Abstract
Base triples are recurrent clusters of three RNA nucleobases interacting edge-to-edge by hydrogen bonding. We find that the central base in almost all triples forms base pairs with the other two bases of the triple, providing a natural way to geometrically classify base triples. Given 12 geometric base pair families defined by the Leontis–Westhof nomenclature, combinatoric enumeration predicts 108 potential geometric base triple families. We searched representative atomic-resolution RNA 3D structures and found instances of 68 of the 108 predicted base triple families. Model building suggests that some of the remaining 40 families may be unlikely to form for steric reasons. We developed an on-line resource that provides exemplars of all base triples observed in the structure database and models for unobserved, predicted triples, grouped by triple family, as well as by three-base combination (http://rna.bgsu.edu/Triples). The classification helps to identify recurrent triple motifs that can substitute for each other while conserving RNA 3D structure, with applications in RNA 3D structure prediction and analysis of RNA sequence evolution.
INTRODUCTION
Base triples are recurrent clusters of three RNA nucleobases interacting edge-to-edge by hydrogen bonding. They occur widely in structured RNA molecules and are found in common, recurrent RNA 3D motifs, including sarcin/ricin loops, kink-turns and C-loops (1–4). In addition, base triples stabilize many RNA tertiary interactions, including hairpin loop–receptor interactions and various ‘A-minor’ motifs (5–7). As an indication of the frequency of base triples in structured RNA molecules, we find ~140 base triples in the 3D structure of bacterial 16S ribosomal RNAs and ~280 in bacterial 23S rRNAs.
Identification of nucleotides forming base triples has proven to be an important tool for modeling the common 3D folding of homologous RNA molecules (8). Bases that participate in triples are generally more constrained in sequence than bases forming only 1 base pair, because mutation of one base of a triple may require changing each of the other two bases in the triple to conserve a functional 3D structure. As early as 1969, Michael Levitt correctly predicted the interaction of nucleotide 9 and the third base pair in the D-stem to form a conserved base triple in the core of tRNA by comparison of aligned tRNA sequences (9). On the basis of this and other tertiary interactions, he built a remarkably accurate 3D model of tRNA. Likewise, in 1990 Michel and Westhof published a highly successful model for the core 3D structure of Group I introns (10), relying largely on a handful of predicted tertiary interactions, most of which also formed base triples, to determine the overall folding topology (11,12).
As of early 2011, 330 distinct atomic-resolution structures containing RNA had been deposited in the PDB/NDB (Supplementary Data S1). With the significant increase in the number of RNA 3D structures reported at atomic resolution (13), the time is ripe for a systematic survey and comprehensive classification of RNA base triples. The systematic compilation of base triples that we present in this article includes structures of all distinct triples found in the current 3D database as well as models of potential triples not yet found. We have organized these data and models in an on-line resource (http://rna.bgsu.edu/Triples) that we anticipate will be useful for comparative sequence analysis of homologous RNA molecules, 3D RNA motif prediction and 3D structure modeling.
Xin and Olson recently provided a compilation of base triples (14) that can be accessed at http://bps.rutgers.edu/atlas/triplet. They organized base triples according to the base combinations forming the triple, grouping together, for example, all AAA triples or all UAC triples. In the classification of Xin and Olson, geometrically similar base triples, which can substitute for each other in homologous structures, end up in different groups, making this approach less useful for bioinformatic applications. In another study, Nagaswamy et al. (15) compiled and named base triples on the basis of the Sanger base pair types (http://prion.bchs.uh.edu/bp_type/bp_structure.html). This approach also separates geometrically similar base triples into different groups. Furthermore, the naming system based on the Sanger base pair types requires keeping track of the numbering of the atoms of each base, which may make it difficult for non-specialists to remember and visualize individual geometries. Finally, it is not clear, using previous classifications, how to predict new base triples, not yet observed in the database.
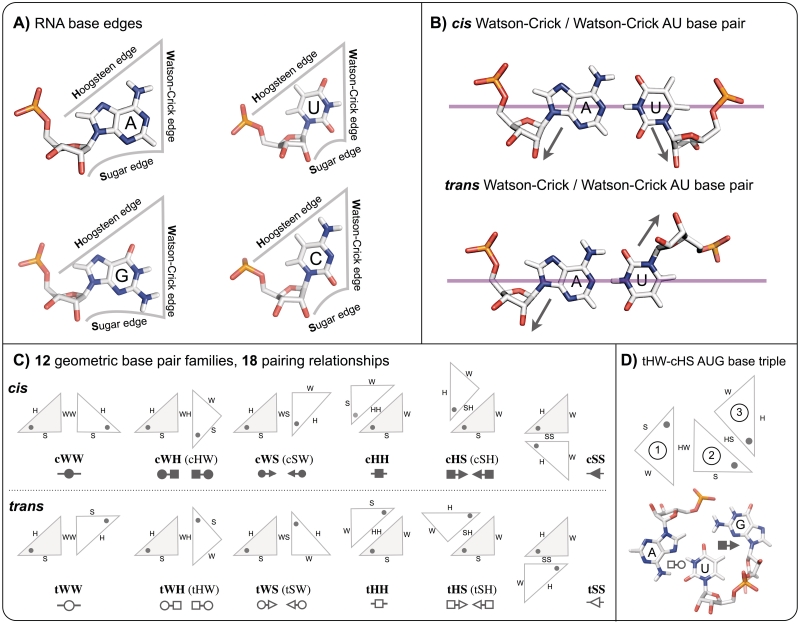
Here, we propose a classification that groups together geometrically similar base triples in ways we find to be most useful for RNA 3D modeling, sequence alignment and phylogenetic analysis. In previous work, we showed that RNA base pairs are conveniently classified according to the interacting edges of the paired bases, the Watson–Crick (W), Hoogsteen (H) and Sugar (S) edges (16). Each unmodified nucleotide, A, G, C or U, presents three edges for H-bonding, as shown in Figure 1A. For each pair of interacting edges, two relative orientations of the glycosidic bonds (cis or trans) are possible (Figure 1B), giving rise to 12 geometric base pair families. These are shown schematically in Figure 1C, using triangles to represent each nucleobase (16,17). Each base pair family is named according to the glycosidic bond orientation and the interacting edges, as previously described in the cited references (16,17). For example, pairing between the Watson–Crick edge of one base and the Hoogsteen edge of a second base with the glycosidic bonds in trans produces a base pair belonging to the trans Watson–Crick/Hoogsteen or ‘tWH’ base pair family. The canonical Watson–Crick base pairs belong to the cis Watson–Crick/Watson–Crick family, abbreviated ‘cWW’. Symbols were proposed for annotating base pairs in 2D diagrams of RNA structure, using circles, squares and triangles to represent the Watson–Crick, Hoogsteen and Sugar edges, respectively. The geometric base pair classification has proven useful in annotating and analyzing RNA 3D structures and understanding RNA sequence variation and evolution (16–19).
Figure 1.
Summary of Leontis/Westhof base pairing classification. (A) Each unmodified RNA nucleotide presents three edges for base pairing interactions, the Hoogsteen (H), Watson–Crick (W) and Sugar (S) edges. Consequently, nucleobases can be conveniently represented by triangles as shown. Note that the sugar edges include the 2′-OH group of the riboses. (B) For each pair of edges, nucleotides can pair in two distinct ways, designated cis and trans, and related by 180° rotation of one nucleotide about the magenta axis that bifurcates the nucleobases perpendicular to the interacting edges. The glycosidic bonds of the nucleotides are on the same side of this axis in the cis configuration, and on opposite sides in the trans configuration (indicated by arrows). (C) Schematic representations of each of the 12 basic base pair families, using triangles to represent each base. Symbols for annotating secondary structures of RNA with non-Watson–Crick base pairs are also provided. The symbols are derived by associating circles with W edges, squares with H edges and triangles with S edges. Filled in symbols represent cis base pairs and open symbols, trans base pairs. Note that the 12 base pair families result in 18 base pairing relations due to the asymmetry of some base pairs. (D) Schematic showing a representative regular base triple, AUG tHW/cHS. The central base (U), numbered base ‘2’, pairs with each of the other two bases of the triple using a distinct base edge. A is base 1 and G is base 3. The triple is named according to the base pairs formed by bases 1 and 2 (tHW in this case) and by bases 2 and 3 (cHS in this case).
Given that base triples are sets of three nucleotides interacting by hydrogen bonding, we investigated whether we can extend the Leontis/Westhof base pair classification to name and classify base triples in a descriptive and comprehensive way (see Figure 1D for an example). Moreover, we anticipated that such an approach could serve to predict additional triples, not yet observed experimentally.
This article addresses three basic questions: (i) how many types of base triples are possible in RNA structures? (ii) How many types are observed in the current database of atomic-resolution 3D structures? (iii) How can base triples best be clustered, classified and named to be most useful in a variety of bioinformatic tasks, including improving methods of aligning homologous RNA sequences and modeling RNA 3D structures using sequence alignments as well as other experimental data?
RESULTS
In the ‘Definition and classification of regular base triple families’ section, we begin by outlining the method we developed to enumerate all potential families of ‘regular’ RNA base triples, which we define as clusters of three RNA nucleobases in which well-defined base pairs form between the central base and each of the other two bases of the triple (Figure 1D). This approach is based on the Leontis/Westhof base pair classification. In the ‘Symbolic searches for base triples using FR3D’ section, we describe the results of searches of a non-redundant (NR) subset of the atomic-resolution RNA structure database to find instances of the predicted geometric families of regular base triples that we carried out using the ‘Find RNA 3D’ (FR3D) program suite, developed in our laboratory (20,21). The observed and predicted instances are presented in comprehensive tables available online (http://rna.bgsu.edu/Triples) as well as in the Supplementary Tables.
In subsequent sections of the ‘Results’, we describe searches we carried out using the new co-planar neighboring relation, which we implemented in FR3D and describe in the Supplementary Data S0, to test the central hypothesis of our approach to classifying RNA base triples. Use of the co-planar relation along with expert manual evaluation of the results identified a very small number of ‘intermediate’ base triples (four distinct types). In these triples, the third base interacts with each of the other two bases by individual hydrogen bonds, without forming full-fledged base pairs with either one alone. In light of the fact that we found very few intermediate base triples, we concluded that almost all base triples are, in fact, regular.
Definition and classification of regular base triple families
Our approach to classifying regular base triple families is based on the Leontis/Westhof classification of RNA base pairs. We note that although there are only 12 base pair families, there are actually 18 pairing relationships (22). Consequently, the base designated ‘base 1’ of a given triple can interact with the central base, designated ‘base 2’ (Figure 1D), by pairing in any of 18 distinct ways, namely cWW (cis Watson–Crick/Watson–Crick), tWW (trans Watson–Crick/Watson–Crick), cWH (cis Watson–Crick/Hoogesteen), tWH (trans Watson–Crick/Hoogesteen), cWS (cis Wastson–Crick/Sugar edge), tWS, cHW, tHW, cHH, tHH, cHS, tHS, cSW, tSW, cSH, tSH, cSS or tSS (Figure 1C). Therefore, base triple families can be grouped into 18 superfamilies. However, since the central base of a regular triple forms two pairs, each base triple family can be designated in two different ways, depending on which of the two component base pairs of the triple is listed first. Thus, each base triple family belongs to two superfamilies. For example, cWW/tHW and tWH/cWW denote the same geometric triple family and this family belongs to both the cWW and the tWH superfamilies. The 18 superfamilies and the triple families that belong to each superfamily are provided in Table 1. Each family is assigned a unique numerical identifier (1 through 108) and appears twice in the table, in two different superfamilies. The italic font indicates the second appearance of each family in Table 1. Cells with gray background in Table 1 indicate triple families for which no instances have yet been observed in the current RNA structure database. We also provide the relative local strand orientations, parallel (P) and anti-parallel (A), for each pair of nucleotides in the triple, in the order, nucleotides 1 and 2, 2 and 3, and 1 and 3. These are the default local strand orientations, which hold when each nucleotide has the anti glycosidic configuration.
Table 1.
The 108 observed and potential geometric base triple families, grouped in 18 triple superfamilies
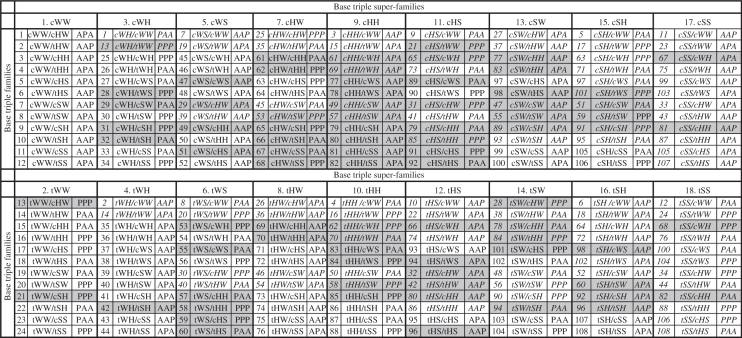 |
Each triple family is assigned a unique number between 1 and 108, but has two names, depending on which component base pair is given first, and thus belongs to two superfamilies. For example, triple family #1 is cWW/cHW or cWH/cWW, depending how the bases are ordered, and belongs to both the cWW and cHW superfamilies (first two columns). For each entry, the chain orientations, parallel (P) or anti-parallel (A), between the 1st and 2nd, 2nd and 3rd and 1st and 3rd nucleotides of the triple are also given, assuming all the nucleobases are in the default anti glycosidic bond configuration. The second appearance of each family in the table is indicated using italic font. Triple families for which instances have not yet been observed in the structure database appear with a gray background.
Symbolic searches for base triples using FR3D
We used FR3D to carry out symbolic searches to find all experimentally observed instances of base triples that can be described as combinations of two (or possibly three) annotated base pairs. We call these ‘regular base triples’. We searched annotations of the NR dataset of RNA-containing PDB structures (described in section 1 of Supplementary Data S0) for all combinations of three bases in which the first and second bases form a given base pair (WC or non-WC), the second and third bases form some type of non-WC base pair and the first and third bases are not stacked. In a small number of cases, the first and third bases also form an annotated pair, but it is not necessary to specify this in the search to obtain these instances. With this procedure, the same triple can be found more than once, so we counted the base triples systematically, to avoid duplication. Moreover, some PDB files have duplicated chains. FR3D identifies these and retains just one instance.
The NR data set we used has 31 617 nucleotides in non-duplicated chains. Of these nucleotides, 25 245 (80%) form at least one base pair, 4773 (15%) are part of at least one base triple, 2368 (7.5%) participate in more than one base pair simultaneously and 146 (0.46%) form three base pairs simultaneously. We find 13 906 base pair instances, 1864 distinct sets of three nucleotides making base triples having exactly two base pairs and 276 base triples with three base pairs, for a total of 2140 base triple instances. The 2140 base triples involve just 4773 nucleotides because many nucleotides participate in more than one base triple. The 146 nucleotides that make three base pairs simultaneously are each involved in three distinct base triples (i.e. forming base quadruples).
In the next sections, we systematically enumerate the regular base triple families by considering each triple superfamily in turn and summarizing the results of database searches we carried out to identify instances of each triple family. Details are compiled in Table 1.
The cWW base triple superfamily
We begin our enumeration of base triple families with base triples in which the first two bases are paired cis Watson–Crick/Watson–Crick (cWW). These triple families belong to the cWW base triple superfamily. We ask how many distinct ways the third base can pair with the second to form a base triple, given that the first two bases are paired cWW. As the second (central) base already interacts with the first base using its Watson–Crick (W) edge, it can pair with the third base using its Hoogsteen (H) or Sugar (S) edges. However, the third base can, in principle, interact with the second base using either its W, H or S edges. Moreover, this interaction can occur with the glycosidic bonds of the second and third bases oriented cis or trans to each other. Thus, there should be 12 different base triple families based on cWW pairing between bases 1 and 2. We group these base triple families into the ‘cWW base triple superfamily’ (superfamily 1 in Table 1) and list them, numbered 1–12, in the first column of the upper part of Table 1. Further details regarding each member of the cWW triple superfamily are provided in Table 2, including base pair symbols introduced in previous work to designate the two component base pairs of each triple family and a schematic representation using triangles to represent the orientations of the bases in the triple (16,18).
Table 2.
Example of a triple superfamily
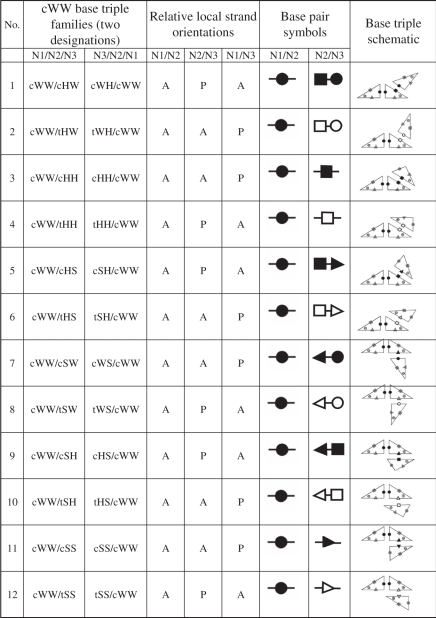 |
The cWW base triple superfamily comprises all geometric base triple families having two bases paired cWW and comprises the most instances. Instances are known for each triple family in the cWW superfamily. The cWW triple families are numbered as in column 1 in Table 1. The alternative names for each triple family are given in columns 2 and 3, and depend on the order in which the nucleotides in the triple are listed. The local strand orientations, parallel (P) or anti-parallel (A) are given in columns 4 through 6. Leontis/Westhof symbols are shown for representing the component base pairs of the triple in columns 7 and 8. In these symbols, circles denote Watson–Crick (W) edges, squares Hoogsteen (H) edges and triangles, Sugar (S) edges, while solid symbols indicate cis base pairs, and open symbols trans base pairs. Finally, column 9 provides schematic representations of each triple family using triangles to represent the bases. Base edges (W, H, S) are labeled with the corresponding symbols (small circles, squares and triangles).
Using the symbolic search capabilities of FR3D, we searched the NR set of RNA-containing PDB structures and found instances of all 12 predicted base triple families of the cWW superfamily. They are shown with white background in the first column of Table 1 to indicate that instances have been observed for each.
The tWW base triple superfamily
The same considerations lead to the prediction that there are potentially 12 families of base triples based on trans Watson–Crick/Watson–Crick (tWW) pairing between the first two bases of the triple. All these triple families are distinct from those in the cWW superfamily; they are numbered 13–24 in Table 1 (superfamily 2, first column, lower section). Using FR3D, we found instances of 10 of these triple families. As for the cWW families, these are shown with white background in Table 1. The two base triple families we did not find in the tWW superfamily are shaded gray in Table 1 (families 13 and 21).
The cWH and tWH base triple superfamilies
Next we considered base triple families in which the first two bases form cis or trans Watson–Crick/Hoogsteen (cWH or tWH) base pairs (triple superfamilies 3 and 4 in Table 1). In these triples, the first base uses its W edge while the second base uses its H edge. Thus, the second base has free W and S edges to pair with the third base. To avoid double counting base triple families already enumerated in the cWW and tWW superfamilies, we exclude triples in which cWW or tWW base pairs form between the second and third bases, and obtain 10 new potential base triple families based on cWH pairing and 10 others based on tWH pairing between the first two bases of the triple. The duplicated triple families are also listed in Table 1, designated with their original numbers and displayed in italic font in Table 1. Using FR3D to carry out symbolic searches, we found instances for seven triple families in the cWH superfamily and 11 families in the tWH superfamily. Note that the cWH and tWH superfamilies also contain 12 base triple families in all, because as noted above, each base triple family belongs to two superfamilies.
The cWS and tWS base triple superfamilies
Superfamilies 5 and 6 comprise base triple families derived by formation of cis or trans Watson–Crick/Sugar-edge (cWS or tWS) base pairs between the first two bases of the triple. In these triples, the second base has free W and H edges for pairing with the third base. Excluding triples in which the second base forms cWW, tWW, cHW or tHW base pairs with the third base, we predict eight new base triple families for the cWS and tWS superfamilies. We found examples for eight families belonging to the cWS superfamily, five of which are new, but only for six families belonging to the tWS superfamily, two of which are new.
The cHW and tHW base triple superfamilies
The cWH and tWH superfamilies do not include base triples in which the first and second bases switch roles, such that the H edge of the first base pairs with the W edge of the second base. Because it is the second base that interacts with the third base, we obtain additional triples by considering families of triples based on cHW and tHW pairing between the first two bases. In the cHW and tHW superfamilies, the second base can pair with the third base using its H or S edges (Table 1, superfamilies 7 and 8). Excluding triples in which the second base forms cHW, tHW, cSW or tSW pairs with the third base, we predict eight new base triple families for the cHW and eight for tHW superfamilies. Using FR3D, we obtained instances representing eight new families, two in the cHW superfamily and six in the tHW superfamily for a total of five observed cHW families and 10 observed tHW families.
The cHH and tHH base triple superfamilies
In these superfamilies (9 and 10 in Table 1), the second base has free W and S edges. Excluding all triples involving the Watson–Crick edge of the second base as well as cSW and tSW pairs between the second and third bases, we predict six additional base triple families for the cHH superfamily and six for the tHH superfamily. There are very few cHH base pairs in the structure database, because it is difficult to form these base pairs without steric clashes between backbone atoms, and so we only find two triple families belonging to the cHH superfamily. However, we do find instances for six tHH base triple families in the database, including three new ones (families 86–88).
The cHS and tHS base triple superfamilies
In superfamilies 11 and 12, the second base can pair with the third base using its W or H edge. Excluding all pairs between the second and third bases except cWS, tWS, cHS and tHS, we obtain four more possible base triple families for each of these triple superfamilies. We found instances for a total of three triple families in the cHS and six in the tHS superfamilies.
The cSW and tSW base triple superfamilies
In the cSW and tSW superfamilies, the second base can interact with the third base using its H or S edges (superfamilies 13 and 14). We predict an additional four possible base triple families in the cSW superfamily and four in the tSW superfamily. Using FR3D, we found instances belonging to six new families for a total of six observed families in the cSW superfamily and seven in the tSW superfamily.
The cSH and tSH base triple superfamilies
Finally, we must consider additional families based on cSH and tSH base pairing between the first and second bases (superfamilies 15 and 16). We found instances for each of the additional four base triplefamilies, which involve cSS and tSS pairing between the second and third bases, for a total of eight triple families in the cSH superfamily and eight in the tSH superfamily.
The cSS and tSS base triple superfamilies
No additional families are obtained by considering cSS or tSS pairing between the first two bases (superfamilies 17 and 18). Triples that involve cSS or tSS are actually common in RNA structures, but were already enumerated in the other triple superfamilies. Combining previous searches, we find instances for 10 triple families belonging to the cSS superfamily and another 10 belonging to the tSS superfamily.
Unobserved base triple families
The symbolic and geometric searches we conducted found instances for 68 of the 108 potential regular base triple families predicted by enumeration. This raises the question why we do not find instances for the other 40 families. One possible explanation is that some of the base triples are so rare that no instances occur in the current atomic-resolution 3D structure database. Another explanation is that some of the triples may simply be impossible to form in RNA for stereo-chemical reasons. We consider each in turn.
We can estimate the expected frequencies of regular base triple families by multiplying the observed frequencies of the component base pairs, obtained in previous work (18) and normalizing. The data are compiled in Table 3, where each entry provides the estimated frequency (upper number in each cell) and the observed frequency (lower number in each cell) of the regular base triple composed of the base pairs given in the column and row headings. Cells with white background indicate observed base triple families, whereas gray background indicates base triple families that have not been observed yet. Three base triple families are observed much more often than expected. The tHW-cHS family occurs 12 times more often than expected, almost certainly because of its central role in the widespread sarcin–ricin and similar motifs. The cHW-cHW family occurs 12 times more often than expected, but this is largely because it forms the well-studied G quadruplex. Each of these families has a small number of average clashes per model. The large discrepancies between observed and estimated frequencies for some families (e.g. tWW-cHH), however, is probably not statistically significant given the very small number of instances in these cases.
Table 3.
Estimated and observed frequencies (%) for regular base triple families
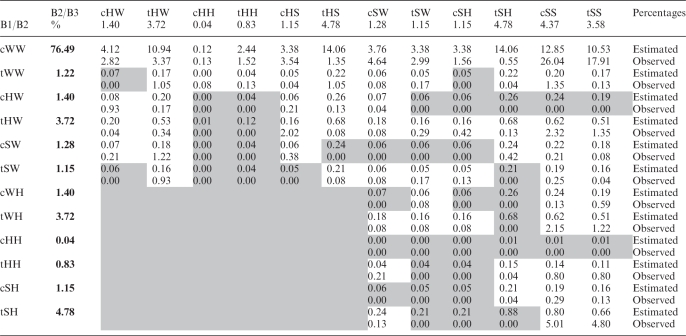 |
Each cell corresponds to the triple family composed of base pairs given in the column and row headers. The estimated frequencies (upper box in each cell) were obtained by multiplying the observed frequencies of the component base pair families (provided in bold font in row 2 and in column 2) and normalizing. Cells with white background indicate base triple families with observed instances. Cells with gray background indicate base triple families for which instances have not yet been observed.
The estimated frequencies for all but 10 of the unobserved base triple families are quite low, 0.12% or less. However, a number of observed base triple families also have low estimated frequencies, so statistical considerations alone probably do not explain the absence of all the unobserved families with low estimated frequencies, or the absence of the remaining 10 unobserved base triple families with relatively high estimated frequencies (0.19–0.88%).
It is also possible that some base triple families are not observed because of steric clashes between the base and/or backbone atoms of the interacting nucleotides. We built 3D models for all possible base combinations of the 40 unobserved base triple families, as described in section 4 of Supplementary Data S0, and examined them for severe steric clashes. We also built models for all unobserved but predicted base combinations in the 68 observed families. A summary of the calculated clash data is provided on the web page http://rna.bgsu.edu/Triples/summary.php. Some, but by no means all, of the triple families with low estimated frequencies exhibit large average numbers of steric clashes in their 3D models, and this may be the definitive factor explaining their absence.
Next we focus attention on the 10 triple families for which no instances are observed yet a significant number is expected statistically. This includes the triple families cSW/tHS, cHW/tSH, cHW/cSS, cHW/tSS, tSW/tSH, cWH/tSH, tWH/tSH, tSH/tSW, tSH/cSH and tSH/tSH. Three of these families, cHW/tSS, cHW/tSH and cSW/tHS, are among the families with the most cumulative clashes and the most average number of clashes per model. It is likely that stereo-chemical reasons preclude these families from forming. Six of the remaining seven families involve the tHS (or tSH) base pair in combination with tSW, tWH, cWH, cSH or tSH. The tHS base pair usually occurs as a local interaction, helping to structure hairpin, internal or junction loops. Frequently, loops containing tHS (tSH) form tertiary interactions, resulting in the triples tSH/cSS and tSH/tSS, which occur more frequently than expected (Table 3). Further work will be needed to understand why the other triples involving tHS (tSH) do not occur, as steric and statistical explanations do not appear to be adequate.
Observed and potential three-base combinations in each base triple family
For each base triple family, there are no more than 43 = 64 possible three-base combinations. One can propose potential base combinations for each base triple family based on the known base pair combinations for bases 1 and 2 and bases 2 and 3. In other words, if base 2 can make the appropriate base pairs with base 1 and 3, then the corresponding base triple may also be possible.
Potential three-base combinations
We devised a general method for generating all potential three-base combinations for each base triple family and obtained a total of 3938 potential three-base combinations. This number is much larger than the 297 distinct three-base combinations, distributed over 68 base triple families that we actually observe in the current database (Supplementary Data S2). We illustrate the method using the cWW/cHW base triple family. We write a 4 × 4 matrix for each component base pair family, entering ‘1’ in cells corresponding to base combinations that form base pairs in the given family and ‘0’ in cells that do not. The corresponding matrices for the cWW and cHW base pair families are shown on the left side of Table 4. Thus, for the cWW family, all base combinations except GG form base pairs, but for the cHW family only 9 of the possible 16 base combinations form base pairs.
Table 4.
Observed and potential three-base combinations in the cWW/cHW base triple family
 |
The left panel shows the 4 × 4 matrices for the two component base pair families (cWW and cHW) of this base triple family. The entries in these matrices designated with ‘1’ and green background are those base combinations that form base pairs in the respective base pair families, cWW and cHW. Base combinations that do not form pairs are designated with ‘0’ and pink background. The right panel shows the derived 16 × 4 matrix representing potential three-base combinations of the cWW/cHW base triple family, computed from the 4 × 4 base pair matrices as described in the text. The base combinations that potentially form base triples are indicated with green background. Pink background indicates those base combinations that are not expected to form triples, because of the absence of one or both of the corresponding base pairs. The entries in the base triple matrix are the numbers of observed instances in the NR data set. Tables for all base triple families are available in Supplementary Data S2.
We use these matrices to generate a 16 × 4 matrix to designate all potential three-base combinations in the corresponding base triple family, as shown for the cWW/cHW base triple family in the right panel of Table 4. Green backgrounds in cells indicate potential base triples (entry = ‘1’) and pink backgrounds indicate base triples anticipated not to exist (entry = ‘0’), based on the non-existence of their component base pairs. This 16 × 4 matrix is actually a 2D representation of a 3D 4 × 4 × 4 matrix (i, j, k). The first four rows of the 2D matrix correspond to the elements (i = 1, j = 1 … 4, k = 1 … 4) of the 3D matrix, the second four rows correspond to the elements (i = 2, j = 1 … 4, k = 1 … 4) of the 3D matrix, and so on. Viewed this way, each element (i, j, k) of the 3D matrix for cWW/cHW is obtained simply by multiplying the (i, j ) element of the cWW matrix by the ( j, k) element of the cHW matrix.
Observed counts of three-base combinations
The right panel of Table 4 also displays the number of observed instances of cWW/cHW base triples in the NR dataset. All actually observed three-base combinations have green background, indicating that they are predicted by this method. However, not all potential triples are observed (blank green cells). In fact, a fairly small number of the potential triples are actually observed (see Supplementary Data S2 for all triple families). Some base combinations are represented by very few instances, others are very common. The most common base combination is cWW/cSS GCA with 341 instances. The cWW superfamily is the most populated superfamily, with 132 distinct base combinations populated by 1576 instances (Table 5).
Table 5.
Base triple families—statistics
| Triple super family |
Occurrences |
|||
|---|---|---|---|---|
| No. | Abbreviation | Potential base combination | Observed base combinations | Total instances |
| 1 | cWW | 519 | 132 | 1576 |
| 2 | tWW | 481 | 27 | 98 |
| 3 | cWH | 320 | 15 | 47 |
| 4 | tWH | 323 | 33 | 149 |
| 5 | cWS | 364 | 9 | 13 |
| 6 | tWS | 320 | 4 | 11 |
| 7 | cHW | 219 | 3 | 8 |
| 8 | tHW | 219 | 17 | 150 |
| 9 | cHH | 103 | 0 | 0 |
| 10 | tHH | 240 | 11 | 39 |
| 11 | cHS | 187 | 1 | 3 |
| 12 | tHS | 133 | 7 | 11 |
| 13 | cSW | 192 | 9 | 16 |
| 14 | tSW | 174 | 6 | 9 |
| 15 | cSH | 80 | 5 | 10 |
| 16 | tSH | 60 | 18 | 233 |
| 17 | cSS | 560 | 78 | 915 |
| 18 | tSS | 298 | 46 | 642 |
For each base triple superfamily, the numbers of three-base combinations forming potential (column 3) and observed (column 4) base triples are listed. The total number of observed instances in the non-redundant RNA structure data set used in this study is given in column 5.
In fact, most predicted base combinations are not observed in the current NR dataset. In some cases, this is probably due to statistics: if the component base pair combinations are rare, then the three-base combination is also likely to be rare. However, we do not expect to observe all the potential base combinations for a given triple family even when the component base pairs are not rare and the appropriate base pairs can form between the central base (base 2) and either of the other two bases (bases 1 and 3), because certain three-base combinations are subject to steric clashes or unfavorable electronic interactions between the bases 1 and 3 of the triple. Conversely, favorable interactions between bases 1 and 3 can stabilize particular three-base combinations. To examine these possibilities, we modeled each of the potential base triples that we do not observe in the structure database and examined the structures for favorable or unfavorable interactions between the bases 1 and 3. We also examined structures of base triples that occur at high or higher than expected frequencies for evidence of favorable interactions.
To generate the 3D models, we added a module to FR3D (20) that identifies the needed base pair exemplars (18) and superposes the bases in each pair that correspond to base 2 of the triple, as described in section 4 of Supplementary Data S0. For example, to generate the 3D model of the UGG cWW/cHW base triple (right panel, Figure 2), the second base of the base pair exemplar for cWW UG was superposed on the first base of the exemplar for cHW GG. The model shows that a steric clash occurs in this triple between O4 of the U (first base of triple) and O6 of the G that is the third base of the triple. This clash explains why this three-base combination is not observed. In the corresponding CGG cWW/cHW base triple, no such clash occurs. Instead a potentially favorable interaction occurs between the electropositive amino group of C and the electronegative carbonyl group of the G (left panel, Figure 2). This triple occurs at high frequency in the database.
Figure 2.
Examples of favorable (A) and unfavorable (B) interactions between the first and third bases of a triple. (A) The exemplar structure of the cWW/cHW CGG base triple which occurs at higher than expected frequency in the structure database. This triple appears to be stabilized by a favorable interaction between the first and third bases of the triple, the H-bond shown with green dotted line between C(N4) and G(O6) of the third base. (B) The steric clash between the first and third base that prevents the cWW/cHW UGG triple from forming. Red dots define clashing van der Waals surfaces.
Expected frequencies of base combinations
We calculated expected occurrence frequencies of three-base combinations for each base triple family by multiplying the occurrence frequencies of the corresponding base combinations of the component base pair families from Ref. (18) and normalizing. The expected base triple frequencies are compared to observed occurrence frequencies in the NR data set for each triple family in Supplementary Data S3.
Table 6 illustrates the use of these data for the cWW/cHW base triple family, taken from Supplementary Data S3. In each cell, the expected and observed frequencies are juxtaposed to identify three-base combinations that occur at significantly higher or lower frequencies than expected. For example, based on the frequencies of the component base pairs, both the cWW/cHW CGG and UGG base triples are expected to have relatively high frequencies. However, only the CGG triple is observed. This prompted us to compare the models shown in Figure 2, to understand why cWW/cHW UGG is not observed, while CGG occurs frequently. The data in Supplementary Data S3 provide the starting point for much further work correlating base triple structure, stability and occurrence frequency.
Table 6.
Estimated and observed frequencies (%) in the cWW/cHW triple family
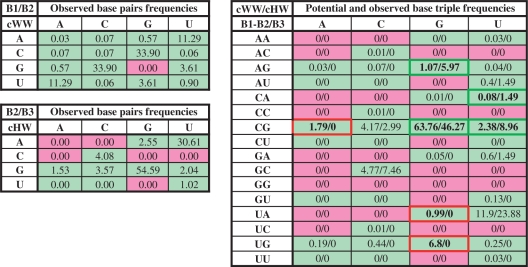 |
The left panel shows the observed frequencies of base combinations in the cWW and cHW base pair families; green and pink background colors indicate base pairs that occur or do not occur, respectively, in the corresponding base pair families. The right panel shows the estimated and observed frequencies for base triples in the cWW/cHW family. Estimated frequencies for each triple combination (first number in each cell) were calculated by multiplying the observed frequencies of the component cWW and cHW base pair combinations and normalizing. Observed frequencies are obtained by normalizing occurrences of cWW/cHW base triples in the NR data set. Green outline and bold font indicates base combinations with high or higher than estimated occurrence frequencies (e.g. CGG) and red outline and bold font indicates those with significantly lower than estimated frequencies (e.g. UGG). As shown in Figure 4, CGG is stabilized by favorable interactions between bases 1 and 3 of the triple, while UGG is destabilized by clashes between bases 1 and 3. Estimated and observed frequencies for all triple families are given in Supplementary Data S3.
Co-planar search for base triples
To test our hypothesis that base triples can be adequately described as combinations of base pairs, we defined a new co-planar neighboring relation between nucleobases in FR3D, to search for possible base triples in the NR data set, making the least number of assumptions about their nature. We define bases as ‘co-planar’ when they lie in the same plane and at a distance conducive to hydrogen bonding, as determined by comparison to 90th percentiles of measures of planarity and interaction distance measured across annotated base pairs. Details are given in section 3 of Supplementary Data S0. Bases that do not meet the strict co-planar criteria may be annotated as near co-planar if they meet somewhat looser criteria.
We searched the NR data set for sets of three RNA nucleobases for which the co-planar or near co-planar relation holds between bases 1 and 2 and between bases 2 and 3, and for which bases 1 and 3 are not stacked. No additional conditions on the sequence or geometry were imposed in this search, and, in particular, no requirement for annotated base pairs was added. This search produced 1317 distinct instances of three bases satisfying the criteria. Of these, 184 instances have three annotated base pairs (i.e. base pairs between bases 1 and 2, between bases 2 and 3, as well as between bases 1 and 3), 731 instances have two annotated base pairs (base pairs between bases 1 and 2 and between bases 2 and 3), 364 instances contain just one annotated base pair and 38 have no annotated base pair at all.
The 915 (184 + 731) instances that comprise two or three annotated base pairs belong to one of the 108 regular base triple families described above and need not be discussed further.
The remaining 402 (364 + 38) instances contain no more than one FR3D-annotated base pair. However, most of these instances do have one or two FR3D-annotated ‘near’ base pairs and therefore represent cases in which the interaction between two of the bases falls just outside the base pair classification limits defined by FR3D. For these cases, we superposed each near base pair with the corresponding base pair exemplar (18) to visually evaluate their structural similarities to annotated pairs and triples. In addition, we conducted geometric searches using FR3D for each of the 402 instances obtained by the co-planar search that comprise no more than one FR3D annotated base pair, to identify structurally similar instances having the same three bases. Manual inspection of the search results showed that 237 of these 402 instances (59%) were also adequately modeled by two known base pairs and thus could also be assigned to one of the regular base triple families already observed.
Manual inspection of the remaining 165 instances revealed that 149 were not triples at all, either because of a lack of edge-to-edge interactions, or, in a small number of cases (ten), due to steric clashes. This left 16 cases of particular interest: four of these 3-nucleotide instances can best be described as belonging to base spirals or ‘2-level bridges’, in which four or more bases interact edge-to-edge but slightly out of plane so that the resulting chain of hydrogen-bonding bases form a spiral with the fourth base stacking on the first base. Such complex base pairing motifs will not be discussed further in this article.
Intermediate base triples
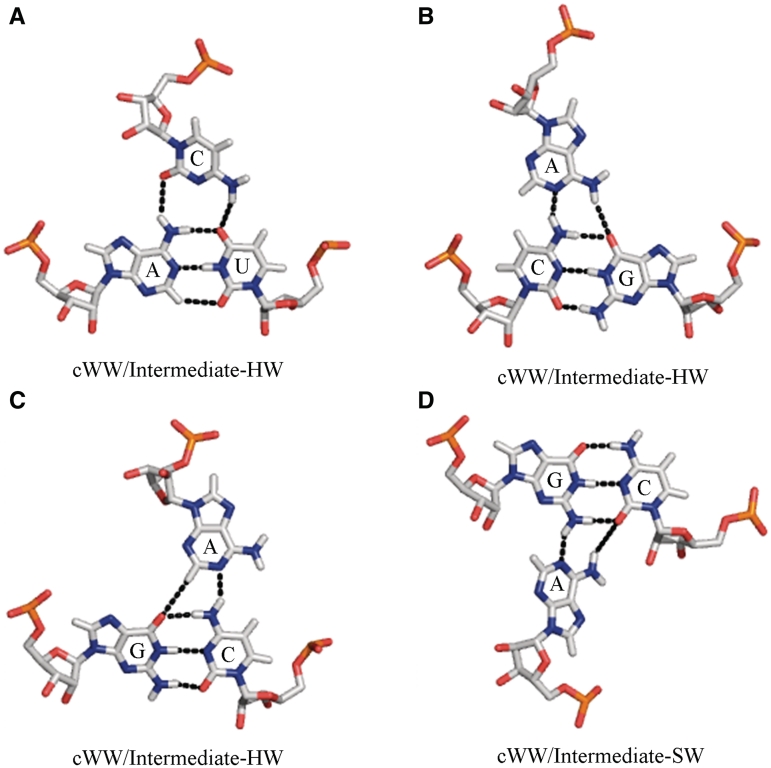
The remaining 12 three-nucleotide instances appear to be genuine ‘intermediate’ base triples, as defined above. Each one involves cWW pairing between bases 1 and 2 of the triple and interaction of the Watson–Crick edge of the third base with atoms belonging to the Hoogsteen or Sugar edges of each of the first two bases, without formation of full base pairs with either one. Four distinct intermediate triples were observed (see Figure 3 and Supplementary Data S4).
Figure 3.
Examples of intermediate base triples in which bases 1 and 2 are cWW paired and the third base interacts with atoms on the adjoining Hoogsteen or Sugar edges of both bases 1 and 2, without forming individual base pairs with either base. (A) Structure of AUC cWW/Intermediate-HW triple A2882/U2836/C2879 from 23S rRNA, PDB file 3I8I (27). (B) Structure of CGA cWW/Intermediate-HW triple G22/C43/A9 from PDB file 2QWY (40). (C) Structure of GCA cWW/Intermediate-HW triple G2580/C2555/A2577 from PDB file 1S72 (42). (D) Structure of GCA cWW/ Intermediate-SW triple G769/C810/A900 from 1J5E (26). In panels (A), (B), and (C) the Watson-Crick edge of the third base interacts with atoms on the Hoogsteen edges of bases 1 and 2, without forming individual base pairs with either one. In panel (D) the Watson-Crick edge of the third base interacts with atoms on the Sugar edges of bases 1 and 2, again without forming individual base pairs with either one.
In the first kind of intermediate base triple, atoms belonging to the Hoogsteen edges of a cWW AU base pair interact with the W edge of C, as shown in Figure 3A. We note that no free-standing CA cWH or CU tWH base pairs have been observed or are expected. Using the co-planar search procedure discussed above, we found two independent instances of this base triple, A2882-U2836-C2879 in Thermus thermophilus 23 S rRNA and A356-U56-C352 in Escherichia coli and T. thermophilus 16 S rRNA (Supplementary Data S4). In these instances, the C(N4) amino group H-bonds with U(O4) and the C(N3) and C(O2) atoms interact with A(N6) amino group, so in each one of these triples the C occupies a position exactly ‘intermediate’ between what it would occupy in a tWH pair with U and in a cWH pair with A.
In the second kind of intermediate base triple, atoms belonging to the Hoogsteen edges of a cWW CG base pair interact with the W edge of A, as shown in Figure 3B. The A(N6) amino group H-bonds with G(O6) and A(N1) interacts with C(N4) so that the triple is ‘intermediate’ between AG tWH and AC cWH. While no free-standing AC cWH base pair has been observed or is expected, AG tWH pairs do exist. However, they are different from what is observed in this base triple. In AG tWH, A(N6) H-bonds with G(N7) and protonated A(N1) H-bonds to G(O6). We found one instance of this intermediate base triple.
The third kind of an intermediate base triple also involves the W edge of an A, but in this case the GC base pair is flipped, as shown in Figure 3C. No free-standing AC tWH base pair has been observed or is expected. While AG cWH base pairs are known, they are different from the interaction observed in this triple, in which A(C2) H-bonds with G(O6) and A(N3) H-bonds with C(N6). We found seven instances of this base triple.
The last case of an ‘intermediate’ base triple involves the W edge of an A interacting with the Sugar edges (minor groove) of a GC base pair, as shown in Figure 3D. We have only observed one instance of this base triple.
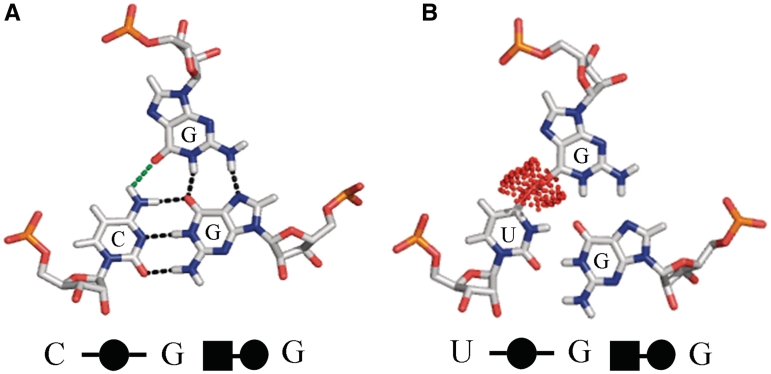
Use of the same edge in pairing to different bases
As mentioned in ‘Symbolic searches for base triples using FR3D’ section, a significant number of triples comprise three base pairs and therefore belong to two different triple families simultaneously. Two types are most prominent, cWW/tSS and tWH/cWS. Generally, when a cWW/tSS base pair forms with an adenosine interacting in the minor groove of the cWW pair, the A forms a tSS pair with the second base of the triple and also a cSS pair with the first base, thus using its sugar edge twice. As there are other cWW/cSS pairs in which the tSS interaction does not form, we classify these triples as cWW/tSS rather than cWW/cSS, and point out that generally there is also a cSS interaction between bases 1 and 3 in the cWW/tSS triples. A second prominent type is the conserved U8-A14-A21 triple in the core of most tRNAs. This triple comprises tWH pairing between U8 and A14 and cWS pairing between A14 A21 pair and is therefore classified tWH/cWS (or cSW/tHW, as it appears in the Base Triple Database). For this, but not all, base combinations in the family, a tSS pair also forms between A21 and U8. As it is not always present, the triple is classified without mentioning the tSS interaction. In a number of other families, we find similar reinforcing interactions between bases 1 and 3 for some base combinations, some of which are fully annotated as base pairs. The proposed classification, however, successfully clusters triples into families according to interactions that occur consistently in all instances.
Database of base triple families
We have prepared an on-line database of observed and modeled base triples, organized in two ways, by geometric triple family and by three-base combination (http://rna.bgsu.edu/Triples). The Base Triple Database provides separate web pages for each base triple family and for each three-base combination. The main page provides access to the individual web pages. To view a particular base triple family, the user clicks on the cell corresponding to that family in the left hand table on the main page. In this table, green colored cells indicate families with observed instances of fully annotated base triples. The number in each cell reports the number of distinct base combinations observed for that family. Yellow cells designate families having no fully annotated instances. These may have near instances, where one or both of the component base pairs are annotated as ‘near’ base pairs by FR3D.
To view the base triple families formed by a particular three-base combination (AAA, AAC, AAG, AAU, etc.), the user clicks on the corresponding cell in the right hand table on the main page. Cells colored green in this table indicate three-base combinations for which some annotated instance has been observed.
On the page corresponding to a given base triple family, the user is presented with a 16 × 4 table with cells corresponding to each of the 64 three-base combinations. The rows correspond to the nucleotides forming the first base pair of the triple. For example, on the page corresponding to the cWW/tHW triple family, the row labeled ‘AU’ contains triples having an AU cWW base pair. The columns correspond to the third base of the triple. Thus, on the cWW/tHW page, the cell corresponding to the row ‘AU’ and column ‘G’ corresponds to the AUG cWW/tHW base triple, which has a UG tHW base pair in addition to the AU cWW base pair.
For each three-base combination for which at least one instance was found having two annotated base pairs in RNA 3D structures, the exemplar base triple is displayed and the cell legend is colored with bright green background. Light green background is used to designate those cases for which the only instances observed have ‘near’ base pairs. In these cases, a 3D model, built as described in the Supplementary Data S0, is displayed. The 3D structures of the near instances are also provided and can be accessed by clicking on the word ‘near’ in the legend of the cell.
The legends of cells corresponding to potential base triples for which no fully annotated or near instances are observed are colored orange and a 3D model is displayed. For base combinations that are not observed and not expected to form, no model is provided and an explanation is provided in the corresponding cell. For example, a cWW/cHW AAA triple is not expected to form because the base combination AA does not form a cHW base pair, and this is stated in the corresponding cell.
For cells that contain instances or models, an interactive 3D structure can be displayed for manipulation by clicking in the check box in the legend. This displays the 3D structure in the Jmol window on the right of the instance table and superposes it on any other triples already displayed to allow comparison. Jmol functions including model manipulation and changes to the display are available. The user can download all exemplar structures as PDB files for further analysis.
Other functions are accessible by clicking the relevant text in the legends of cells. Clicking on the letters of the three-base combination links the user to the corresponding table displaying all triples formed by that combination. Clicking on the number of instances allows one to superpose the structures of all the instances of that base triple in the Jmol window to compare them and assess the degree of variation among them.
The number of clashes calculated using the Probe program (23) is displayed in the legend as clickable text for all exemplars and models. Clicking on the number of clashes brings up a KiNG (24) applet with all-atom contacts shown as dots. Clashes are calculated between base atoms and between base atoms and sugar–phosphate atoms. All models are displayed, regardless of the number of clashes, to allow users to download, refine and evaluate them. The fact that a model is shown in the database is not a statement that an instance exists. The fact that a large number of clashes are calculated for the model is not a statement that it cannot exist.
An interactive gallery showing all known cases of intermediate base triples is accessible from the front page of the triple website.
New versions of the database will be released on a regular basis. All releases can be accessed independently to facilitate referencing. The version of the database described in this article is 1.4. A user guide to the website is accessible from the front page.
Covariation of base triples in the ribosome
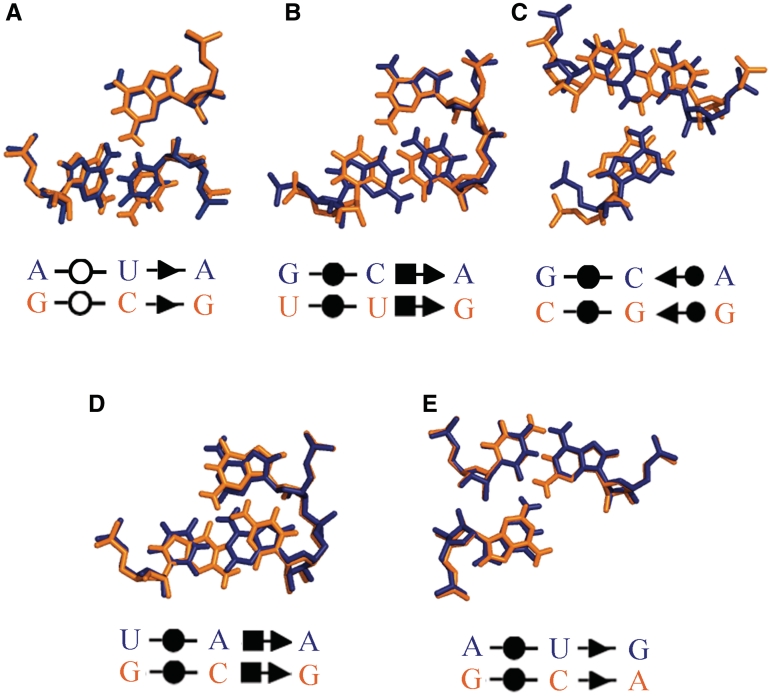
To assess the degree of conservation of base triples in homologous RNA molecules, we identified base triples formed by equivalent nucleotides in the 16S and 23S rRNAs of E. coli and T. thermophilus, for which high-quality 3D structures exist. FR3D annotates 124 regular base triples in E. coli 16S rRNA (25) and 141 in T. thermophilus 16S rRNA (26), of which 91 occur at equivalent positions in the two 16S rRNA structures and belong to the same base triple families. Of these 91 structurally conserved triples, 81 (89%) are also conserved in sequence, having the same three-base combination as well as the same base triple family. FR3D annotates 279 base triples in E. coli 23S rRNA (25) and 267 in T. thermophilus 23S rRNA (27) of which 199 occur at equivalent positions in the two 23S rRNA structures and belong to the same base triple family. Of these 199 structurally conserved triples, 167 (85%) are also conserved in sequence. Among the 42 corresponding base triples from 16S and 23S rRNA that differ in sequence in the E. coli and T. thermophilus structures, most differ in one or two bases, but a few differ at all three bases. We compared five of these by superposition (Figure 4). As shown in Figure 4, the corresponding triples are very similar in structure. This analysis indicates that base substitutions in homologous structures preserve the geometric base triple families we have identified and is consistent with our previous finding that 98% of corresponding base pairs in bacterial ribosomes, including Watson–Crick and non-WC base pairs, are structurally conserved, i.e. isosteric or near isosteric (18). We note that the core triple found by Levitt in tRNA involving A9 also differs in all three bases. He noted in his 1969 paper that when position 9 is G, the third base pair of the D-stem is G12/C24 and when position 9 is A, this pair is U12/A24, and on this basis predicted a triple base interaction between positions 9, 12 and 24 (9). In fact, we find that GCA and UAA are two of the most common variants of the cWW/tHH base triple family, and most instances are from tRNAs.
Figure 4.
Examples of covariation of conserved base triples observed at corresponding positions in E. coli (blue) and T. thermophilus (orange) from 23S rRNA (Upper row) and 16S rRNA (Lower row). Upper row (A): Conserved tWW/cSS base triple in 23S rRNA nucleotides 430/234/219, which is AUA in E.c. and GCG in T. th. (B): Conserved cWW/cHS base triple in 23S rRNA position 757/740/739, which is GCA in E.c. and UUG in T. th. (C): Conserved cWW/cSW base triple in 23S rRNA position 418/409/226, which is GCA in E.c. and CGG in T. th. Lower row (D): Conserved cWW/cHS base triple in 16S rRNA position 644/595/596, which is UAA in E.c. and GCG in T. th. (E): Conserved cWW/cSS base triple in 16S rRNA position 1326/1311/1268, which is AUG in E. coli and GCA in T. th. Structures were taken from the following PDB files: E. coli 23S rRNA, 2QBE (25); E. coli 16S rRNA, 2QAN (25) T. th. 23S rRNA, 3I8I (27) and T. th. 16S rRNA, 1J5E (26).
Effects of modified nucleotides on triple formation
We have also examined base triples formed by modified nucleotides, although these triples are not included in the current Base Triple Database. There is increasing evidence for the presence of modified nucleotides in a wide variety of RNAs (28), in addition to tRNA (29,30) and rRNA (31). The MODOMICS and RNA Modification Databases currently list over 100 modified nucleotides, most of which are rare or only occur in tRNA (32,33). A total of 92 different modifications have been found in tRNA and a recent survey showed that on average ~12% of tRNA residues are modified (34,35).
We extracted all clusters of three neighboring nucleotides comprising at least one modified nucleotide from our NR data set and examined each manually for possible base triples. Given the high percentage of modified bases that tRNAs contain, we also manually examined all tRNA structures in the PDB for triples involving modified bases. We found that a relatively small number of base triples contain modified nucleotides. The results are provided as Supplementary Data S5. No base triples were observed involving highly modified bases. All the base triples involving modified bases that we found belong to triple families that were already identified and, with the exception of one instance, all the base combinations were also observed with unmodified bases. The only new instance involves the non-planar dihydro-uridine (D) modification, forming a CGD tWW/cSW base triple (PDB file 1SER).The corresponding triple with unmodified U has not been observed.
The most complex modifications occur in the anti-codon loop of tRNA, especially at positions 34 and 37. Extensive experimental work shows that these modifications pre-structure the anti-codon for specific interaction with the intended codon sequence(s) by reducing the available conformational space and restricting dynamics (29). It should therefore not be surprising that these bases are not observed forming base triples, at least in biologically relevant structures.
In 1999, Helm et al. provided the first direct evidence for the role of modified nucleotides in RNA folding (36). They used chemical and enzymatic structure probing to compare the folding of native human mitochondrial tRNALys, which folds into the expected cloverleaf structure in vitro, with the folding of the completely unmodified, in vitro transcript of this tRNA and that of a chimeric synthetic version that contained m1A9 as the sole modified base. They found that the unmodified in vitro transcript folds into an extended hairpin rather than the native cloverleaf structure, with unmodified A9 forming a Watson–Crick base pair with U64, which normally is part of the T-stem. The chimeric tRNA containing m1A9, however, correctly folds into the expected cloverleaf structure. These results indicate that methylation of N1 on the Watson–Crick edge of A9 guides correct folding of the tRNA by preventing incorrect pairing with U64. As the Hoogsteen edge of A9 is still available for pairing, it is able to form the conserved base triple predicted by Levitt with the third base pair of the D-stem, which is U12/A23. In our proposed nomenclature, this triple belongs to the cWW/tHH triple family.
Interestingly, the base triples that form the core, conserved tertiary interactions in tRNA frequently involve one or more modified bases. Given the experimental precedents noted above, these modifications, which almost always involve methylation on a specific base edge, thus effectively blocking base pairing on that edge, evidently function to restrict, rather than expand, the kinds of base pairs, and therefore, base triples, that the base can form. For example, methylation of purine N7 or pyrimidine C5 blocks pairing on the Hoogsteen edge, while methylation of purine N1 or pyrimidine N3 blocks pairing on the Watson–Crick edge, and methylation of 2′-O or the G-N2 probably modulates the kinds of Sugar-edge interactions a nucleotide can form. In addition, base methylations are likely to modulate stacking interactions.
Base triples and RNA motifs
Base triples are components or submotifs of recurrent, modular RNA motifs. These motifs have varied functions: they may mediate tertiary interactions in the same RNA, or bind other RNAs, proteins or small molecules. The same motif can have multiple functions in different contexts (37–39). RNA motifs also form as the result of RNA tertiary interactions. In fact, the most common base triples mediate long-range RNA tertiary interactions: The base triples cWW/cSS, cWW/tSS, cWW/cSW, cWW/tSW, tSH/cSS and tSH/tSS are components of tertiary interactions involving the minor groove (6). Less frequent but also important are tertiary interactions involving the Hoogsteen edge. Long-range triples resulting from tertiary interactions at the Hoogsteen edge include cWW/tHH (as in the tRNA 9-12-23 triple), cWW/cHW and cWW/tHW. Another example is the tWW/tHW triple that forms when the GAAA loop binds to the cognate loop receptor (7,41). In contrast, the cWW/cHS and tHW/cHS triples usually occur within internal loops and do not involve a long-range tertiary interaction.
DISCUSSION
With respect to our original goals, we find that although it is not yet possible to state how many different base triples are possible in biological RNA molecules, we are able to provide a fairly accurate upper limit to the number of types of triples (i.e. geometric base triple families) and the number of base combinations within each family that can form stable triples. The fact that almost all observed triples are also predicted by the proposed classification scheme provides a strong argument for its soundness as a conceptual framework for organizing base triples. The small number of exceptions, i.e. observed triples that are not predicted starting from known base pairs, nonetheless fit within a modified framework. They are all intermediate cases in which a cWW base pair forms between two bases of the triple, while the third base interacts in an intermediate geometry with the aligned edges (Hoogsteen or Sugar edges) of the first two bases, without fully base pairing with either base, as discussed above and illustrated in Figure 3.
Furthermore, the fact that almost all three-base combinations that are predicted to not form base triples also are not observed to do so, provides further support for the proposed geometric base triple classification. The only exceptions are the small number of intermediate cases just discussed.
A prominent outcome of our analysis is the prediction of a large number of possible base triples that are not observed. A significant, but relatively small number of these appear to be precluded from forming by steric clashes between the first and third base. In addition, within most families, individual examples of base combinations that cannot form due to steric clashes are evident by inspection. An example was provided in Figure 2. However, for most predicted but not observed three-base combinations, obvious steric clashes are not apparent.
Many of these predicted three-base combinations may not yet be observed in structures, because of the limited nature of the RNA 3D structure database. Therefore, we anticipate that new base triples will be found as new structures are solved at atomic resolution. FR3D now provides a data pipeline for analyzing new structures to identify and classify new base triples and to update the Base Triple Database automatically. Another source of experimental evidence for identifying as yet unobserved base triples is homologous sequence alignments. If a base triple is conserved with regard to geometric triple family in the 3D structures of the distantly related E. coli and T. thermophilus 16S or 23S rRNAs, it is also likely to be conserved in other bacterial species. We are currently developing data pipelines to apply this approach to identify new base triples in structurally annotated sequence alignments of ribosomal RNAs. Preliminary work provides evidence that a significant increase in the number of base triple combinations will be obtained in this way.
Steric and statistical considerations may not be the only factors responsible for the large variations observed in the occurrence frequencies of different three-base combinations within a given base triple family. The example of CGG, the most frequent three-base combination in the cWW/cHW family (Figure 2) suggests the importance of favorable interactions between bases 1 and 3 in stabilizing the triple. Conversely, we anticipate that unfavorable electronic interactions, even in the absence of obvious steric clashes, may destabilize particular three-base combinations. These considerations indicate that detailed energetic analysis using high-level quantum chemical calculations can be expected to play an important role in deepening our understanding of the most frequently occurring base triples, on the one hand, and a significant fraction of the large number of predicted but not observed triple combinations, on the other. We plan to maintain and update the on-line resource that we have developed to facilitate further work on base triples. The database can be studied to select observed, or unobserved but predicted, base triples of special interest for detailed analysis. The models provided can be downloaded and studied further with modeling tools to optimize geometries and calculate and compare intrinsic interaction energies.
CONCLUSION
In summary, we have carried out an exhaustive analysis of observed and potential base triples in atomic-resolution RNA 3D structures deposited in PDB/NDB. We undertook a concerted and systematic effort to test our model for classifying base triples. We thoroughly searched the structure database by solely geometric means for sets of three co-planar bases, without regard to precomputed base pair annotations. We carefully examined each of the instances obtained to find new base triples, not previously classified according to our model. This approach yielded a very limited number (four) of new, intermediate types of triples and effectively validated the generality of the proposed classification.
We have organized the triple data into an on-line electronic resource that includes exemplars of observed instances and 3D models where instances are not available. Because of the dynamic nature of the experimental RNA structure database, and the fact that many predicted base triples are not yet observed, we plan to update the Base Triple Database periodically as new atomic-resolution structures are solved, to identify new, previously unobserved instances of triples and to recalculate occurrence frequencies using updated non-redundant data sets, so as to maintain its usefulness to the community.
We suggest that the proposed geometric classification of RNA base triples provides a robust framework for organizing these data in ways that are useful for RNA 3D structure prediction and modeling, RNA sequence alignment and phylogenetic analysis. All base triples belonging to the same triple family are geometrically similar, having all three glycosidic bonds located and oriented in the same way in 3D space. This explains the high degree of conservation of base triple families revealed by comparison of the 3D structures of the rRNAs of the distantly related bacteria E. coli and T. thermophilus and provides additional strong support for the validity of the proposed classification.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online: Supplementary Methods, Supplementary Data 0–5, Supplementary References [13,18,20,21,23,24,43,44].
FUNDING
National Institutes of Health (grant numbers 1R01GM085328-01A1 to C.L.Z. and N.B.L. and 2R15GM055898-05 to N.B.L.). Funding for open access charge: National Institutes of Health.
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGMENTS
The authors thank the reviewers for careful reading of the article and suggestions for improving the article, especially for the suggestion to include modified nucleotides in the analysis. A.S.A. thanks her parents for giving her a chance to pursue her graduate education in the United States.
REFERENCES
- 1.Leontis NB, Westhof E. A common motif organizes the structure of multi-helix loops in 16 S and 23 S ribosomal RNAs. J. Mol. Biol. 1998;283:571–583. doi: 10.1006/jmbi.1998.2106. [DOI] [PubMed] [Google Scholar]
- 2.Szewczak AA, Moore PB. The sarcin/ricin loop, a modular RNA. J. Mol. Biol. 1995;247:81–98. doi: 10.1006/jmbi.1994.0124. [DOI] [PubMed] [Google Scholar]
- 3.Klein DJ, Schmeing TM, Moore PB, Steitz TA. The kink-turn: a new RNA secondary structure motif. Embo J. 2001;20:4214–4221. doi: 10.1093/emboj/20.15.4214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lescoute A, Leontis NB, Massire C, Westhof E. Recurrent structural RNA motifs, Isostericity Matrices and sequence alignments. Nucleic Acids Res. 2005;33:2395–2409. doi: 10.1093/nar/gki535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Doherty EA, Batey RT, Masquida B, Doudna JA. A universal mode of helix packing in RNA. Nat. Struct. Biol. 2001;8:339–343. doi: 10.1038/86221. [DOI] [PubMed] [Google Scholar]
- 6.Nissen P, Ippolito JA, Ban N, Moore PB, Steitz TA. RNA tertiary interactions in the large ribosomal subunit: the A-minor motif. Proc. Natl Acad. Sci. USA. 2001;98:4899–4903. doi: 10.1073/pnas.081082398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cate JH, Gooding AR, Podell E, Zhou K, Golden BL, Kundrot CE, Cech TR, Doudna JA. Crystal structure of a group I ribozyme domain: principles of RNA packing. Science. 1996;273:1678–1685. doi: 10.1126/science.273.5282.1678. [DOI] [PubMed] [Google Scholar]
- 8.Michel F, Costa M, Massire C, Westhof E. Modeling RNA tertiary structure from patterns of sequence variation. Methods Enzymol. 2000;317:491–510. doi: 10.1016/s0076-6879(00)17031-4. [DOI] [PubMed] [Google Scholar]
- 9.Levitt M. Detailed molecular model for transfer ribonucleic acid. Nature. 1969;224:759–763. doi: 10.1038/224759a0. [DOI] [PubMed] [Google Scholar]
- 10.Michel F, Westhof E. Modeling of the 3-dimensional architecture of group-I catalytic introns based on comparative sequence-analysis. J. Mol. Biol. 1990;216:585–610. doi: 10.1016/0022-2836(90)90386-Z. [DOI] [PubMed] [Google Scholar]
- 11.Michel F, Hanna M, Green R, Bartel DP, Szostak JW. The guanosine binding site of the Tetrahymena ribozyme. Nature. 1989;342:391–395. doi: 10.1038/342391a0. [DOI] [PubMed] [Google Scholar]
- 12.Michel F, Ellington AD, Couture S, Szostak JW. Phylogenetic and genetic evidence for base-triples in the catalytic domain of group I introns. Nature. 1990;347:578–580. doi: 10.1038/347578a0. [DOI] [PubMed] [Google Scholar]
- 13.Deshpande N, Addess KJ, Bluhm WF, Merino-Ott JC, Townsend-Merino W, Zhang Q, Knezevich C, Xie L, Chen L, Feng ZK, et al. The RCSB Protein Data Bank: a redesigned query system and relational database based on the mmCIF schema. Nucleic Acids Res. 2005;33:D233–D237. doi: 10.1093/nar/gki057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Xin Y, Olson WK. BPS: a database of RNA base-pair structures. Nucleic Acids Res. 2009;37:D83–88. doi: 10.1093/nar/gkn676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nagaswamy U, Voss N, Zhang ZD, Fox GE. Database of non-canonical base pairs found in known RNA structures. Nucleic Acids Res. 2000;28:375–376. doi: 10.1093/nar/28.1.375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Leontis NB, Westhof E. Geometric nomenclature and classification of RNA base pairs. RNA. 2001;7:499–512. doi: 10.1017/s1355838201002515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Leontis NB, Stombaugh J, Westhof E. The non-Watson-Crick base pairs and their associated isostericity matrices. Nucleic Acids Res. 2002;30:3497–3531. doi: 10.1093/nar/gkf481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Stombaugh J, Zirbel CL, Westhof E, Leontis NB. Frequency and isostericity of RNA base pairs. Nucleic Acids Res. 2009;37:2294–2312. doi: 10.1093/nar/gkp011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cruz JA, Westhof E. The dynamic landscapes of RNA architecture. Cell. 2009;136:604–609. doi: 10.1016/j.cell.2009.02.003. [DOI] [PubMed] [Google Scholar]
- 20.Sarver M, Zirbel CL, Stombaugh J, Mokdad A, Leontis NB. FR3D: finding local and composite recurrent structural motifs in RNA 3D structures. J. Math. Biol. 2008;56:215–252. doi: 10.1007/s00285-007-0110-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Petrov AI, Zirbel CL, Leontis NB. WebFR3D–a server for finding, aligning and analyzing recurrent RNA 3D motifs. Nucleic Acids Res. 2011;39:W50–W55. doi: 10.1093/nar/gkr249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hoendorf R, Batchelor C, Bittner T, Dumontier M, Eilbeck K, Knight R, Mungall CJ, Richardson JS, Stombaugh J, Westhof E, et al. The RNA Ontology (RNAO): an ontology for integrating RNA sequence and structure data. Appl. Ontol. 2011;6:53–89. [Google Scholar]
- 23.Word JM, Lovell SC, LaBean TH, Taylor HC, Zalis ME, Presley BK, Richardson JS, Richardson DC. Visualizing and quantifying molecular goodness-of-fit: small-probe contact dots with explicit hydrogen atoms. J. Mol. Biol. 1999;285:1711–1733. doi: 10.1006/jmbi.1998.2400. [DOI] [PubMed] [Google Scholar]
- 24.Chen VB, Davis IW, Richardson DC. KiNG (Kinemage, Next Generation): a versatile interactive molecular and scientific visualization program. Protein Sci. 2009;18:2403–2409. doi: 10.1002/pro.250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Borovinskaya MA, Pai RD, Zhang W, Schuwirth BS, Holton JM, Hirokawa G, Kaji H, Kaji A, Cate JHD. Structural basis for aminoglycoside inhibition of bacterial ribosome recycling. Nat. Struct. Mol. Biol. 2007;14:727–732. doi: 10.1038/nsmb1271. [DOI] [PubMed] [Google Scholar]
- 26.Wimberly BT, Brodersen DE, Clemons WM, Morgan-Warren RJ, Carter AP, Vonrhein C, Hartsch T, Ramakrishnan V. Structure of the 30S ribosomal subunit. Nature. 2000;407:327–339. doi: 10.1038/35030006. [DOI] [PubMed] [Google Scholar]
- 27.Jenner LB, Demeshkina N, Yusupova G, Yusupov M. Structural aspects of messenger RNA reading frame maintenance by the ribosome. Nat. Struct. Mol. Biol. 2010;17:555–560. doi: 10.1038/nsmb.1790. [DOI] [PubMed] [Google Scholar]
- 28.Karijolich J, Yu YT. Spliceosomal snRNA modifications and their function. RNA Biol. 2010;7:192–204. doi: 10.4161/rna.7.2.11207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Agris PF, Vendeix FA, Graham WD. tRNA's wobble decoding of the genome: 40 years of modification. J. Mol. Biol. 2007;366:1–13. doi: 10.1016/j.jmb.2006.11.046. [DOI] [PubMed] [Google Scholar]
- 30.Phizicky EM, Hopper AK. tRNA biology charges to the front. Genes Dev. 2010;24:1832–1860. doi: 10.1101/gad.1956510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chow CS, Lamichhane TN, Mahto SK. Expanding the nucleotide repertoire of the ribosome with post-transcriptional modifications. ACS Chem. Biol. 2007;2:610–619. doi: 10.1021/cb7001494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Czerwoniec A, Dunin-Horkawicz S, Purta E, Kaminska KH, Kasprzak JM, Bujnicki JM, Grosjean H, Rother K. MODOMICS: a database of RNA modification pathways. 2008 update. Nucleic Acids Res. 2009;37:D118–D121. doi: 10.1093/nar/gkn710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Cantara WA, Crain PF, Rozenski J, McCloskey JA, Harris KA, Zhang X, Vendeix FA, Fabris D, Agris PF. The RNA Modification Database, RNAMDB: 2011 update. Nucleic Acids Res. 2011;39:D195–D201. doi: 10.1093/nar/gkq1028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sprinzl M, Vassilenko KS. Compilation of tRNA sequences and sequences of tRNA genes. Nucleic Acids Res. 2005;33:D139–D140. doi: 10.1093/nar/gki012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Phizicky EM, Alfonzo JD. Do all modifications benefit all tRNAs? FEBS Lett. 2010;584:265–271. doi: 10.1016/j.febslet.2009.11.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Helm M, Giege R, Florentz C. A Watson-Crick base-pair-disrupting methyl group (m1A9) is sufficient for cloverleaf folding of human mitochondrial tRNALys. Biochemistry. 1999;38:13338–13346. doi: 10.1021/bi991061g. [DOI] [PubMed] [Google Scholar]
- 37.Leontis NB, Lescoute A, Westhof E. The building blocks and motifs of RNA architecture. Curr. Opin. Struct. Biol. 2006;16:279–287. doi: 10.1016/j.sbi.2006.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Leontis NB, Stombaugh J, Westhof E. Motif prediction in ribosomal RNAs Lessons and prospects for automated motif prediction in homologous RNA molecules. Biochimie. 2002;84:961–973. doi: 10.1016/s0300-9084(02)01463-3. [DOI] [PubMed] [Google Scholar]
- 39.Leontis NB, Westhof E. Analysis of RNA motifs. Curr. Opin. Struct. Biol. 2003;13:300–308. doi: 10.1016/s0959-440x(03)00076-9. [DOI] [PubMed] [Google Scholar]
- 40.Gilbert SD, Rambo RP, Van Tyne D, Batey RT. Structure of the SAM-II riboswitch bound to S-adenosylmethionine. Nat. Struct. Mol. Biol. 2008;15:177–182. doi: 10.1038/nsmb.1371. [DOI] [PubMed] [Google Scholar]
- 41.Cate JH, Gooding AR, Podell E, Zhou K, Golden BL, Szewczak AA, Kundrot CE, Cech TR, Doudna JA. RNA tertiary structure mediation by adenosine platforms. Science. 1996;273:1696–1699. doi: 10.1126/science.273.5282.1696. [DOI] [PubMed] [Google Scholar]
- 42.Klein DJ, Moore PB, Steitz TA. The roles of ribosomal proteins in the structure assembly, and evolution of the large ribosomal subunit. J. Mol. Biol. 2004;340:141–177. doi: 10.1016/j.jmb.2004.03.076. [DOI] [PubMed] [Google Scholar]
- 43.Guex N, Peitsch MC. SWISS-MODEL and the Swiss-PdbViewer: an environment for comparative protein modeling. Electrophoresis. 1997;18:2714–2723. doi: 10.1002/elps.1150181505. [DOI] [PubMed] [Google Scholar]
- 44.DeLano WL. The PyMOL User's Manual. San Carlos: DeLano Scientific; 2002. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.