Abstract
Cell cycle-dependent gene expression is often controlled on the transcriptional level. Genes like cyclin B, CDC2 and CDC25C are regulated by cell cycle-dependent element (CDE) and cell cycle genes homology region (CHR) promoter elements mainly through repression in G0/G1. It had been suggested that E2F4 binding to CDE sites is central to transcriptional regulation. However, some promoters are only controlled by a CHR. We identify the DREAM complex binding to the CHR of mouse and human cyclin B2 promoters in G0. Association of DREAM and cell cycle-dependent regulation is abrogated when the CHR is mutated. Although E2f4 is part of the complex, a CDE is not essential but can enhance binding of DREAM. We show that the CHR element is not only necessary for repression of gene transcription in G0/G1, but also for activation in S, G2 and M phases. In proliferating cells, the B-myb-containing MMB complex binds the CHR of both promoters independently of the CDE. Bioinformatic analyses identify many genes which contain conserved CHR elements in promoters binding the DREAM complex. With Ube2c as an example from that screen, we show that inverse CHR sites are functional promoter elements that can bind DREAM and MMB. Our findings indicate that the CHR is central to DREAM/MMB-dependent transcriptional control during the cell cycle.
INTRODUCTION
The expression of many genes that play a central role in the cell cycle is regulated on the transcriptional level. They are characterized by a cyclic expression during different phases of the cell cycle. Genes expressed at the G1/S transition are often regulated by complexes formed by E2F transcription factors, their dimerization partners DP1 or DP2, and the pocket proteins pRB, p130 or p107 (1–3). While the regulation of these S phase genes is well understood, many open questions remain for the regulation of genes with a maximal expression in late S, G2 or M phases. Many of these genes like cyclin B, CDC25C, CDC2, CKS1 and aurora kinase B are repressed in the early phases of the cell cycle by unknown mechanisms. However, the promoters of these genes appear to share some common features. Most strikingly, phylogenetically conserved cell cycle-dependent element (CDE) and cell cycle genes homology region (CHR) sites and CCAAT-boxes can often be found close to the transcription start in the promoters of such genes. While CDE and CHR mediate transcriptional repression in early cell cycle phases, the CCAAT-boxes are necessary for transcriptional activation (4).
The CDE was first observed by in vivo footprinting in the human CDC25C promoter to be protected during G0 (5). Mutation of this element and subsequent analysis of the promoter in reporter assays revealed that the CDE is important for transcriptional repression in early cell cycle phases. Soon after this observation, another element, then named the CHR, was discovered by sequence comparison and functional analysis of the CDC25C, cyclin A2 and CDC2 promoters (6). The CHR consensus resembles the sequence TTTGAA. Its mutation leads to a loss of transcriptional repression in G0 and G1 phases as observed for the CDE site. Further analysis provided evidence that the CDE and the CHR always appear in close proximity with a spacer of four nucleotides. The sequence of the CDE is rich in guanine and cytosine and is related to the TTGGCGC E2F-binding consensus. Consistent with a similarity of CDE and E2F sites, binding of E2F and pocket proteins was shown to several CDE-regulated promoters, e.g. CDC2, cyclin A2 and aurora kinase B (6–9).
However, a similarity between E2F- and CDE-dependent regulations would not explain the necessity of CHRs in CDE/CHR-controlled genes. Assuming that protein binding is required for CHR function, one would speculate that CDE and CHR elements cooperate in protein binding. Consistently, it was observed for the CDC2 promoter that E2F4 binding to the CDE is abolished after mutating the CHR (6,10). This result may be explained by proteins associated with the CHR being essential for E2F4 binding to the CDE. Several groups tried to identify the factors binding to CHRs responsible for regulating different genes. For the cyclin A2 promoter, a protein named CHF has been observed to bind to the CHR in EMSAs (11). Another factor that was called CDF-1 was found to associate with the CDE/CHR elements in CDC25C and CCNA2 promoters (12,13). However, cloning or further characterization of any of these factors was not accomplished.
The link between CDE and CHR regulation and protein binding becomes more puzzling when genes like mouse Cdc25C, human cyclin B1, cyclin B2 and PLK1 are considered. Their promoters have been shown by sequence comparison and mutation analyses to be controlled by a single CHR without additional CDE or E2F site (14–18). Thus, central questions remain open as to which proteins are binding to the CDE and/or the CHR elements and how these proteins control transcriptional repression of genes in the early cell cycle phases.
Here, we observe that the DREAM (DP, RB-like, E2F4 and MuvB) complex binds to the CHR of mouse and human cyclin B2 promoters in G0. Mutation of the CHR in both promoters results in a complete loss of DREAM complex binding and deregulation of the promoters in the cell cycle. Although E2F4 is part of the complex in G0 and early G1 phase (19–21), an E2F-like CDE element is not essential for DREAM binding. However, the CDE in the mouse cyclin B2 promoter can support binding of the DREAM complex. Surprisingly, we find that the CHR is not only necessary for suppression of promoter activity, but also for activation. In proliferating cells, the B-myb-containing MMB (Myb-MuvB) complex binds to the CHR of both promoters independently of the CDE. Additionally, bioinformatic analyses of experimental data identify many more genes which bind the DREAM complex and contain evolutionary conserved CHR elements in their promoters. We prove that this approach is applicable for the identification of promoters with functional CHR elements by showing that the mouse Ube2c promoter binds the DREAM and MMB complexes through an inverse CHR that is essential for the regulation of cell cycle-dependent expression. Our findings indicate that the CHR is a central element in many promoters mediating transcriptional regulation in the cell cycle by the DREAM and MMB complexes.
MATERIALS AND METHODS
Cell culture
NIH3T3 and F9 cells obtained from DSMZ (Braunschweig, Germany) were grown in Dulbecco's modified Eagle's medium (DMEM, Lonza, Basel, Switzerland) supplemented with 10% fetal calf serum (FCS) and penicillin/streptomycin. Cells were maintained at 37°C and 10% CO2. F9 cells were grown on 0.1% gelatin-coated dishes.
For synchronization in G0 phase, NIH3T3 cells were either density arrested for 3–4 days or arrested by serum starvation (DMEM with 0% FCS) for 60 h. To analyze cells during their passage through the cell cycle, they were restimulated with 20% FCS in DMEM after the serum-deprivation phase.
Stably transfected NIH3T3 cell lines were created by transfecting promoter–reporter constructs based on the pGL4.14-Hygro vector (Promega, Madison, WI, USA) and selection with hygromycin (PAA, Linz, Austria) at a concentration of 500 µg/ml.
For SILAC analysis, NIH3T3 cells were grown in DMEM minus arginine and lysine (Thermo Fisher Scientific, Rockford, IL, USA) supplemented with 10% FCS and penicillin/streptomycin (PAA, Linz, Austria). To produce the ‘light’ media, l-arginine–HCl and l-lysine–HCl were added to a final concentration of 84 µg/ml and 146 µg/ml, respectively, whereas 13C6, 15N4 l-arginine–HCl and 13C6 l-lysine–HCl (Thermo Fisher Scientific) were applied at the same concentrations to obtain the ‘heavy’ medium.
Plasmids and DNA probes
The human cyclin B2 promoter (nt −903 to +1 relative to translational start, designated as hCCNB2-long) has been described before (17). hCCNB2 wt, hCCNB2 ‘CDE'mut, hCCNB2 CHRmut and hCCNB2 ‘CDE’/CHRmut were cloned in the pGL4.10 vector. hCCNB2-short was produced by PCR amplification creating promoter fragments with a size of 263 nt (nucleotides −262 to +1) followed by ligation into the pGL4.10 or pGL4.14 vectors (Table 1).
Table 1.
Mutations introduced in the CDE and CHR elements of human and mouse cyclin B2 promoters
 |
The mouse cyclin B2 promoter with a size of 1190 bp (nt −1189 to +1, named mCcnb2-long) and the mouse Ube2c promoter (nt −478 to −3) were amplified from genomic NIH3T3 cell DNA and cloned in the pGL4.10 vector. mCcnb2-short was produced by PCR amplification creating promoter fragments with a size of 210 nt (nt −209 to +1) and subsequent ligation in pGL4.10 or pGL4.14 vectors. Mutations were introduced with the QuikChange site-directed mutagenesis kit (Agilent Technologies, Santa Clara, CA, USA) (Table 1). Plasmids used for B-myb-knockdown (sh-Bmyb1, sh-Bmyb2) were kind gifts from Kenneth Boheler (22). DNA probes for affinity purification with the same sequence as hCCNB2-short and mCcnb2-short and truncated variants of mCcnb2-short without CCAAT-boxes (nt −130 to +1) were obtained by PCR using a biotinylated primer for labeling the 3′-end (Invitrogen, Carlsbad, CA, USA). As a negative control, a fragment of the mouse Gapdhs promoter (nt −169 to +12 relative to the transcription start site) was used. Sequences of primers are available in the Supplementary Methods.
DNA affinity purification
For DNA affinity purification of protein complexes in G0 cells, NIH3T3 cells were grown to confluence and density arrested for three additional days. F9 cells were grown to 80–90% confluence. Cells were washed with PBS, scraped and centrifuged. Nuclei were isolated by incubation for 10 min in buffer A [10 mM HEPES–KOH, pH 7.9, 1.5 mM MgCl2, complete EDTA-free protease inhibitors and phosphatase inhibitors (Roche, Basel, Switzerland), 10 mM KCl, 0.5 mM DTT] on ice and centrifugation at 4000g for 10 min. Nuclei were lysed by incubation for 20 min on ice in buffer C [20 mM HEPES/KOH (pH 7.9), 1.5 mm MgCl2, 0.2 mm EDTA, 450 mM NaCl, complete EDTA-free protease inhibitors]. Nuclei were disrupted by using a syringe with a 23 gauge needle. Cells were drawn from the sample tube into the syringe and the contents were ejected back into the sample tube for five times. Extracts were centrifuged at 20 000g at 4°C to remove debris. After that, NaCl concentration was adjusted to 150 mM by dilution with buffer C (without NaCl). Protein concentration in the nuclear extract was determined by the Bradford assay (Thermo Fisher Scientific). Sonicated salmon sperm DNA was added at a final concentration of 100 µg/ml and incubated at room temperature for 15 min. After that, extracts were aliquoted in 1.5 ml Eppendorf tubes (extract with 1 mg protein per tube). One microgram of biotinylated DNA probe was added to the aliquots and extracts were incubated for 30 min at room temperature. Afterwards, 50 µl of magnetic streptavidin bead suspension (Miltenyi, Bergisch Gladbach, Germany) were added and the mixture was incubated for additional 5 min. Proteins binding to the probes were isolated using the μMACS Streptavidin Kit (Miltenyi). Washing was performed six times with 200 µl buffer C supplemented with 150 mM NaCl. For the detection of MMB complex proteins, the washing buffer was supplemented with 0.5% NP40. Proteins were eluted in two steps with 20 µl and 60 µl elution buffer at 95°C. Twenty microliters of the eluates and 15 µg nuclear extract (input) were subjected to sodium dodecyl sulfate–olyacrylamide gel electrophoresis (SDS–PAGE) and western blot.
SDS–PAGE and western blot
SDS–PAGE and western blot were performed following standard protocols (23). For detection of DREAM complex components, the following antibodies were applied: E2F4 (C-20, Santa Cruz Biotechnology, Santa Cruz, CA, USA, 1:1000 dilution), Tfdp1 (Ab6, Thermo Scientific, 1:400 dilution), p130 (C-20, Santa Cruz Biotech., 1:1000 dilution), p107 (C-18, Santa Cruz Biotech., 1:1000 dilution), Lin9, Lin37, Lin52, Lin54 (1:1000 dilution) (19) and Rbbp4 (ab79416, Abcam, Cambridge, UK, 1:5000 dilution). Nfya was detected with NF-YA (G-2) monoclonal antibody (Santa Cruz Biotech., 1:1000 dilution). The BMyb LX015.1 monoclonal antibody was a kind gift from Roger Watson (24).
SILAC
For SILAC analysis, NIH3T3 cells were grown in ‘heavy’ or ‘light’ medium as described above. Nuclear extracts of density-arrested cells were prepared and DNA affinity purification with the wild-type mouse cyclin B2 probe and the CDE/CHR mutant probe was performed as described above. Four milligrams of nuclear extract were subjected to affinity purification with magnetic streptavidin beads (Miltenyi). Columns were washed 10 times with 200 µl buffer C. Equal volumes of eluates from affinity purifications with wild-type and mutant probe were combined and separated on a 12% SDS–PAGE. After separation, the gel was stained with Coomassie Imperial Protein Stain (Thermo Fisher Scientific) and 2 mm slices were excised.
Mass spectrometry and data analysis
Tryptic digest of proteins and nanoLC-MS/MS experiments were done as described previously (25). In brief, tryptic peptides were separated by a reversed-phase capillary liquid chromatography system (Eksigent 2D nanoflow LC, Axel Semrau GmbH, Sprockhövel, Germany) connected to an LTQ-Orbitrap XL mass spectrometer (Thermo Scientific). Mass spectra were acquired in a data-dependent mode with one MS survey scan (with a resolution of 60 000) in the Orbitrap and MS/MS scans of the five most intense precursor ions in the LTQ. The MS survey range was m/z 350–1500. The dynamic exclusion time (for precursor ions) was set to 120 s and automatic gain control was set to 3 × 106 and 20 000 for Orbitrap-MS and LTQ-MS/MS scans, respectively.
Identification and quantification of proteins were carried out with version 1.0.12.31 of the MaxQuant software package (26). Generated peak lists (msm files) were submitted to a MASCOT search engine (version 2.2, Matrix Science Ltd, London, UK) and searched against an IPI mouse protein database (version 3.52). The mass tolerance of precursor and sequence ions was set to 7 ppm and 0.35 Da, respectively. Methionine oxidation and the acrylamide modification of cysteine were used as variable modifications. False discovery rates were <1% based on matches to reversed sequences in the concatenated target-decoy database. Proteins were considered if at least two sequenced peptides were quantified.
Transfections and luciferase assays
For measuring cell cycle-dependent promoter activity with luciferase reporter assays, NIH3T3 cells were plated in 12-well plates (23 000 cells per well). After 24 h, cells were transfected with pGL4 reporter constructs [transfection mixture per well: 0.2 µg promoter reporter plasmid (pGL4.10), 0.05 µg Renilla luciferase plasmid (pGL4.70), 1.5 µl GeneJuice (Merck, Darmstadt, Germany), 60 µl DMEM without FCS]. Twenty-four hours after transfection, cells were starved for 60 h by exchanging growth medium with 10% FCS to DMEM with 0% FCS. Cells were restimulated for cell cycle re-entry with 20% FCS in DMEM and collected at given time points by adding 200 µl luciferase lysis buffer (Promega). Cells were stored at −80 C and luciferase activity was measured with the Dual-Luciferase Reporter Assay system (Promega) following the manufacturer's recommendations. Relative light units (RLUs) were calculated by normalizing firefly luciferase activity to Renilla luciferase activity. Subsequently, RLUs for promoter reporters for each time point were divided by the RLUs from pGL4.10 empty vector transfections to compensate for changes in the luciferase expression caused by factors binding to cryptic sites in the vector backbone, thereby changing the expression of firefly luciferase reporter independently from the promoter.
F9 cells (40 000 per well in 24-well plates) were transfected with 1 µl Fugene HD (Roche, Basel, Switzerland), 0.2 µg promoter reporter plasmid (pGL4.10) and 0.02 µg Renilla luciferase plasmid (pGL4.70). For B-myb knockdown experiments, NIH3T3 cells (12 000 per well in 24-well plates) were transfected with 1 µl Fugene HD (Roche), 0.1 µg promoter reporter plasmid (pGL4.10), 0.2 µg pSuper construct and 0.02 µg Renilla luciferase plasmid (pGL4.70). Cells were collected 48 h after transfection.
FACS analysis
Cells were fixed overnight at 4°C in one volume PBS/1 mM EDTA and three volumes of absolute ethanol. DNA was stained with Hoechst 33343 (Invitrogen) at a final concentration of 10 µg/ml for 15 min at 37°C. DNA content per cell was measured by flow cytometry on a LSR II instrument (Becton Dickinson, Franklin Lakes, NJ, USA). Data analysis was done with BD FACSDiva 6.1 and WinMDI 2.9 software.
Chromatin immunoprecipitations
Density-arrested NIH3T3 cells were fixed in 1% formaldehyde for 10 min. ChIPs were performed as described earlier (16,17,27). The following antibodies were used to precipitate DREAM complex components: E2F4 (C-20, Santa Cruz Biotech.), p130 (C-20, Santa Cruz Biotech.) and Lin9 (19). A non-related rabbit antibody was used as a control for non-specific signals. For all precipitations, 1 µg of antibody and 25 µl of Protein G Dynabead suspension (Invitrogen) were used. qPCR was carried out with the QuantiTect SYBR Green PCR kit (QIAGEN, Hilden, Germany). All PCR results were normalized to input controls. ChIP primer sequences can be obtained upon request.
Bioinformatic identification of CHR elements
The 817 genomic regions in human reference genome build hg17 (http://genome.ucsc.edu) that were bound by LIN9, p130 and E2F4 in G0 phase were searched for all annotated transcription start sites whose [−200, +200] bp promoter windows were completely contained within the regions. We found promoters associated with a total of 792 unique HGNC official gene symbols. A corresponding set of 792 genes was chosen at random out of the set of all genes annotated in build hg17. All further analysis was carried out on all promoters contained in the DREAM-binding set as well as all [−200, +200] promoters in the entire genome associated with the randomized gene set. MATCH was used to find all experimentally validated CHR motifs (TTTGAA, TTTAAA, TAGGAA and CTTGAA) in the promoters (28). The PhastCons 17-vertebrate conservation score (29) was then used to filter the sites with a posterior probability cutoff of 95% for at least five bases and a minimum score of 85% for the sixth. A gene was counted as a ‘CHR-containing gene’ if it had at least one conserved CHR site in any of its associated promoter windows. Significance of the overlap between DREAM-binding promoters and CHR-containing genes was computed using Fisher's exact test.
RESULTS
Components of the DREAM complex bind to the CDE/CHR sites of the mouse cyclin B2 promoter
In an earlier report, we had shown that the CDE/CHR element in the mouse cyclin B2 gene is essential for mediating repression in early cell cycle phases (30). We intended to identify the proteins binding to these sites in G0. To this end, we applied a mass spectrometry-based approach employing the method of stable isotope labeling with amino acids in cell culture (SILAC). After density arrest in G0, DNA affinity purification was performed with mouse NIH3T3 nuclear cell extracts using biotinylated DNA probes for the wild-type promoter when cells grown in ‘heavy’ medium and a CDE/CHR double-mutant promoter probe for extracts from cells grown in ‘light’ medium (Table 1). Eluates with DNA-binding complexes were mixed, the sample resolved by SDS–PAGE and the resulting lane cut into equal segments that were then subjected to trypsin digestion. Subsequently, tandem MS/MS analysis was performed for identification and relative quantification of proteins which bind to the wild-type probe compared with the CDE/CHR-mutant promoter (Figure 1). A high ratio of signals for 13C versus 12C and 15N versus 14N containing tryptic peptides indicated proteins to bind to the cyclin B2 promoter in G0 requiring the intact CDE and CHR elements.
Figure 1.
Strategy for the identification of proteins binding to the CDE/CHR tandem element of the mouse cyclin B2 promoter. NIH3T3 cells were labeled by cultivation in ‘heavy’ medium lacking natural Lys and Arg amino acids supplemented with 13C6 lysine and 13C6, 15N4 arginine or regular ‘light’ medium (12C-Lys/12C, 14N-Arg). After density arrest in G0, nuclear extracts were prepared. DNA affinity purification was performed with biotinylated cyclin B2 wild-type and CDE/CHR mutant probes. Precipitated proteins were separated by SDS–PAGE, digested with trypsin and analyzed by mass spectrometry.
Nine proteins were enriched more than 4-fold at the wild-type promoter when compared with the mutant probe (Supplementary Table S1). Interestingly, seven of these nine proteins are members of the DREAM complex which is known to be involved in the regulation of several cell cycle-regulated genes. In G0, the mammalian DREAM complex consists of E2F4, DP1, p130/p107 and the MuvB core proteins LIN9, LIN37, LIN52, LIN54 and RBBP4 (19). Six out of that group of proteins were observed to be enriched at the wild-type probe more than 7-fold (Table 2). Rbbp4 was found to bind to both the wild-type and the mutant probe. Interestingly, Rbbp4 enrichment at the wild-type promoter was only ∼2-fold and thereby considerably less than the other DREAM components. Tryptic peptides of Lin52 and Dp1 could not be detected in the screen (Supplementary Table S2). Thus, we show that the DREAM complex, that was shown to repress cell cycle genes in G0/G1 (19), binds to the CDE/CHR element of the mouse cyclin B2 promoter in G0.
Table 2.
Identification of components of the DREAM complex binding to the CDE/CHR element in the mouse cyclin B2 promoter
| Protein | Unique peptides sequenced | Ratio wt/mut |
|---|---|---|
| Lin9 | 5 | 27.3 |
| p130 | 9 | 21.9 |
| Lin37 | 2 | 16.4 |
| Lin54 | 7 | 12.2 |
| E2f4 | 6 | 10.6 |
| p107 | 4 | 7.2 |
After DNA affinity purification of proteins from stable isotope-labeled nuclear extracts, relative protein abundance was measured by mass spectrometry. Proteins are enriched at the wild-type mouse cyclin B2 promoter in comparison to a promoter harboring a CDE/CHR double mutation. The enrichment factor of proteins binding to the wild-type probe is shown as well as the number of unique peptide sequences.
The CHR is essential for recruiting the DREAM complex to the cyclin B2 promoter in vitro and in vivo
After applying a DNA probe mutated in both the CDE and CHR sites for identification of DREAM binding to the mouse cyclin B2 gene, we were wondering if both elements are required for the DREAM complex to associate with the cyclin B2 promoter. Since E2F4 is an essential component of the complex and the DNA sequence of the CDE is similar to a canonical E2F-binding site, one would assume that E2F4 binds to the CDE. However, it was shown that the human in contrast to the mouse cyclin B2 promoter does not hold a CDE next to the CHR (17,18). Therefore, comparing protein binding to the mouse and human cyclin B2 promoters should yield individual contributions of the CDE and the CHR to DREAM binding.
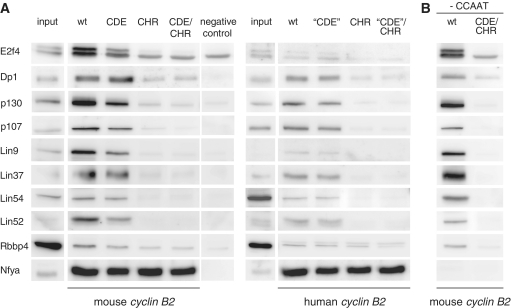
We performed DNA affinity purification with biotinylated human and mouse wild-type and mutated (CDE, CHR and CDE/CHR) cyclin B2-promoter probes (Table 1) from nuclear extracts of density-arrested NIH3T3 cells followed by western blot analysis. A fragment of the mouse Gapdhs promoter served as a negative control. Using antibodies against DREAM proteins, we observed that all complex components were enriched at both the human and mouse wild-type cyclin B2 promoters (Figure 2A). Employing this DNA pull-down western approach, we detected E2f4, p107, p130, Lin9, Lin37, Lin54 and Rbbp4 proteins that we had previously identified by mass spectrometry. Furthermore, DREAM complex components Lin52 and Dp1, which were not observed in the SILAC experiment, also bound to the mouse and human cyclin B2 promoters. Thus, we could identify all known proteins of the mammalian DREAM complex bound to the wild-type human and mouse cyclin B2 promoters.
Figure 2.
In vitro analysis of DREAM protein binding to cyclin B2 wild-type or mutant promoters. Nuclear extracts of density-arrested NIH3T3 cells were employed for DNA affinity purification using biotinylated human or mouse cyclin B2 promoters. (A) Binding was tested to wild-type, CDE, CHR or CDE/CHR mutant DNA probes by western blot analysis. As a negative control, a fragment of the Gapdhs promoter was used. As a protein binding to all cyclin B2 probes containing CCAAT-boxes, Nfya was detected. (B) Binding of the DREAM complex to the cyclin B2 promoters is independent of CCAAT-boxes. A truncated fragment of the mouse cyclin B2 promoter ending upstream of the CDE lacking the CCAAT-boxes was assayed for DREAM binding in comparison to a probe of the same length but with mutated CDE and CHR elements.
Given the ability of the DREAM complex to bind to the mouse and human cyclin B2 probes, we tested the contribution of CDE and CHR sites to DREAM binding relative to each other and to the wild-type probes. Although the human cyclin B2 promoter does not contain a functional CDE as shown by sequence comparisons and mutation analyses (17,18), the DREAM complex can nevertheless bind to it. We observed that the DREAM components are enriched to a higher degree on the mouse cyclin B2 probe than on the human probe when compared with input (Figure 2A). To rule out that proteins from mouse cell extracts preferentially bind to the mouse compared with the human probe, we looked for binding of DREAM complex components to human and mouse cyclin B2 promoters in density-arrested human foreskin fibroblasts. Also human DREAM proteins bound more avidly to the mouse probe than to the human promoter (data not shown).
When probes of the mouse cyclin B2 promoter with a mutated CDE were used, a slight reduction of DREAM protein binding could be observed. In contrast, binding of DREAM proteins to the probe ‘CDE’ was essentially the same as that to the wild-type promoter (Figure 2A). The probe ‘CDE’ was created on the basis of human cyclin B2 by mutating the nucleotides corresponding to the position of the CDE in the mouse promoter.
Interestingly, mutation of the CHR in both human and mouse probes led to a loss of binding of DREAM complex components to the cyclin B2 promoters. Double-mutation of both CDE and CHR elements did not show a further reduction in binding of all proteins analyzed. Only Rbbp4 was still binding to the probes with mutated CHR to almost the same degree as to the wild-type probe, which is in agreement with the data from the MS/MS analyses (Supplementary Table S2). Binding of the nuclear transcription factor-Y subunit alpha (Nfya), that had been shown to bind to CCAAT-boxes of the cyclin B2 promoter (31), is independent of CDE or CHR mutations and served here as a positive control for equal pull-down efficiency (Figure 2A). Taken together, these data demonstrate that the CHR is essential for binding of the DREAM complex to the cyclin B2 promoters. The CDE in the mouse promoter enhances binding of the DREAM complex, but is not indispensable for interaction of the complex with the DNA. Consistently, after mutation of the CDE in the cyclin B2 promoter slightly reduced protein binding was observed (Figure 2A).
In addition to the in vitro DREAM binding as assayed by pull-down western approach, we tested in vivo binding of the DREAM proteins to the CDE and CHR elements by chromatin immunoprecipitation (ChIP). To this end, we stably transfected NIH3T3 cells with luciferase reporter plasmids containing the mCcnb2-short and hCCNB2-short promoter fragments as wild-type, CDE, CHR or CDE/CHR mutants (Table 1). These constructs contained the same DNA sequences that we had used for probes in the pull-down assays. Nuclear extracts were collected from cells that were density arrested and ChIPs were performed for E2f4, p130 and Lin9 binding to the stably transfected promoters. For all three proteins, maximal binding appeared at both wild-type promoters (Figure 3). When the CDE in the mouse cyclin B2 promoter and the corresponding region in the human promoter were mutated, a slight decrease of E2f4, p130 and Lin9 binding was observed. In contrast, binding of all proteins to both promoters was reduced to background level when the CHR was mutated. In vivo binding was not reduced further with the CDE/CHR double mutation in the promoters (Figure 3). Interestingly, the ChIP signals at the mouse wild-type promoter appeared to be stronger in comparison to the human promoter, consistent with the higher affinity of DREAM proteins to the mouse compared with the human promoter in the pull-down western experiments (Figures 2 and 3).
Figure 3.
Chromatin immunoprecipitation analysis of DREAM proteins binding to (A) mouse or (B) human cyclin B2 promoters in vivo. NIH3T3 cells were stably transfected with wild-type, CDE, CHR or CDE/CHR mutant promoter constructs. Nuclear extracts of density-arrested cells were prepared and ChIPs were performed with antibodies targeting E2f4, p130 and Lin9. As a negative control, a non-targeting rabbit antibody was used. All signals are given relative to the input.
Therefore, regardless of the experimental approach, the data strongly suggest that the DREAM complex is binding to the cyclin B2 promoter primarily via the CHR. A CDE can support the binding, but is not essential for the interaction of DREAM proteins with the DNA.
The CHR element participates in transcriptional activation and repression of cyclin B2 promoters
With the DREAM complex binding to cyclin B2 promoters mostly through the CHR, we wished to have a more precise look at the function of this site in regulating promoter activity. In two earlier reports, we had identified the CHR as the dominant element for cell cycle-dependent repression in cyclin B2 promoters (17,30). However, we did not search for a contribution to transcriptional activation. Additionally, in experiments published earlier, the promoter constructs increased activity later in the cell cycle even when the CHR was mutated (17,30). As an important technical note, we found that pGL3 luciferase reporter vectors were responsible for a good part of the regulation that could not be attributed to a particular promoter element. Another change to earlier experiments was that we have employed the hCCNB2-short and mCcnb2-short fragments for DNA affinity purification and ChIP experiments. These promoter fragments were much shorter than the segments employed for the original cyclin B2 reporter assays (17,30). Since CCAAT-boxes and CDE/CHR elements are present in the short constructs, we assumed that these promoter fragments are able to mediate cell cycle-dependent regulation in the same manner as the long variants. However, experimental validation of this assumption was still necessary. To address these questions, we cloned both the long and short fragments of the mouse and human promoters into pGL4 series plasmids, which are largely devoid of cryptic transcription factor binding sites that were masking the real promoter activity in their predecessors. All four wild-type and mutant promoter–reporter constructs were assayed in serum-starved cells following their activity after serum stimulation throughout the subsequent cell cycle (Figure 4).
Figure 4.
Reporter activities of human and mouse cyclin B2 wild-type and mutant promoters. NIH3T3 cells were transfected with the wild-type (wt) and mutant (CDE, CHR, CDE/CHR) promoter-luciferase reporter constructs (A) mouse Ccnb2 long, (B) mouse Ccnb2 short, (C) human CCNB2 long and (D) human CCNB2 short. Cells were arrested in G0 by serum starvation and stimulated to re-enter the cell cycle by addition of FCS to the media. Cells were collected every 3 h and luciferase activity was measured. (E) To determine cell cycle distribution of cell populations at the different time points, the DNA in the cells was stained with Hoechst 33342 and fluorescence was measured by flow cytometry.
Expression from wild-type human and mouse cyclin B2 promoters was found at background level in G0 cells (Figure 4). Transcription was activated at the beginning of S phase at around 15 h after serum restimulation. Expression of the reporters reached a maximum after 27 h in G2/M. Importantly, the expression patterns from wild-type promoters reflect cyclin B2 mRNA expression from endogenous genes (Supplementary Figure S1). Regulation during the cell cycle of long and short promoter constructs was nearly identical, indicating that short cyclin B2 promoter fragments including merely CCAAT-boxes and CDE/CHR elements are sufficient for cell cycle-dependent expression. Cell cycle-dependent activity of the short wild-type mouse cyclin B2 promoter activity increased 30-fold when expression of the peak in G2/M was compared with G0. The increase in activity between G0 and G2/M phase was found to be higher in the mouse (mCcnb2 short, 30-fold; mCcnb2 long, 42-fold) than in the human promoters (hCCNB2 short, 12-fold; hCCNB2 long, 13-fold).
We compared the activity of the cyclin B2 promoters with mutations in the CDE, CHR or both elements. Mutation of the CDE sites in the mouse cyclin B2 promoter constructs resulted in a loss of repression of ∼2-fold for Ccnb2 long and 3-fold for Ccnb2 short in G0 and early G1 phase. In contrast, mutation of the corresponding ‘CDE’ region in the human promoter constructs essentially did not alter the activity of the promoters in the early cell cycle phases. These differences in the role of the CDE region in mouse and human cyclin B2 promoters are consistent with published observations (17,30). Mutation of the CDE regions in all four promoters did not change timing of increase in activity beginning at 15 h. In contrast, the reporters with CHR mutations or CDE/CHR double mutations lost cell cycle-dependent regulation (Figure 4). Higher activities were observed in G0 and G1. Furthermore, promoter activity of the CHR and double mutants fluctuated <2-fold during all phases of the cell cycle. Interestingly, no substantial increase of promoter activity occurred in S, G2 or M phases beyond the activity already reached in G0, leaving the activity of CHR mutant promoters in G2/M consistently below the maximum activity reached for the corresponding wild-type constructs. Interestingly, this suggests that about a 3-fold activation is contributed by CHR elements and the factors binding to them. Taken together, the luciferase reporter assays revealed that the CHR is a central element necessary for repression as well as activation of human and mouse cyclin B2 promoters, whereas the CDE in the mouse gene is only of minor importance for repression and the human promoter does not contain a functional CDE.
Sequence and position of CHR and CCAAT-box elements in cyclin B2 promoters are phylogenetically well conserved
We observed that the minimal cyclin B2 promoter with lengths of 210 bp (mouse) or 263 bp (human) gave the same regulation as much longer promoter fragments (Figure 4) similar to the mRNA expression from the endogenous cyclin B2 gene (Supplementary Figure S1). Thus, the minimal promoter should hold all relevant transcription factor-binding sites. We compared nucleotide sequences of cyclin B2 genes from 26 mammalian species just upstream from their translational start sites using the UCSC Genome Browser (32). Only four elements were identified as perfectly conserved in their nucleotide sequence in all cyclin B2 promoters compared: the three CCAAT-boxes and the CHR. Interestingly, the sequence of the mouse CDE was not observed in many of the other mammalian genes (Supplementary Figure S2). This lack of phylogenetical conservation of the CDE and the non-existence of canonical E2F-sites support the conclusion that cell cycle-dependent regulation and binding of the DREAM complex can occur exclusively through a CHR.
The CCAAT-boxes had already been shown to be necessary for basal activity of the promoter (17,31,33). Also the distance of ∼33 bp between two neighboring CCAAT-boxes and the distance of the CCAAT-boxes to the CHR is highly conserved (Supplementary Figure S2). In addition, no other highly conserved elements could be identified upstream of the 200 bp minimal promoter in a region of 800 nt upstream of the translational start site. In general, the data suggest that transcription of cyclin B2 is mainly controlled by three CCAAT-boxes and the CHR.
CCAAT-boxes are not required for binding of the DREAM complex to the CHR
Three CCAAT-boxes and the CCAAT-binding factors Nfya, Nfyb and Nfyc have been implicated in the regulation of the cyclin B2 promoter before (17,31). In addition, CCAAT-boxes are the only well-conserved elements besides the CHR. Therefore, we tested if CCAAT-boxes are necessary for binding of DREAM. To address this question, we created biotinylated probes of the mouse cyclin B2 promoter with a length of 130 nt ending just upstream of the CDE element. These probes included the CDE/CHR element but not the CCAAT-boxes (Ccnb2–CCAAT, Supplementary Figure S2). Furthermore, we amplified another probe of the same length, but with mutated CDE and CHR elements. Using these probes for pull-down western experiments with nuclear extracts of density-arrested NIH3T3 cells, we were able to compare protein binding to probes with or without CCAAT-boxes. Binding of Nfya is lost in the probe lacking CCAAT-boxes. However, all the analyzed DREAM proteins are still bound to the mouse cyclin B2 promoter even in the absence of CCAAT-boxes (Figure 2B). This experiment indicates that there is no direct cooperation between the CHR element and the CCAAT-boxes in recruiting the DREAM complex. The CHR is the only phylogenetically well-conserved element that is essential for binding of DREAM.
The CHR in the cyclin B2 promoter is required for binding of the B-myb-containing MMB complex
It was shown earlier that the composition of the DREAM complex changes during the cell cycle. In early cell cycle phases, the complex includes E2F4/DP1 together with p130/p107. This complex seems to have repressing functions. Later in the cell cycle, a shift in composition from E2F4/DP1 and p130/p107 to B-MYB is observed. Together with the MuvB core proteins, B-MYB forms the MMB (MYB-MuvB) complex that participates in the activation of various genes (19–21,34,35). B-MYB is essential for activation of many genes expressed in S, G2 and M phases (10,22,35). Since we were able to show that the CHR participates in repression as well as in activation of cyclin B2, we were wondering if MMB can bind to the CHR. We performed pull-down assays applying the same probes as used for the purification of the repressing DREAM complex. However, we employed extracts from proliferating NIH3T3 cells instead of density-arrested cells since B-myb is not expressed (36,37) and the MMB complex cannot be detected in G0 cells (19). Western-blot analyses showed that B-myb binds to mouse and human cyclin B2 promoters with similar affinity (Figure 5A). Mutation of the CHR leads to a loss of B-myb binding to both promoters. Mutation of the CDE region in both genes does not affect binding of B-myb. Apparently, binding of B-myb to cyclin B2 promoters is independent of CDE sites, but dependent on CHR elements. Since in a lysate from proliferating cells both the DREAM and MMB complexes are present, it is not possible to discriminate if detected proteins of the MuvB core are part of DREAM or MMB. To overcome this problem, we employed F9 embryonal carcinoma cells derived from a mouse testicular teratocarcinoma. F9 cells were shown not to form the DREAM complex in any cell cycle phase, but to carry the MMB complex (34). Thus, in these cells DREAM-dependent repression of promoters through CHR elements should not be possible. We then performed DNA affinity purification with nuclear extracts of F9 cells to find out if B-myb binds to the cyclin B2 promoters together with other components of the MMB complex. Indeed, proteins of the MuvB core bind to the wild-type cyclin B2 probes as well as to the probes with mutated CDE (Figure 5B). Consistent with the notion that F9 cells lack DREAM, E2f4 and p130 were clearly detectable in the nuclear extracts, but did not bind to any of the DNA probes which again proves that DREAM does not form in F9 cells and that protein binding to the DNA probes in our assay is highly specific. To evaluate if binding of MMB to the CHR is indeed necessary for the maximal activity of the cyclin B2 promoters, we transfected NIH3T3 cells with wild-type cyclin B2 luciferase reporters or constructs with mutated CHR elements together with two different plasmids expressing shRNAs targeting B-myb. The activity of the wild-type cyclin B2 promoters is clearly reduced when B-myb is knocked down relative to a control that was cotransfected with a non-targeting GFP-shRNA construct. This effect was completely abolished when the CHR was mutated (Figure 5C). Taken together, these experiments demonstrate that the MMB complex binds to the CHR independently of a CDE and that this interaction is necessary for full activation of the cyclin B2 promoter.
Figure 5.
The MMB complex binds and activates the cyclin B2 promoters through the CHR. (A) Nuclear extracts of proliferating NIH3T3 cells were employed for DNA affinity purification with biotinylated DNA probes of the human and mouse cyclin B2 promoters. Protein binding to wild-type, CDE, CHR or CDE/CHR mutant promoter fragments was tested by western blot analysis. As a protein binding to all cyclin B2 probes containing CCAAT-boxes, Nfya was detected. As a negative control, a fragment of the Gapdhs promoter was used. (B) Binding of the MMB complex to the mouse and human cyclin B2 DNA probes was assayed with DNA affinity purification of proteins derived from F9 cell nuclear extracts followed by western blot. Note that the DREAM complex components E2f4 and p130 do not bind to the probes. (C) To determine the effect of B-myb knockdown on the activity of mouse and human cyclin B2 promoters, NIH3T3 cells were transfected with the reporter constructs mouse Ccnb2 short and human CCNB2 short (wild-type or CHR mutant) together with vectors expressing shRNAs targeting B-myb (sh-Bmyb 1, sh-Bmyb 2). A shRNA construct expressing a non-targeting GFP-shRNA served as a negative control (sh-GFP). Luciferase activities were measured 48 h after transfection.
Promoters binding the DREAM complex often contain CHR elements that are conserved in vertebrate genomes
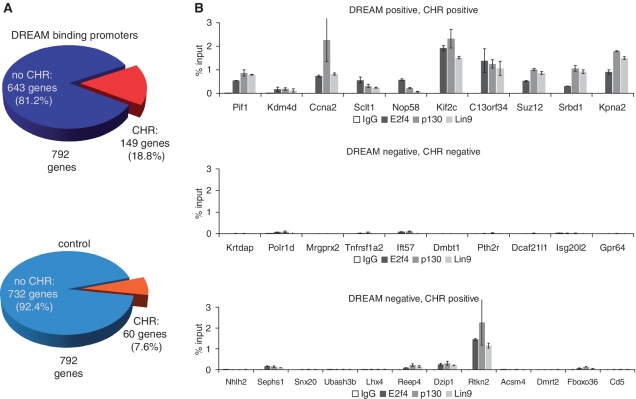
The data on cyclin B2 regulation suggest a central role for the CHR element in cell cycle-dependent transcription. To date, roughly a dozen promoters have been experimentally validated to be regulated through CHRs or CDE/CHR tandem sites (4). We wondered if DREAM binding to CHR promoters is limited to the cyclin B2 gene or if it is observed in a larger number of promoters. The most commonly reported CHR corresponds to the consensus TTTGAA. Another functional CHR with the sequence TTTAAA was identified in the cyclin B1 promoter. The mouse B-myb promoter possesses a CHR with the sequence TAGGAA and the human cyclin A2 gene contains a functional CHR with the sequence CTTGAA [reviewed in (4)]. Using these four verified CHR sites as a basis, we searched promoters described to bind DREAM components. To this end, we employed a dataset of DNA fragments that were shown by ChIP-chip assays to bind DREAM proteins in resting cells (19). The dataset comprised 817 DNA fragments observed to bind E2F4, p130 and LIN9 in G0. We searched these ChIP-chip fragments for transcription start sites identifying promoters of 792 genes annotated in the HGNC database. We searched for subgroups of these promoters, which possess evolutionary conserved CHR sites located in a region of 200 bp upstream or downstream from transcriptional start sites in the human genome. To qualify as a conserved CHR, a site had to have a >95% conservation score for five out of six nucleotides and the sixth score had to be >85% when the human sequence was compared with 16 other vertebrate genomes. We identified 149 genes matching all requirements (Supplementary Table S2) which accounts for 18.8% of all DREAM-binding promoters (Figure 6A). Interestingly, 76% of the identified genes contain conserved CHR elements with the sequence TTTGAA (or inverse TTCAAA). A similar percentage of all experimentally validated CHRs have the same nucleotide sequence (4). This indicates that the CHRs with the sequence TTTGAA are likely the most common sites among the four used for the search.
Figure 6.
In silico identification of CHR elements in promoters of genes bound by components of the DREAM complex. (A) The 792 promoters have been identified to bind the DREAM complex in ChIP-Seq assays (19). 149 of these genes (18.8%) possess CHR elements that are located in a distance of 200 bp relative to the transcription start site and that are phylogenetically conserved in vertebrates. As a control, a set of 792 randomly selected genes was assayed using the same parameters. In this control group, 60 conserved CHR elements were identified (7.6%). Using the Fisher's exact test, the overlap between DREAM-binding genes and CHR-containing genes appears to be highly significant (P = 4.8 × 10−11). (B) Randomly selected genes from subgroups of identified genes were further tested by chromatin immunoprecipitation. Nuclear extracts from density-arrested NIH3T3 cells were prepared and ChIPs were performed with antibodies targeting E2f4, p130 and Lin9. As a negative control, a non-targeting rabbit antibody was used (IgG). All signals are given relative to the input.
As a negative control, we screened a set of randomly selected genes for CHRs following the same protocol. Only 60 out of 792 genes were found to hold conserved CHR elements (Figure 6A). Using the Fisher's exact test, the overlap between DREAM-binding genes and CHR-containing genes appears to be highly significant (P = 4.8 × 10−11). To further substantiate these findings, we performed ChIP analyses with antibodies targeting E2f4, p130 and Lin9 using extracts from density-arrested NIH3T3 cells. We randomly selected genes from the following subgroups: (i) genes that bind DREAM and were identified to possess conserved CHR elements (DREAM positive, CHR positive); (ii) genes from the control group that have not been identified to bind DREAM and which do not contain CHRs (DREAM negative, CHR negative); and (iii) genes from the control group that have not been identified to bind DREAM but possess evolutionary conserved CHR elements (DREAM negative, CHR positive). For the genes with CHR elements, we designed primers for the amplification of DNA fragments that completely overlap with the CHR regions. A possible binding of DREAM components to CHR-negative promoters was tested with primers for the amplification of fragments that overlap with the transcription start. As predicted, we show binding of E2f4, p130 and Lin9 to all selected promoters from the subgroup of DREAM-positive and CHR-positive genes and no binding of these proteins to the genes of the control group lacking a CHR element (Figure 6B). One even more fascinating question was whether one would detect DREAM proteins at the promoters that have not been identified in the original screen to bind DREAM, but possess conserved CHRs. In fact, while 8 out of the 12 randomly selected genes were negative for binding of DREAM components, one (Rtkn2) was clearly positive for E2f4, p130 and Lin9 binding. Furthermore, a weak but detectable binding of these DREAM proteins to other three genes (Sephs1, Reep4, Dzip1) could be shown as well (Figure 6B). Therefore, the number of promoters that bind DREAM proteins and that possess evolutionary conserved CHR elements will be even higher than the 149 genes we have identified in our screen with the employed parameters.
Taken together, these observations suggest that the CHR is a highly conserved element in many promoters that are regulated by the DREAM complex.
An inverse CHR in the Ube2c promoter mediates cell cycle-dependent transcription and binding of the DREAM and MMB complexes
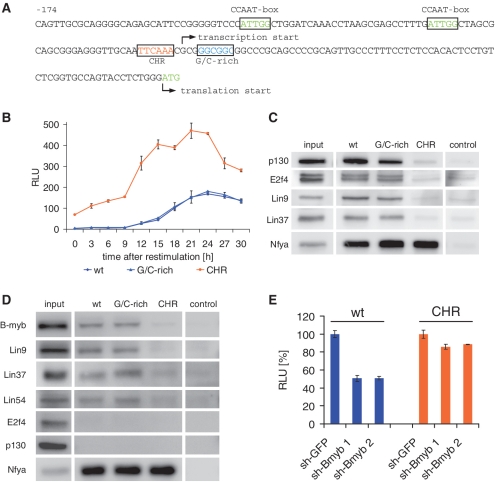
One of the genes that were identified in the screen of DREAM-binding promoters to contain a CHR element is the ubiquitin-conjugating enzyme Ube2c gene (Supplementary Table S2). Ube2c is an important regulator of the cell cycle. It is required for the destruction of mitotic cyclins and for exit from mitosis (38,39). Like cyclin B2, expression of Ube2c is regulated on the transcriptional level with a maximal expression in G2/M (Supplementary Figure S1) (40,41). The sequence of the CHR in the Ube2c promoter follows the consensus TTTGAA. However, this element differs from previously tested CHRs in that its orientation is inverse (Figure 7A). To find out if such CHR elements predicted by the bioinformatic screen are also functional in cell cycle-dependent regulation and DREAM/MMB binding, we tested a fragment of the mouse Ube2c promoter (nucleotides −478 to −3 relative to the translation start) for transcriptional regulation in reporter assays. In serum-starved NIH3T3 cells, activity of the promoter is low and starts to increase at 12 h after serum restimulation reaching maximal expression 24 h after restimulation (45-fold activation) (Figure 7B). The activity of a construct with a mutated CHR is 19-fold higher than the wild-type reporter in the serum-starved cells (0 h time point) indicating a strong loss of repression for the mutant. However, in contrast to the cyclin B2 constructs with mutated CHRs that do not show any cell cycle-dependent regulation, the Ube2c CHR mutant is still activated ∼7-fold in a cell cycle-dependent manner. This change in activity could be caused by other activating transcription factors regulating the Ube2c promoter independently of the CHR. In contrast to the cyclin B2 promoter, several highly conserved regions in addition to the CHR may serve as potential binding sites for additional regulators (Supplementary Figure S3). We also searched for potential CDE sites close to the CHR. Upstream of the CHR, the nucleotide composition does not follow the CDE consensus. However, the region downstream of the CHR consists only of guanines and cytosines and displays a sequence similar to CDE sites. Mutation of this G/C-rich region did not alter cell cycle-dependent transcription (Figure 7B). Thus, the G/C-rich element cannot be marked as a CDE.
Figure 7.
Confirmation of a functional CHR element binding DREAM and MMB in the mouse Ube2c promoter predicted through bioinformatic analyses. (A) The inverse CHR is located close to the transcription start and in proximity to two inverse CCAAT-boxes. The region downstream of the CHR is rich in guanines and cytosines (blue, G/C-rich). (B) Activities of the wild-type Ube2c promoter, a CHR mutant and a promoter with mutated G/C-rich region were tested in luciferase reporter assays in serum-starved and restimulated NIH3T3. (C) Binding of the DREAM complex to Ube2c probes was assayed with DNA affinity purifications using nuclear extracts of density-arrested NIH3T3 cells followed by western blot. (D) The analogous experiment with nuclear extracts from proliferating F9 cells was performed to test binding of the MMB complex to the Ube2c promoter. (E) The effect of B-myb knockdown on Ube2c promoter reporter constructs was tested with luciferase assays in proliferating NIH3T3 cells. All experiments were essentially performed as shown for cyclin B2 (Figures 2, 4 and 5).
Binding of the DREAM and MMB components to the promoter fragment was shown by DNA affinity purification followed by western blot (Figure 7C and D). Matching with the functional data employing the cyclin B promoter, binding of DREAM and MMB proteins is disrupted when the CHR is mutated. Mutation of the G/C-rich downstream region does not affect binding of the DREAM and MMB complexes. Knockdown of B-myb leads to a 2-fold reduction of the Ube2c wild-type promoter activity and to minor changes in the activity of the CHR mutant (Figure 7E).
Taken together, these experiments constitute verification for the ability to identify functional CHR elements by a bioinformatic approach. Furthermore, we provide evidence that inverse CHR elements are functional and can bind the DREAM and MMB complexes.
DISCUSSION
Transcriptional regulation of genes that are differentially expressed during the cell cycle is a central mechanism of cell cycle control. The observation that some genes whose transcription is repressed in G0 and G1 but is activated in S, G2 and M phases are controlled by CDE and CHR elements was made more than a decade ago (5–7). However, protein binding to the elements and the general mechanism of CDE/CHR-dependent transcriptional regulation still remain to be elucidated.
Here, we provide evidence that protein complexes named DREAM and MMB bind to the promoter of human and mouse cyclin B2 genes as well as to the mouse Ube2c promoter. We find that DREAM is the long sought-after complex binding to the CHR in resting cells.
E2F- and pRB-related proteins had been described as components of a complex that was first identified in Drosophila and named dREAM (Drosophila RBF, dE2F2 and dMyb-interacting proteins) (42). Later, the ortholog complex, containing E2F4, DP1, p130 and p107 as essential components, has been identified as the mammalian DREAM complex. In addition to these proteins, this complex consists of RBBP4 and the MuvB-like LIN proteins LIN9, LIN37, LIN52 and LIN54 that form the MuvB-core of DREAM (19,20). By ChIP, components of human DREAM were shown to bind to many promoters of genes that are differentially expressed during the cell cycle (19,20). The DREAM complex binds to these promoters in G0 and early G1 and is necessary for repression of transcription. When a cell progresses through the cell cycle, E2F4/DP1 and p107/p130 appear to be released from the DREAM complex, which then incorporates B-MYB. As this complex consists of the MuvB-core and B-MYB, it is designated as the MMB (MYB-MuvB) complex that activates gene expression in S phase (19–21,34,35,43).
Here, we show that DREAM binding to cyclin B2 promoters in G0 is dependent on an intact CHR since mutation of this element results in a loss of DREAM association in vitro and in vivo. Interestingly, six of the nine nuclear proteins identified by mass spectrometry to be enriched >4-fold at the wild-type probe are components of DREAM. Of the remaining three proteins, Snd1, Cand1 and Cfl1, only Snd1 is a factor that has been implicated in transcriptional regulation (44–47). However, in pull-down western experiments we were unable to corroborate the observation that Snd1 binds to the CDE or CHR sites of the cyclin B2 promoter. Therefore, it seems likely that DREAM constitutes the main complex binding to the CHR of the cyclin B2 promoter in G0.
Interestingly, while mutation of the CDE/CHR tandem element resulted in dissociation of E2F4, p107, p130, Lin9, Lin37 and Lin54 from the cyclin B2 promoter (factors for differential binding from 7.2 to 27.3), the amount of bound Rbbp4 changed only to a small degree (factor 1.9) (Supplementary Table S1). A possible explanation for the lack of stoichiometric loss of binding is that Rbbp4 is not only part of the DREAM complex, but can also bind directly to histones (48) or is additionally a component of other chromatin remodeling complexes like the NuRD and Sin3 complexes (49,50). These complexes are able to bind either to transcription factors contacting the DNA or directly to methylated DNA. Many proteins that are part of both complexes like Mta1, Mta2, Mbd3, Chd4, Hdac1, Hdac3, Gatad2b, Sin3a and Sap18 could be identified by mass spectrometry to bind to the mouse cyclin B2 promoter (Supplementary Table S1). Also binding of these complex components to wild-type versus CDE/CHR mutant cyclin B2 probes only differed to a small extend. Since the basal cyclin B2 promoter is rich in GC sequences, it is likely that the NuRD complex including Rbbp4 is able to bind to the probes independent from the CDE/CHR. It remains open which function, in addition to its DREAM association, binding of Rbbp4 to complexes like NuRD and Sin3 may have in transcriptional regulation of the cyclin B2 promoter. Furthermore, it is not known what impact binding of NuRD and Sin3 complexes to the promoter has on cyclin B2 expression.
After finding that DREAM binds the cyclin B2 promoter preferentially through the CHR and not through the CDE as a potential E2F4-binding site, the question comes up which protein in this complex directly contacts the DNA. Recently, LIN54 was shown to bind to DNA in a sequence-specific manner (51). Two sites were proposed for LIN54 binding in the human CDC2 promoter. One of the binding elements overlaps with the CHR, whereas the other one is located further upstream in the promoter and is an overlap of a potential B-MYB-binding site with a CHR-like element. It was suggested that an E2F4/p130-containing DREAM complex binds to the CDE/CHR region in quiescent cells, whereas in S phase after the shift from E2F4/p130 to B-MYB, the then formed MMB complex would bind to the upstream B-MYB/CHR-like site (51). However, we tested the upstream CHR-like site in the human CDC2 promoter for function in reporter luciferase assays and found that mutation of this element did neither change cell cycle-dependent regulation nor general promoter activity significantly. Observations from the report show that LIN54 cannot easily be produced in recombinant form to be tested for in vitro binding in EMSAs (51). If binding of LIN54 to a non-functional CHR-like site is similar to its affinity to the established functional CHR, one may suggest awaiting the results from more experiments to either establish that LIN54 binds CHRs or identify some other DREAM component to establish the contact with the DNA in CDE/CHR-controlled promoters.
Another important issue related to DREAM binding is the relative contribution of CDE and CHR elements. The human cyclin B2 promoter does not have a functional CDE and DREAM binding is exclusively dependent on the CHR (Figures 2 and 3). Mutation of the CDE in the mouse cyclin B2 promoter led to a small deregulation of cell cycle-dependent transcriptional control caused by a 2- to 3-fold increase of activity over background in G0 and early G1 phase. In the same experiment, the wild-type mouse cyclin B2 promoter yielded a regulation of about 30- to 40-fold when activity is measured in G2/M relative to reporter activity in G0 and early G1 (Figure 4). Comparing DREAM binding of the CDE mutant with binding to the wild-type mouse promoter shows a slightly reduced protein binding to the mutant promoter (Figure 2A). In the case of the human cyclin B2 promoter with a lack of a functional CDE, mutation of the corresponding region does not influence promoter activity in G0/G1 and DREAM binding is also not altered in pull-down assays. Furthermore, the small functional contribution of the CDE in the mouse cyclin B2 gene matches well with the observation of the intact CDE supporting binding of the DREAM complex to the CHR. Taken together, the data from functional assays are consistent with DREAM-binding results.
Interestingly, cyclin B2 CHR mutant reporter activity does not reach the maximum in G2 seen with the wild-type constructs, which would be expected if cell cycle-dependent regulation would only be a result of repression early in the cell cycle. Although a substantial part of general promoter activity is likely due to NF-Y activating the genes through the CCAAT-boxes (4), our current observation leads to the conclusion that full activation of the promoter is not possible when the CHR is mutated (Figure 4). This loss of activation for the short mCcnb2 construct is ∼3-fold. Taken together, the data prove not only a strong repressive function for the CHR, but also a smaller but detectable role in the activation of cyclin B2 promoters. As we were able to show that DREAM can bind to the CHR without the necessity of the interaction of E2f4 with an E2f-binding site, we wondered if the MMB complex might bind to the same sequence without a direct interaction of B-myb with a Myb-binding site. This hypothesis would be consistent with the detected activation of the cyclin B2 promoters through the CHR.
Prompted by these indicators, we tested B-myb binding. B-myb was detected at the human and mouse cyclin B2 promoters in pull-down experiments using extracts from proliferating NIH3T3 cells (Figure 5A). In both promoters, binding depends only on a functional CHR. Mutation of the CDE region does not reduce binding of B-myb. Because the CHR is not related to canonical Myb-binding sites, it is likely that B-myb is recruited to the CHR by components of the MuvB-core. As it is not possible to discriminate if components of the MuvB-core are parts of DREAM or MMB, we made use of F9 cells that were shown not to possess the DREAM complex even in early G1 (34). Indeed, we were able to show that B-myb together with other MMB proteins binds to the CHR elements of cyclin B2 promoters independently of the CDE (Figure 5B). A reduction of activity of the wild-type promoters but not of promoters with mutated CHR elements after B-myb knockdown (Figure 5C) further support the hypothesis that MMB can activate cyclin B2 expression by binding to the CHR. It is highly likely that the effect is directly mediated by an interaction of MMB with the cyclin B2 promoter through the CHR and not by a shift in cell cycle phases after B-myb knockdown, because RNAi of B-myb provokes a partial arrest of cells in G2/M (52,34), which would lead to an increase and not to a reduction of cyclin B2 promoter activity due to the cell cycle shift. Consistent with these observations, it was shown by microarray analyses that a knockdown of the MMB component Lin9 in F9 cells also leads to a 2-fold reduction of cyclin B2 mRNA (34).
These results provide a first link between the CHR and activation of promoters in S, G2 and M phases. At first glance, activation through CHR sites in the later phases of the cell cycle stands in contrast to the original observation made by in vivo footprinting that CDE/CHR sites are not occupied by protein when the promoters get activated (6). However, clear-cut results by these footprints can only be obtained for the CDE sites carrying several guanines. CHRs mostly contain only one guanine or none at all. Interestingly, an adenine gave a weak signal while the only guanine in the cyclin A promoter did not yield a differential footprint during the cell cycle (6). Taken together with our observations, this may lead to a model by which CDE/CHR tandem elements are occupied in G0/G1 by DREAM, while the CDE is free of proteins and only the CHR is occupied by MMB when the genes become expressed.
Observations made analyzing the survivin gene hinted to a classical binding of B-myb to B-myb consensus sites. Knight et al. propose that four different Myb-binding sites are necessary for binding of B-myb and Lin9 to the survivin promoter. Mutation of each site resulted in loss of protein binding as well as a decrease in promoter activity (34). Schmit et al. have shown that several potential Myb-binding sites are not necessary for binding DREAM components from S phase extracts to the cdc2 promoter. Furthermore, they observed that the activating DREAM complex seems to bind a different site in the promoter than the repressing complex (51). One may speculate that in some promoters the DREAM complex is recruited to the DNA through E2F4 or B-myb to E2F/CDE sites or Myb-binding sites, respectively, whereas in a group of cyclin B2-like genes the CHR is sufficient to recruit DREAM through other components of the complex. This could explain DREAM recruitment to promoters that do not contain CHR elements (Figure 6). Taken together, it appears that different mechanisms mediate regulation of genes by the DREAM complex even if promoters share common features like CDEs, CHRs and CCAAT-boxes. In addition, other transcription factors like FoxM1, ZNF143, c-Met and c-Myb have been shown to participate at least in part in activating genes expressed in S, G2 and M phases (53–56). Activation by transcription factors that do not bind through the CHR could be an explanation why the Ube2c promoter with a mutated CHR still shows a moderate cell cycle-dependent regulation in synchronized NIH3T3 cells (Figure 7B). Indeed, it had been shown that transcription of the Ube2c gene is activated by the factors EWS/FLI1, cdc42, c-MYC and v-ABL (40). Our current model for CHR-dependent regulation is that transcriptional repression of CHR-containing promoters may be a general mechanism, whereas activation in S, G2 and M phases may be regulated by a complex interplay of different transcription factors that could vary for many subgroups of cell cycle-regulated genes. This model could also explain why most genes differ in their exact timing of expression during the cell cycle.
With a bioinformatic approach, we were able to identify a subgroup of DREAM complex-binding promoters containing phylogenetically conserved CHR elements close to the transcriptional start site. With 18.8% of genes conforming the selection criteria from the group of all DREAM complex-binding promoters, CHR-containing promoters are strongly enriched in comparison to the control group.
We used a very stringent approach to identify promoters containing CHR elements to reduce the number of false calls. Only four published CHR-sequences, namely TTTGAA, TTTAAA, TAGGAA and CTTGAA (6,57,58) were used as templates for the analyses. Interestingly, the CHR with the sequence TTTGAA, which is mostly found in functionally characterized CDE/CHR-regulated promoters to date, with its inverse counterpart TTCAAA are most frequently observed with about two-thirds of the hits among the four sequences used for the bioinformatic search. More importantly, all previously described experimentally validated CHR elements appear in the list of CHR-containing promoters (Supplementary Table S2). In addition, the predicted inverse CHR in the Ube2c promoter was shown to be necessary for cell cycle-dependent regulation and DREAM/MMB binding (Figure 7). This indicates that the parameters are adequate also for the identification of further previously unknown CHR elements. However, we cannot rule out that some of the newly identified CHR elements are not functional or are part of DNA elements binding other transcription factors than DREAM proteins. Furthermore, it is likely that there are additional CHR elements in promoters binding the DREAM complex that have not been identified in our screen since we used only four experimentally verified CHR sequences for the analysis. Recent observations indicate that additional functional CHRs with variant CHR sequences exist which would have to be included in a more complex search in the future (our unpublished data). Furthermore, we were able to show that promoters from the control group that have not been detected in the original screen as DREAM-binding proteins (19) indeed bind the DREAM complex. However, such genes like Reep4, Dzip1 and Rtkn2 were only identified among the genes that possess evolutionary conserved CHR sequences, which again suggests a correlation between DREAM-binding and CHR elements (Figure 6B). Taken together, there will be even more promoters binding DREAM that may contain CHRs than the genes we uncovered by the current analyses. Even though the bioinformatic analyses have not detected every functional CHR, the data strongly suggest that CHR elements play an important role in binding of the DREAM complex in many promoters.
In summary, we identify the DREAM complex to bind mainly through the CHR to mammalian cyclin B2 promoters in G0. In the human cyclin B2 promoter, binding of E2F4 to the E2F-related CDE site is not required for DREAM function. However, a functional CDE as observed in the mouse cyclin B2 promoter is able to enhance binding of DREAM proteins. The CHR element, and in association with it likely also the DREAM proteins, confers repression in G0 and participates in the activation of CDE/CHR promoters in later cell cycle phases. Binding of B-myb to the CHR of the cyclin B2 promoter could provide a link to the activation in S, G2 and M phases. Bioinformatic analyses suggest that many more genes binding the DREAM complex in their promoters possess phylogenetically conserved CHR elements. These findings indicate that the CHR is a central element in transcriptional regulation by the DREAM and MMB complexes.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online: Supplementary Tables 1–3, Supplementary Figures 1–3, Supplementary Methods.
FUNDING
Public Health Service grants (P01CA050661, RO1CA93804, and R01CA63113 to J.A.D.); DOD New Investigator Award (W81XWH-08-1-0048 to L.L.). Bundesministerium für Bildung und Forschung (BMBF) through grants by the Interdisciplinary Center for Clinical Research (IZKF) at the University of Leipzig and the Deutsche Forschungsgemeinschaft (DFG) through grants (SPP 314, EN 218/6-1 and 6-2 to K.E.). Funding for open access charge: Waived by Oxford University Press.
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
The authors are indebted to Carola Koschke and Andrea Rothe for expert technical assistance, Andreas Lösche and Kathrin Jäger at the IZKF Leipzig core unit for performing FACS analyses. We thank Roger Watson and Kenneth Boheler for the kind gifts of the B-myb antibody and B-myb short hairpin constructs, respectively. Author contributions: G.A.M. and K.E. conceived the experiments. M.S. and E.K. performed quantitative MS/MS for the SILAC experiment; M.S., E.K. and G.A.M. analyzed the data. M.P. performed the bioinformatic analyses. M.F. performed parts of the Ube2c experiments. G.A.M. and M.Q. performed all other experiments. L.L. and J.A.D. contributed essential reagents and discussed experiments. G.A.M., J.A.D. and K.E., including segments provided by E.K. and M.P., wrote the article. All authors read and approved the final article.
REFERENCES
- 1.DeGregori J. The genetics of the E2F family of transcription factors: shared functions and unique roles. Biochim. Biophys. Acta. 2002;1602:131–150. doi: 10.1016/s0304-419x(02)00051-3. [DOI] [PubMed] [Google Scholar]
- 2.Stevaux O, Dyson NJ. A revised picture of the E2F transcriptional network and RB function. Curr. Opin. Cell Biol. 2002;14:684–691. doi: 10.1016/s0955-0674(02)00388-5. [DOI] [PubMed] [Google Scholar]
- 3.Trimarchi JM, Lees JA. Sibling rivalry in the E2F family. Nat. Rev. Mol. Cell Biol. 2002;3:11–20. doi: 10.1038/nrm714. [DOI] [PubMed] [Google Scholar]
- 4.Müller GA, Engeland K. The central role of CDE/CHR promoter elements in the regulation of cell cycle-dependent gene transcription. FEBS J. 2010;277:877–893. doi: 10.1111/j.1742-4658.2009.07508.x. [DOI] [PubMed] [Google Scholar]
- 5.Lucibello FC, Truss M, Zwicker J, Ehlert F, Beato M, Müller R. Periodic cdc25C transcription is mediated by a novel cell cycle- regulated repressor element (CDE) EMBO J. 1995;14:132–142. doi: 10.1002/j.1460-2075.1995.tb06983.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zwicker J, Lucibello FC, Wolfraim LA, Gross C, Truss M, Engeland K, Müller R. Cell cycle regulation of the cyclin A, cdc25C and cdc2 genes is based on a common mechanism of transcriptional repression. EMBO J. 1995;14:4514–4522. doi: 10.1002/j.1460-2075.1995.tb00130.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tommasi S, Pfeifer GP. In vivo structure of the human cdc2 promoter: release of a p130-E2F-4 complex from sequences immediately upstream of the transcription initiation site coincides with induction of cdc2 expression. Mol. Cell. Biol. 1995;15:6901–6913. doi: 10.1128/mcb.15.12.6901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Schulze A, Zerfass K, Spitkovsky D, Middendorp S, Berges J, Helin K, Jansen-Durr P, Henglein B. Cell cycle regulation of the cyclin A gene promoter is mediated by a variant E2F site. Proc. Natl Acad. Sci. USA. 1995;92:11264–11268. doi: 10.1073/pnas.92.24.11264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kimura M, Uchida C, Takano Y, Kitagawa M, Okano Y. Cell cycle-dependent regulation of the human aurora B promoter. Biochem. Biophys. Res. Commun. 2004;316:930–936. doi: 10.1016/j.bbrc.2004.01.178. [DOI] [PubMed] [Google Scholar]
- 10.Zhu W, Giangrande PH, Nevins JR. E2Fs link the control of G1/S and G2/M transcription. EMBO J. 2004;23:4615–4626. doi: 10.1038/sj.emboj.7600459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Philips A, Chambeyron S, Lamb N, Vie A, Blanchard JM. CHF: a novel factor binding to cyclin A CHR corepressor element. Oncogene. 1999;18:6222–6232. doi: 10.1038/sj.onc.1203017. [DOI] [PubMed] [Google Scholar]
- 12.Liu N, Lucibello FC, Engeland K, Müller R. A new model of cell cycle-regulated transcription: repression of the cyclin A promoter by CDF-1 and anti-repression by E2F. Oncogene. 1998;16:2957–2963. doi: 10.1038/sj.onc.1201838. [DOI] [PubMed] [Google Scholar]
- 13.Liu N, Lucibello FC, Korner K, Wolfraim LA, Zwicker J, Müller R. CDF-1, a novel E2F-unrelated factor, interacts with cell cycle- regulated repressor elements in multiple promoters. Nucleic Acids Res. 1997;25:4915–4920. doi: 10.1093/nar/25.24.4915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Haugwitz U, Wasner M, Wiedmann M, Spiesbach K, Rother K, Mössner J, Engeland K. A single cell cycle genes homology region (CHR) controls cell cycle-dependent transcription of the cdc25C phosphatase gene and is able to cooperate with E2F or Sp1/3 sites. Nucleic Acids Res. 2002;30:1967–1976. doi: 10.1093/nar/30.9.1967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Katula KS, Wright KL, Paul H, Surman DR, Nuckolls FJ, Smith JW, Ting JP, Yates J, Cogswell JP. Cyclin-dependent kinase activation and S-phase induction of the cyclin B1 gene are linked through the CCAAT elements. Cell Growth Differ. 1997;8:811–820. [PubMed] [Google Scholar]
- 16.Wasner M, Tschöp K, Spiesbach K, Haugwitz U, Johne C, Mössner J, Mantovani R, Engeland K. Cyclin B1 transcription is enhanced by the p300 coactivator and regulated during the cell cycle by a CHR-dependent repression mechanism. FEBS Lett. 2003;536:66–70. doi: 10.1016/s0014-5793(03)00028-0. [DOI] [PubMed] [Google Scholar]
- 17.Wasner M, Haugwitz U, Reinhard W, Tschöp K, Spiesbach K, Lorenz J, Mössner J, Engeland K. Three CCAAT-boxes and a single cell cycle genes homology region (CHR) are the major regulating sites for transcription from the human cyclin B2 promoter. Gene. 2003;312:225–237. doi: 10.1016/s0378-1119(03)00618-8. [DOI] [PubMed] [Google Scholar]
- 18.Müller GA, Heissig F, Engeland K. Chimpanzee, orangutan, mouse, and human cell cycle promoters exempt CCAAT boxes and CHR elements from interspecies differences. Mol. Biol. Evol. 2007;24:814–826. doi: 10.1093/molbev/msl210. [DOI] [PubMed] [Google Scholar]
- 19.Litovchick L, Sadasivam S, Florens L, Zhu X, Swanson SK, Velmurugan S, Chen R, Washburn MP, Liu XS, DeCaprio JA. Evolutionarily conserved multisubunit RBL2/p130 and E2F4 protein complex represses human cell cycle-dependent genes in quiescence. Mol. Cell. 2007;26:539–551. doi: 10.1016/j.molcel.2007.04.015. [DOI] [PubMed] [Google Scholar]
- 20.Schmit F, Korenjak M, Mannefeld M, Schmitt K, Franke C, von EB, Gagrica S, Hanel F, Brehm A, Gaubatz S. LINC, a human complex that is related to pRB-containing complexes in invertebrates regulates the expression of G2/M genes. Cell Cycle. 2007;6:1903–1913. doi: 10.4161/cc.6.15.4512. [DOI] [PubMed] [Google Scholar]
- 21.Pilkinton M, Sandoval R, Colamonici OR. Mammalian Mip/LIN-9 interacts with either the p107, p130/E2F4 repressor complex or B-Myb in a cell cycle-phase-dependent context distinct from the Drosophila dREAM complex. Oncogene. 2007;26:7535–7543. doi: 10.1038/sj.onc.1210562. [DOI] [PubMed] [Google Scholar]
- 22.Tarasov KV, Tarasova YS, Tam WL, Riordon DR, Elliott ST, Kania G, Li J, Yamanaka S, Crider DG, Testa G, et al. B-MYB is essential for normal cell cycle progression and chromosomal stability of embryonic stem cells. PLoS One. 2008;3:e2478. doi: 10.1371/journal.pone.0002478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kirschner RD, Sänger K, Müller GA, Engeland K. Transcriptional activation of the tumor suppressor and differentiation gene S100A2 by a novel p63-binding site. Nucleic Acids Res. 2008;36:2969–2980. doi: 10.1093/nar/gkn132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tavner F, Frampton J, Watson RJ. Targeting an E2F site in the mouse genome prevents promoter silencing in quiescent and post-mitotic cells. Oncogene. 2007;26:2727–2735. doi: 10.1038/sj.onc.1210087. [DOI] [PubMed] [Google Scholar]
- 25.Lange S, Sylvester M, Schumann M, Freund C, Krause E. Identification of phosphorylation-dependent interaction partners of the adapter protein ADAP using quantitative mass spectrometry: SILAC vs (18)O-labeling. J. Proteome Res. 2010;9:4113–4122. doi: 10.1021/pr1003054. [DOI] [PubMed] [Google Scholar]
- 26.Cox J, Mann M. MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification. Nat. Biotechnol. 2008;26:1367–1372. doi: 10.1038/nbt.1511. [DOI] [PubMed] [Google Scholar]
- 27.Friedrich M, Böhlig L, Kirschner RD, Engeland K, Hauschildt S. Identification of two regulatory binding sites which confer myotube specific expression of the mono-ADP-ribosyltransferase ART1 gene. BMC. Mol. Biol. 2008;9:91. doi: 10.1186/1471-2199-9-91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kel AE, Gossling E, Reuter I, Cheremushkin E, Kel-Margoulis OV, Wingender E. MATCH: A tool for searching transcription factor binding sites in DNA sequences. Nucleic Acids Res. 2003;31:3576–3579. doi: 10.1093/nar/gkg585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Siepel A, Bejerano G, Pedersen JS, Hinrichs AS, Hou M, Rosenbloom K, Clawson H, Spieth J, Hillier LW, Richards S, et al. Evolutionarily conserved elements in vertebrate, insect, worm, and yeast genomes. Genome Res. 2005;15:1034–1050. doi: 10.1101/gr.3715005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lange-zu Dohna C, Brandeis M, Berr F, Mössner J, Engeland K. A CDE/CHR-tandem element regulates cell cycle-dependent repression of cyclin B2 transcription. FEBS Lett. 2000;484:77–81. doi: 10.1016/s0014-5793(00)02133-5. [DOI] [PubMed] [Google Scholar]
- 31.Bolognese F, Wasner M, Lange-zu Dohna C, Gurtner A, Ronchi A, Muller H, Manni I, Mössner J, Piaggio G, Mantovani R, et al. The cyclin B2 promoter depends on NF-Y, a trimer whose CCAAT- binding activity is cell-cycle regulated. Oncogene. 1999;18:1845–1853. doi: 10.1038/sj.onc.1202494. [DOI] [PubMed] [Google Scholar]
- 32.Kent WJ, Sugnet CW, Furey TS, Roskin KM, Pringle TH, Zahler AM, Haussler D. The human genome browser at UCSC. Genome Res. 2002;12:996–1006. doi: 10.1101/gr.229102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Salsi V, Caretti G, Wasner M, Reinhard W, Haugwitz U, Engeland K, Mantovani R. Interactions between p300 and multiple NF-Y trimers govern cyclin B2 promoter function. J. Biol. Chem. 2003;278:6642–6650. doi: 10.1074/jbc.M210065200. [DOI] [PubMed] [Google Scholar]
- 34.Knight AS, Notaridou M, Watson RJ. A Lin-9 complex is recruited by B-Myb to activate transcription of G2/M genes in undifferentiated embryonal carcinoma cells. Oncogene. 2009;28:1737–1747. doi: 10.1038/onc.2009.22. [DOI] [PubMed] [Google Scholar]
- 35.Osterloh L, von EB, Schmit F, Rein L, Hubner D, Samans B, Hauser S, Gaubatz S. The human synMuv-like protein LIN-9 is required for transcription of G2/M genes and for entry into mitosis. EMBO J. 2007;26:144–157. doi: 10.1038/sj.emboj.7601478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lam EW, Watson RJ. An E2F-binding site mediates cell-cycle regulated repression of mouse B- myb transcription. EMBO J. 1993;12:2705–2713. doi: 10.1002/j.1460-2075.1993.tb05932.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lam EW, Bennett JD, Watson RJ. Cell-cycle regulation of human B-myb transcription. Gene. 1995;160:277–281. doi: 10.1016/0378-1119(95)00184-8. [DOI] [PubMed] [Google Scholar]
- 38.Hershko A, Ganoth D, Sudakin V, Dahan A, Cohen LH, Luca FC, Ruderman JV, Eytan E. Components of a system that ligates cyclin to ubiquitin and their regulation by the protein kinase cdc2. J. Biol. Chem. 1994;269:4940–4946. [PubMed] [Google Scholar]
- 39.Townsley FM, Aristarkhov A, Beck S, Hershko A, Ruderman JV. Dominant-negative cyclin-selective ubiquitin carrier protein E2-C/UbcH10 blocks cells in metaphase. Proc. Natl Acad. Sci. USA. 1997;94:2362–2367. doi: 10.1073/pnas.94.6.2362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Arvand A, Bastians H, Welford SM, Thompson AD, Ruderman JV, Denny CT. EWS/FLI1 up regulates mE2-C, a cyclin-selective ubiquitin conjugating enzyme involved in cyclin B destruction. Oncogene. 1998;17:2039–2045. doi: 10.1038/sj.onc.1202129. [DOI] [PubMed] [Google Scholar]
- 41.Yamanaka A, Hatakeyama S, Kominami K, Kitagawa M, Matsumoto M, Nakayama K. Cell cycle-dependent expression of mammalian E2-C regulated by the anaphase-promoting complex/cyclosome. Mol. Biol. Cell. 2000;11:2821–2831. doi: 10.1091/mbc.11.8.2821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Korenjak M, Taylor-Harding B, Binne UK, Satterlee JS, Stevaux O, Aasland R, White-Cooper H, Dyson N, Brehm A. Native E2F/RBF complexes contain Myb-interacting proteins and repress transcription of developmentally controlled E2F target genes. Cell. 2004;119:181–193. doi: 10.1016/j.cell.2004.09.034. [DOI] [PubMed] [Google Scholar]
- 43.Litovchick L, Florens LA, Swanson SK, Washburn MP, DeCaprio JA. DYRK1A protein kinase promotes quiescence and senescence through DREAM complex assembly. Genes Dev. 2011;25:801–813. doi: 10.1101/gad.2034211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Tong X, Drapkin R, Yalamanchili R, Mosialos G, Kieff E. The Epstein-Barr virus nuclear protein 2 acidic domain forms a complex with a novel cellular coactivator that can interact with TFIIE. Mol. Cell Biol. 1995;15:4735–4744. doi: 10.1128/mcb.15.9.4735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Yang J, Aittomaki S, Pesu M, Carter K, Saarinen J, Kalkkinen N, Kieff E, Silvennoinen O. Identification of p100 as a coactivator for STAT6 that bridges STAT6 with RNA polymerase II. EMBO J. 2002;21:4950–4958. doi: 10.1093/emboj/cdf463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Solan NJ, Miyoshi H, Carmona EM, Bren GD, Paya CV. RelB cellular regulation and transcriptional activity are regulated by p100. J. Biol. Chem. 2002;277:1405–1418. doi: 10.1074/jbc.M109619200. [DOI] [PubMed] [Google Scholar]
- 47.Paukku K, Yang J, Silvennoinen O. Tudor and nuclease-like domains containing protein p100 function as coactivators for signal transducer and activator of transcription 5. Mol. Endocrinol. 2003;17:1805–1814. doi: 10.1210/me.2002-0256. [DOI] [PubMed] [Google Scholar]
- 48.Yoon HG, Chan DW, Huang ZQ, Li J, Fondell JD, Qin J, Wong J. Purification and functional characterization of the human N-CoR complex: the roles of HDAC3, TBL1 and TBLR1. EMBO J. 2003;22:1336–1346. doi: 10.1093/emboj/cdg120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zhang Y, LeRoy G, Seelig HP, Lane WS, Reinberg D. The dermatomyositis-specific autoantigen Mi2 is a component of a complex containing histone deacetylase and nucleosome remodeling activities. Cell. 1998;95:279–289. doi: 10.1016/s0092-8674(00)81758-4. [DOI] [PubMed] [Google Scholar]
- 50.Zhang Y, Ng HH, Erdjument-Bromage H, Tempst P, Bird A, Reinberg D. Analysis of the NuRD subunits reveals a histone deacetylase core complex and a connection with DNA methylation. Genes Dev. 1999;13:1924–1935. doi: 10.1101/gad.13.15.1924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Schmit F, Cremer S, Gaubatz S. LIN54 is an essential core subunit of the DREAM/LINC complex that binds to the cdc2 promoter in a sequence-specific manner. FEBS J. 2009;276:5703–5716. doi: 10.1111/j.1742-4658.2009.07261.x. [DOI] [PubMed] [Google Scholar]
- 52.Santilli G, Schwab R, Watson R, Ebert C, Aronow BJ, Sala A. Temperature-dependent modification and activation of B-MYB: implications for cell survival. J. Biol. Chem. 2005;280:15628–15634. doi: 10.1074/jbc.M411747200. [DOI] [PubMed] [Google Scholar]
- 53.Laoukili J, Kooistra MR, Bras A, Kauw J, Kerkhoven RM, Morrison A, Clevers H, Medema RH. FoxM1 is required for execution of the mitotic programme and chromosome stability. Nat. Cell Biol. 2005;7:126–136. doi: 10.1038/ncb1217. [DOI] [PubMed] [Google Scholar]
- 54.Izumi H, Wakasugi T, Shimajiri S, Tanimoto A, Sasaguri Y, Kashiwagi E, Yasuniwa Y, Akiyama M, Han B, Wu Y, et al. Role of ZNF143 in tumor growth through transcriptional regulation of DNA replication and cell-cycle-associated genes. Cancer Sci. 2010;101:2538–2545. doi: 10.1111/j.1349-7006.2010.01725.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Factor VM, Seo D, Ishikawa T, Kaposi-Novak P, Marquardt JU, Andersen JB, Conner EA, Thorgeirsson SS. Loss of c-Met disrupts gene expression program required for G2/M progression during liver regeneration in mice. PLoS One. 2010;5 doi: 10.1371/journal.pone.0012739. pii: e12739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Dang CV. c-Myc target genes involved in cell growth, apoptosis, and metabolism. Mol. Cell. Biol. 1999;19:1–11. doi: 10.1128/mcb.19.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Bennett JD, Farlie PG, Watson RJ. E2F binding is required but not sufficient for repression of B- myb transcription in quiescent fibroblasts. Oncogene. 1996;13:1073–1082. [PubMed] [Google Scholar]
- 58.Liu N, Lucibello FC, Zwicker J, Engeland K, Müller R. Cell cycle-regulated repression of B-myb transcription: cooperation of an E2F site with a contiguous corepressor element. Nucleic Acids Res. 1996;24:2905–2910. doi: 10.1093/nar/24.15.2905. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.