing the target context (bold letters); the methylated base is highlighted in red. Error bars represent the standard error of the ratio of means and is calculated as described in the ‘Materials and Methods’ section.
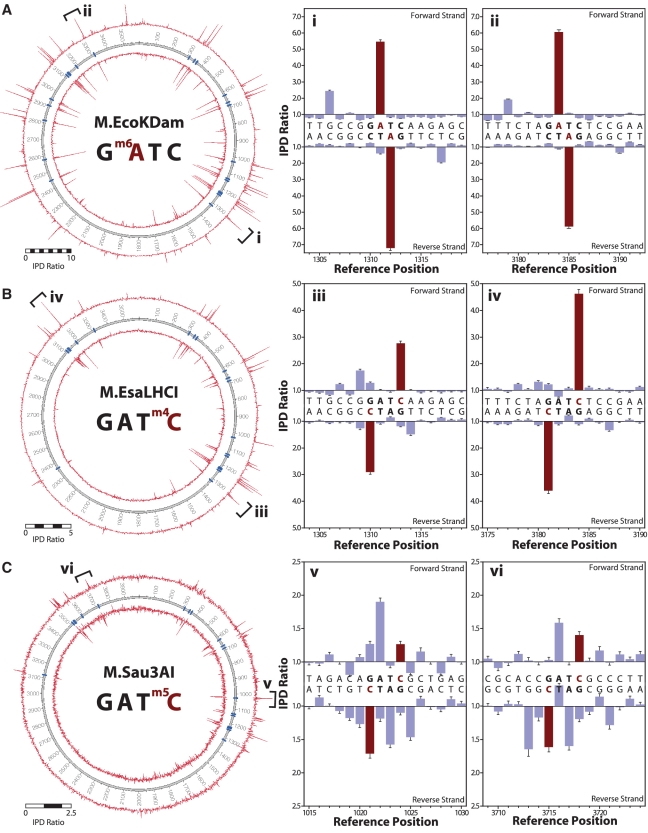
For the sequence context 5′-GATC-3′, MTases exist that can impart all three methylation types: M.EcoKDam (m6A), M.EsaLHCI (m4C) and M.Sau3AI (m5C) (Figure 4). For the dam MTase in E. coli (M.EcoKDam), we obtained data consistent with previously observed kinetic signals for m6A (28) (Figure 4A), showing large IPD ratios at m6A template positions. In addition, we observed a secondary peak at the +5 position relative to the modified base, likely due to the intimate contact of the DNA polymerase with the nascent double-stranded DNA, and its dynamic sensitivity to slight structural perturbations caused by the presence of the methyl group (28,34). Such secondary peaks enhance the ability to detect and discriminate different base modifications.
Figure 4.
MTase specificities determined from SMRT sequencing. The sequence context 5′-GATC-3′ is methylated by (A) M.EcoKDam (m6A), (B) M.EsaLHCI (m4C) and (C) M.Sau3AI (m5C). The left panel shows IPD ratio data for both strands over the entire plasmid, with the inner and outer circles representing the reverse and forward DNA template strands, respectively. Template positions are indicated by the numbered track middle circle, with blue markers denoting occurrences of MTase target sequence contexts. The right panels show IPD ratios for two representative template positions containing the target context (bold letters), the methylated base is highlighted in red. Error bars represent the standard error of the ratio of means and is calculated as described in the ‘Materials and Methods’ section.
Figure 4B shows the methylation activity of M.EsaLHCI that adds a methyl group to the N4 position of cytosine in the 5′-GATC-3′ sequence context. Encountering m4C DNA base modifications in the template, the DNA polymerase slows significantly when incorporating a G nucleotide opposite the m4C base, resulting in IPDs at m4C positions ∼3- to 5-fold higher for sequences with 4mC compared with unmodified control sequences. To our knowledge, this study represents the first demonstration of directly sequencing the m4C base modification.
The third common base modification in bacteria, m5C, is imparted on the 5′-GATC-3′ sequence context by the MTase M.Sau3AI (Figure 4C). Whereas the methyl groups in m6A and m4C are directly involved in base pairing, the m5C methyl group is not and is instead positioned in the major groove of the nascent double-stranded DNA which has few direct contacts with the DNA polymerase. As a result, m5C modifications cause more subtle perturbations of the DNA which in turn result in smaller effects on the DNA polymerase dynamics (28). While under the current sequencing conditions, the IPD ratios are therefore smaller and spread across multiple bases surrounding the modification, the majority of m5C positions could be distinguished from unmodified cytosine based on polymerase kinetics, with each of the MTase target sites showing IPD ratio peaks above background.
Determination of methylation patterns of DNA MTases with unknown specificities
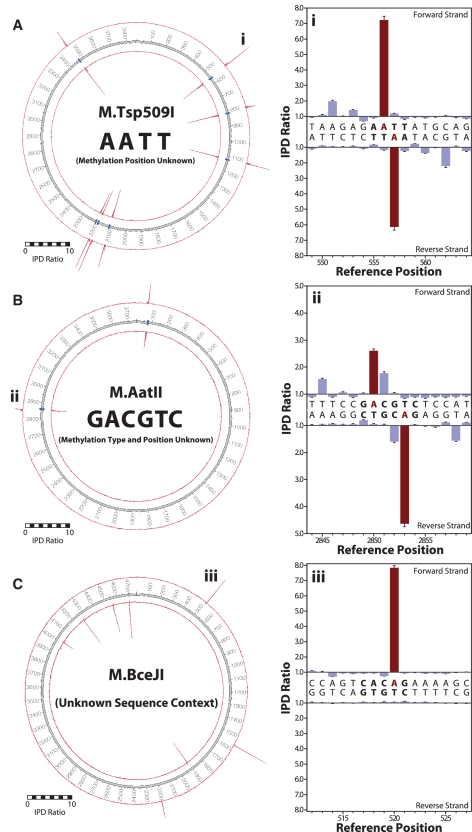
We applied our method to determine the methylation patterns of several DNA MTases for which the position and/or type of methylation is not known (Figure 5). The first example, M.Tsp509I, was selected from the category of MTases for which the target sequence context and the type of methylation was available, but the exact site of methylation was undetermined. Taking advantage of the base resolution of methylation detection during SMRT sequencing, we determined that it is the inner adenine base of the sequence context 5′-AATT-3′ that was methylated to m6A (Figure 5A), as the IPD ratio peak was observed at this position, as well as the secondary peak at position +5 (see above).
Figure 5.
Determination of unknown MTase specificities. Base-resolved MTase target specificities were resolved for (A) M.Tsp509I as 5′-A m6A TT-3′, (B) M.AatII as 5′-G m6A CGTC-3′ and (C) M.BceJI as 5′-CAC m6A G-3′. The left panel shows IPD ratio data for both strands over the entire plasmid, with the inner and outer circles representing the reverse and forward DNA template strands, respectively. Template positions are indicated by the numbered track middle circle, with blue markers denoting occurrences of MTase target sequence contexts (if known). The right panels show IPD ratios for two representative template positions containing the target context (bold letters), the methylated base is highlighted in red. Error bars represent the standard error of the ratio of means and is calculated as described in the ‘Materials and Methods’ section.
The second example of determining the specificity of an MTase was from the category of MTases where the target sequence context was available, but both the type and position of methylation were unknown. We sequenced a plasmid expressing M.AatII (Figure 5B), known to methylate the target sequence 5′-GACGTC-3′ either at m4C or m6A, and determined that this enzyme methylates the adenine, but not the cytosines, again directly visible from the strong signals at the A position and the secondary peak at +5.
The third example was M.BceJI, an MTase for which neither the target sequence, nor the type and position of methylation were known (Figure 5C). This MTase is part of a putative Type III restriction system from Burkholderia cenocepacia strain J2315 that is difficult to transform. Type III REases rarely give complete digests making it difficult to deduce their specificity. Since the specificity-defining sequence is in the MTase protein, this means that the cognate MTases have previously been employed to determine specificity, but the approach needed is quite tedious (18). Kinetic signatures obtained from SMRT sequencing the plasmid expressing this Type III MTase were observed at every occurrence of the sequence context 5′-CACAG-3′ and at no other positions. For each occurrence, the second adenine position displayed kinetic signals of methylation to m6A (Supplementary Figure S4). Because the target sequence is non-palindromic and does not contain adenosine in the reverse complement, the methylation occurs only on one strand as is typical for Type III MTases (35). The presence of m6A in the 5′-CACAG-3′ sequence context was further validated by inhibition of digestion of an overlapping sequence by the methyl-sensitive ScaI restriction endonuclease (Supplementary Figure S5).
Some MTases display off-target activities
While the majority of MTases displayed kinetic signals present exclusively at their cognate sites indicating strict context specificities, we observed additional, off-target kinetic signals for some MTases. For example, a global analysis of M.EcoKDam modified DNA shows numerous smaller kinetic signals in addition to the 5′-GATC-3′ cognate site, suggesting partial methylation at off-target sites (Figure 4A, left panel). These off-target signals were found to encompass sequence contexts differing from the target context by one base (Figure 6). For M.EcoKDam, the 5′-GACC-3′ off-target context was most affected, followed by detectable signals for 5′-HATC-3′ (H = A, C or T) and 5′-GATT-3′. For example, the inset of Figure 6 shows IPD ratio data for the off-target 5′-GACC-3′ sequence: the lower average IPD ratio value is suggestive of partial methylation and in this case, methylation only occurs on one strand because there is no adenine in the reverse complementary sequence (5′-GGTC-3′). This example also exhibits a secondary IPD ratio peak at the +5 position which is commonly observed with m6A modifications in similar sequence contexts. The partial methylation of an off-target 5′-GACC-3′ site was confirmed by showing ∼50% inhibition of SalI cleavage at an overlapping 5′-GTCGACC-3′ sequence (Supplementary Figure S6).
Figure 6.
Sequence context specificity of M.EcoKDam. Kinetic signals for the target 5′-GATC-3′ and all 1-base neighboring sequence contexts containing adenine at the second position are compared. The values plotted represent average IPD ratios of all occurrences of each sequence context (error bars are s.e.m.). The inset shows an example for a methylation signal at the off-target 5′-GACC-3′ sequence context. Error bars represent the standard error of the ratio of means and is calculated as described in the ‘Materials and Methods’ section.
DISCUSSION
Thanks to the availability of a large number of cloned DNA methyltransferase genes that are expressed in an E. coli background devoid of other DNA methylation, it has been possible to prepare a series of pure plasmid DNAs each of which carries methylation signatures for just one methyltransferase. Using a few well-characterized methyltransferases as controls, we have been able to confirm their methylation specificity using the SMRT sequencing approach and find that in addition to obtaining interpretable signals for m6A and m5C as reported previously (28), it is also possible to detect m4C. This means that the three common types of methylation typically found in bacterial and archaeal genomes can be unambiguously distinguished. Furthermore, when a suitable number of sites are present, both the type of methylation and a unique recognition sequence for the methyltransferase can be assigned. By applying this same technique to several methyltransferases with either unknown types and/or positions of methylation or in one case, both an unknown type of methylation and an unknown recognition sequence, we have been able to assign specificity to the M.Tsp509I methyltransferase, the M.AatII methyltransferase and a new methyltransferase, M.BceJI, encoded by ORF 3494 in the B. cenocepacia genome. The latter is of particular interest because it is part of a Type III RM system and traditionally it has been quite difficult to obtain the recognition sequences for these enzymes. The SMRT approach described here is well-suited for that purpose and if applied at a whole bacterial genome level, should also be suitable to obtain recognition sequences for Type I RM systems, which typically have been even more difficult to determine than the Type III systems.
Since most bacterial genomes contain many RM systems of all types, it seems likely that genome sequencing using the SMRT approach should, in many cases, allow the direct discovery of recognition sequences for the methyltransferases encoded by those genomes. Already, for all fully-sequenced genomes, REBASE (4) contains a theoretical analysis of the sequences and predictions of the methyltransferase genes present in those genomes. In many cases, the recognition sequences of those methyltransferases can be inferred on the basis of sequence similarity and these predictions are included in the database. Confirmation of many of these predictions should now be possible by SMRT sequencing, while in addition, the recognition sequences for many unassigned methyltransferases should also be obtainable. For all genomic sequences that become newly determined by the SMRT approach, the raw data will include both methylation status, as well as just the raw sequence, a considerable bonus in terms of data acquisition.
In previous studies of DNA methyltransferase specificity, there has been no easy way to discover the fidelity of the methyltransferase to find out whether it matches the fidelity of the cognate restriction enzyme. In the few cases where it has been possible to check this, the enzymes have shown the same degree of specificity of recognition as the restriction enzymes, but in at least one case, some promiscuity has been noted (36). It was thus of great interest to look at the promiscuity of the methyltransferases used in this study. For the Dam methyltransferase of E. coli (M.EcoKDam recognizing GATC) we found that, surprisingly, there was rather more off-target methylation than we anticipated. Given the role of the Dam methyltransferase in the initiation of DNA synthesis, one might have expected a high degree of fidelity since this would seem to be a critical function and many GATC sites are found close to the origin. It appears that the initiation of DNA replication is not affected if additional methyl groups are present at non-cognate sites. Dam methyltransferase can affect transcription when GATC sites are methylated [reviewed earlier (5)]. The other known function of the Dam methyltransferase in damage repair would not be expected to be affected by off-target methylation. Whereas it is possible that the observed off-target signals are caused by mutation in the MTase genes present as a smaller fraction in the samples, we have not observed such minority species from SMRT sequencing, with a current detection limit of ∼2–3% (30).
It should be noted though that for most of the methyltransferases that we studied, the fidelity was actually quite high and relatively few off-target signals were found. This might be viewed as a little surprising since there is no clear biological selection mechanism that would ensure the accuracy of the methyltransferase component of an RM system. This is in contrast to the restriction enzyme itself where there is a strong selection for the cognate site since off-target cleavage known as star activity could easily cause damage to host genomes. In this context, it is worth noting that many restriction enzymes do show unwanted star activity (37) and it would be of considerable interest to check whether the methyltransferases that accompany these promiscuous restriction enzymes show a similar promiscuity in their recognition, thereby protecting the organism against the potentially deleterious action of its restriction enzyme.
We are greatly encouraged by the results of the studies described here, and plan to undertake a more systematic analysis of whole genome sequences, both with a view to examining the levels of DNA methylation in these genomes and possibly assigning specificity to the methyltransferases and correlating it with the predicted methyltransferase genes in the genome. Such studies will also provide the significant quantities of data needed to enable exact quantitation of the degree of partial methylation and to define the precise sequences that lead to off-site methylation. By judicious analysis of methylation patterns, SMRT sequencing could also prove extremely useful in permitting the correct reassembly of individual genomes from complex metagenomic sequences.
ACCESSION NUMBER
Genbank accession number JN569339.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online: Supplementary Tables 1 and 2, Supplementary Figures 1–6 and Supplementary File 1.
FUNDING
National Institutes of Health (grants 1RC2HG005618-01; National Human Genome Research Institute (NHGRI) and 1RC2GM092602-01; National Institute of General Medical Sciences (NIGMS)). Funding for open access charge: Internal funds of Pacific Biosciences.
Conflict of interest statement. T.A.C., A.O.K., K.E.S., M.B. and J.K. are full-time employees at Pacific Biosciences, a company commercializing single-molecule, real-time nucleic acid sequencing technologies. I.A.M., R.D.M., A.F. and R.J.R. are full-time employees of New England Biolabs, a company that sells research reagents such as DNA methyltransferases.
Supplementary Material
ACKNOWLEDGEMENTS
We would like to thank L. Mazzola and N. Badger at the NEB sequencing core facility, D. Heiter, B. Jack, K. Lunnen, J. Samuelson, S.-Y. Xu and Z. Zhu for providing genomic or plasmid DNA samples containing MTase genes, and S.W. Turner, K. Luong, J. Bullard, J. Lee, J. Bingham, E. Schadt, O. Banerjee, and D. Webster for helpful discussions. We thank Dr. E. Mahenthiralingam for the gift of Burkholderia cepacia J2315 DNA.
REFERENCES
- 1.Malone T, Blumenthal RM, Cheng X. Structure-guided analysis reveals nine sequence motifs conserved among DNA amino-methyltransferases, and suggests a catalytic mechanism for these enzymes. J. Mol. Biol. 1995;253:618–632. doi: 10.1006/jmbi.1995.0577. [DOI] [PubMed] [Google Scholar]
- 2.Kumar S, Cheng X, Klimasauskas S, Mi S, Posfai J, Roberts RJ, Wilson GG. The DNA (cytosine-5) methyltransferases. Nucleic Acids Res. 1994;22:1–10. doi: 10.1093/nar/22.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wu JC, Santi DV. Kinetic and catalytic mechanism of HhaI methyltransferase. J. Biol. Chem. 1987;262:4778–4786. [PubMed] [Google Scholar]
- 4.Roberts RJ, Vincze T, Posfai J, Macelis D. REBASE–a database for DNA restriction and modification: enzymes, genes and genomes. Nucleic Acids Res. 2010;38:D234–D236. doi: 10.1093/nar/gkp874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Marinus MG, Casadesus J. Roles of DNA adenine methylation in host-pathogen interactions: mismatch repair, transcriptional regulation, and more. FEMS Microbiol. Rev. 2009;33:488–503. doi: 10.1111/j.1574-6976.2008.00159.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cheng X, Blumenthal RM. Mammalian DNA methyltransferases: a structural perspective. Structure. 2008;16:341–350. doi: 10.1016/j.str.2008.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bestor TH. The DNA methyltransferases of mammals. Hum. Mol. Genet. 2000;9:2395–2402. doi: 10.1093/hmg/9.16.2395. [DOI] [PubMed] [Google Scholar]
- 8.Fuks F. DNA methylation and histone modifications: teaming up to silence genes. Curr. Opin. Genet. Dev. 2005;15:490–495. doi: 10.1016/j.gde.2005.08.002. [DOI] [PubMed] [Google Scholar]
- 9.Okamoto I, Otte AP, Allis CD, Reinberg D, Heard E. Epigenetic dynamics of imprinted X inactivation during early mouse development. Science. 2004;303:644–649. doi: 10.1126/science.1092727. [DOI] [PubMed] [Google Scholar]
- 10.Surani MA. Reprogramming of genome function through epigenetic inheritance. Nature. 2001;414:122–128. doi: 10.1038/35102186. [DOI] [PubMed] [Google Scholar]
- 11.Penn NW, Suwalski R, O'Riley C, Bojanowski K, Yura R. The presence of 5-hydroxymethylcytosine in animal deoxyribonucleic acid. Biochem. J. 1972;126:781–790. doi: 10.1042/bj1260781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kriaucionis S, Heintz N. The nuclear DNA base 5-hydroxymethylcytosine is present in Purkinje neurons and the brain. Science. 2009;324:929–930. doi: 10.1126/science.1169786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tahiliani M, Koh KP, Shen Y, Pastor WA, Bandukwala H, Brudno Y, Agarwal S, Iyer LM, Liu DR, Aravind L, et al. Conversion of 5-methylcytosine to 5-hydroxymethylcytosine in mammalian DNA by MLL partner TET1. Science. 2009;324:930–935. doi: 10.1126/science.1170116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dunn DB, Smith JD. The occurrence of 6-methylaminopurine in deoxyribonucleic acids. Biochem. J. 1958;68:627–636. doi: 10.1042/bj0680627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Janulaitis A, Klimasauskas S, Petrusyte M, Butkus V. Cytosine modification in DNA by BcnI methylase yields N4-methylcytosine. FEBS Lett. 1983;161:131–134. doi: 10.1016/0014-5793(83)80745-5. [DOI] [PubMed] [Google Scholar]
- 16.Kong H, Lin LF, Porter N, Stickel S, Byrd D, Posfai J, Roberts RJ. Functional analysis of putative restriction-modification system genes in the Helicobacter pylori J99 genome. Nucleic Acids Res. 2000;28:3216–3223. doi: 10.1093/nar/28.17.3216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bitinaite J, Maneliene Z, Menkevicius S, Klimasauskas S, Butkus V, Janulaitis A. Alw26I, Eco31I and Esp3I–type IIs methyltransferases modifying cytosine and adenine in complementary strands of the target DNA. Nucleic Acids Res. 1992;20:4981–4985. doi: 10.1093/nar/20.19.4981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bachi B, Reiser J, Pirrotta V. Methylation and cleavage sequences of the EcoP1 restriction-modification enzyme. J. Mol. Biol. 1979;128:143–163. doi: 10.1016/0022-2836(79)90123-2. [DOI] [PubMed] [Google Scholar]
- 19.Bart A, van Passel MW, van Amsterdam K, van der Ende A. Direct detection of methylation in genomic DNA. Nucleic Acids Res. 2005;33:e124. doi: 10.1093/nar/gni121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Singer-Sam J, Grant M, LeBon JM, Okuyama K, Chapman V, Monk M, Riggs AD. Use of a HpaII-polymerase chain reaction assay to study DNA methylation in the Pgk-1 CpG island of mouse embryos at the time of X-chromosome inactivation. Mol. Cell. Biol. 1990;10:4987–4989. doi: 10.1128/mcb.10.9.4987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Frommer M, McDonald LE, Millar DS, Collis CM, Watt F, Grigg GW, Molloy PL, Paul CL. A genomic sequencing protocol that yields a positive display of 5-methylcytosine residues in individual DNA strands. Proc. Natl Acad. Sci. USA. 1992;89:1827–1831. doi: 10.1073/pnas.89.5.1827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Herman JG, Graff JR, Myohanen S, Nelkin BD, Baylin SB. Methylation-specific PCR: a novel PCR assay for methylation status of CpG islands. Proc. Natl Acad. Sci. USA. 1996;93:9821–9826. doi: 10.1073/pnas.93.18.9821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lister R, O'Malley RC, Tonti-Filippini J, Gregory BD, Berry CC, Millar AH, Ecker JR. Highly integrated single-base resolution maps of the epigenome in Arabidopsis. Cell. 2008;133:523–536. doi: 10.1016/j.cell.2008.03.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lister R, Pelizzola M, Dowen RH, Hawkins RD, Hon G, Tonti-Filippini J, Nery JR, Lee L, Ye Z, Ngo QM, et al. Human DNA methylomes at base resolution show widespread epigenomic differences. Nature. 2009;462:315–322. doi: 10.1038/nature08514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kinney SM, Chin HG, Vaisvila R, Bitinaite J, Zheng Y, Esteve PO, Feng S, Stroud H, Jacobsen SE, Pradhan S. Tissue specific distribution and dynamic changes of 5-hydroxymethylcytosine in mammalian genome. J. Biol. Chem. 2011;286:24685–24693. doi: 10.1074/jbc.M110.217083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Song CX, Szulwach KE, Fu Y, Dai Q, Yi C, Li X, Li Y, Chen CH, Zhang W, Jian X, et al. Selective chemical labeling reveals the genome-wide distribution of 5-hydroxymethylcytosine. Nat. Biotechnol. 2011;29:68–72. doi: 10.1038/nbt.1732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cohen-Karni D, Xu D, Apone L, Fomenkov A, Sun Z, Davis PJ, Morey-Kinney SR, Yamada-Mabuchi M, Xu SY, Davis T, et al. The MspJI family of modification-dependent restriction endonucleases for epigenetic studies. Proc. Natl Acad. Sci. USA. 2011;108:11040–11045. doi: 10.1073/pnas.1018448108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Flusberg BA, Webster DR, Lee JH, Travers KJ, Olivares EC, Clark TA, Korlach J, Turner SW. Direct detection of DNA methylation during single-molecule, real-time sequencing. Nat. Methods. 2010;7:461–465. doi: 10.1038/nmeth.1459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Eid J, Fehr A, Gray J, Luong K, Lyle J, Otto G, Peluso P, Rank D, Baybayan P, Bettman B, et al. Real-time DNA sequencing from single polymerase molecules. Science. 2009;323:133–138. doi: 10.1126/science.1162986. [DOI] [PubMed] [Google Scholar]
- 30.Travers KJ, Chin CS, Rank DR, Eid JS, Turner SW. A flexible and efficient template format for circular consensus sequencing and SNP detection. Nucleic Acids Res. 2010;38:e159. doi: 10.1093/nar/gkq543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chin CS, Sorenson J, Harris JB, Robins WP, Charles RC, Jean-Charles RR, Bullard J, Webster DR, Kasarskis A, Peluso P, et al. The origin of the Haitian cholera outbreak strain. N. Engl. J. Med. 2011;364:33–42. doi: 10.1056/NEJMoa1012928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Korlach J, Bjornson KP, Chaudhuri BP, Cicero RL, Flusberg BA, Gray JJ, Holden D, Saxena R, Wegener J, Turner SW. Real-time DNA sequencing from single polymerase molecules. Methods Enzymol. 2010;472:431–455. doi: 10.1016/S0076-6879(10)72001-2. [DOI] [PubMed] [Google Scholar]
- 33.Krzywinski M, Schein J, Birol I, Connors J, Gascoyne R, Horsman D, Jones SJ, Marra MA. Circos: an information aesthetic for comparative genomics. Genome Res. 2009;19:1639–1645. doi: 10.1101/gr.092759.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kamtekar S, Berman AJ, Wang J, Lazaro JM, de Vega M, Blanco L, Salas M, Steitz TA. Insights into strand displacement and processivity from the crystal structure of the protein-primed DNA polymerase of bacteriophage phi29. Mol. Cell. 2004;16:609–618. doi: 10.1016/j.molcel.2004.10.019. [DOI] [PubMed] [Google Scholar]
- 35.Madhusoodanan UK, Rao DN. Diversity of DNA methyltransferases that recognize asymmetric target sequences. Crit. Rev. Biochem. Mol. Biol. 2010;45:125–145. doi: 10.3109/10409231003628007. [DOI] [PubMed] [Google Scholar]
- 36.Woodbury CP, Jr, Downey RL, von Hippel PH. DNA site recognition and overmethylation by the Eco RI methylase. J. Biol. Chem. 1980;255:11526–11533. [PubMed] [Google Scholar]
- 37.Wei H, Therrien C, Blanchard A, Guan S, Zhu Z. The Fidelity Index provides a systematic quantitation of star activity of DNA restriction endonucleases. Nucleic Acids Res. 2008;36:e50. doi: 10.1093/nar/gkn182. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.