Abstract
DNA decatenation mediated by Topoisomerase II is required to separate the interlinked sister chromatids post-replication. SGS1, a yeast homolog of the human RecQ family of helicases interacts with Topoisomerase II and plays a role in chromosome segregation, but this functional interaction has yet to be identified in higher organisms. Here, we report a physical and functional interaction of Topoisomerase IIα with RECQL5, one of five mammalian RecQ helicases, during DNA replication. Direct interaction of RECQL5 with Topoisomerase IIα stimulates the decatenation activity of Topoisomerase IIα. Consistent with these observations, RECQL5 co-localizes with Topoisomerase IIα during S-phase of the cell cycle. Moreover, cells with stable depletions of RECQL5 display a slow proliferation rate, a G2/M cell cycle arrest and late S-phase cycling defects. Metaphase spreads generated from RECQL5-depleted cells exhibit undercondensed and entangled chromosomes. Further, RECQL5-depleted cells activate a G2/M checkpoint and undergo apoptosis. These phenotypes are similar to those observed when Topoisomerase II catalytic activity is inhibited. These results reveal an important role for RECQL5 in the maintenance of genomic stability and a new insight into the decatenation process.
INTRODUCTION
The RecQ family of helicases has been called the ‘guardians of the genome’ because they play critical roles in maintaining genome integrity. In general, the loss of function of RecQ helicases is thought to increase the mutational load in the cell. Mutations in at least three of the five RecQ helicases, Werner (WRN), Bloom (BLM) and RECQL4 in humans give rise to segmental premature aging phenotypes and cancer predisposition (1). RecQ helicases are characterized by the presence of a DExH and a RecQ-Ct domain (2). RECQL5 is one of the least characterized human RecQ helicases with a C-terminal region that shares no homology with the other family members. There are at least three different isoforms of RECQL5 resulting from alternative RNA splicing. The largest splice variant, RECQL5β (hereafter referred to as RECQL5), localizes to the nucleus and possesses DNA helicase activity (3,4). It has also been shown to harbor an intrinsic strand annealing activity, strand exchange activity and promote branch migration of Holliday junctions (5).
RECQL5 interacts with the MRN complex, Topoisomerase IIIα and β and RNA polymerase II (4,6–8). RECQL5s role in replication has been documented by its co-localization to PCNA during S-phase after hydroxyurea (HU) and UV stress (9). Work from our laboratory demonstrated that RECQL5 interacts with FEN1 and stimulates FEN1 cleavage activity on a variety of DNA substrates that are proposed intermediates in DNA replication (10). Further Recql5 knockout mouse embryonic fibroblasts are sensitive to camptothecin and to thymidine-induced replication stress (11,12). Evidence for a role in homologous recombination (HR) comes from studies in knockout mice where RECQL5 displaced Rad51 from single strand DNA and disrupted Rad51 presynaptic filament formation (13). Recql5 knockout mice are also highly cancer prone and the embryonic fibroblasts cells display elevated levels of sister chromatid exchanges (SCE) (13,14). Interestingly, loss of RecQ5 in Drosophila was shown to cause mitotic defects and more recent work demonstrated that lack of RecQ5 in Drosophila syncytial embryos induced the formation of anaphase bridges (15,16).
Segregation of intertwined chromosomes immediately after replication along with condensation of chromosomes and centromere separation requires the activity of a type II topoisomerase. Topoisomerase IIα is a highly conserved nuclear enzyme which plays a role in relieving topological stress in cells. It catalyzes the transient breaking of both strands of a duplex DNA, which then allows another intact duplex to pass through the break, and rejoining of two strands of cut duplex DNA, thus altering the topology of DNA (17). Topoisomerase IIα is the molecular target for many clinically useful anti-neoplastic drugs (18,19). Studies have also implicated Topoisomerase IIα as a potential prognostic marker in tumors (20–22).
Interestingly, SGS1, the RecQ homolog in yeast, interacts with Topoisomerase II to faithfully segregate chromosomes (23). Though BLM interacts with Topoisomerase IIα, no mammalian RecQ helicase has been shown to influence the decatenation activity of Topoisomerase IIα (24). In this report, we demonstrate the interaction between RECQL5 and Topoisomerase IIα that serves to stimulate the decatenation of DNA by Topoisomerase IIα and we show the co-localization of the two proteins during late S-phase. We describe a Topoisomerase IIα dysfunction in RECQL5-depleted cells, leading to a modest G2/M arrest and the formation of undercondensed and entangled chromosomes. We show that the stimulation of Topoisomerase IIα decatenation activity is RECQL5 specific; thus identifying RECQL5 as a functional homolog of yeast SGS1. Further, RECQL5 depletion in cells leads to increased apoptosis. Based on this work, we propose a model for how RECQL5 may serve to maintain genome integrity.
MATERIALS AND METHODS
Cell lines
HeLa, U2OS, WI38 and HCT116 were purchased from ATCC and grown according to ATCC protocols. HEK 293T (ATCC) used for generating lentivirus was cultured in DMEM media supplemented with 10% Hyclone characterized FBS.
Generation of lentivirus and stable knockdown cells
pLKO.1 vector harboring shRNA constructs targeting human Recql5 was obtained from Sigma Aldrich. The sequences used were shRECQL5-1 targeting the 3′-UTR CCGGGCCTTGTGTTTAGACCTGGATCTCGAGATCCAGGTCTAAACACAAGGCTTTTTG and shRECQL5-2 5′- CCGGCCCTAAAGGTACGAGTAAGTTCTCGAGAACTTACTCGTACCTTTAGGGTTTTTG-3′ targeting the CDS. shRNA construct expressing scrambled sequence was purchased from Addgene (Deposited by the Sabatini lab). Second-generation VSV-G pseudotyped lentiviruses were generated by transient co-transfection of HEK 293T cells with a three-plasmid combination. Briefly, one 50–60% confluent 10 cm dish with 293T cells was transfected using FuGENE 6 (Roche) with 5 µg lentiviral vector, 2.5 µg pCMV ΔR8.2 (Addgene, deposited by the Weinberg lab) and 2.5 µg pCMV VSV-G (Addgene, deposited by the Weinberg lab). Supernatants were collected 48 h after transfection, pooled, filtered through a 0.45-μm filter, flash frozen and stored at −80°C. For lentiviral transduction, 2 ×105 cells were seeded in 10 cm culture plates and transduced the following day with appropriate lentivirus. Forty-eight hours post transduction, the cells were split and selected with puromycin at 2 µg/ml. The cells were maintained in selection media.
RECQL5 antibody generation
A fragment of Recql5 gene corresponding to amino acids 813–963 was PCR amplified and cloned into pTXB1 plasmid (New England Biolabs). The resulting construct was sequenced and transformed into BL21(DE3) CodonPlus RIPL Escherichia coli strain (Stratagene). Bacteria were grown to OD600= 0.7 in LB medium supplemented with 100 μg/ml ampicillin and 50 µg/ml chloramphenicol. Protein expression was induced with 0.2 mM IPTG for 3 h at 37°C. Cells were collected and protein purification performed on chitin resin (New England Biolabs) according to manufacturer's instructions. Purified C-terminal fragment of RECQL5 was sent to Convance Inc. (Denver, PA, USA) for polyclonal antibody production in rabbits. Antibodies were affinity-purified using AminoLink Plus Immobilization Kit (Thermo Scientific).
Growth assays
Scrambled or RECQL5 knockdowns were counted and plated 96 h after transduction and 48 h post selection into eighteen dishes, 104 cells/dish. Three dishes were harvested every 24-h post plating and counted using a Coulter counter. The first set of three harvested on Day 1 were used to normalize as plating controls.
Flow cytometry
To measure cell cycle status, cells were harvested by trypsinization, and were combined with the media from the cell cultures containing floating (mitotic) cells to ensure that the analysis was not biased by excluding M-phase events. After a brief centrifugation, the cell pellets were resuspended in 0.5 ml of FBS and frozen at −70°C. They were stained for cell cycle analysis using the Nuclear Isolation Medium (NIM) procedure (25). This detergent-based one-step method [50 μg/ml propidium iodide (PI) in the presence of RNAse] produces nuclei that have DNA histograms with tight CVs (<1% for lymphocytes), thus yielding more precise cell cycle measurements. The percentage of cells in the G0/G1, S and G2+M phases of the cell cycle were deconvoluted using Multicycle Software (Phoenix Flow Systems, San Diego, CA, USA) and corrected for the effects of debris and doublets by software algorithms (25,26).
To quantitate the percentage of cells directly undergoing DNA synthesis (S-phase), cells were exposed to freshly made 10 μM 5-bromodeoxyuridine (BrdU; Sigma Chemical Co., St Louis, MO, USA) for 30 min, harvested by centrifugation at 4°C, washed twice with cold phosphate-buffered saline (PBS), fixed with 10 ml of cold 70% ethanol at −20°C and stored at this temperature until analysis. For bivariate analysis of BrdU incorporation across the cell cycle, fixed cells were processed following the procedure described by Donahue et al. (27) in which pepsin and acid treatments are used for high resolution BrdU analysis. Incorporated BrdU was detected using a fluorescein (FITC)-conjugated antibody to BrdU (clone B44, BD Biosciences, San Jose, CA, USA) and counterstained with 10 μg/ml PI before flow cytometric analysis. Histograms consisting of 25 000–50 000 cells were acquired on a LSRII (BD Biosciences). In some experiments, cells were incubated with 10 μM BrdU for 24 h. This procedure provides a dynamic approach to access cell division history across one cell cycle (27). Notably the absence of daughter G0/1 cells with diminished BrdU content indicates either diminished time to traverse the cell cycle or block which can be identified on the basis of DNA content and also provides an estimate of the number of quiescent cells in non-G1 phase compartments (i.e. those that fail to take up BrdU in a 24-h time period).
Metaphase spreads and telomeric FISH
RECQL5 depleted and scrambled control U20S cells were treated with 200 μl of 0.5% colcemid for 3 h. Cells were then harvested and immediately incubated in 75 mM KCl for 20 min in 37°C, followed by fixation in ice-cold (3:1) methanol and glacial acetic acid. Metaphase spreads were then made by dropping the cells onto a glass slide. The spreads were stained using 4,6-diamidino-2-phenylindole (DAPI) as described previously (28). For telomeric FISH, metaphase spreads generated above were hybridized with a Cy3-labeled PNA (CCCTAA)3 C-telomere probe (0.3 mg/ml, Panagene) and counterstained with DAPI. Images were captured using CytovisionTM software (Applied Imaging Corp.) on a fluorescence microscope (Axio2; Carl Zeiss, Germany).
Immunofluorescence
Exponentially growing cells were plated on Lab-Tek II chambered glass slides (Thermo-Fisher Scientific) and treated with 2 mM HU (Sigma) for 16 h or 9 μM R03306 for 20 h. HU containing media was then replaced by DMEM and cells were fixed at the time points indicated in text. Cells were fixed and treated with antibodies as described previously (29). The following antibodies were used as described in the ‘Results’ section- Primary: RECQL5 (1:200, rabbit), Topoisomerase IIα (Santa Cruz Biotechnology; 1:200, mouse). Secondary: Donkey anti-rabbit AlexaFluor 488 at 1:1000 dilution and donkey anti-mouse AlexaFluor 647, 1:1000 dilutions. Images were captured with Nikon Eclipse TE2000 confocal microscope and analyzed using Volocity-5 software (PerkinElmer). Co-localization channels (positive product of the difference from the mean (PDM) channels) were generated to highlight the co-localized foci.
DNA decatenation assay
Nuclear extracts were prepared as previously described (30). Recombinant RECQL5 protein was overproduced as a fusion protein with an intein–chitin-binding domain (CBD) self-cleaving affinity tag in E. coli BL21(DE3) CodonPlus RIPL strain (Stratagene), and purified as previously described (31). Recombinant Topoisomerase IIα was commercially available as a Topoisomerase IIα assay kit from VAXRON. Kinetoplast DNA (kDNA) was prepared from Crithidia fasciculata as described previously (32).
DNA decatenation assays were performed as suggested by the manufacturer but using C. fasciculata kDNA. Briefly, varying amounts of nuclear extracts from scrambled or RECQL5-depleted cells (0.5, 1.0 and 2.0 μg) were incubated with 100 ng of kDNA substrate in 10 μl reaction buffer (50 mM Tris–HCl pH 8.0, 100 mM NaCl, 10 mM MgCl2, 2 mM ATP and 1 mM DTT) at 37°C for 15 min. The reactions were terminated by adding SDS and proteinase K and further incubation at 37°C for 30 min. The entire reaction mixtures were separated on 1% agarose gel containing ethidium bromide (0.5 μg/ml). Results were analyzed by ImageQuant version 5.2 (GE Healthcare). To examine functional interaction, Topoisomerase IIα (20 fmol) was incubated with recombinant RECQL5 (0.2, 1.0, 2.0, 10, 20 fmols). Reaction was performed as described above.
Decatenation activities of the nuclear extracts were quantified by measuring the decatenated kDNA band intensity, and shown as relative decatenation (100% for 2.0 μg of scrambled nuclear extracts). Decatenation activities of recombinant proteins were quantified using the same procedure and shown as relative decatenation (one for Topoisomerase IIα alone).
Helicase assay
RECQL5 (8 fmols) was incubated with 1 fmole of a forked duplex substrate (22-bp with a 3′ overhang of 15-bp and labeled on the 5′ end) in 20 μl reaction buffer containing 20 mM Tris pH 7.4, 20 mM NaCl, 25 mM KCl, 2.5 mM MgCl2, 2.5 mM ATP, 1 mM DTT, 100 μg/ml BSA and 5% glycerol at 37°C for 20 min. Increasing amounts of Topoisomerase IIα (8, 40, 80 fmols) were added into the reaction mixtures. Reactions were terminated by addition of 10 μl of stop dye (50 mM EDTA, 38% glycerol, 0.9% SDS, 0.05% bromophenol blue and 0.05% xylene cyanol) and products were separated on 8% native polyacrylamide gel. Results were visualized using a Typhoon phosphorimager (GE Health Sciences) and analyzed by ImageQuant version 5.2 (GE Healthcare).
Western blots
Cells were lysed in RIPA buffer (50 mM Tris–HCl pH 8.0, 150 mM NaCl, 0.1% SDS and 0.5% sodium deoxycholate) in the presence of protease and phosphatase inhibitors (Roche). Protein concentrations of the extracts were determined using Bradford protein assay kit (Bio-rad). Thirty or 50 μg of total protein from each sample was applied onto precast 4–12% SDS–polyacrylamine gel (Invitrogen) and transferred to PVDF membrane (Invitrogen). The following antibodies were used: rabbit-anti RECQL5 generated in house, mouse-anti Topoisomerase IIα (Santa Cruz Biotechnology; 1:1000), mouse-anti PARP1 (Santa Cruz Biotechnology; 1:1000), anti-Chk1 (Santa Cruz Biotechnology; 1:1000), anti-Chk2 (Santa Cruz Biotechnology; 1:1000), anti-phospho Chk2 thr 68 (Santa Cruz Biotechnology; 1:1000), anti-phospho ATM ser 1981 (Santa Cruz Biotechnology; 1:1000), rabbit-anti cleaved PARP1 (Cell Signaling; 1:1000), anti-phospho Chk1 Ser 345 (Abcam; 1:1000), anti-ATM (Abcam; 1:1000), rabbit anti-lamin A/C (Sigma; 1:10 000) and mouse-anti β-Actin (Sigma;1:10 000).
Quantitative RT–PCR
The extent of lentiviral-mediated Recql5 mRNA reduction was measured using real-time PCR analysis conducted using RNA extracted from cells 3 days after infection. cDNA was made from total RNA using MultiScribe™ reverse transcriptase (Roche). A commercial TAQMAN™ probe (Applied Biosystems) for Recql5 (Hs01005415_m1) was used to measure the Recql5 mRNA transcripts. A probe for the house-keeping gene Gapdh (Hs00266705_g1) was used as an internal loading control. All samples were run on a 7900T fast real-time PCR system (Applied Biosystems).
Immunoprecipitations
Cells were washed in ice-cold PBS and lysed in a lysis buffer [50 mM Tris–HCl (pH 7.4), 150 mM NaCl, 1% Triton X-100, 2 mM EDTA] containing Complete Protease Inhibitor Cocktail Tablets (Roche), 1 mM phenylmethylsulfonyl fluoride (PMSF) and 50 μg/ml ethidium bromide for 30 min on ice. The suspension was centrifuged at 14 000 g for 20 min at 4°C. Lysates were pre-cleared with 40 μl slurry of protein A/G-agarose beads (Thermo Sciences) for 1 h at 4°C. Extracts (500 μg each) were incubated with either 4 μg of 0.2 mg/ml rabbit anti-RECQL5, 4 μg of rabbit anti-Topoisomerase IIα antibody (Santa Cruz Biotechnology) or 4 μg of rabbit IgG (Santa Cruz Biotechnology) as a negative control, overnight at 4°C. Immunocomplex was captured by 80 μl of protein A/G beads slurry for 4 h at 4°C, followed by multiple washes. Bound proteins were eluted from beads by boiling in 2× reducing loading dye for 5 min and were analyzed by 4–15 % Tris–Glycine SDS-PAGE. The mouse anti-Topoisomerase IIα antibody (Santa Cruz Biotechnology; 1:1000), rabbit anti–RECQL5 antibody (1:1000) generated in house and anti-mouse HRP conjugated antibody (Amersham; 1:10 000) were used in western Blot analysis following ECL chemiluminescent analysis (Amersham). For detection of RECQL5 protein, rabbit IgG TrueBlot antibody (eBioscience) was used.
RESULTS
RECQL5 depletion is associated with slow proliferation rate, arrest in G2/M phase and late S-phase cycling defects
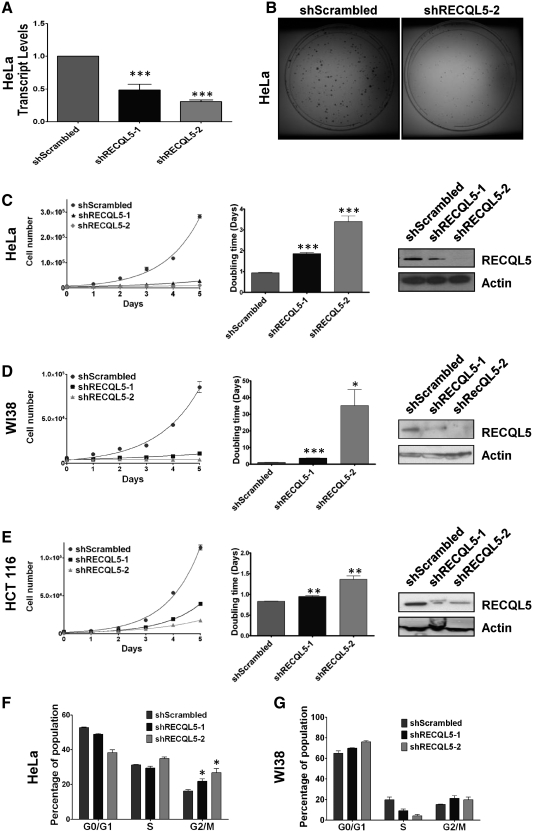
In order to characterize the role of RECQL5 in human cells, RECQL5 was depleted by using lentivirus packaged with plasmids stably expressing a short hairpin (sh)RNA targeted to the 3′-UTR (shRECQL5-1) or the coding sequence (shRECQL5-2). Using quantitative PCR, we measured the Recql5 mRNA and it was reduced by ∼50% with the shRECQL5-1 and >70% with the shRECQL5-2 compared to the control cells (Figure 1A). Upon reduction of the endogenous RECQL5 protein levels, the knockdown cells failed to proliferate at the same rate as their scrambled shRNA counterparts as evidenced by the size of the colonies formed by these cells (Figure 1B). RECQL5 protein levels in HeLa cells were significantly reduced after 2 days of selection as compared to cells transduced with the control (shScrambled) RNA (Figure 1C). Additionally, measurement of the proliferation rate indicated that by Day 10 post lentiviral transduction, control cells grew nearly 7-fold faster than cells depleted using shRECQL5-1, and nearly 15-fold faster than cells depleted using shRECQL5-2 (Figure 1C), suggesting that this phenotype was dose dependent. Similar results were observed in WI38 and HCT116 cells stably expressing RECQL5 targeting shRNAs, indicating that the results were not cell type specific (Figure 1D and E). The rate of cell doubling also appeared to correlate, in a dose-dependent manner, with the RECQL5 expression levels (Figure 1C–E).
Figure 1.
RECQL5 depletion is associated with slow proliferation rate, arrest in G2/M phase, and late S-phase cycling defects. (A) Quantitative RT–PCR analysis of the Recql5 mRNA levels normalized to endogenous GAPDH, in HeLa shScrambled, shRECQL5-1 (n = 3) P = 0.0006 and shRECQL5-2 (n = 3) P = 0.0002, Student's t-test. (B) Methylene blue stained colonies of HeLa shScrambled and HeLa shRECQL5-2. Growth assays were performed with (C) HeLa, (D) WI38 or (E) HCT116 using shScrambled, shRECQL5-1 and shRECQL5-2 as described in ‘Materials and Methods’ section. Error bars represent ±SD, n = 3. The non-linear fit was calculated and R square (Y = Y0*ek*X) values are 0.989 for shScrambled, 0.982 for shRECQL5-1 and 0.7416 for shRECQL5-2 in HeLa, 0.983 for shScrambled, 0.927 for shRECQL5-1 and 0.25 for shRECQL5-2 for WI38 experiments and 0.995 for shScrambled, 0.993 for shRECQL5-1 and 0.976 for shRECQL5-2 in HCT 116 experiments. Graphical representation of the doubling time calculated as an average of two independent growth assays. Error bars represent ±SD, n = 6 for each sample, each day; ***P < 0.0005, **P < 0.005, *P < 0.05, Student's t-test. Western blot showing reduced expression of RECQL5 in (C) HeLa, (D) WI38 or (E) HCT116 whole-cell lysates 96 h following transduction with lentivirus harboring shRECQL5-1, shRECQL5-2 RNA compared to control shScrambled RNA. Equal loading was confirmed by probing with anti-Actin antibody. Quantification of PI staining of asynchronous populations in (F) HeLa and (G) WI38 shScrambled and RECQL5-depleted cells. Error bars represent ±SD, n = 2; *P = 0.0359 between HeLa shScrambled and sh RECQL5-1 and *P = 0.0262 between HeLa shScrambled and shRECQL5-2, Student's t-test.
To further investigate the cellular proliferation defect after RECQL5 depletion, cell cycle analysis of asynchronous cultures was performed. Our results indicated that depletion of RECQL5 caused a modest accumulation of cells in the G2/M phase of the cell cycle, observed in both HeLa (Figure 1F) and in WI38 cells (Figure 1G). We also observed a decrease in G0/G1 phase in RECQL5-depleted HeLa cells and a decrease in S-phase in RECQL5-depleted WI38 cells.
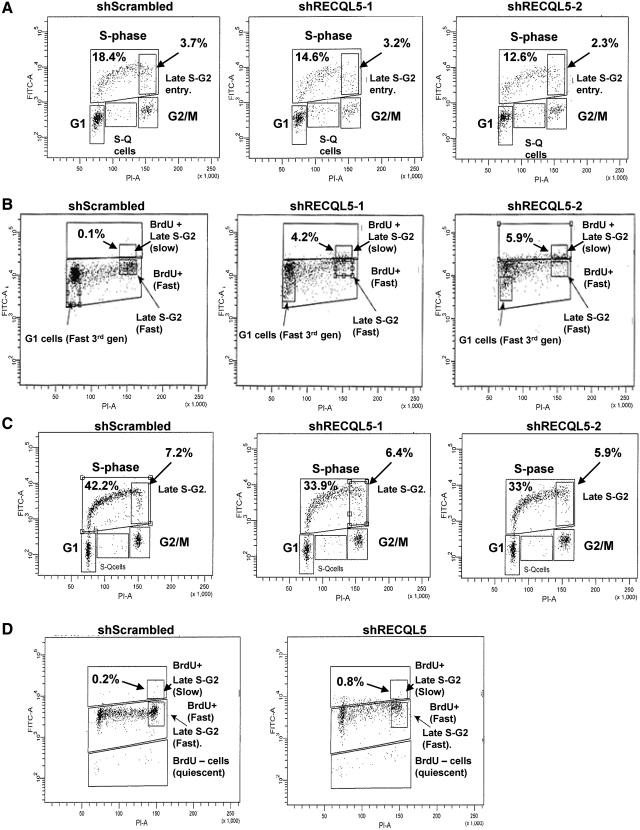
Next, we checked for the cycling of the cells during S-phase using BrdU incorporation, for either 30 min or 24 h, followed by BrdU content analysis across the cell cycle. The 30 min pulse experiments showed an expected decrease in the BrdU incorporation in both HeLa (Figure 2A, 18.4% for control versus 14.6% and 12.6% for RECQL5 shRNA treated) and in HCT116 cells (Figure 2C, 42.2% for control versus 33.9% and 33% for RECQL5 shRNA treated). However, significantly noticeable was the decrease in the overall BrdU positive 4N population in the RECQL5 knockdowns, as compared to the scrambled, suggesting a loss of actively replicating cells at the 4N stage. This was clearly evidenced in the 24 h pulse (Figure 2B and D), where the 4N populations were arrested in the first cell cycle round in the knockdowns when compared to the scrambled (compare quantified BrdU+ late S-G2 populations in Figures 2B and 2D). The RECQL5 knockdown cells finished duplicating their DNA, but they were slower to continue through the 4N stage to the second and subsequent generations. Consistent with these observations, the BrdU intensity in the shScrambled cells was lower than the RECQL5 deficient cells indicating the increased rate of proliferation of the shScrambled cells because BrdU incorporation is halved with every cell cycle. Combining these results with the accumulation of cells at the G2/M phase, we concluded that RECQL5 deficient cells were slower to traverse the G2/M cell cycle post-replication.
Figure 2.
RECQL5 depletion is associated with late S-phase cycling defects. Representative flow cytometry images of (A and B) HeLa and (C and D) HCT116 scrambled and RECQL5 knockdown cells analyzed for the ability to incorporate bromodeoxyuridine (BrdU) and plotted against DNA content in (A and C) a 30 min BrdU pulse and (B and D) 24 h pulse. For panels A and C, the fraction of BrdU positive cells are denoted in the upper left. The late S-G2 of the BrdU positive cells subpopulation, boxed area, is also quantified in the upper right. For panels B and D, the first generation slow late S-G2 population is boxed and quantified.
Stable RECQL5 depletion causes metaphase chromosome defects
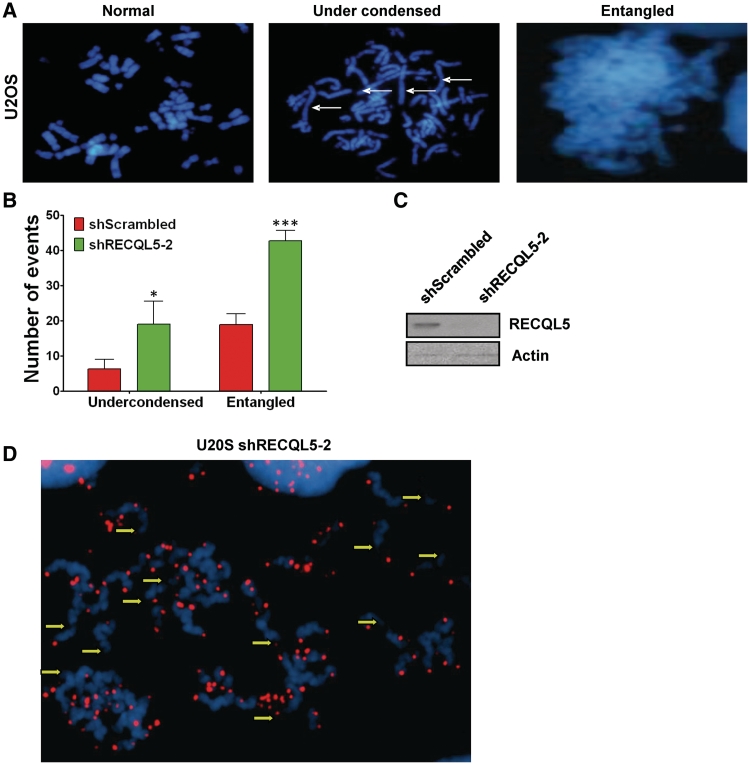
To identify whether RECQL5-depleted cells have a post-replication defect, we generated metaphase spreads from U2OS and WI38 cells expressing shRECQL5-2 or shScrambled RNA. Previous studies in Drosophila have indicated that loss of RecQ5 leads to mitotic defects (15). Strikingly, metaphase spreads from RECQL5 knockdown cells generated nearly 5-fold more undercondensed chromosomes and more than double the number of entangled chromosomes relative to the scrambled controls (Figure 3A–C). In order to confirm that our observation of undercondensed chromosomes was not due to end-to-end chromosome cohesion, we labeled the metaphases with a telomeric probe and were able to identify the chromosomes as separate (Figure 3D). However, not all the telomeres were labeled and DAPI staining was diffused. We attributed the absence of some telomeric signals to the decondensation of DNA, which could account for the diffused pattern of DAPI staining (Figure 3D, indicated by the yellow arrows). Similar results were obtained in WI38 cells after stable depletion of RECQL5 (Supplementary Table S1).
Figure 3.
Stable RECQL5 depletion causes metaphase chromosome defects. (A) Metaphase spreads of U2OS cells after RECQL5 depletion. Two observed phenotypes, undercondensation and entanglement of chromosomes, are shown. (B) Quantification of metaphase spreads from shScrambled and shRECQL5-2 depleted U20S cells. Results are an average of two independent biological experiments. A minimum of 50 metaphase spreads per cell line were analyzed. Error bars represent ±SD, *P = 0.0349, ***P = 0.0006, Student's t-test (C) Western blot showing reduced expression of RECQL5 in U2OS whole-cell lysates 96 h following transduction with lentivirus harboring shRECQL5-2 RNA compared to shScrambled RNA. Equal loading was confirmed by probing with anti-Actin antibody. (D) Fluorescence in situ hybridization in metaphase spreads of U2OS shRECQL5-2 using a telomeric probe. Yellow arrows indicate ends of undercondensed chromatids.
RECQL5 interacts with Topoisomerase IIα
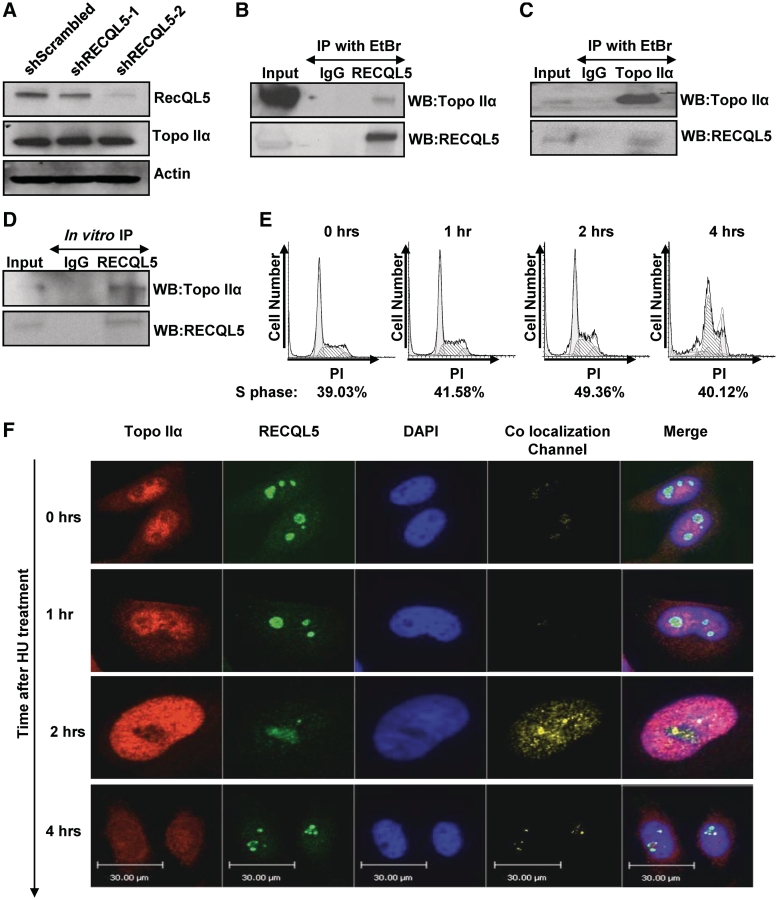
The aberrant condensation of mitotic chromatids has previously been described in cells treated with Topoisomerase II catalytic inhibitors, e.g.: ICRF-193 (33). Additionally, a depletion of Topoisomerase IIα resulted in accumulation of cells in G2/M similar to the one we observed after stable knockdown of RECQL5 (34). Given similar cellular phenotypes, we hypothesized that depletion of RECQL5 generates a Topoisomerase IIα dysfunction. Previously, it has been reported that RECQL5 interacts with RNA polymerase II and represses transcription (35). We therefore examined if changes in RECQL5 levels could result in the modulation of Topoisomerase IIα levels in the cells. Western blot analysis showed that Topoisomerase IIα was expressed at a similar level in RECQL5-depleted and in control cells (Figure 4A). Next, we determined if there was any physical interaction between RECQL5 and Topoisomerase IIα. Immunoprecipitation of endogenous RECQL5 from HeLa cells pulled down Topoisomerase IIα indicating the presence of RECQL5 and Topoisomerase IIα in a complex (Figure 4B). Reciprocal immunoprecipitation of Topoisomerase IIα precipitated RECQL5 as well (Figure 4C). In order to determine whether these proteins interact directly, we performed an in vitro immunoprecipitation of purified RECQL5 with purified Topoisomerase IIα. This experiment demonstrated the possibility of a direct binding of Topoisomerase IIα to RECQL5 (Figure 4D).
Figure 4.
RECQL5 physically interacts with Topoisomerase IIα during S-phase. (A) Western blot of whole cell lysates from HeLa shScrambled, shRECQL5-1 and shRECQL5-2 cells, probed with indicated antibodies. Equal loading was confirmed by probing with anti-Actin antibody. (B) Immunoprecipitation of RECQL5 and (C) Topoisomerase IIα from HeLa whole cell extracts and probed with indicated antibodies. (D) Immunoprecipitation of RECQL5 from a mixture of RECQL5 and Topoisomerase IIα recombinant proteins and probed with indicated antibodies. (E) Representative flow cytometry histograms of HeLa cells analyzed after 2 mM hydroxyurea treatment and released for the time indicated. DNA content stained with PI was plotted against cell number. (F) Confocal images of representative HeLa cells fixed and stained for RECQL5 and Topoisomerase IIα after 2 mM hydroxyurea treatment and then released for the time indicated. Staining protocol is described in ‘Materials and Methods’ section. Images are pseudo colored: green-RECQL5, red-Topoisomerase IIα and blue-DAPI stain of nucleus, with yellow areas indicating the co-localization. The co-localization channel was generated by the Volocity software. Co-localization was determined by the following equation: (Xi −Xmean) (Yi −Ymean) where Xi is the intensity of the voxel for the Red Fluorescence Channel and Yi is the intensity of the voxel for the Green Fluorescence channel (62). The fifth panel is the merge of all three channels.
Since we found that only a small fraction of Topoisomerase IIα interacted with RECQL5, we next inquired whether the direct interaction between RECQL5 and Topoisomerase IIα was cell cycle dependent. It has been previously reported that a type II topoisomerase is required, among other processes, for replication termination (17). The expression of Topoisomerase IIα fluctuates during the cell cycle and peaks during the replicative phase of growth (36). To examine at what stage of the cell cycle RECQL5 and Topoisomerase IIα were acting in concert, we arrested cells in S-phase using a sublethal dose of HU (37). Cells were then released and the co-localization of RECQL5 and Topoisomerase IIα was followed by co-immunofluorescence at the indicated time points (Figure 4F). A cell cycle profile for each time point was analyzed (Figure 4E). Immediately after release from HU, there was low detectable co-localization (as shown by the intensity of the yellow signal under co-localization channel). With increasing time, the co-localization increased and reached a maximum 2-h post release (Figure 4F). The observed co-localization across the nucleus suggests that the two proteins interact around mid-S-phase when the parental DNA strands are being duplicated. The interaction persisted through S-phase and until 4 h after HU release. This was further confirmed by a co-immunoprecipitation experiment, where we observed increased interaction of Topoisomerase IIα with RECQL5 as the cells passed through S-phase (Supplementary Figure 1A and B). These results were consistent with our previous data where we observed a late S-phase cycling defect that may lead to entangled and undercondensed chromosomes.
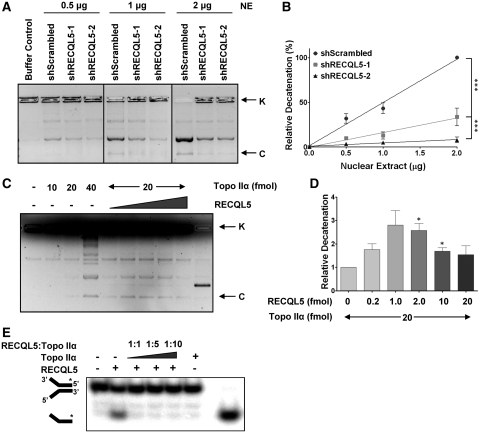
RECQL5 stimulates Topoisomerase IIα decatenation activity
Topoisomerase IIα is indispensible for the decatenation of intertwined sister chromatids after replication and before mitosis (38). Since we identified a high incidence of entangled and undercondensed chromosomes in the metaphase spreads of the RECQL5-depleted cells and a physical interaction between RECQL5 and Topoisomerase IIα, we tested the possibility that RECQL5 could modulate the decatenation activity of Topoisomerase IIα. Comparison of the decatenation activity within knockdown cells to their scrambled counterparts indicated that nuclear extracts of HeLa cells depleted of RECQL5 displayed decreased decatenation of kinetoplast DNA (kDNA) as compared to the scrambled shRNA treated controls (Figure 5A). To measure decatenation activity quantitatively, we used varying amounts of nuclear extracts in the in vitro decatenation assays. Quantification of the relative decatenation revealed that there is a correlation between the RECQL5 levels and the decatenation activity of the nuclear extracts (Figure 5B). Similar results were observed with WI38 (Supplementary Figure 2A and B) indicating that this is not cell type dependent. Interestingly, RECQL5 directly stimulated Topoisomerase IIα catalyzed decatenation of kDNA in vitro, as evidenced by the increase in the decatenated DNA band intensity with increasing RECQL5 (Figure 5C and D). The stimulation was not as profound as the results observed with the cell extracts. This could be because in a cellular context, other protein(s) may influence the activity along with RECQL5 in a complex or perhaps a post-translational modification in RECQL5 stimulates its activity.
Figure 5.
RECQL5 stimulates Topoisomerase IIα catalyzed DNA decatenation. (A) Decatenation assay carried out with nuclear extracts of HeLa shScrambled, shRECQL5-1 and shRECQL5-2 at indicated concentrations. (B) Quantification of the decatenation was performed by measuring the intensity of the decatenated bands. Error bars represent ±SD, n = 3. ***P < 0.0001, f-test (C) DNA decatenation assays were carried out as described in ‘Materials and Methods’ section using recombinant Topoisomerase IIα with recombinant RECQL5 added in increasing amounts as indicated. K-kinetoplast DNA and C-covalently closed minicircles. (D) Quantification of the decatenation was performed by measuring the intensity of the decatenated bands. Error bars represent ±SD, n = 3. *P < 0.01, Student's t-test. (E) Helicase assay using purified RECQL5 and increasing quantity of Topoisomerase IIα.
RECQL5 is a DNA helicase which associates with the replication machinery and accumulates at sites of stalled replication forks (6,11). We tested whether Topoisomerase IIα could influence the helicase function of RECQL5, and observed that the presence of Topoisomerase IIα strongly inhibited the helicase activity of RECQL5 (Figure 5E). This could be attributed to competition for the DNA substrate. It could also be a mechanism of a functional co-regulation of the two proteins; in the presence of supercoils, the helicase activity of RECQL5 could be modulated temporarily by Topoisomerase IIα to accommodate the decatenation activity. In an analogous manner, previous studies indicated that Werner ATPase activity was inhibited by Topoisomerase I (39).
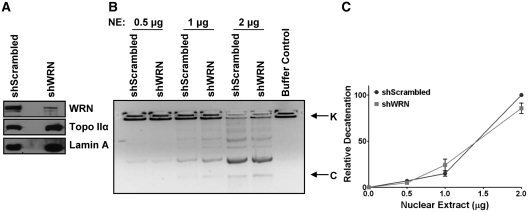
RECQL5, but not WRN, can stimulate Topoisomerase IIα mediated DNA decatenation
RecQ helicase family members WRN, BLM and RECQL5, have been shown to interact with type I topoisomerases (4,39,40). Recently, it was shown that BLM helicase and Topoisomerase IIα interact to maintain genomic integrity (24). Although this interaction served to stimulate BLM helicase activity, BLM helicase was not reported to stimulate DNA decatenation (24). Since Werner helicase has been suggested to play a role in DNA decatenation, nuclear extracts were prepared from U2OS cells stably depleted of WRN and used in the in vitro decatenation assay (41,42). Our results indicate that WRN depletion does not influence the DNA decatenation activity of Topoisomerase IIα in cell extracts (Figure 6A–C). Thus stimulation of Topoisomerase IIα decatenation activity seems to be specific to RECQL5.
Figure 6.
RECQL5, but not WRN, can stimulate Topoisomerase IIα mediated DNA decatenation. (A) Western blot showing reduced expression of WRN in nuclear extracts of U2OS shWRN cells compared to control shScrambled cells. Equal loading was confirmed by probing with a Lamin A specific antibody. (B) Decatenation assay carried out with the nuclear extracts of U2OS shScrambled and shWRN at indicated concentrations. (C) Quantification of the decatenation was performed by measuring the intensity of the decatenated bands. Error bars represent ±SD, n = 3.
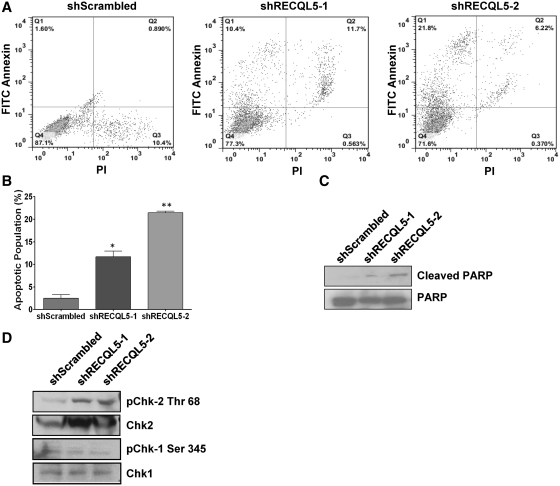
Depletion of RECQL5 increases apoptosis and activates a G2/M checkpoint
Topoisomerase II activity is required to separate replicated daughter DNA strands and a failure to do so results in cell death (43). Similarly prolonged treatment of cells with ICRF193 causes apoptosis, suggesting that the cells are sensitive to the catalytic inhibition of Topoisomerase IIα activity (43,44). We observed that stable depletion of RECQL5 in human cells decreased their survival in culture. In fact, most cells failed to survive after 2 weeks of RECQL5 depletion and those that did had reversed to normal RECQL5 levels. In order to measure the level of apoptosis in the RECQL5 knockdowns, we stained the cells with antibodies to annexin and performed flow cytometric analysis. Our data showed a dose-dependent increase in the percentage of apoptotic cells in RECQL5-depleted HeLa cells. This increase ranged from nearly 6-fold in the shRECQL5-1 to nearly 14-fold in the shRECQL5-2 silenced cells (Figure 7A and B). Similar results were observed in WI38 (Supplementary Figure S3A and B) suggesting that this result was independent of cell type. Consistent with the above data, the ratio of cleaved PARP to total PARP was significantly higher in the RECQL5-depleted cells as compared to the control scrambled cells (Figure 7C).
Figure 7.
Depletion of RECQL5 increases apoptosis and activates a G2/M checkpoint. (A) Dot plots of FITC-Annexin versus PI two parameter flow cytometry of HeLa shScrambled, shRECQL5-1 and shRECQL5-2 cells. (B) Quantification of Annexin positive and PI negative cells in HeLa. Error bars represent ±SD, n = 2; *P = 0.02, **P = 0.002, Student's t-test. (C and D) Western blot of whole cell lysates from HeLa shScrambled, shRECQL5-1 and shRECQL5-2 cells probed with indicated antibodies.
It has previously been reported that inhibition of Topoisomerase IIα catalytic activity or DNA decatenation in HeLa cells can activate a DNA damage-independent checkpoint (45). Although controversial, the checkpoint has been shown to activate Chk2 and be independent of Chk1 phosphorylation (43,44). We therefore checked the levels of phospho-Chk2 and -Chk1 in RECQL5-depleted cells. Our results indicated increased levels of Chk2 phosphorylated at threonine 68, but not Chk1 phosphorylated at serine 345 in the RECQL5-depleted cells as compared to the scrambled controls (Figure7D). This observation was consistent with our cell cycle data where we observed accumulation of cells in G2/M phase and the lack of cells passing the 4N stage of the cell cycle, as the activation of Chk2 at the G2/M phase triggers increased apoptosis.
DISCUSSION
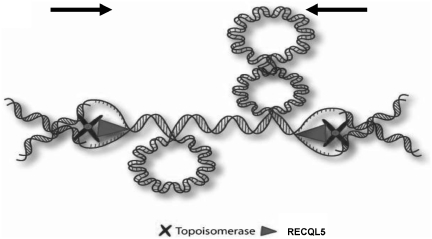
Based on the data presented here, we propose a model (Figure 8) in which the interaction between RECQL5 and Topoisomerase IIα plays a putative role in resolving torsional and replicative stress associated with termination of replication (when the opposing replication forks approach each other). In this model, RECQL5 helicase could resolve the steric constraints that arise due to the formation of positive supercoils during convergence of the opposite replication forks on a linear chromosome. We hypothesize that by unwinding the duplex DNA and converting positive supercoils into interlocked catenates, RECQL5 would provide access for Topoisomerase IIα, which could then resolve the daughter strands. This model is similar to previous models suggesting that a helicase and a type II topoisomerase working together could co-operate in separation of the sister strands (46). Yeast SGS1 is proposed to interact with Topoisomerase II in a similar manner and participate in faithful chromosome segregation (23).
Figure 8.
Model to demonstrate the interaction of RECQL5 and Topoisomerase IIα. Model to demonstrate the interaction of RECQL5 and Topoisomerase IIα functioning to separate daughter chromatids at the end of replication. RECQL5 serves to unwind the parental DNA at the converging replication forks with Topoisomerase IIα behind functioning to decatenate newly formed daughter molecules. The helicase activity is then inhibited till Topoisomerase II performs the decatenation.
Our work also demonstrates a ‘division of labor’ that takes place in the overlapping functional roles of the RecQ helicases. Mammals have evolved to have five different RecQ helicases whereas bacteria and yeast express one (47). Yeast SGS1 has been suggested to be a functional homolog of human WRN and BLM helicases (1,48,49). By virtue of its ability to stimulate Topoisomerase IIα-mediated decatenation, we identify RECQL5 as a mammalian functional counterpart of SGS1. BLM and RECQL5 have been shown to disrupt RAD51 filament formation, and both interact with Topoisomerase IIIα (4,14,40,50). The present study identifies the interaction of RECQL5 and Topoisomerase IIα during replication whereas the interaction between BLM and Topoisomerase IIα has been identified to be highest during mitosis (24). Another distinguishing element of the BLM–Topoisomerase IIα interaction is that Topoisomerase IIα stimulates the helicase activity of BLM whereas BLM was unable to stimulate the decatenation by Topoisomerase IIα (24). This is in stark contrast to what we report here for the RECQL5-Topoisomerase IIα interaction. It could be possible that the two sets of interactions could follow one another, highlighting the importance of RecQ helicases in the maintenance of genomic integrity during the various stages of cell cycle. In this work, we also show that Werner does not influence Topoisomerase IIα-mediated DNA decatenation. Therefore, the ability to affect decatenation appears to be RECQL5 specific.
The functional demarcation is also seen among the RecQ helicases through their interaction with Topoisomerase IIIα. While BLM forms a part of a multiprotein complex and is involved in the dissolution of the sister chromatids, no such function has yet been attributed to the RECQL5-Topoisomerase IIIα interaction (51–53). This could in part explain why RECQL5 and BLM have non-redundant roles in the suppression of crossovers during replication (14).
Recql5 knockout mice display chromosomal abnormalities including a high incidence of sister chromatid exchanges (13). SCE are believed to arise as process of recombination that occurs after double-strand breakage in sister chromatids (54). Though RECQL5 promotes strand exchange on DNA structures that mimic stalled forks, it has been suggested that the frequency of SCE depends on the size of the replicon rather than stalling of replication forks (9,55). Previous reports also suggest that recombinational processes take place in generation of SCEs which could involve the participation of type II topoisomerase (55,56). Indeed, inhibition of Topoisomerase II by ICRF-193 has been demonstrated to increase SCE levels (57). In the absence of RECQL5, it is quite possible that an impaired Topoisomerase IIα bound to DNA could be converted to double strand breaks. This could explain in part the accumulation of H2AX foci observed when RECQL5 is depleted in cells (13); in turn, this could lead to formation of tumors as observed in Recql5 knockout mice (13).
RecQ5 loss in Drosophila causes mitotic defects and more recently was found to induce the formation of anaphase bridges in syncytial embryos (15,16). The role of Topoisomerase IIα in the formation of anaphase bridges is well documented (58). It is therefore likely that a decreased interaction between RECQL5 and Topoisomerase IIα could contribute to the formation of anaphase bridges (59). Moreover, previous work has also demonstrated the link between Topoisomerase IIα activity in the separation of telomeric ends at late S-phase and the occurrence of mitotic abnormalities similar to those described here (60,61). We also observed the interaction of RECQL5 and Topoisomerase IIα during mid-late S-phase and have documented mitotic abnormalities as a consequence of RECQL5 knockdown. The differences in the length of the telomeric ends in mice and humans could perhaps explain why the Recql5 knockout mice do not display the phenotypes we have observed.
In conclusion, this study identifies a novel physical and functional interaction between human RECQL5 and Topoisomerase IIα. This interaction is likely to be an important mechanism by which RECQL5 and Topoisomerase IIα coordinately enhance the stability of eukaryotic chromosomes.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online: Supplementary Table 1, Supplementary Figures 1–4.
FUNDING
This work was supported by the National Institutes of Health Intramural Program of the National Institute on Aging (Z01-AG000726-18). Funding for open access charge: National Institutes of Health.
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
The authors would like to thank Dr Pavel Janscak for the human RECQL5 expression construct and Dr Theresa Shapiro for the kinetoplast DNA. The authors would like to thank Dr Chandrika Canugovi for her help with the model rendition. The authors appreciate the technical assistance of Dr Ramesh Subramanyam (LMBI) with the annexin staining. The authors would like to thank Dr Brian Berquist, Dr Avvaru Suhasini, Dr Morten Scheibye-Knudsen and Dr Ian Hickson (Faculty of Health Sciences, University of Copenhagen) for critically reading this manuscript. M.R. and V.B. conceived and designed the study. M.R., T.T., I.R., A.G., R.W., A.M., T.K. and P.S. performed the experiments and analyzed the data. M.R., D.C. and V.B. interpreted the data. M.R. wrote the manuscript with comments from D.C., T.K. and V.B.
REFERENCES
- 1.Bohr VA. Rising from the RecQ-age: the role of human RecQ helicases in genome maintenance. Trends Biochem. Sci. 2008;33:609–620. doi: 10.1016/j.tibs.2008.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bernstein DA, Keck JL. Domain mapping of Escherichia coli RecQ defines the roles of conserved N- and C-terminal regions in the RecQ family. Nucleic Acids Res. 2003;31:2778–2785. doi: 10.1093/nar/gkg376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sekelsky JJ, Brodsky MH, Rubin GM, Hawley RS. Drosophila and human RecQ5 exist in different isoforms generated by alternative splicing. Nucleic Acids Res. 1999;27:3762–3769. doi: 10.1093/nar/27.18.3762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Shimamoto A, Nishikawa K, Kitao S, Furuichi Y. Human RecQ5beta, a large isomer of RecQ5 DNA helicase, localizes in the nucleoplasm and interacts with topoisomerases 3alpha and 3beta. Nucleic Acids Res. 2000;28:1647–1655. doi: 10.1093/nar/28.7.1647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Garcia PL, Liu Y, Jiricny J, West SC, Janscak P. Human RECQ5beta, a protein with DNA helicase and strand-annealing activities in a single polypeptide. EMBO J. 2004;23:2882–2891. doi: 10.1038/sj.emboj.7600301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zheng L, Kanagaraj R, Mihaljevic B, Schwendener S, Sartori AA, Gerrits B, Shevelev I, Janscak P. MRE11 complex links RECQ5 helicase to sites of DNA damage. Nucleic Acids Res. 2009;37:2645–2657. doi: 10.1093/nar/gkp147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Aygun O, Svejstrup J, Liu Y. A RECQ5-RNA polymerase II association identified by targeted proteomic analysis of human chromatin. Proc. Natl Acad. Sci. USA. 2008;105:8580–8584. doi: 10.1073/pnas.0804424105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Izumikawa K, Yanagida M, Hayano T, Tachikawa H, Komatsu W, Shimamoto A, Futami K, Furuichi Y, Shinkawa T, Yamauchi Y, et al. Association of human DNA helicase RecQ5beta with RNA polymerase II and its possible role in transcription. Biochem. J. 2008;413:505–516. doi: 10.1042/BJ20071392. [DOI] [PubMed] [Google Scholar]
- 9.Kanagaraj R, Saydam N, Garcia PL, Zheng L, Janscak P. Human RECQ5beta helicase promotes strand exchange on synthetic DNA structures resembling a stalled replication fork. Nucleic Acids Res. 2006;34:5217–5231. doi: 10.1093/nar/gkl677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Speina E, Dawut L, Hedayati M, Wang Z, May A, Schwendener S, Janscak P, Croteau DL, Bohr VA. Human RECQL5beta stimulates flap endonuclease 1. Nucleic Acids Res. 2010;38:2904–2916. doi: 10.1093/nar/gkp1217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Blundred R, Myers K, Helleday T, Goldman AS, Bryant HE. Human RECQL5 overcomes thymidine-induced replication stress. DNA Repair (Amst) 2010;9:964–975. doi: 10.1016/j.dnarep.2010.06.009. [DOI] [PubMed] [Google Scholar]
- 12.Hu Y, Lu X, Zhou G, Barnes EL, Luo G. Recql5 plays an important role in DNA replication and cell survival after camptothecin treatment. Mol. Biol. Cell. 2009;20:114–123. doi: 10.1091/mbc.E08-06-0565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hu Y, Raynard S, Sehorn MG, Lu X, Bussen W, Zheng L, Stark JM, Barnes EL, Chi P, Janscak P, et al. RECQL5/Recql5 helicase regulates homologous recombination and suppresses tumor formation via disruption of Rad51 presynaptic filaments. Genes Dev. 2007;21:3073–3084. doi: 10.1101/gad.1609107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hu Y, Lu X, Barnes E, Yan M, Lou H, Luo G. Recql5 and Blm RecQ DNA helicases have nonredundant roles in suppressing crossovers. Mol. Cell Biol. 2005;25:3431–3442. doi: 10.1128/MCB.25.9.3431-3442.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nakayama M, Yamaguchi S, Sagisu Y, Sakurai H, Ito F, Kawasaki K. Loss of RecQ5 leads to spontaneous mitotic defects and chromosomal aberrations in Drosophila melanogaster. DNA Repair (Amst) 2009;8:232–241. doi: 10.1016/j.dnarep.2008.10.007. [DOI] [PubMed] [Google Scholar]
- 16.Sakurai H, Okado M, Ito F, Kawasaki K. Anaphase DNA bridges induced by lack of RecQ5 in Drosophila syncytial embryos. FEBS Lett. 2011;585:1923–1928. doi: 10.1016/j.febslet.2011.04.074. [DOI] [PubMed] [Google Scholar]
- 17.Wang JC. Cellular roles of DNA topoisomerases: a molecular perspective. Nat. Rev. Mol. Cell Biol. 2002;3:430–440. doi: 10.1038/nrm831. [DOI] [PubMed] [Google Scholar]
- 18.Hande KR. Clinical applications of anticancer drugs targeted to topoisomerase II. Biochim. Biophys. Acta. 1998;1400:173–184. doi: 10.1016/s0167-4781(98)00134-1. [DOI] [PubMed] [Google Scholar]
- 19.Li TK, Liu LF. Tumor cell death induced by topoisomerase-targeting drugs. Annu. Rev. Pharmacol. Toxicol. 2001;41:53–77. doi: 10.1146/annurev.pharmtox.41.1.53. [DOI] [PubMed] [Google Scholar]
- 20.Jarvinen TA, Kononen J, Pelto-Huikko M, Isola J. Expression of topoisomerase IIalpha is associated with rapid cell proliferation, aneuploidy, and c-erbB2 overexpression in breast cancer. Am. J. Pathol. 1996;148:2073–2082. [PMC free article] [PubMed] [Google Scholar]
- 21.Jarvinen TA, Holli K, Kuukasjarvi T, Isola JJ. Predictive value of topoisomerase IIalpha and other prognostic factors for epirubicin chemotherapy in advanced breast cancer. Br. J. Cancer. 1998;77:2267–2273. doi: 10.1038/bjc.1998.377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sandri MI, Hochhauser D, Ayton P, Camplejohn RC, Whitehouse R, Turley H, Gatter K, Hickson ID, Harris AL. Differential expression of the topoisomerase II alpha and beta genes in human breast cancers. Br. J. Cancer. 1996;73:1518–1524. doi: 10.1038/bjc.1996.286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Watt PM, Louis EJ, Borts RH, Hickson ID. Sgs1: a eukaryotic homolog of E. coli RecQ that interacts with topoisomerase II in vivo and is required for faithful chromosome segregation. Cell. 1995;81:253–260. doi: 10.1016/0092-8674(95)90335-6. [DOI] [PubMed] [Google Scholar]
- 24.Russell B, Bhattacharyya S, Keirsey J, Sandy A, Grierson P, Perchiniak E, Kavecansky J, Acharya S, Groden J. Chromosome breakage is regulated by the interaction of the BLM helicase and topoisomerase II{alpha} Cancer Res. 2011;71:561–571. doi: 10.1158/0008-5472.CAN-10-1727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wersto RP, Chrest FJ, Leary JF, Morris C, Stetler-Stevenson MA, Gabrielson E. Doublet discrimination in DNA cell-cycle analysis. Cytometry. 2001;46:296–306. doi: 10.1002/cyto.1171. [DOI] [PubMed] [Google Scholar]
- 26.Wersto RP, Stetler-Stevenson M. Debris compensation of DNA histograms and its effect on S-phase analysis. Cytometry. 1995;20:43–52. doi: 10.1002/cyto.990200108. [DOI] [PubMed] [Google Scholar]
- 27.Donahue RE, Sorrentino BP, Hawley RG, An DS, Chen IS, Wersto RP. Fibronectin fragment CH-296 inhibits apoptosis and enhances ex vivo gene transfer by murine retrovirus and human lentivirus vectors independent of viral tropism in nonhuman primate CD34+ cells. Mol. Ther. 2001;3:359–367. doi: 10.1006/mthe.2001.0269. [DOI] [PubMed] [Google Scholar]
- 28.Zijlmans JM, Martens UM, Poon SS, Raap AK, Tanke HJ, Ward RK, Lansdorp PM. Telomeres in the mouse have large inter-chromosomal variations in the number of T2AG3 repeats. Proc. Natl Acad. Sci. USA. 1997;94:7423–7428. doi: 10.1073/pnas.94.14.7423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rossi ML, Ghosh AK, Kulikowicz T, Croteau DL, Bohr VA. Conserved helicase domain of human RecQ4 is required for strand annealing-independent DNA unwinding. DNA Repair (Amst) 2010;9:796–804. doi: 10.1016/j.dnarep.2010.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Dignam JD, Lebovitz RM, Roeder RG. Accurate transcription initiation by RNA polymerase II in a soluble extract from isolated mammalian nuclei. Nucleic Acids Res. 1983;11:1475–1489. doi: 10.1093/nar/11.5.1475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Janscak P, Garcia PL, Hamburger F, Makuta Y, Shiraishi K, Imai Y, Ikeda H, Bickle TA. Characterization and mutational analysis of the RecQ core of the bloom syndrome protein. J. Mol. Biol. 2003;330:29–42. doi: 10.1016/s0022-2836(03)00534-5. [DOI] [PubMed] [Google Scholar]
- 32.Kulikowicz T, Shapiro TA. Distinct genes encode type II Topoisomerases for the nucleus and mitochondrion in the protozoan parasite Trypanosoma brucei. J. Biol. Chem. 2006;281:3048–3056. doi: 10.1074/jbc.M505977200. [DOI] [PubMed] [Google Scholar]
- 33.Ishida R, Sato M, Narita T, Utsumi KR, Nishimoto T, Morita T, Nagata H, Andoh T. Inhibition of DNA topoisomerase II by ICRF-193 induces polyploidization by uncoupling chromosome dynamics from other cell cycle events. J. Cell Biol. 1994;126:1341–1351. doi: 10.1083/jcb.126.6.1341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bower JJ, Karaca GF, Zhou Y, Simpson DA, Cordeiro-Stone M, Kaufmann WK. Topoisomerase IIalpha maintains genomic stability through decatenation G(2) checkpoint signaling. Oncogene. 2010;29:4787–4799. doi: 10.1038/onc.2010.232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Aygun O, Xu X, Liu Y, Takahashi H, Kong SE, Conaway RC, Conaway JW, Svejstrup JQ. Direct inhibition of RNA polymerase II transcription by RECQL5. J. Biol. Chem. 2009;284:23197–23203. doi: 10.1074/jbc.M109.015750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Goswami PC, Roti Roti JL, Hunt CR. The cell cycle-coupled expression of topoisomerase IIalpha during S phase is regulated by mRNA stability and is disrupted by heat shock or ionizing radiation. Mol. Cell Biol. 1996;16:1500–1508. doi: 10.1128/mcb.16.4.1500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Adams RL, Lindsay JG. Hydroxyurea reversal of inhibition and use as a cell-synchronizing agent. J. Biol. Chem. 1967;242:1314–1317. [PubMed] [Google Scholar]
- 38.DiNardo S, Voelkel K, Sternglanz R. DNA topoisomerase II mutant of Saccharomyces cerevisiae: topoisomerase II is required for segregation of daughter molecules at the termination of DNA replication. Proc. Natl Acad. Sci. USA. 1984;81:2616–2620. doi: 10.1073/pnas.81.9.2616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Laine JP, Opresko PL, Indig FE, Harrigan JA, von KC, Bohr VA. Werner protein stimulates topoisomerase I DNA relaxation activity. Cancer Res. 2003;63:7136–7146. [PubMed] [Google Scholar]
- 40.Wu L, Davies SL, North PS, Goulaouic H, Riou JF, Turley H, Gatter KC, Hickson ID. The Bloom's syndrome gene product interacts with topoisomerase III. J. Biol. Chem. 2000;275:9636–9644. doi: 10.1074/jbc.275.13.9636. [DOI] [PubMed] [Google Scholar]
- 41.Franchitto A, Oshima J, Pichierri P. The G2-phase decatenation checkpoint is defective in Werner syndrome cells. Cancer Res. 2003;63:3289–3295. [PubMed] [Google Scholar]
- 42.Opresko PL, Otterlei M, Graakjaer J, Bruheim P, Dawut L, Kolvraa S, May A, Seidman MM, Bohr VA. The Werner syndrome helicase and exonuclease cooperate to resolve telomeric D loops in a manner regulated by TRF1 and TRF2. Mol. Cell. 2004;14:763–774. doi: 10.1016/j.molcel.2004.05.023. [DOI] [PubMed] [Google Scholar]
- 43.Akimitsu N, Kamura K, Tone S, Sakaguchi A, Kikuchi A, Hamamoto H, Sekimizu K. Induction of apoptosis by depletion of DNA topoisomerase IIalpha in mammalian cells. Biochem. Biophys. Res. Commun. 2003;307:301–307. doi: 10.1016/s0006-291x(03)01169-0. [DOI] [PubMed] [Google Scholar]
- 44.Park I, Avraham HK. Cell cycle-dependent DNA damage signaling induced by ICRF-193 involves ATM, ATR, CHK2, and BRCA1. Exp. Cell Res. 2006;312:1996–2008. doi: 10.1016/j.yexcr.2006.02.029. [DOI] [PubMed] [Google Scholar]
- 45.Deming PB, Cistulli CA, Zhao H, Graves PR, Piwnica-Worms H, Paules RS, Downes CS, Kaufmann WK. The human decatenation checkpoint. Proc. Natl Acad. Sci. USA. 2001;98:12044–12049. doi: 10.1073/pnas.221430898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Duguet M. When helicase and topoisomerase meet! J. Cell Sci. 1997;110(Pt 12):1345–1350. doi: 10.1242/jcs.110.12.1345. [DOI] [PubMed] [Google Scholar]
- 47.Hickson ID. RecQ helicases: caretakers of the genome. Nat. Rev. Cancer. 2003;3:169–178. doi: 10.1038/nrc1012. [DOI] [PubMed] [Google Scholar]
- 48.Versini G, Comet I, Wu M, Hoopes L, Schwob E, Pasero P. The yeast Sgs1 helicase is differentially required for genomic and ribosomal DNA replication. EMBO J. 2003;22:1939–1949. doi: 10.1093/emboj/cdg180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Spillare EA, Wang XW, von KC, Bohr VA, Hickson ID, Harris CC. Redundancy of DNA helicases in p53-mediated apoptosis. Oncogene. 2006;25:2119–2123. doi: 10.1038/sj.onc.1209242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Bugreev DV, Yu X, Egelman EH, Mazin AV. Novel pro- and anti-recombination activities of the Bloom's syndrome helicase. Genes Dev. 2007;21:3085–3094. doi: 10.1101/gad.1609007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Bussen W, Raynard S, Busygina V, Singh AK, Sung P. Holliday junction processing activity of the BLM-Topo IIIalpha-BLAP75 complex. J. Biol. Chem. 2007;282:31484–31492. doi: 10.1074/jbc.M706116200. [DOI] [PubMed] [Google Scholar]
- 52.Singh TR, Ali AM, Busygina V, Raynard S, Fan Q, Du CH, Andreassen PR, Sung P, Meetei AR. BLAP18/RMI2, a novel OB-fold-containing protein, is an essential component of the Bloom helicase-double Holliday junction dissolvasome. Genes Dev. 2008;22:2856–2868. doi: 10.1101/gad.1725108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Xu D, Guo R, Sobeck A, Bachrati CZ, Yang J, Enomoto T, Brown GW, Hoatlin ME, Hickson ID, Wang W. RMI, a new OB-fold complex essential for Bloom syndrome protein to maintain genome stability. Genes Dev. 2008;22:2843–2855. doi: 10.1101/gad.1708608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wolff S. Sister chromatid exchange. Annu. Rev. Genet. 1977;11:183–201. doi: 10.1146/annurev.ge.11.120177.001151. [DOI] [PubMed] [Google Scholar]
- 55.Cleaver JE. Correlations between sister chromatid exchange frequencies and replicon sizes. A model for the mechanism of SCE production. Exp. Cell Res. 1981;136:27–30. doi: 10.1016/0014-4827(81)90034-3. [DOI] [PubMed] [Google Scholar]
- 56.Painter RB. A replication model for sister-chromatid exchange. Mutat. Res. 1980;70:337–341. doi: 10.1016/0027-5107(80)90023-8. [DOI] [PubMed] [Google Scholar]
- 57.Dominguez I, Pastor N, Mateos S, Cortes F. Testing the SCE mechanism with non-poisoning topoisomerase II inhibitors. Mutat. Res. 2001;497:71–79. doi: 10.1016/s1383-5718(01)00241-8. [DOI] [PubMed] [Google Scholar]
- 58.Spence JM, Phua HH, Mills W, Carpenter AJ, Porter AC, Farr CJ. Depletion of topoisomerase IIalpha leads to shortening of the metaphase interkinetochore distance and abnormal persistence of PICH-coated anaphase threads. J. Cell Sci. 2007;120:3952–3964. doi: 10.1242/jcs.013730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Laulier C, Cheng A, Stark JM. The relative efficiency of homology-directed repair has distinct effects on proper anaphase chromosome separation. Nucleic Acids Res. 2011;39:5935–5944. doi: 10.1093/nar/gkr187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Stewenius Y, Gorunova L, Jonson T, Larsson N, Hoglund M, Mandahl N, Mertens F, Mitelman F, Gisselsson D. Structural and numerical chromosome changes in colon cancer develop through telomere-mediated anaphase bridges, not through mitotic multipolarity. Proc. Natl Acad. Sci. USA. 2005;102:5541–5546. doi: 10.1073/pnas.0408454102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Stewenius Y, Jin Y, Ora I, de KJ, Bras J, Frigyesi A, Alumets J, Sandstedt B, Meeker AK, Gisselsson D. Defective chromosome segregation and telomere dysfunction in aggressive Wilms' tumors. Clin. Cancer Res. 2007;13:6593–6602. doi: 10.1158/1078-0432.CCR-07-1081. [DOI] [PubMed] [Google Scholar]
- 62.Barlow AL, Macleod A, Noppen S, Sanderson J, Guerin CJ. Colocalization analysis in fluorescence micrographs: verification of a more accurate calculation of pearson's correlation coefficient. Microsc. Microanal. 2010;16:710–724. doi: 10.1017/S143192761009389X. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.