Abstract
The pluripotency factor Lin28 is a highly conserved protein comprising a unique combination of RNA-binding motifs, an N-terminal cold-shock domain and a C-terminal region containing two retroviral-type CCHC zinc-binding domains. An important function of Lin28 is to inhibit the biogenesis of the let-7 family of microRNAs through a direct interaction with let-7 precursors. Here, we systematically characterize the determinants of the interaction between Lin28 and pre-let-7g by investigating the effect of protein and RNA mutations on in vitro binding. We determine that Lin28 binds with high affinity to the extended loop of pre-let-7g and that its C-terminal domain contributes predominantly to the affinity of this interaction. We uncover remarkable similarities between this C-terminal domain and the NCp7 protein of HIV-1, not only in terms of primary structure but also in their modes of RNA binding. This NCp7-like domain of Lin28 recognizes a G-rich bulge within pre-let-7g, which is adjacent to one of the Dicer cleavage sites. We hypothesize that the NCp7-like domain initiates RNA binding and partially unfolds the RNA. This partial unfolding would then enable multiple copies of Lin28 to bind the extended loop of pre-let-7g and protect the RNA from cleavage by the pre-microRNA processing enzyme Dicer.
INTRODUCTION
MicroRNAs (miRNAs) are short single-stranded RNAs of ∼22 nt found in virus, plant and animal species that act as post-transcriptional regulators of mRNA expression [for recent reviews, see (1–4)]. They are generated from a longer RNA, the primary transcript (pri-miRNA), by a multi-step process. The pri-miRNA is first cleaved by the microprocessor complex containing the endonuclease Drosha and the double-stranded RNA-binding protein DGCR8 to produce a 60–70 nts RNA hairpin known as the precursor miRNA (pre-miRNA). After being exported to the cytoplasm, the pre-miRNA is further cleaved by the endonuclease Dicer to form a ∼22-nt dsRNA. The single-stranded mature miRNA is then loaded into the RNA-induced silencing complex to regulate its target mRNAs.
miRNAs play important roles in cell differentiation (5–7), and, in mammals, several miRNAs have been shown to act as oncogenes and tumor suppressors [reviewed in (8–13)]. Among those playing a role as tumor suppressors, the let-7 family of miRNAs have been extensively characterized, and are known inhibitors of oncogenes such as RAS, MYC, HMGA2, and cyclin D1 (10). The let-7 miRNAs are often present in multiple copies in a single genome, with the mature let-7 being highly conserved across species. In human and mouse, there are 10 mature let-7 family sequences (let-7a, let-7b, etc.) produced from 13 precursors.
Although levels of let-7 pri-miRNAs are controlled by transcription factors, post-transcriptional regulation is critical in determining the levels of mature let-7 miRNAs (14–18). Recent studies in embryonic cells have highlighted the importance of Lin28 in post-transcriptional regulation of the let-7 family of miRNAs, where it acts as a selective inhibitor of let-7 miRNAs maturation (19–21). The various members of the let-7 family are not affected to the same degree by Lin28, with let-7a, let-7d and let-7g being among the most affected. Several mechanisms have been proposed to explain the Lin28 inhibition of let-7 biogenesis. Lin28 was shown to interfere with the Drosha cleavage of pri-let-7 (16,19,21) and with the cleavage of pre-let-7 by Dicer (22,23). In addition, Lin28 was shown to induce the uridylylation of pre-let-7 by the recruitment of TUT4 (Zcchc11), which leads to its degradation (22,24–26). Although the relative importance of these mechanisms in vivo has not been clearly established (27), they all involve the formation of a complex between Lin28 and the immature forms of the let-7 miRNA.
Lin28 is a highly conserved protein of 209 amino acids known to be an important pluripotency factor (28), and its role in pluripotency is likely related to its function in let-7 biogenesis (19,29). Lin28 contains a unique set of RNA-binding motifs (30,31); an N-terminal cold shock domain (CSD) and a C-terminal region composed of two CCHC-type zinc-binding domains [ZBDs; (30)]. CSDs are found in several RNA- and DNA-binding proteins (32), whereas the CCHC-type ZBDs are most commonly found in retroviral nucleocapsid proteins, such as the NCp7 protein from HIV-1 (33). Although Lin28 has been shown to regulate the stability and translation of selected mRNAs (34–37), it plays a central role in regulating levels of mature let-7.
Several in vivo and in vitro studies have sought to characterize the interaction between pre-let-7 and Lin28 (19,20,23,24,38). It was demonstrated that both the CSD and the ZBDs of Lin28 are necessary for pre-let-7g binding in vitro and maturation inhibition in vivo (20). As determined by in vitro binding assays, Lin28 binds the extended terminal loop of pre-let-7g (20,38). Mutation of a conserved cytosine in this loop was shown to reduced its in vitro affinity for Lin28 (20). A G-rich sequence at the 5′-end of the pre-let-7g terminal loop was found to be strongly protected from ribonuclease cleavage by Lin28 (38). In addition, mutations of a few conserved nucleotides in the terminal loop make the immature miRNA resistant to Lin28 inhibition in P19 embryonal carcinona extract (19). Lin28 also binds the extended terminal loop of pre-let-7a-2, and the sequence composing the mature miRNA (let-7a) can compete with pre-let-7a-2 binding for Lin28 (23). Moreover, a four-nucleotide 5′-GGAG-3' sequence important for Lin28 binding and its uridylylation by Zcchc11 was identified at the 3′-end of the terminal loop region of pre-let-7a-1 (24). Although several studies have contributed to establish that the RNA-binding domains of Lin28 are important for recognition of the extended terminal loop of pre-let-7, the key determinants of this interaction have not been systematically defined.
In this work, we used electrophoretic mobility shift assay (EMSA), ribonuclease protection assay, in-line probing and NMR spectroscopy with purified molecules to map the interaction between the pre-let-7g RNA and the Lin28 protein from mouse. We determine that the C-terminal domain of Lin28 contributes predominantly to the high-affinity interaction with pre-let-7g and its sequence is very similar to the NCp7 protein of HIV-1. We also uncover several similarities in terms of RNA binding between NCp7 and the C-terminal domain of Lin28.
MATERIALS AND METHODS
Plasmids
Lin28 expression vectors are derived from pGEX4T (GE Healthcare) and were constructed using the Lin28 cDNA from Mus musculus (Open Biosystems BC068304). Vectors for RNA transcription were derived either from the pARiBo1 plasmid or the pRSA-VS plasmid (39). Mutant vectors were prepared using the Strategene QuikChangeII site-directed mutagenesis method or by standard cloning of restriction fragments. All plasmids created for this study were verified by DNA sequencing.
RNA preparation for biochemical characterization and NMR studies
Most RNAs used here were transcribed in vitro as ARiBo-tagged precursors and purified by batch affinity purification (39,40). In one case (TL-let-7g), the RNA was synthesized in vitro as a precursor with a VS ribozyme substrate at its 3′-end and purified as described previously (41). For biochemical characterization (gel-shift, footprinting and in-line probing), the RNAs were [5′-32P]-labeled and further purified by 20% denaturing gel electrophoresis (42). For NMR studies, the RNAs were concentrated and exchanged with an Amicon Ultra-15 3000 NMWL (Millipore) in NMR buffer (10 mM d18-HEPES at pH 6.4, 50 mM NaCl, 0.05 mM NaN3 and 10% D2O).
Protein expression and purification for biochemical characterization
Lin28 and related mutants were expressed in Escherichia coli strain BL21 cells (Stratagene). The bacterial cultures were grown in LB medium at 37°C and induced with 1 mM isopropyl-β-d-thiogalactopyranoside (IPTG) for 4 h at 30°C. The cells were harvested by centrifugation and resuspended in binding buffer [25 mM Tris, pH 8.0, 1 M NaCl, 0.1% NP-40 alternative (Calbiochem) and 1 mM DTT] supplemented with Complete EDTA-free protease inhibitor (Roche) and 10 U/ml of DNase I recombinant RNase-free (Roche). The cells were lysed by French press and centrifuged at 100 000g for 1 h at 4°C. The supernatant was incubated for 1 h at 4°C with GSH-Sepharose 4B resin (GE Healthcare). After incubation, the resin was washed three times with the binding buffer and three times with the S7 buffer (50 mM Tris, pH 8.0, 50 mM NaCl, 5 mM CaCl2 and 1 mM DTT). The washed resin was resuspended in S7 buffer and incubated overnight at room temperature with 5 U/ml of Nuclease S7 (Roche). The resin was subsequently washed three times with S7 buffer and incubated 1 h at room temperature with 100 U of thrombin (Calbiochem). The eluted protein was dialyzed 4 h at 4°C in 2 l of 20 mM Tris, pH 8.0, 2 M NaCl, 1 mM DTT and overnight at 4°C in 20 mM Tris, pH 8.0, 1 M urea, 200 mM NaCl and 1 mM DTT. The dialyzed protein was loaded on an SP-Sepharose high-performance column (GE Healthcare) equilibrated with FPLC-A (20 mM Tris, pH 8.0 and 1 mM DTT). The protein was eluted from the column using a gradient (from 0% to 100% over 525 mL) of FPLC-B (20 mM Tris, pH 8.0, 2 M NaCl and 1 mM DTT). The fractions containing the protein were combined, concentrated with an Amicon Ultra-15 3000 NMWL (Millipore) and dialyzed in storage buffer (100 mM Tris, pH 7.6, 100 mM NaCl, 20% glycerol and 2 mM DTT). The NCp7 protein was expressed and purified as described previously (43). All proteins purified for this study were verified by mass spectrometry.
Protein expression and purification for NMR studies
For NMR studies, uniform 15N- and 15N/13C labeling was obtained by growing the cells in minimal media containing 15N-labeled NH4Cl and 13C6-glucose as the sole sources of nitrogen and carbon, respectively. Protein purification was conducted as described above, but with the following modifications. The selected fractions from the SP-Sepharose column were dialyzed in 5% acetic acid, concentrated on a rotary evaporator and purified on a Vydac C4 reverse-phase HPLC column using an acetonitrile gradient (from 15% to 35% over 335 ml) in 0.05% TFA. After HPLC purification, the proteins were refolded in the presence of zinc, as described previously (44).
Electrophoretic mobility shift assay
For EMSA, the 32P-labeled RNA was first heated and snap cooled (heated 2 min at 95°C and snap-cooled on ice for 5 min) to promote hairpin formation. The protein samples were diluted in EMSA buffer (50 mM Tris, pH 7.6, 50 mM NaCl, 10% glycerol, 0.05% NP-40 alternative and 2 mM DTT) and their concentrations were adjusted to span from 0.02× to 50× of the estimated Kd. The binding reactions (20 ml) were initiated by mixing 1 pM of 32P-labeled RNA with the diluted proteins and incubated at 4°C for 30 min. For each Kd determination, ∼14 binding reactions were loaded directly on an 8% native polyacrylamide gel (37.5:1 polyacrylamide/bisacrylamide) and run in Tris–Glycine buffer (25 mM Tris–Base and 200 mM glycine) at 200 V for 2 h with active water cooling in the cold room. The gels were fixed in 50% methanol and 10% acetic acid for 1 h, washed 15 min in 30% ethanol, quickly rinsed with H2O and exposed overnight to a storage phosphor screen (Bio-Rad). The 32P-labeled RNA was visualized with a Bio-Rad Molecular Imager FX densitometer, and band intensities were quantified using the QuantityOne software (version 4.6.6 from Bio-Rad). The fraction of bound RNA was plotted against protein concentration, and the data were fitted to the one-site binding equation or to the Hill equation (only for the two cases in Table 1) by nonlinear regression analysis within the Origin 7 SR4 version 7.0552 software (OriginLab, MA, USA). For each protein–RNA complex, at least three independent Kd determination experiments were performed. The reported Kd's and their errors are, respectively, the average values and the standard deviations from these multiple experiments.
Table 1.
Dissociation constants (Kd in nM) of different domains of Lin28 for various pre-let-7g constructs
| RNA | Lin281–209 | Lin2839–112 | Lin28119–180 |
|---|---|---|---|
| Pre-let-7g | 0.15 ± 0.04 | 41 ± 10 | 0.6 ± 0.1 |
| n = 2.7 ± 0.5a | |||
| Duplex let-7g GNRA | 9 ± 3 | >200 | n.b.b |
| TL-let-7g | 0.13 ± 0.02 | 126 ± 41 | 1.3 ± 0.3 |
| n = 2.9 ± 0.7a |
aIn these cases, the Hill equation was used to derived the Kd values.
bNo specific binding observed. The gel mobility shift assays display smearing and multiple shifts, indicating non-specific binding.
In-line probing assay
In-line probing assays were performed as described previously (45). The 32P-labeled RNA was visualized with a Bio-Rad Molecular Imager FX densitometer, and band intensities were quantified using the Image Lab software (version 3.0 from Bio-Rad).
RNA footprinting assay
For RNase footprinting, the 32P-labeled RNA was first heated and snap cooled to promote hairpin formation. The Lin28119–180 protein was diluted at various concentrations in EMSA buffer. The protein was first incubated with 1 nM of 32P-labeled RNA (10 ml total volume) for 30 min at 4°C. Then, 1 U of T1 ribonuclease from Aspergillus oryzae (Sigma) was added and the incubation continued for 15 min at 4°C. The reaction was stopped by the addition of Precipitation/Inactivation buffer (Ambion), incubation for 15 min at −20°C and centrifugation at 16 000g for 15 min. The RNA pellet was dissolved in Gel loading buffer II (Ambion), loaded on a 20% polyacrylamide/7 M urea sequencing gel and run at 1900 V for 5 h. The sequencing gel was exposed 2 h to a storage phosphor screen (Bio-Rad). The 32P-labeled RNA bands were visualized and quantified as for the EMSA assay.
NMR Spectroscopy
For NMR studies, the following samples were prepared in NMR buffer: 1.0 mM 15N-labeled Lin28136–180; 1.3 mM 15N-labeled Lin28119–180; 1.1 mM 13C/15N-labeled Lin28119–180; 0.1 mM TL-let-7g:15N-labeled Lin28119–180; 0.1 mM Δbulge TL-let-7g:15N-labeled Lin28119–180; 0.1 mM G34AG35A TL-let-7g:15N-labeled Lin28119–180; 1.0 mM TL-let-7g:15N-labeled Lin28119–180; and 1.3 mM TL-let-7g:13C/15N-labeled Lin28119–180. For the TL-let-7g:Lin28119–180 complexes, the samples were prepared by titration of Lin28119–180 into a TL-let-7g sample. All NMR experiments were collected on Varian UnityINOVA 500 and 600 MHz spectrometers equipped with a pulse-field gradient unit and an actively shielded z gradient probe (either a room-temperature probe or a cryogenic probe). The backbone resonances (1H, 15N and 13C) of Lin28119–180 in the free and TL-let-7g-bound form were assigned using the following NMR experiments collected at 35°C: two-dimensional (2D) 1H-15N HSQC (46); three-dimensional (3D) HNCACB (47–49); and 3D (HB)CBCA(CO)NNH (48,49). 1H, 13C and 15N chemical shifts were referenced to an external standard of 2,2-dimethyl-2-silapentane-5-sulfonic acid (DSS) at 0.00 ppm (50). NMR data were processed with NMRPipe/NMRDraw (51) and analyzed with NMRView (52).
RESULTS
Lin28 recognizes the terminal loop of pre-let-7g with its C-terminal domain providing the most important energetic contribution
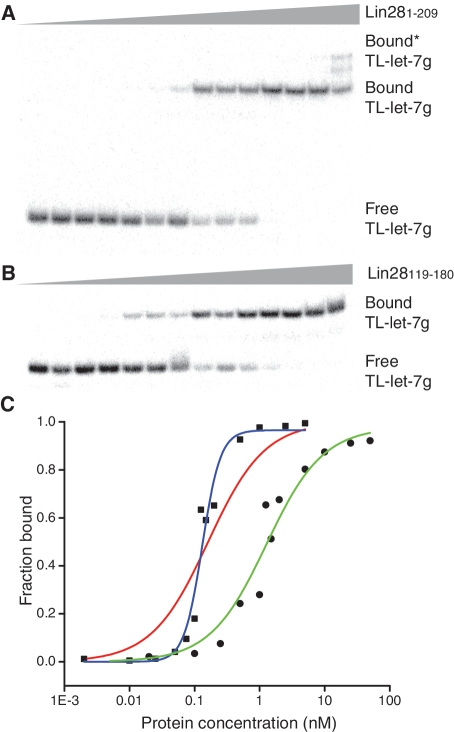
To identify the domain(s) of Lin28 important for binding the let-7g precursor miRNA (pre-let-7g), we used EMSAs with purified recombinant proteins and in vitro transcribed RNAs (Figure 1). It was previously established that Lin28 recognizes pre-let-7g and its terminal loop with a similar affinity [Kd of 1–2 µM; (20,38)]. Thus, we initiated our study by determining the Kd of full-length Lin28 (Lin281–209) for the terminal loop of pre-let-7g (TL-let-7g; Figure 1D). We attempted to fit the data (Figure 2A) using the classical one-site binding equation, but the fit was rather poor (Figure 2C, red line). Using the Hill equation, a much better fit was obtained (Figure 2C, blue line), from which we derived a Kd of 0.13 ± 0.02 nM with a Hill coefficient of 2.9 ± 0.7 (Table 1). These results clearly demonstrate that Lin28 binds the terminal loop of pre-let-7g with much higher affinity than previously reported (20,38).
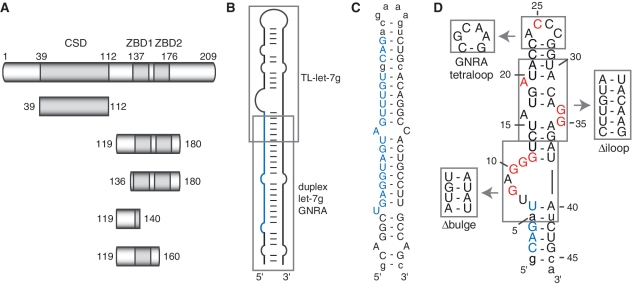
Figure 1.
The Lin28 protein, pre-let-7g RNA and related sequences used in this study. (A) Schematic representation of the primary structures of Lin28 and deletion fragments. The gray boxes delineate sequences of known RNA-binding motifs: a cold shock domain (CSD) and a pair of retroviral–type CCHC zinc-binding domains (ZBD1 and ZBD2). (B) Schematic representation of pre-let-7g, indicating the regions (gray boxes) from which TL-let-7g and duplex let-7g GNRA were derived. (C and D) Primary and secondary structures of the (C) duplex let-7g GNRA and (D) TL-let-7g. Nucleotides within the mature miRNA sequence are in blue and non-natural nucleotides are shown in lowercase. In (D), site-specific mutations of TL-let-7g are in red and regions that were replaced by alternative structured elements are boxed.
Figure 2.
EMSA of TL-let-7g with Lin281–209 and Lin28119–180. (A) Typical EMSA performed with 1 pM of 5′-[32P]-labeled TL-let-7g and increasing concentrations of Lin281–209 (0.0, 0.002, 0.010, 0.025, 0.050, 0.075, 0.10, 0.15, 0.20, 0.50, 1.0, 2.5 and 5.0 nM). (B) Typical EMSA for TL-let-7g and increasing concentrations of Lin28119–180 (0.0, 0.02, 0.10, 0.25, 0.50, 0.75, 1.0, 1.25, 1.5, 1.75, 2.0, 5.0,10, 25 and 50 nM). (C) The bound fraction of RNA is plotted against the total concentration of protein. The data for binding of TL-let-7g to Lin281–209 (squares) are fitted to both the one site binding equation (red line; Kd = 0.2 nM) and the Hill equation (blue line; Kd = 0.13 nM). The data for binding of TL-let-7g to Lin28119–180 (dots) are fitted to the one site binding equation (green line; Kd = 1.3 nM).
Given this unexpected result, we verified the binding of Lin28 to the full-length pre-let-7g. We obtained a Kd of 0.15 ± 0.04 nM with a Hill coefficient of 2.7 ± 0.5 for this interaction (Table 1), confirming that pre-let-7g and its terminal loop have similar affinities for Lin28, as previously established (20,38). As a control, we measured the binding for Lin28 to an RNA that contains only the miRNA stem of pre-let-7g (duplex let-7g GNRA; Figure 1C) and obtained a 60-fold lower Kd (Kd = 9 ± 3 nM), further supporting that the terminal loop is the main determinant of Lin28 binding to pre-let-7g.
Since Lin28 contains two different RNA-binding domains, we determined the affinity of Lin28 fragments containing either the CSD (Lin2839–112) or the two ZBDs (Lin28119–180) for pre-let-7g, TL-let-7g and duplex let-7g GNRA (Figure 1). For the N-terminal Lin2839–112 fragment comprising the CSD (Figure 1A), Kd values of 41 ± 10 nM and 126 ± 41 nM were obtained for pre-let-7g and TL-let-7g, respectively, indicating significantly weaker affinity (<250-fold) compared to the full protein. For the duplex let-7g GNRA, only a minimum Kd value could be obtained (>250 nM), because of aggregation of the Lin2839–112 domain detected at concentrations higher than 250 nM. In contrast, the Lin28119–180 fragment containing the ZBDs displays only slightly lower affinities than full-length Lin28 toward pre-let-7g and TL-let-7g, with Kd values of 0.6 ± 0.1 nM and 1.3 ± 0.3 nM, respectively (Table 1, Figure 2B and C). These binding data for Lin28119–180 can be fitted to a classical one-site binding equation (Figure 2C and Table 1). For the duplex-let-7g GNRA, no specific binding could be observed with Lin28119–180. Thus, compared to the full-length protein, the C-terminal Lin28119–180 fragment containing the two ZBDs displays similar specificity toward RNAs derived from pre-let-7g. Furthermore, the affinity between Lin28119–180 and TL-let-7g is only 10-fold weaker than between the full-length Lin28 protein and pre-let-7g, indicating that Lin28119–180 and TL-let-7g encompass the main determinants of the Lin28/pre-let-7g interaction.
Similarities between the C-terminal domain of Lin28 and the HIV-1 NCp7 protein
It has been previously noted that the C-terminal domain of Lin28 contains two ZBDs similar to those found in viral nucleocapsid proteins (30). Given the importance of this RNA-binding domain for pre-let-7g binding, we searched for proteins containing a similar domain in the Swiss-Prot database using BLAST (53). The nucleocapsid proteins from simian and human immunodeficiency viruses give the highest scores after Lin28 proteins from different species. The sequence alignment between the well-characterized HIV-1 nucleocapsid NCp7 and metazoan Lin28 sequences indicates significant similarities in the ZBDs, and also, surprisingly, in the N-terminal KR-rich domain (Figure 3). The percentage of sequence similarity to HIV-1 NCp7 is relatively high for both murine (45%) and human Lin28123–180 (45%). All three proteins contain identical zinc-chelating amino acids (CCHC) and spacing between these residues in the two ZBDs. In addition, the spacing between the two ZBDs is similar with seven residues in NCp7 of HIV-1 and eight residues in the Lin28 sequences (Figure 3). Furthermore, this sequence similarity also involves several of the NCp7 residues from the KR-rich domain and the two ZBDs that contribute to the RNA-binding interface as observed in the NMR structures of RNA/NCp7 complexes (33,54).
Figure 3.
Sequence similarity between the HIV-1 NCp7 and the C-terminal domain of Lin28. The sequences of HIV-1 NCp7 and Lin28 from Mus musculus (mmu), Homo sapiens (hsa), Gallus gallus (gga), Xenopus laevis (xla) and Danio rerio (dre) were aligned using ClustalW2 (70). A consensus sequence is given with the standard one-letter code in capital letters for amino acids, as well as the following notation: a, aromatic; h, hydrophobic; p, polar; +, positively charged. The schematic representation of NCp7 highlights the domains that contribute to RNA binding: an N-terminal KR-rich domain and two zinc-binding domains (ZBD1 and ZBD2). The residues of NCp7 in red and blue make direct contact with zinc and RNA, respectively (33,54). Those residues that could play an equivalent role in Lin28 are similarly colored.
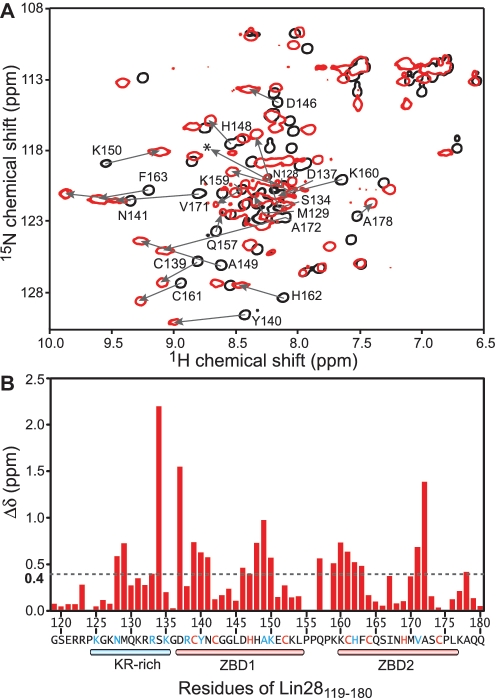
To further investigate the binding of Lin28119–180 to TL-let-7g, we performed NMR chemical shift perturbation experiments. In both the free and bound forms, Lin28119–180 displays a well-dispersed 1H-15N HSQC spectrum (Figure 4A), indicating that both adopt a stable homogenous conformation in solution. Analysis of the 1H and 15N chemical shifts reveals that 21 of the 56 amino acid residues analyzed display significant chemical shift differences between the free and RNA-bound form (Figure 4B; Δδ > 0.4 ppm). When mapped onto the primary structure of Lin28119–180, the residues showing significant chemical shift differences are found in the KR-rich domain, both ZBDs, as well as in the linker between ZBD1 and ZBD2. These results indicate that all these domains participate in RNA binding either by direct contact or through conformational rearrangement of the protein.
Figure 4.
(A) Superposition of 2D 1H-15N HSQC spectra of 1 mM 13C/15N-labeled Lin28119–180 in the free form (black) and bound to 1 mM TL-let-7g (red). The signals from the free form that display a significant chemical shift change (Δδ > 0.4 ppm) as a result of RNA binding are annotated and the change is illustrated with an arrow. A very weak signal for D137 in the complex is indicated by a star. (B) Histogram displaying the differences in chemical shifts (Δδ in ppm ± 0.03 ppm) observed after the addition of a molar equivalent of TL-let-7g to 1 mM 13C/15N-labeled Lin28119–180. The chemical shift differences (Δδ) were calculated according to the formula Δδ = [(ΔHN)2 + (0.17ΔNH)2]1/2.
To determine if the Lin28119–180 fragment could be shortened while maintaining its affinity for the terminal loop of pre-let-7g, we generated several N-terminal and C-terminal deletions (Figure 1A). Of the three deletion fragments that were expressed and purified (Lin28136–180, Lin28119–140 and Lin28119–160), none binds TL-let-7g with high affinity (Kd > 5 µM; Table 2), and thus Lin28119–180 constitutes the minimal domain required for TL-let-7 g binding. It is particularly striking that removal of the first 17 amino acids encompassing the KR-rich domain is as detrimental to binding as removal of one or two ZBDs. To insure that the absence of binding with Lin28136–180 is not due to protein misfolding, we compared the 1H-15N HSQC spectrum of this fragment with that of Lin28119–180 (Supplementary Figure S1). The chemical shift similarity between these two spectra indicates that the ZBDs adopt a similar fold in Lin28136–180 and Lin28119–180. In addition, the importance of the KR-rich domain was further investigated using a mutant of Lin28119–180 in which all lysines and arginines of the KR-rich domain are mutated (KR- with mutations R122A, R123A, K125A, K127A, K131A, R132A, R133A and K135G). As expected, Lin28119–180 (KR−) does not bind TL-let-7g with high affinity (Kd > 5 µM; Table 2). Interestingly, we found that the NCp7 protein of HIV-1 binds with the same affinity to TL-let-7g (1.1 ± 0.3 nM) as Lin28119–180 (Table 2). Thus, in addition to sharing sequence similarity with NCp7, Lin28119–180 also uses both its KR-rich and ZBDs for RNA recognition and binds with the same affinity as NCp7 to TL-let-7g. To emphasize these similarities with NCp7, we defined Lin28119–180 as the NCp7-like domain of Lin28.
Table 2.
Dissociation constants (Kd in nM) of mutants of the NCp7-like domain of Lin28 (Lin28119–180) for the TL-let-7g RNA
| Protein | Kd (nM) |
|---|---|
| Lin28119–180 | 1.3 ± 0.3 |
| Lin28136–180 | >5000 |
| Lin28119–140 | >2500 |
| Lin28119–160 | >5000 |
| Lin28119–180 (KR−) | >5000 |
| HIV–1 NCp7 | 1.1 ± 0.3 |
Global mapping of the interaction site using ribonuclease protection assay
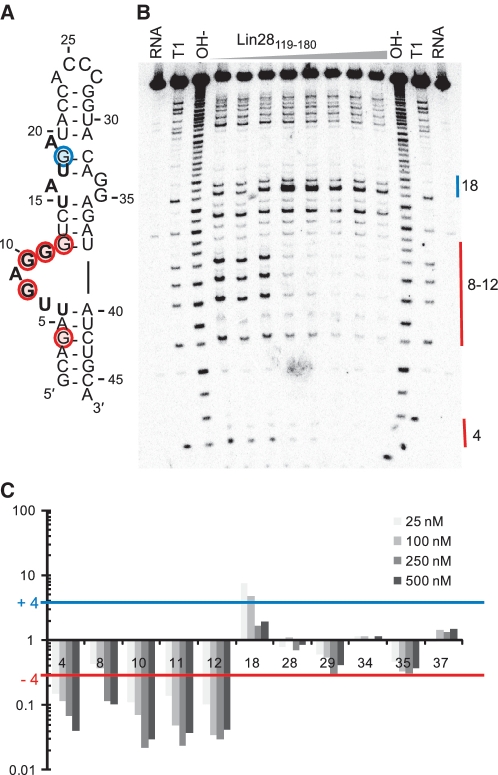
A ribonuclease protection assay was used to identify the region(s) of TL-let-7g interacting with the NCp7-like domain. As a first step, in-line probing (Supplementary Figure S2) was performed to establish the secondary structure of TL-let-7g. These results confirm that the free TL-let-7g adopts a hairpin conformation, with dynamic residues in the G-rich bulge (residues 7–11), the adjoining internal loop (residues 16–19 and 32–35) and the hairpin loop (residues 24–27; Figure 5A). For the ribonuclease protection assay, RNase T1 was selected because it shares the specificity of ZBDs for single-stranded guanines (55). The results of nuclease mapping in the presence of increasing concentration of Lin28119–180 (Figure 5) clearly demonstrate that only the G residues within the G-rich bulge (G8, G10 and G11) and adjacent stems (G4 and G12) are protected by Lin28119–180. Only one G residue (G18) becomes more accessible in the presence of Lin28119–180, most likely resulting from destabilization of the predicted G18–C12 base pair (see Discussion section). Thus, the ribonuclease protection assay indicates that the G-rich bulge is the main binding site for the NCp7-like domain and that the adjoining internal loop is also affected by binding.
Figure 5.
Footprint analysis of TL-let-7g with RNase T1. (A) Secondary structure of TL-let-7g with the mapping of in-line probing and T1 footprinting data. Residues that are the most susceptible to spontaneous cleavage through in-line attack are in bold (Supplementary Figure S2), and residues that experience a significant reduction (Ip/I0 = −4) or enhancement (Ip/I0 = +4) of T1 cleavage in the presence of Lin28119–180 are shaded in red and blue, respectively. (B) Typical RNA footprinting gel of TL-let-7g in the absence and presence of Lin28119–180 (at concentrations of 0, 1, 5, 25, 100, 250, 500 and 1000 nM). Lanes with input TL-let-7g (RNA), an alkaline hydrolysis ladder (OH−) and a T1 hydrolysis ladder (T1) are also included. (C) Histogram of normalized band sensitivity (Ip/I0, where Ip and I0 are, respectively, the intensity in the presence and absence of protein) for T1 cleaveage of each guanine obtained at 25–500 nM Lin28119–180.
Detailed mapping of the pre-let-7g determinants for Lin28119–180 binding
Next, we performed an exhaustive EMSA analysis of TL-let-7g mutants to identify the RNA determinants of the high-affinity interaction between TL-let-7g and Lin28119–180 (Kd = 1.3 nM; Table 3). Since NCp7 and several other ZBDs specifically recognize single-stranded nucleic acids (33,54), several mutants of the loop regions of TL-let-7g (Figure 1D) were investigated. Replacement of the ACCC hairpin loop by a stable GNRA tetraloop (GCAA) increases the Kd by a factor of 3, and the punctual C25A mutation increases the Kd by a factor of 1.6. Deletion of unpaired nucleotides in the internal loop to create a stable stem (Figure 1D; Δiloop) causes a 5.3-fold increase in the Kd compared with the wild-type RNA. Similar changes in affinity are also observed with simultaneous mutations of three unpaired nucleotides from the internal loop (A19C/G34A/G35A: 6.2-fold Kd increase) or from the combined effect of two related mutations (1.2- and 3.8-fold Kd increase for the A19C and G34A/G35A mutants, respectively). Thus, the internal loop of TL-let-7g makes a minor contribution to Lin28119–180 binding, but the hairpin loop appears to be replaceable.
Table 3.
Dissociation constants (Kd in nM) of the NCp7-like domain of Lin28 (Lin28119–180) for various mutants of the TL-let-7g RNA
| TL-let-7g RNA | Kd (nM) | Kd/[Kd (wt)]a |
|---|---|---|
| Wild type (wt) | 1.3 ± 0.3 | 1 |
| GNRA tetraloop | 4 ± 1 | 3.1 |
| C25A | 2.1 ± 0.5 | 1.6 |
| Δiloop | 6.9 ± 0.7 | 5.3 |
| A19C | 1.6 ± 0.6 | 1.2 |
| G34A G35A | 5 ± 1 | 3.8 |
| A19C G34A G35A | 8 ± 2 | 6.2 |
| Δbulge | n.b.b | n.b.b |
| G8C G10C G11A G12A | n.b.b | n.b.b |
| G8C | 1.1 ± 0 1 | 0.9 |
| G10C | 9 ± 2 | 6.9 |
| G11A | 3 ± 1 | 2.3 |
| G12A | 25 ± 7 | 19 |
| G10C G12A | 18 ± 5 | 14 |
aThe Kd/[Kd (wt)] is the ratio of the Kd obtained for the mutant TL-let-7g over the Kd obtained for the wild-type TL-let-7g RNA.
bNo specific binding observed. The gel mobility shift assays display smearing and multiple shifts, indicating non-specific binding.
Several EMSA results indicate that the G-rich bulge contributes significantly to Lin28119–180 binding. First, deletion of the G-rich bulge (Figure 1D; Δbulge) has a substantial effect since no specific binding could be observed with this mutant at a protein concentration as high as 5 µM. Similarly, no specific binding could be observed for a mutant in which all guanines at the bulge were mutated (G8C/G10C/G11A/G12A). To identify the guanine residues of the G-rich bulge that are important for Lin28119–180 binding, each guanine was individually mutated and the Kd was determined by EMSA (Table 3). Although the G8C and G11A mutations have a negligible effect on binding, the G10C or G12A mutations cause 7- and 19-fold increases in Kd, respectively, which represent the largest changes observed in this study for single nucleotide mutations. Surprisingly, the double G10C/G12A mutation does not completely abolish specific binding, but instead has a similar effect on binding as the single G12A mutation, suggesting that the remaining guanine residues (G8 and G11) may contribute to binding in the double mutant (G10C/G12A). Thus, it appears that G10 and G12 are key residues for the recognition, but that other G residues may also contribute to the affinity of the G-rich bulge for Lin28119–180. The G-rich bulge clearly represents the main RNA determinant of the high-affinity interaction with Lin28119–180, although the internal loop of TL-let-7g makes a minor contribution to binding.
To provide additional evidence for the importance of the G-rich bulge, we performed NMR studies of Lin28119–180 in complex with TL-let-7g mutants (Supplementary Figure S3). As expected, the 1H-15N HSQC spectrum of the G34AG35A/Lin28119–180 complex is almost identical to that of the TL-let-7g/Lin28119–180 complex, particularly for residues from the two ZBDs. In contrast, the 1H-15N HSQC spectrum of the Δbulge/Lin28119–180 complex indicates that, at the high concentration (0.1 mM) used for these NMR studies, Lin28119–180 interacts with the Δbulge mutant, but in a different manner than observed for the wild-type TL-let-7 g. Thus, these NMR results confirm that the G-rich bulge is the main determinant for high-affinity binding of Lin28119–180 to TL-let-7g.
Given the importance of the G-rich bulge for Lin28119–180 binding, we examined the sequences of all known pre-let-7g RNAs (Supplementary Figure S4). We find that the sequences of the G-rich bulge and adjacent residues are highly conserved in mammals, birds and amphibians and fit the consensus sequence U*AGGGU (* is A, C or G), with the UGAGGGU sequence found in mouse and human being the most common.
DISCUSSION
In this work, we systematically identified the key determinants of the interaction between pre-let-7g and Lin28 using highly-purified proteins and RNAs. A surprising result is the high affinity (Kd of 0.15 nM) measured for Lin28 binding to pre-let-7g, given that a Kd of 1–2 µM has previously been reported for this interaction (20,38). One important factor that may explain the higher affinity measured in this study is the absence of RNA competitor in the binding buffer. We also took great care of removing RNA contaminants during the purification of the Lin28 proteins and found it necessary to use the S7 nuclease. Nevertheless, our results are in general agreement with previous studies, which established that the extended terminal loop of pre-let-7g is the binding site for Lin28 (20,38).
The EMSA data for the binding of full-length Lin28 to pre-let-7g and TL-let-7g could be fitted well by using the Hill equation, but not the classical one-site binding equation (Figure 2C). The Hill coefficients of ∼2.8 indicate that these interactions involve a minimum of three binding sites for Lin28 on the target RNA, and most likely reflect positive cooperativity for these binding events (56). The concept that Lin28 can bind multiple sites on the RNA is further supported by supershifts observed at higher protein concentration (>5 nM; Figure 2A). Since the Hill coefficients obtained for TL-let-7g and pre-let-7g are essentially identical, we propose that full-length Lin28 can cooperatively bind a minimum of three sites in the extended terminal loop of pre-let-7g. This ability of Lin28 to cooperatively bind multiple sites is not observed with the isolated NCp7-like domain and could not be identified from our binding data with the isolated CSD due to severe aggregation problems, as previously reported (31). The CSD of Lin28 is similar to bacterial cold shock proteins (30), and other members of this family of proteins are reported to display cooperativity and weak specificity (57). For example, the RNA chaperone CspA is known to destabilize RNA secondary structure by cooperatively binding to single-stranded regions with low sequence specificity (58). Thus, although the CSD of Lin28 does not contribute significantly to the affinity of Lin28 to pre-let-7g, it may mediate cooperative binding in the context of the full-length protein.
The C-terminal domain of Lin28 displays remarkable similarities with the NCp7 protein of HIV-1. The sequence alignment between the NCp7 protein and metazoan Lin28 proteins shows a high degree of similarity, which involves several residues from the KR-rich domain and the two ZBDs of NCp7. Several of these residues have been shown to contribute to RNA binding in NMR structures of NCp7 bound to RNA hairpins derived from the HIV-1 Ψ site (33,54). Here, both NMR and mutational studies confirmed that the KR-rich domain and both ZBDs of Lin28 participate in TL-let-7g binding. In particular, truncation of the KR-rich domain or mutations of K/R residues within the KR-rich domain of Lin28119–180 abrogate high-affinity binding to TL-let-7g. Similarly, truncation of the KR-rich domain from HIV-1 NCp7 was previously shown to prevent the specific binding of NCp7 to its Ψ-site RNA target (59).
The similarity between Lin28119–180 and the NCp7 protein also extends to their binding affinity and specificity. Indeed, both Lin28119–180 and NCp7 display low nanomolar affinities toward their specific RNA targets (60,61) and specifically recognize a G-rich single-stranded region. Furthermore, we find that Lin28119–180 and NCp7 bind TL-let-7g with similar affinities under the same conditions. Lin28119–180 preferentially binds the G10-X-G12 unit of the G-rich bulge, but may also bind G8-X-X-G11 when G10 and G12 are simultaneously mutated. Our results are compatible with a recent study in which Lin28 was found to strongly protect residues G8, G10, G11 and G12 of the G-rich bulge of pre-let-7g from ribonuclease cleavage (38). The sequence at the G-rich bulge of pre-let-7g is highly conserved in mammals (Supplementary Figure S4), and sequence similarity was also found in this region for most human and mouse let-7 family members (19,20,24). Thus, it is likely that this region is important for Lin28 binding to other pre-let-7 miRNAs. NCp7 from HIV-1 was also reported to bind exposed guanines with its two ZBDs (33,54,60,62,63). Its target sequences in the Ψ-site RNA are generally located in a hairpin loop, where they form a G-X-G motif, but exceptions such as the binding to the 1 × 3 internal loop of SL1 demonstrate flexibility in RNA recognition by NCp7 (60,63). This flexibility in target sequence recognition may be inherent to the adaptive nature of the NCp7 motif and may explain, in part, why Lin28 binds a wide variety of mRNA targets in addition to let-7 precursors (35,36,64,65). Lin28119–180 possibly recognizes its RNA target in the same way that HIV-1 NCp7 binds hairpin loops in the HIV Ψ RNA (33,54). In this model, the two ZBDs of Lin28 would each bind an exposed guanine in the G-rich bulge and the KR-rich domain would bind an adjacent stem or enlarged major groove. Interaction of Lin28119–180 with a domain adjacent to the G-rich bulge is consistent with our mutational studies, which indicate that the internal loop of TL-let-7g makes a small contribution to binding. Given all these similarities between NCp7 and Lin28119–180, it is tempting to speculate on a role for HIV-1 NCp7 in let-7 biogenesis and a common origin for NCp7 and the C-terminal domain of Lin28. The latter is inconsistent with a classical evolutionary model that does not include viruses, but agrees with an alternative model in which viruses play an important role in cellular evolution (66,67).
The high-affinity of Lin28 toward TL-let-7g is mostly due to its C-terminal NCp7-like domain, which has been shown to be required for several functional aspects of Lin28. It was shown to be required for Lin28 processing inhibition of pre-let-7g in vivo (20), its localization to P-bodies (31), its specific binding to let-7 precursors (20,24) and for the Lin28-mediated uridylylation of pre-let-7-a-1 by TUT4 (22). However, the NCp7-like domain is not sufficient for Lin28 function, as previously demonstrated with two Lin28 homologs, Lin28B and Lin28B-S, which are overexpressed in human hepatocellular carcinoma and in several cancer lines (68). The Lin28B-S preserves the NCp7-like domain but contains a truncation of the cold-shock domain. It has been shown that the overexpression of Lin28B-S does not induce cancer cell proliferation in contrast to what is observed with Lin28B (68). In addition, Lin28B-S does not inhibit the processing of pri-let-7g like Lin28 and Lin28B (21). Thus, although the NCp7-like domain of Lin28 likely contributes to its in vivo function through high-affinity and specific binding to the terminal loop of pre-let-7g, the cold-shock domain is required for Lin28 to function as an effective oncogene and inhibitor of let-7 biogenesis.
Given its high-affinity for pre-let-7g, the NCp7-like domain may be responsible for the initial targeting of pre-let-7g. After this initial binding event, partial unfolding of the terminal loop of pre-let-7g would make it more accessible for binding multiple copies of Lin28. Both the NCp7 domain and the CSD have been previously described as RNA chaperones (58,69) and could contribute to making the terminal loop more accessible. In agreement with this role of the NCp7-like domain in Lin28, its binding to TL-let-7g makes G18 of the internal loop more accessible to ribonuclease cleavage. The high-affinity and specificity of the NCp7-like domain for the G-rich bulge may also allow Lin28 to bind its RNA target in an orderly fashion to insure that important functional regions of the RNA are protected from binding of miRNA processing enzymes. The G-rich bulge is directly adjacent to one of the Dicer processing site. Thus, it is likely that Lin28 binding at the G-rich bulge protects the pre-let-7g RNA from Dicer cleavage at this site, likely by both steric hindrance and destabilization of the stem region near the G-rich bulge (38).
Our mutagenesis, T1 footprinting and NMR data all indicate that the G-rich bulge of TL-let-7g is the main determinant for high-affinity binding to Lin28119–180. In an apparent contradiction with our results, a previous report identified a different G-rich region of pre-let-7 (GGAG residues 34–37 of the internal loop in Figure 5A) to be important for Lin28 binding (24). Both these G-rich regions are highly conserved in mammalian let-7g, and similar G-rich regions are found both at the 5′-end and 3′-end of the terminal loop in most members of the human and mouse let-7 family (19,20,24). Thus, it is likely that both G-rich regions in pre-let-7 miRNAs are important for binding full-length Lin28. For example, initial binding of Lin28 to the G-rich bulge may expose internal loop residues and allow Lin28 binding to the other G-rich region. Alternatively, the levels of Lin28 and other cellular factors may affect binding of Lin28 to the two G-rich regions. Interestingly, both hnRNP A1 and KSRP specifically bind G-rich sequences within the terminal loop of pre-let-7a1 (17,18) and may regulate Lin28 binding at these sites in some pre-let-7 members. Clearly, further studies are needed to more precisely determine how each G-rich region contributes with cellular factors to regulate pre-let-7 biogenesis.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online: Supplementary Figures S1–4, Supplementary References [71–73].
FUNDING
The National Sciences and Engineering Research Council of Canada (NSERC) (to P.L. and J.G.O.). A.D. holds a Ph.D. scholarship from the Canadian Institutes for Health Research. J.B. holds an MSc scholarship from the Université de Montréal. P.L. holds a Canada Research Chair in Structural Biology and Engineering of RNA. Funding for open access charge: National Sciences and Engineering Research Council of Canada.
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
The authors thank Dominique Chaussé, Geneviève Di Tomasso and Alexis Rompré-Brodeur for sample preparation.
REFERENCES
- 1.Bartel DP. MicroRNAs: target recognition and regulatory functions. Cell. 2009;136:215–233. doi: 10.1016/j.cell.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Winter J, Jung S, Keller S, Gregory RI, Diederichs S. Many roads to maturity: microRNA biogenesis pathways and their regulation. Nat. Cell. Biol. 2009;11:228–234. doi: 10.1038/ncb0309-228. [DOI] [PubMed] [Google Scholar]
- 3.Filipowicz W, Bhattacharyya SN, Sonenberg N. Mechanisms of post-transcriptional regulation by microRNAs: are the answers in sight? Nat. Rev. Genet. 2008;9:102–114. doi: 10.1038/nrg2290. [DOI] [PubMed] [Google Scholar]
- 4.Kim VN, Han J, Siomi MC. Biogenesis of small RNAs in animals. Nat. Rev. Mol. Cell Biol. 2009;10:126–139. doi: 10.1038/nrm2632. [DOI] [PubMed] [Google Scholar]
- 5.Zhao Y, Ransom JF, Li A, Vedantham V, von Drehle M, Muth AN, Tsuchihashi T, McManus MT, Schwartz RJ, Srivastava D. Dysregulation of cardiogenesis, cardiac conduction, and cell cycle in mice lacking miRNA-1-2. Cell. 2007;129:303–317. doi: 10.1016/j.cell.2007.03.030. [DOI] [PubMed] [Google Scholar]
- 6.Peter ME. Let-7 and miR-200 microRNAs: guardians against pluripotency and cancer progression. Cell Cycle. 2009;8:843–852. doi: 10.4161/cc.8.6.7907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ambros V. MicroRNAs and developmental timing. Curr. Opin. Genet. Dev. 2011;21:511–517. doi: 10.1016/j.gde.2011.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Esquela-Kerscher A, Slack FJ. Oncomirs—microRNAs with a role in cancer. Nat. Rev. Cancer. 2006;6:259–269. doi: 10.1038/nrc1840. [DOI] [PubMed] [Google Scholar]
- 9.Shi XB, Tepper CG, deVere White RW. Cancerous miRNAs and their regulation. Cell Cycle. 2008;7:1529–1538. doi: 10.4161/cc.7.11.5977. [DOI] [PubMed] [Google Scholar]
- 10.Roush S, Slack FJ. The let-7 family of microRNAs. Trends Cell Biol. 2008;18:505–516. doi: 10.1016/j.tcb.2008.07.007. [DOI] [PubMed] [Google Scholar]
- 11.Bussing I, Slack FJ, Grosshans H. Let-7 microRNAs in development, stem cells and cancer. Trends Mol. Med. 2008;14:400–409. doi: 10.1016/j.molmed.2008.07.001. [DOI] [PubMed] [Google Scholar]
- 12.Visone R, Croce CM. MiRNAs and cancer. Am. J. Pathol. 2009;174:1131–1138. doi: 10.2353/ajpath.2009.080794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.O'Day E, Lal A. MicroRNAs and their target gene networks in breast cancer. Breast Cancer Res. 2010;12:201. doi: 10.1186/bcr2484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Thomson JM, Newman M, Parker JS, Morin-Kensicki EM, Wright T, Hammond SM. Extensive post-transcriptional regulation of microRNAs and its implications for cancer. Genes Dev. 2006;20:2202–2207. doi: 10.1101/gad.1444406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wulczyn FG, Smirnova L, Rybak A, Brandt C, Kwidzinski E, Ninnemann O, Strehle M, Seiler A, Schumacher S, Nitsch R. Post-transcriptional regulation of the let-7 microRNA during neural cell specification. FASEB J. 2007;21:415–426. doi: 10.1096/fj.06-6130com. [DOI] [PubMed] [Google Scholar]
- 16.Van Wynsberghe PM, Kai ZS, Massirer KB, Burton VH, Yeo GW, Pasquinelli AE. LIN-28 co-transcriptionally binds primary let-7 to regulate miRNA maturation in Caenorhabditis elegans. Nat. Struct. Mol. Biol. 2011;18:302–308. doi: 10.1038/nsmb.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Michlewski G, Caceres JF. Antagonistic role of hnRNP A1 and KSRP in the regulation of let-7a biogenesis. Nat. Struct. Mol. Biol. 2010;17:1011–1018. doi: 10.1038/nsmb.1874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Trabucchi M, Briata P, Garcia-Mayoral M, Haase AD, Filipowicz W, Ramos A, Gherzi R, Rosenfeld MG. The RNA-binding protein KSRP promotes the biogenesis of a subset of microRNAs. Nature. 2009;459:1010–1014. doi: 10.1038/nature08025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Newman MA, Thomson JM, Hammond SM. Lin-28 interaction with the Let-7 precursor loop mediates regulated microRNA processing. RNA. 2008;14:1539–1549. doi: 10.1261/rna.1155108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Piskounova E, Viswanathan SR, Janas M, LaPierre RJ, Daley GQ, Sliz P, Gregory RI. Determinants of microRNA processing inhibition by the developmentally regulated RNA-binding protein Lin28. J. Biol. Chem. 2008;283:21310–21314. doi: 10.1074/jbc.C800108200. [DOI] [PubMed] [Google Scholar]
- 21.Viswanathan SR, Daley GQ, Gregory RI. Selective blockade of microRNA processing by Lin28. Science. 2008;320:97–100. doi: 10.1126/science.1154040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Heo I, Joo C, Cho J, Ha M, Han J, Kim VN. Lin28 mediates the terminal uridylation of let-7 precursor MicroRNA. Mol. Cell. 2008;32:276–284. doi: 10.1016/j.molcel.2008.09.014. [DOI] [PubMed] [Google Scholar]
- 23.Rybak A, Fuchs H, Smirnova L, Brandt C, Pohl EE, Nitsch R, Wulczyn FG. A feedback loop comprising lin-28 and let-7 controls pre-let-7 maturation during neural stem-cell commitment. Nat. Cell Biol. 2008;10:987–993. doi: 10.1038/ncb1759. [DOI] [PubMed] [Google Scholar]
- 24.Heo I, Joo C, Kim YK, Ha M, Yoon MJ, Cho J, Yeom KH, Han J, Kim VN. TUT4 in concert with Lin28 suppresses microRNA biogenesis through pre-microRNA uridylation. Cell. 2009;138:696–708. doi: 10.1016/j.cell.2009.08.002. [DOI] [PubMed] [Google Scholar]
- 25.Hagan JP, Piskounova E, Gregory RI. Lin28 recruits the TUTase Zcchc11 to inhibit let-7 maturation in mouse embryonic stem cells. Nat. Struct. Mol. Biol. 2009;16:1021–1025. doi: 10.1038/nsmb.1676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lehrbach NJ, Armisen J, Lightfoot HL, Murfitt KJ, Bugaut A, Balasubramanian S, Miska EA. LIN-28 and the poly(U) polymerase PUP-2 regulate let-7 microRNA processing in Caenorhabditis elegans. Nat. Struct. Mol. Biol. 2009;16:1016–1020. doi: 10.1038/nsmb.1675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Newman MA, Hammond SM. Lin-28: an early embryonic sentinel that blocks Let-7 biogenesis. Int. J. Biochem. Cell Biol. 2010;42:1330–1333. doi: 10.1016/j.biocel.2009.02.023. [DOI] [PubMed] [Google Scholar]
- 28.Yu J, Vodyanik MA, Smuga-Otto K, Antosiewicz-Bourget J, Frane JL, Tian S, Nie J, Jonsdottir GA, Ruotti V, Stewart R, et al. Induced pluripotent stem cell lines derived from human somatic cells. Science. 2007;318:1917–1920. doi: 10.1126/science.1151526. [DOI] [PubMed] [Google Scholar]
- 29.Viswanathan SR, Daley GQ. Lin28: a microRNA regulator with a macro role. Cell. 2010;140:445–449. doi: 10.1016/j.cell.2010.02.007. [DOI] [PubMed] [Google Scholar]
- 30.Moss EG, Lee RC, Ambros V. The cold shock domain protein LIN-28 controls developmental timing in C. elegans and is regulated by the lin-4 RNA. Cell. 1997;88:637–646. doi: 10.1016/s0092-8674(00)81906-6. [DOI] [PubMed] [Google Scholar]
- 31.Balzer E, Moss EG. Localization of the developmental timing regulator Lin28 to mRNP complexes, P-bodies and stress granules. RNA Biol. 2007;4:16–25. doi: 10.4161/rna.4.1.4364. [DOI] [PubMed] [Google Scholar]
- 32.Landsman D. RNP-1, an RNA-binding motif is conserved in the DNA-binding cold shock domain. Nucleic Acids Res. 1992;20:2861–2864. doi: 10.1093/nar/20.11.2861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.De Guzman RN, Wu ZR, Stalling CC, Pappalardo L, Borer PN, Summers MF. Structure of the HIV-1 nucleocapsid protein bound to the SL3 psi-RNA recognition element. Science. 1998;279:384–388. doi: 10.1126/science.279.5349.384. [DOI] [PubMed] [Google Scholar]
- 34.Polesskaya A, Cuvellier S, Naguibneva I, Duquet A, Moss EG, Harel-Bellan A. Lin-28 binds IGF-2 mRNA and participates in skeletal myogenesis by increasing translation efficiency. Genes Dev. 2007;21:1125–1138. doi: 10.1101/gad.415007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Xu B, Huang Y. Histone H2a mRNA interacts with Lin28 and contains a Lin28-dependent posttranscriptional regulatory element. Nucleic Acids Res. 2009;37:4256–4263. doi: 10.1093/nar/gkp372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Xu B, Zhang K, Huang Y. Lin28 modulates cell growth and associates with a subset of cell cycle regulator mRNAs in mouse embryonic stem cells. RNA. 2009;15:357–361. doi: 10.1261/rna.1368009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Qiu C, Ma Y, Wang J, Peng S, Huang Y. Lin28-mediated post-transcriptional regulation of Oct4 expression in human embryonic stem cells. Nucleic Acids Res. 2010;38:1240–1248. doi: 10.1093/nar/gkp1071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lightfoot HL, Bugaut A, Armisen J, Lehrbach NJ, Miska EA, Balasubramanian S. A LIN28-dependent structural change in pre-let-7g directly inhibits dicer processing. Biochemistry. 2011;50:7514–7521. doi: 10.1021/bi200851d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Di Tomasso G, Lampron P, Dagenais P, Omichinski JG, Legault P. The ARiBo tag: a reliable tool for affinity purification of RNAs under native conditions. Nucleic Acids Res. 2011;39:e18. doi: 10.1093/nar/gkq1084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Di Tomasso G, Dagenais P, Desjardins A, Rompré-Brodeur A, Delfosse V, Legault P. Affinity purification of RNA using an ARiBo tag. Methods Mol. Biol. 2012 doi: 10.1007/978-1-62703-113-4_11. In Press. [DOI] [PubMed] [Google Scholar]
- 41.Delfosse V, Bouchard P, Bonneau E, Dagenais P, Lemay JF, Lafontaine DA, Legault P. Riboswitch structure: an internal residue mimicking the purine ligand. Nucleic Acids Res. 2010;38:2057–2068. doi: 10.1093/nar/gkp1080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bouchard P, Lacroix-Labonté J, Desjardins G, Lampron P, Lisi V, Lemieux S, Major F, Legault P. Role of SLV in SLI substrate recognition by the Neurospora VS ribozyme. RNA. 2008;14:736–748. doi: 10.1261/rna.824308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Jenkins LM, Byrd JC, Hara T, Srivastava P, Mazur SJ, Stahl SJ, Inman JK, Appella E, Omichinski JG, Legault P. Studies on the mechanism of inactivation of the HIV-1 nucleocapsid protein NCp7 with 2-mercaptobenzamide thioesters. J. Med. Chem. 2005;48:2847–2858. doi: 10.1021/jm0492195. [DOI] [PubMed] [Google Scholar]
- 44.Omichinski JG, Clore GM, Schaad O, Felsenfeld G, Trainor C, Appella E, Stahl SJ, Gronenborn AM. NMR structure of a specific DNA complex of Zn-containing DNA binding domain of GATA-1. Science. 1993;261:438–446. doi: 10.1126/science.8332909. [DOI] [PubMed] [Google Scholar]
- 45.Regulski EE, Breaker RR. In-line probing analysis of riboswitches. Methods Mol. Biol. 2008;419:53–67. doi: 10.1007/978-1-59745-033-1_4. [DOI] [PubMed] [Google Scholar]
- 46.Kay L, Keifer P, Saarinen T. Pure absorption gradient enhanced heteronuclear single quantum correlation spectroscopy with improved sensitivity. J. Am. Chem. Soc. 1992;114:10663–10665. [Google Scholar]
- 47.Wittekind M, Mueller L. HNCACB, a high-sensitivity 3D NMR experiment to correlate amide-proton and nitrogen resonances with the alpha- and beta-carbon resonances in proteins. J. Magn. Reson. Series B. 1993;101:201–205. [Google Scholar]
- 48.Grzesiek S, Dobeli H, Gentz R, Garotta G, Labhardt AM, Bax A. 1H, 13C, and 15N NMR backbone assignments and secondary structure of human interferon-gamma. Biochemistry. 1992;31:8180–8190. doi: 10.1021/bi00150a009. [DOI] [PubMed] [Google Scholar]
- 49.Muhandiram DR, Kay LE. Gradient-enhanced triple-resonance three-dimensional NMR experiments with improved sensitivity. J. Magn. Reson. Series B. 1994;103:203–216. [Google Scholar]
- 50.Wishart DS, Bigam CG, Yao J, Dyson HJ, Oldfield E, Markley JL, Sykes BD. 1H, 13C, 15N chemical shift referencing in biomolecular NMR. J. Biomol. NMR. 1995;6:135–140. doi: 10.1007/BF00211777. [DOI] [PubMed] [Google Scholar]
- 51.Delaglio F, Grzesiek S, Vuister GW, Zhu G, Pfeifer J, Bax A. NMRPipe: a multidimensional spectral processing system based on UNIX pipes. J. Biomol. NMR. 1995;6:277–293. doi: 10.1007/BF00197809. [DOI] [PubMed] [Google Scholar]
- 52.Johnson BA, Blevins RA. NMR view: a computer program for the visualization and analysis of NMR data. J. Biomol. NMR. 1994;4:603–614. doi: 10.1007/BF00404272. [DOI] [PubMed] [Google Scholar]
- 53.Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Amarasinghe GK, De Guzman RN, Turner RB, Chancellor KJ, Wu ZR, Summers MF. NMR structure of the HIV-1 nucleocapsid protein bound to stem-loop SL2 of the psi-RNA packaging signal. Implications for genome recognition. J. Mol. Biol. 2000;301:491–511. doi: 10.1006/jmbi.2000.3979. [DOI] [PubMed] [Google Scholar]
- 55.Steyaert J. A decade of protein engineering on ribonuclease T1-atomic dissection of the enzyme-substrate interactions. Eur. J. Biochem. 1997;247:1–11. doi: 10.1111/j.1432-1033.1997.t01-1-00001.x. [DOI] [PubMed] [Google Scholar]
- 56.Weiss JN. The Hill equation revisited: uses and misuses. FASEB J. 1997;11:835–841. [PubMed] [Google Scholar]
- 57.Ermolenko DN, Makhatadze GI. Bacterial cold-shock proteins. Cell Mol. Life Sci. 2002;59:1902–1913. doi: 10.1007/PL00012513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Jiang W, Hou Y, Inouye M. CspA, the major cold-shock protein of Escherichia coli, is an RNA chaperone. J. Biol. Chem. 1997;272:196–202. doi: 10.1074/jbc.272.1.196. [DOI] [PubMed] [Google Scholar]
- 59.Dannull J, Surovoy A, Jung G, Moelling K. Specific binding of HIV-1 nucleocapsid protein to PSI RNA in vitro requires N-terminal zinc finger and flanking basic amino acid residues. EMBO J. 1994;13:1525–1533. doi: 10.1002/j.1460-2075.1994.tb06414.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Shubsda MF, Paoletti AC, Hudson BS, Borer PN. Affinities of packaging domain loops in HIV-1 RNA for the nucleocapsid protein. Biochemistry. 2002;41:5276–5282. doi: 10.1021/bi016045+. [DOI] [PubMed] [Google Scholar]
- 61.Paoletti AC, Shubsda MF, Hudson BS, Borer PN. Affinities of the nucleocapsid protein for variants of SL3 RNA in HIV-1. Biochemistry. 2002;41:15423–15428. doi: 10.1021/bi026307n. [DOI] [PubMed] [Google Scholar]
- 62.Fisher RJ, Rein A, Fivash M, Urbaneja MA, Casas-Finet JR, Medaglia M, Henderson LE. Sequence-specific binding of human immunodeficiency virus type 1 nucleocapsid protein to short oligonucleotides. J. Virol. 1998;72:1902–1909. doi: 10.1128/jvi.72.3.1902-1909.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Yuan Y, Kerwood DJ, Paoletti AC, Shubsda MF, Borer PN. Stem of SL1 RNA in HIV-1: structure and nucleocapsid protein binding for a 1 × 3 internal loop. Biochemistry. 2003;42:5259–5269. doi: 10.1021/bi034084a. [DOI] [PubMed] [Google Scholar]
- 64.Jin J, Jing W, Lei XX, Feng C, Peng S, Boris-Lawrie K, Huang Y. Evidence that Lin28 stimulates translation by recruiting RNA helicase A to polysomes. Nucleic Acids Res. 2011;39:3724–3734. doi: 10.1093/nar/gkq1350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Peng S, Chen LL, Lei XX, Yang L, Lin H, Carmichael GG, Huang Y. Genome-wide studies reveal that lin28 enhances the translation of genes important for growth and survival of human embryonic stem cells. Stem Cells. 2011;29:496–504. doi: 10.1002/stem.591. [DOI] [PubMed] [Google Scholar]
- 66.Claverie JM. Viruses take center stage in cellular evolution. Genome Biol. 2006;7:110. doi: 10.1186/gb-2006-7-6-110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Bandea CI. The origin and evolution of viruses as molecular organisms. Nat. Precedings. 2009 npre.2009.3886.2001. [Google Scholar]
- 68.Guo Y, Chen Y, Ito H, Watanabe A, Ge X, Kodama T, Aburatani H. Identification and characterization of lin-28 homolog B (LIN28B) in human hepatocellular carcinoma. Gene. 2006;384:51–61. doi: 10.1016/j.gene.2006.07.011. [DOI] [PubMed] [Google Scholar]
- 69.Tsuchihashi Z, Khosla M, Herschlag D. Protein enhancement of hammerhead ribozyme catalysis. Science. 1993;262:99–102. doi: 10.1126/science.7692597. [DOI] [PubMed] [Google Scholar]
- 70.Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, McWilliam H, Valentin F, Wallace IM, Wilm A, Lopez R, et al. Clustal W and Clustal X version 2.0. Bioinformatics. 2007;23:2947–2948. doi: 10.1093/bioinformatics/btm404. [DOI] [PubMed] [Google Scholar]
- 71.Zuker M. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 2003;31:3406–3415. doi: 10.1093/nar/gkg595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Parisien M, Major F. The MC-Fold and MC-Sym pipeline infers RNA structure from sequence data. Nature. 2008;452:51–55. doi: 10.1038/nature06684. [DOI] [PubMed] [Google Scholar]
- 73.Notredame C, Higgins DG, Heringa J. T-Coffee: a novel method for fast and accurate multiple sequence alignment. J. Mol. Biol. 2000;302:205–217. doi: 10.1006/jmbi.2000.4042. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.