Abstract
We recently reported that a DNA catalyst (deoxyribozyme) can site-specifically hydrolyze DNA on the minutes time scale. Sequence specificity is provided by Watson-Crick base pairing between the DNA substrate and two oligonucleotide binding arms that flank the 40-nt catalytic region of the deoxyribozyme. The DNA catalyst from our recent in vitro selection effort, 10MD5, can cleave a single-stranded DNA substrate sequence with the aid of Zn2+ and Mn2+ cofactors, as long as the substrate cleavage site encompasses the four particular nucleotides ATG^T. Thus, 10MD5 can cleave only 1 out of every 256 (44) arbitrarily chosen DNA sites, which is rather poor substrate sequence tolerance. In this study, we demonstrated substantially broader generality of deoxyribozymes for site-specific DNA hydrolysis. New selection experiments were performed, revealing the optimality of presenting only one or two unpaired DNA substrate nucleotides to the N40 DNA catalytic region. Comprehensive selections were then performed, including in some cases a key selection pressure to cleave the substrate at a predetermined site. These efforts led to identification of numerous new DNA-hydrolyzing deoxyribozymes, many of which require merely two particular nucleotide identities at the cleavage site (e.g. T^G), while retaining Watson-Crick sequence generality beyond those nucleotides along with useful cleavage rates. These findings establish experimentally that broadly sequence-tolerant and site-specific deoxyribozymes are readily identified for hydrolysis of single-stranded DNA.
INTRODUCTION
Deoxyribozymes are particular DNA sequences with catalytic activity (1,2). First reported in 1994 for RNA cleavage (3), which has been their most widely explored activity (4,5), DNA catalysts have been identified by in vitro selection (6,7) for a variety of chemical reactions (8). We recently reported that deoxyribozymes have the unexpected ability to hydrolyze phosphodiester linkages in single-stranded DNA (ssDNA) substrates, forming 5′-phosphate and 3′-hydroxyl products and achieving 1012 rate enhancement with the aid of Zn2+ and Mn2+ cofactors (9,10). This catalysis contrasts with that of Cu2+-dependent DNA-cleaving deoxyribozymes, which oxidatively degrade their DNA substrates (11–13). In follow-up efforts, we explored important practical aspects of our best initial DNA-hydrolyzing deoxyribozyme, named 10MD5. We found that 10MD5 and sequence variants compromise among key attributes such as the ability to tolerate different pH values and the ability to cleave site-specifically at a particular phosphodiester linkage (14). We also found that a modest set of mutations in 10MD5 obviated its Mn2+ requirement, allowing Zn2+ to be used as the sole divalent metal ion cofactor (15).
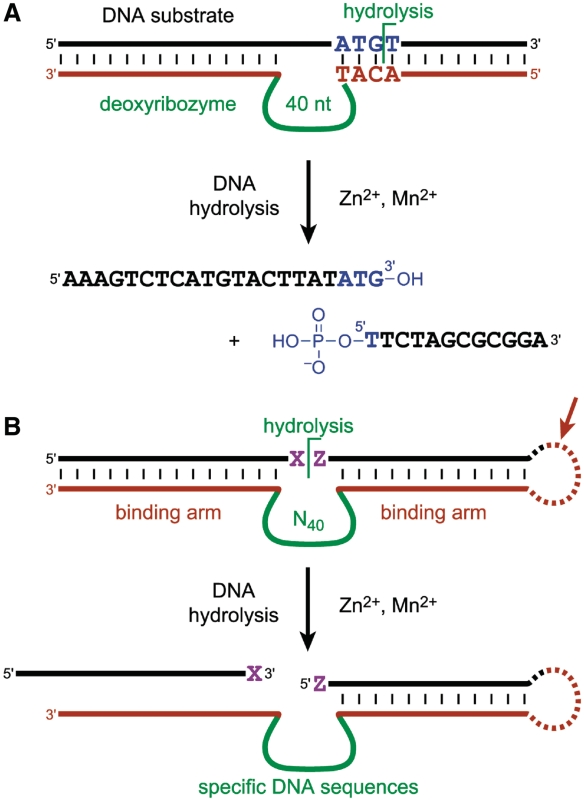
One important attribute of any DNA hydrolysis catalyst (or small set of these catalysts) is the ability to tolerate a reasonably broad range of substrate sequences, such that a new catalyst does not have to be evolved for each new substrate sequence. For currently unknown mechanistic reasons, the 10MD5 deoxyribozyme requires four particular nucleotides in its DNA substrate at the cleavage site, ATG^T, where the hydrolysis products are one oligonucleotide terminating with ATG (3′-hydroxyl) and a second oligonucleotide terminating with T (5′-phosphate) (9,14). All other nucleotides of the DNA substrate may have essentially any identity, as long as Watson-Crick base pairing interactions are maintained between the substrate and the two binding arms of the deoxyribozyme (Figure 1A). This Watson-Crick requirement to bind its substrate means that 10MD5 is inherently highly sequence-specific, unlike many model nuclease systems for which little or no sequence specificity is achieved (16–23). However, any arbitrarily chosen DNA substrate sequence has only 1 in 44 = 256 chance of satisfying 10MD5's ATG^T requirement at the intended cleavage site. Therefore, the 40-nt catalytic region of 10MD5 cannot be relocated within appropriate binding arms and expected to function for most DNA substrates. In other words, 10MD5 is not broadly general in terms of its substrate sequence tolerance.
Figure 1.
The 10MD5 deoxyribozyme, and identification by in vitro selection of new DNA-hydrolyzing deoxyribozymes. (A) 10MD5 and interactions with its DNA substrate (9). Sequence specificity is inherently provided by the Watson-Crick base pairs between deoxyribozyme and substrate. However, the 4-nt ATG^T requirement at the cleavage site means that 10MD5 is not broadly tolerant of different substrate sequences. (B) In vitro selection strategy. The loop on the right side (arrow) enables the selection process but is dispensable for catalysis; its removal enables in trans (intermolecular) assays. The two DNA substrate nucleotides immediately flanking the cleavage site are denoted here generically as X^Z. DNA-catalyzed hydrolysis is sought at or near the XZ region of the substrate.
In this study, we sought to determine if broadly sequence-tolerant DNA-hydrolyzing deoxyribozymes can be identified. The discovery of 10MD5, with its 4-nt ATG^T requirement, still leaves entirely open the question of whether more sequence-tolerant deoxyribozymes exist and can be identified by in vitro selection (Figure 1B). That is, new selection experiments are required to establish whether or not the catalytic regions of DNA-hydrolyzing deoxyribozymes are always so specific for particular cleavage-site sequences such that generality for DNA hydrolysis cannot be achieved. To set, in advance, a reasonable threshold for ‘general sequence tolerance’, we considered what is known to be achievable by RNA-cleaving deoxyribozymes, where RNA cleavage by transesterification is a much easier reaction to catalyze than is DNA cleavage by hydrolysis. Santoro and Joyce reported the 10–23 and 8–17 deoxyribozymes, which cleave RNA while imposing rather modest 2-nt sequence requirements of R^Y (R = purine, Y = pyrimidine) and A^G, respectively (24). Follow-up work has shown that the 8–17 motif is especially versatile, enabling cleavage of nearly any RNA dinucleotide junction sequence using a suitable 8–17-like variant (5,25,26).
For the present efforts, we adopted the benchmark that a 2-nt requirement (analogous to A^G for the parent 8–17 RNA-cleaving deoxyribozyme; see X^Z in Figure 1B) is a sensible threshold for declaring useful sequence generality of a DNA-hydrolyzing deoxyribozyme. If any particular deoxyribozyme requires only two specific nucleotides such as A^G immediately at the cleavage site of its DNA substrate, whereas the flanking substrate nucleotides require (at most) Watson-Crick base pairing to freely chosen deoxyribozyme binding arms, then a collection of merely 42 = 16 such catalysts would suffice to enable hydrolysis of any arbitrarily chosen ssDNA substrate sequence at a desired and wholly predetermined site. It should be emphasized that because DNA hydrolysis is a much more difficult reaction than RNA cleavage by transesterification [e.g. uncatalyzed half-lives of 30 million years for DNA (27) versus 10 years for RNA (28)], the known 2-nt requirements of RNA-cleaving deoxyribozymes do not automatically imply that DNA-hydrolyzing deoxyribozymes are also capable of such short cleavage-site requirements. More extensive contacts by a deoxyribozyme with a DNA substrate could be required to enable the more challenging DNA hydrolysis reaction.
With this threshold for sequence tolerance thus chosen in advance, we performed several sets of in vitro selection experiments. We first aimed to determine the optimal number of unpaired DNA substrate nucleotides to present for hydrolysis while achieving sequence tolerance by the resulting DNA catalysts. We then sought numerous individual deoxyribozymes that catalyze site-specific DNA hydrolysis while being compatible with the 2-nt cleavage site threshold. The results establish that broadly general deoxyribozymes for ssDNA hydrolysis are indeed readily identified via a suitable selection strategy, aided by a key selection pressure to enforce hydrolysis at a predetermined cleavage site within the substrate. These findings not only experimentally establish this broad generality but also reveal an initial set of deoxyribozymes that together enable cleavage of any N^G site in a DNA substrate (i.e. one-fourth of all possible N^N sites), including one deoxyribozyme fully optimized by reselection for efficient hydrolysis at any T^G site. We anticipate that comprehensive follow-up experiments should allow the discovery of a complete set of deoxyribozymes that enable site-specific hydrolysis of essentially any arbitrarily chosen ssDNA substrate, in analogy to what has already been achieved for the very different—and considerably easier to catalyze—reaction of DNA-catalyzed RNA cleavage (5,25,26).
MATERIALS AND METHODS
Oligonucleotides and in vitro selection
All oligonucleotides were prepared by solid-phase synthesis at Integrated DNA Technologies (Coralville, IA) and purified by denaturing 20% or 8% PAGE. To avoid chelation of Zn2+ by adventitious EDTA, all substrates and deoxyribozymes used in kinetic assays were extracted from gels using TN buffer (10 mM Tris, pH 8.0, 300 mM NaCl) lacking EDTA and precipitated with ethanol. The in vitro selection experiments were performed essentially as described previously, using 200 pmol (∼1014 molecules) in each initial round of selection (29,30), where sequence space encompasses 440 ≈ 1024 possibilities (10−10 sampling). Reselection experiments were performed with 25% partially randomized pools, which were prepared using phosphoramidite mixtures as described previously (14,31).
The DNA substrate sequences during the selection process were 5′-TAATACGACTCACTAT-(unpaired)-GAAGAGATGGCGACgga-3′, where the unpaired nucleotides differed for each specific selection experiment (e.g. Figure 2). These are the ‘parent’ (original) substrate sequences, whereas the nucleotides in the binding arms (i.e. all of the nucleotides except those designated as unpaired) were different in the substrates with systematic sequence changes as described in the Figure 2 caption. The 3′-terminal gga residues were RNA rather than DNA to enable ligation of the substrate to the deoxyribozyme pool strand by T4 RNA ligase during each selection round. The DNA substrate sequences used for cleavage assays were the same as those used during selection, except the 3′-terminal –Cgga was replaced with –CGGA (i.e. all DNA).
Figure 2.
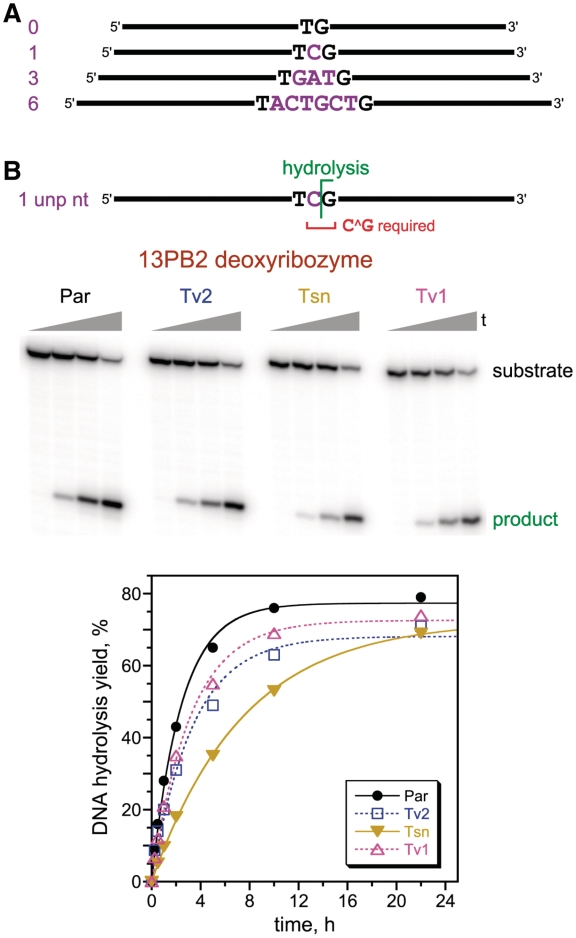
Selections with DNA substrates that have 0, 1, 3, or 6 unpaired nucleotides between the two duplex binding arms. (A) Substrates, showing the unpaired nucleotides (purple). The complete sequences of the binding arms (black) are provided in the Experimental Section. Compare to Figure 1B for structural context of the substrate. The corresponding selections were designated PA–PD. (B) Single-turnover in trans assays of the 13PB2 deoxyribozyme, which was identified from the selection with 1 unpaired nucleotide. 13PB2 requires only C^G at the cleavage site and forms 3′-hydroxyl + 5′-phosphate termini. On the PAGE image are shown representative time points at t = 30 s, 15 min, 2 h and 22 h. The activity is maintained well when all nucleotides of the DNA substrate outside of the cleavage-site C^G are changed from the parent (Par) sequence via systematic changes: transitions (Tsn: A↔G and T↔C); transversions-1 (Tv1: A↔C and G↔T); transversions-2 (Tv2: A↔T and G↔C). All nucleotides in the deoxyribozyme binding arms (except opposite the unchanged C^G of the substrate) were covaried to maintain Watson-Crick base pairing with the substrate. kobs values (h−1): Par 0.41, Tv2 0.30, Tsn 0.13, Tv1 0.32. The 13PB2 sequence is given in Figure 3. 13PB2 also has moderate activity with G^G rather than C^G substrates (Supplementary Figure S3).
The deoxyribozyme pool strand for the selections designated PA–PD and RQ–RT was 5′-CGAAGTCGCCATCTCTTC-N40-ATAGTGAGTCGTATTA-3′, where the two underlined regions denote the substrate binding arms. The deoxyribozyme pool strand for the RV–RY and VH–VL selections (as well as the corresponding reselection, YR, in the case of VJ) was 5′-CGAAGTCGCCATCTCTTC-N40-ATAGTGAGTCGTATTAAGCTGATCCTGATGG-3′. In all instances, the 5′-CGAA was replaced with 5′-CC for individual deoxyribozymes prepared by solid-phase synthesis and used in the single-turnover in trans kinetic assays. For the shorter pool strands (PA–PD and RQ–RT), the two PCR primers used during selection were 5′-CGAAGTCGCCATCTCTTC-3′ (5′-phosphorylated to enable ligation by T4 RNA ligase) and 5′-(AAC)4XTATTACGACTCACTAT-3′ (where X denotes Glen Spacer 18, which is a PEG spacer that stops extension by Taq polymerase and leads to a size difference between the two PCR product strands). For the longer pool strands (RV–RY and VH–VL), the two PCR primers used during selection were 5′-CGAAGTCGCCATCTCTTC-3′ (5′-phosphorylated) and 5′-(AAC)4XCCATCAGGATCAGCT-3′. It was empirically found that the longer pool strands led to more efficient PCR and earlier appearance of catalytic activity, which is why the later selection efforts used the longer pools.
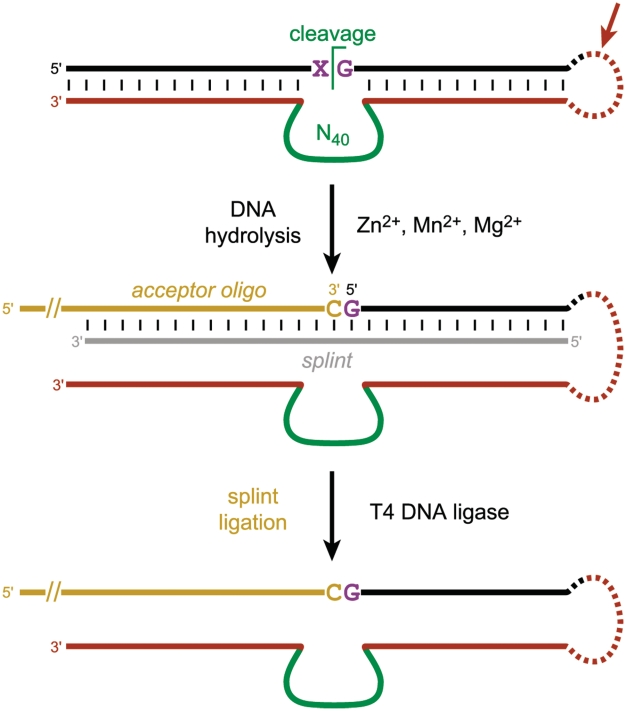
For the selections that incorporated splint ligation pressure (VH–VL and YR; Figure 5), the splint ligation step was performed as follows. The selection sample was precipitated with ethanol and redissolved in 17 µl total volume containing 5 mM Tris, pH 7.5, 15 mM NaCl and 0.1 mM EDTA along with 50 pmol of splint oligonucleotide and 75 pmol of acceptor oligonucleotide. The sample was annealed by heating at 95°C for 3 min and cooling on ice for 5 min. The sample was then brought to 20 µl total volume containing 1× of T4 DNA ligase buffer and one unit of T4 DNA ligase (Fermentas), incubated for 1 h at 37°C, and separated by 8% PAGE. The splint was 5′-TTCGTCCCAGCGGTAGAGAAC-(C)-GCGCTGTCTGATGCGGC-3′, where the unpaired (C) was complementary to the single unpaired 5′-G that remained on the DNA substrate after DNA-catalyzed hydrolysis at the X^G position depicted in Figure 5. The acceptor oligonucleotide for the splint ligation reaction was 5′-(AAC)5AAGGCCGCATCAGACAGCGC-3′, where the underlined portion was complementary to the 3′-end of the splint.
Figure 5.
Selection process that includes splint ligation pressure for DNA-catalyzed DNA hydrolysis at a particular predetermined site (X^G, where four different selections were performed for X = one of C, T, A, or G). The hydrolysis products are 3′-hydroxyl + 5′-phosphate products, as required by T4 DNA ligase. Note that the acceptor oligonucleotide, which has a 3′-hydroxyl for joining to the 5′-phosphate of the G nucleotide, has a long 5′-extension that leads to a large upward PAGE shift upon splint ligation. For nucleotide details, see the Experimental Section.
Cleavage assays
The DNA hydrolysis (cleavage) assays using individual deoxyribozymes were performed under single-turnover in trans conditions with the procedure described previously (14). The final incubation conditions were 70 mM HEPES, pH 7.5, 1 mM ZnCl2, 20 mM MnCl2, 40 mM MgCl2 and 150 mM NaCl at 37°C, with 10 nM 5′-32P-radiolabeled DNA substrate and 1 µM deoxyribozyme (1:100) in 20 µl total volume. Samples were separated by 20% PAGE and quantified with a PhosphorImager. Values of kobs and final yield were obtained by fitting the data directly to first-order kinetics.
Mass spectrometry assays to validate product identities
Samples for mass spectrometry were prepared using 100 pmol of substrate and 200 pmol of deoxyribozyme in 50 µl reaction volume using the procedure described previously (9). All MALDI mass spectra were obtained in the mass spectrometry laboratory of the UIUC School of Chemical Sciences. For detailed tables of mass spectrometry data, see Supplementary Table S1.
RESULTS
Establishing the optimal number of unpaired DNA substrate nucleotides for DNA-hydrolyzing deoxyribozymes
The initial DNA-hydrolyzing deoxyribozyme named 10MD5 was identified by in vitro selection using a substrate that incorporated a tripeptide moiety between two DNA segments, each of which was Watson-Crick base-paired to one of two deoxyribozyme binding arms that surrounded an N40 catalytic region (9). Although the original goal of that previous study was peptide-cleaving deoxyribozymes, 10MD5 can equally well cleave a substrate that is solely DNA. For the present experiments that seek DNA-catalyzed DNA hydrolysis from the outset, we initially chose a set of four all-DNA substrates that differ only in the number of unpaired nucleotides presented to the N40 region (Figure 2A). In our efforts with RNA ligase deoxyribozymes, we have anecdotally observed that the presence of unpaired substrate nucleotides during the selection process often leads to specific identity requirements at these nucleotides by the resulting deoxyribozymes, likely because the single-stranded portion of the substrate can interact more intimately than can duplex DNA with the deoxyribozyme's catalytic region (29,32). Here we used DNA substrates that present 0, 1, 3, or 6 unpaired nucleotides to the N40 region. In vitro selection was performed via our standard approach, which depends on PAGE shift after DNA-catalyzed DNA cleavage (29). In each case, the incubation conditions during the key DNA-hydrolyzing selection step were 70 mM HEPES, pH 7.5, 1 mM ZnCl2, 20 mM MnCl2, 40 mM MgCl2 and 150 mM NaCl at 37°C for 14 h. (Both Zn2+ and Mn2+ were included during all selections and activity assays in this report; studying the detailed metal ion and pH requirements of all new deoxyribozymes will require comprehensive efforts that are beyond the scope of this study.) PAGE shift was used to separate deoxyribozymes that cleave their covalently attached DNA substrate at a location approximately in the middle of the substrate, as shown in Figure 1B. As was the case for 10MD5 (9) and many of our other DNA catalysts (32), the resulting deoxyribozymes are anticipated to function in trans (intermolecularly), without any covalent attachment to the substrate.
In the selection experiment with the 0 unpaired nt DNA substrate, the resulting individual deoxyribozymes (Supplementary Figure S1) appeared largely to catalyze deglycosylation followed by strand cleavage via β-elimination, rather than hydrolysis, and some product formation by oxidative degradation cannot be ruled out (Supplementary Figure S2) (33). In contrast, the other three selections led to the desired DNA hydrolysis, with no evidence for deglycosylation. The selection experiment with the 1 unpaired nt substrate provided the 13PB2 deoxyribozyme (Figure 3), which has 70–80% hydrolysis yield in 22 h (Figure 2B). 13PB2 requires only C^G at the cleavage site, although substantial activity is also achieved with G^G (Supplementary Figure S3); all substrate nucleotides other than C^G can be varied freely, as long as Watson-Crick base-pairing is maintained with the deoxyribozyme binding arms. Hydrolytic cleavage by 13PB2 of its DNA substrate to leave 3′-hydroxyl + 5′-phosphate termini was verified by mass spectrometry (Supplementary Table S1). The selection experiment with the 3 unpaired nt substrate led to DNA-hydrolyzing deoxyribozymes that were much slower than 13PB2 (Supplementary Figure S4) and required specific identities at more than four nucleotides (data not shown). Finally, the experiment with the 6 unpaired nt substrate provided deoxyribozymes (Supplementary Figure S5) that demanded specific identities at more than five nucleotides (data not shown). From all of these data, we concluded that substrates with one and possibly two unpaired nucleotides should be examined more comprehensively in subsequent selection experiments.
Figure 3.
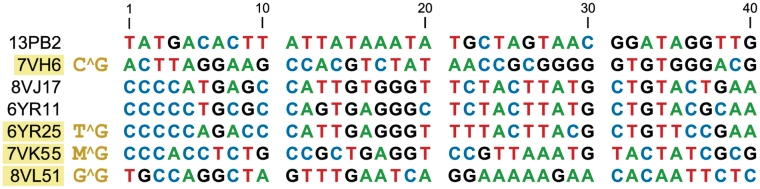
Sequences of the initially random (N40) catalytic regions of key deoxyribozymes described in this report. To create each functional deoxyribozyme, the catalytic region is flanked by two Watson-Crick binding arms that are simply covaried with the relevant substrate sequence. For 13PB2, which requires C^G at the cleavage site, the G is base-paired to the last nucleotide of the corresponding binding arm. For all of the other deoxyribozymes, the two X^G nucleotides at the cleavage site are both unpaired, whereas all other substrate nucleotides are base-paired to the binding arms. The four deoxyribozymes that are useful for each of the four possible X^G sites are highlighted in yellow (7VK55 cleaves either A^G or C^G, where M = A or C).
Three deoxyribozymes from the selection effort with the six unpaired nucleotide substrate hydrolyzed their DNA substrate relatively rapidly, with kobs = 0.3–1.2 min−1 (Supplementary Figure S5). The value of 1.2 min−1 for the 13PD1 deoxyribozyme (Supplementary Figure S1), which was not improved upon partial (25%) randomization and reselection for even faster rate (data not shown), corresponds to a rate enhancement of 3 × 1013 over the uncatalyzed reaction, To our knowledge, this is the highest rate enhancement ever reported for a DNA catalyst. The previous highest rate enhancement was 1 × 1012, for 10MD5 with kobs = 0.05 min−1 (9).
Systematic selections with DNA substrates that have one or two unpaired nucleotides
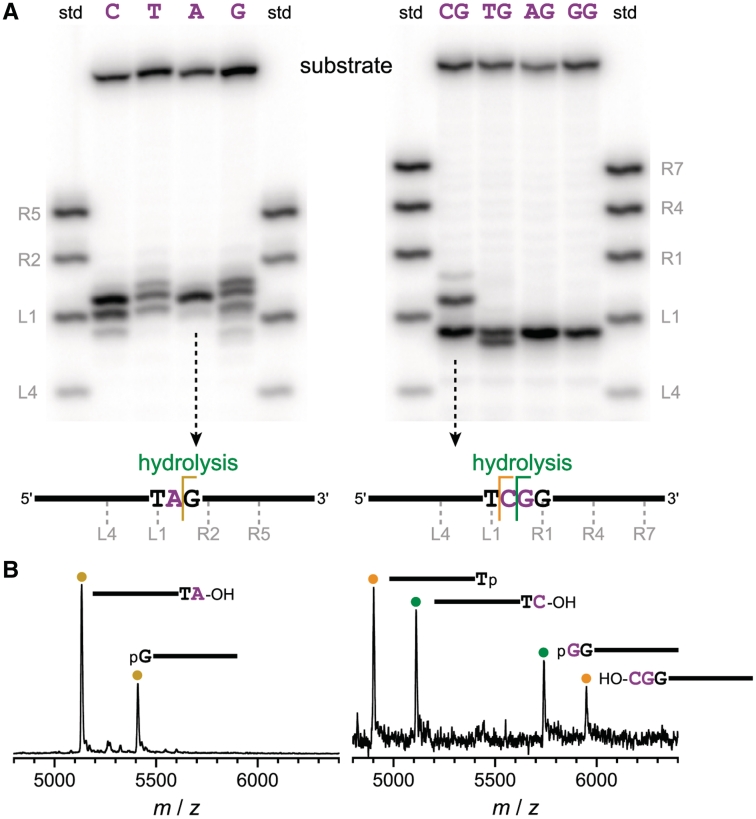
We next performed two sets of four selection experiments, one set with each of the four possible single unpaired nucleotides, and another set with each of the four possible unpaired XG dinucleotides (where X = one of C, T, A, or G; the experiment with a single unpaired C was essentially a replication of the effort described above). The immediate goal of these experiments was to determine the range of hydrolysis site locations in each resulting collection of deoxyribozymes. For all eight of these selections, cleavage by the uncloned pools was observed at various positions clustered near the middle of the substrate (Figure 4). Some of the deoxyribozymes led to 3′-hydroxyl + 5′-phosphate termini whereas others formed 3′-phosphate + 5′-hydroxyl, as assessed by MALDI mass spectrometry (see representative data in Figure 4B); arguably the former combination is more desirable, because of compatibility with the termini formed by conventional restriction enzymes. In addition, for several of the selections with one unpaired nucleotide, a small amount of substrate cleavage by deglycosylation and β-elimination was observed by the uncloned pool, along with hydrolytic cleavage (Supplementary Figure S6). Taken together, these findings indicated that for systematic development of a full collection of DNA-hydrolyzing deoxyribozymes, a selection pressure must be developed such that the resulting catalysts hydrolyze the substrate at a predetermined site rather than anywhere within a rather large region. Ideally, this selection pressure should also ensure that the catalysts uniformly provide 3′-hydroxyl + 5′-phosphate hydrolysis products.
Figure 4.
Outcomes of systematic selections with DNA substrates that have one or two unpaired nucleotides, as revealed by single-turnover in trans assays of uncloned selection pools. For substrates with one unpaired nucleotide X, selections were designated RV–RY; in cis uncloned pool yields for round 7 were 37, 27, 37 and 18%. For substrates with two unpaired nucleotides XG, selections were designated RQ–RT; in cis uncloned pool yields for round 8 were 44, 43, 56 and 28%. (A) PAGE assays using 5′-32P-radiolabeled substrates and standards. The unpaired nucleotide(s) of the substrate are shown above each lane. The 3′-OH standards are each designated with their 3′-terminal nucleotide, where L1 is the first paired nucleotide immediately 5′ of the unpaired nucleotide(s) and R1 is the first paired nucleotide immediately 3′ of the unpaired nucleotide(s). (B) Representative MALDI mass spectrometry data for the two indicated uncloned pools. Each assigned peak has observed mass within 0.05% of the calculated value (data not shown). DNA hydrolysis was predominant in each uncloned pool, although a small amount of deglycosylation-induced cleavage was also evident in some cases. For example, the selection with a single unpaired G gave a mixture of hydrolysis and deglycosylation products, as assessed by mass spectrometry (Supplementary Figure S6).
Selections with pressure for predetermined cleavage site and 5′-phosphate product
The necessary selection pressure was achieved by adding an additional step to each round of the selection process (Figure 5). This step was performed immediately after the deoxyribozyme pool was allowed to cleave the attached DNA substrate at any central position. To the product mixture was added a DNA splint, an acceptor oligonucleotide (with 3′-OH), and T4 DNA ligase. Only those deoxyribozymes that had created a 5′-phosphate terminus directly at the predetermined cleavage site would undergo ligation with the acceptor and therefore migrate much more slowly on PAGE (34) (even more slowly than the original uncleaved pool-substrate conjugate, due to a lengthy 5′-extension included on the acceptor oligonucleotide). This enhanced selection procedure incorporating the splint ligation pressure was performed with each of the four substrates that have XG unpaired nucleotides, with selection pressure that is selective for hydrolyzing between the unpaired nucleotides X^G (i.e. products X-OH-3′ and 5′-pG).
Each of these four selection experiments gave strong DNA hydrolysis activity, with 40–50% cleavage by rounds 7–8. Individual deoxyribozymes from each of the four selections were cloned and assayed, focusing explicitly on identifying those clones that broadly tolerate different substrate sequences. For this purpose, clones that appeared substantially active with the original (parent) substrate sequence in the preliminary activity screen were sequenced and immediately prepared by solid-phase synthesis with systematic sequence changes at all binding arm nucleotides. The substrates for these deoxyribozymes had corresponding Watson-Crick changes at all nucleotides except the unpaired X^G between the two binding arms. As a summary of extensive characterization results, the ensuing assays revealed numerous deoxyribozymes that had at most a two-nucleotide (X^G) substrate requirement, thereby achieving the generality threshold established in the Introduction. The outcome was slightly different for each of the four X^G selection experiments; a summary of the key findings is provided here. In all cases, hydrolytic cleavage was validated by MALDI mass spectrometry (Supplementary Table S1). No deglycosylation-induced cleavage [also forming a 5′-phosphate terminus (33)] was observed, although the splint ligation selection pressure would have permitted that outcome. Sequences of key deoxyribozymes are collected in Figure 3.
C^G selection
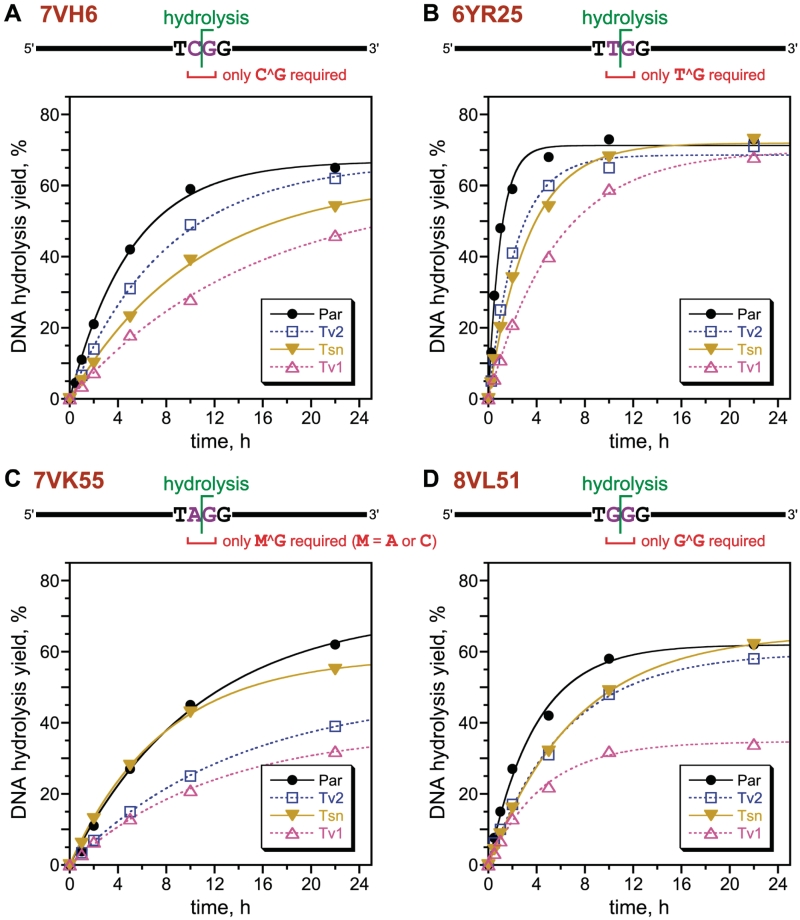
Eight unique deoxyribozyme sequences from round 7 (47% in cis uncloned pool activity) were identified through the initial screening process, and four of these were examined in more detail after solid-phase synthesis. One of the four, 7VH6, had 45–65% single-turnover in trans cleavage yield in 22 h with each of four systematically varied substrates that collectively changed every nucleotide to each possible identity except for the unaltered C^G directly at the cleavage site (Figure 6A; see Supplementary Figure S7 for PAGE images). When, separately, the C^G nucleotides were themselves changed, little or no DNA hydrolysis activity by 7VH6 was observed (Supplementary Figure S8A). Therefore, 7VH6 requires C^G in its substrate at the cleavage site but works well with any nucleotides in the surrounding sequences.
Figure 6.
Assays of deoxyribozymes from the four selections that included the splint ligation pressure of Figure 5. Each DNA substrate had two unpaired nucleotides X^G, where X = one of C, T, A, or G, and is cleaved to form 3′-hydroxyl + 5′-phosphate termini. As observed for 13PB2 (Figure 2B), the single-turnover in trans catalytic activities were maintained for each deoxyribozyme when all nucleotides of the DNA substrate outside of the required cleavage-site X^G (Supplementary Figure S8) were changed systematically. See Supplementary Figure S7 for accompanying PAGE images. For 7VK55, C^G is tolerated at least as well as A^G (Supplementary Figure S9). Sequences of these deoxyribozymes are in Figure 3. kobs values (h−1) were as follows. (A) 7VH6: Par 0.20, Tv2 0.13, Tsn 0.095, Tv1 0.067. (B) 6YR25: Par 1.0, Tv2 0.43, Tsn 0.30, Tv1 0.18. (C) 7VK55: Par 0.094, Tv2 0.073, Tsn 0.13, Tv1 0.083. (D) 8LV51: Par 0.26, Tv2 0.16, Tsn 0.14, Tv1 0.22.
T^G selection
Ten unique sequences from round 8 (40% in cis uncloned pool yield) emerged after screening, and six were prepared by solid-phase synthesis. One of these six, 8VJ17, worked nearly equally well with each of the four systematically varied substrates, albeit with only 15–25% single-turnover in trans cleavage yield in 22 h (data not shown). 8VJ17 was partially (25%) randomized and subjected to six rounds of reselection, leading to 49% in cis uncloned pool yield; 28 individual sequences differed from 8VJ17 itself by 4–9 nt. Six of these variants were assayed in more detail; two of them, 6YR11 and 6YR25, retained very high substrate generality outside of T^G and furthermore had substantially improved 65–75% single-turnover in trans cleavage yield in 22 h (Figure 6B; kobs = 1.0 h−1 for 6YR25 with the parent substrate sequence). Both of these deoxyribozymes required T^G directly at the cleavage site (Supplementary Figure S8B).
A^G selection
Twenty-four unique sequences were found from round 7 (44% in cis uncloned pool yield), of which 15 were made by solid-phase synthesis and examined further. One deoxyribozyme, 7VK55, provided 30–65% single-turnover in trans cleavage yield in 22 h with all four substrate sequence variants that retained A^G at the cleavage site (Figure 6C). Moreover, activity was even higher using the parent substrate sequence that has C^G in place of A^G at the cleavage site (Supplementary Figure S8C), and this high activity was retained with C^G substrates for which the surrounding nucleotides were systematically varied (Supplementary Figure S9). Therefore, 7VK55 is general for sites that are either A^G or C^G, i.e. M^G (where M is the IUB code for A or C).
G^G selection
The screening process for this final selection experiment led to 17 unique sequences from round 8 (44% in cis uncloned pool yield), and 12 of these were studied further. The 8VL51 deoxyribozyme cleaved the G^G substrates with 35–66% single-turnover in trans yield in 22 h (Figure 6D) and required G^G directly at the cleavage site (Supplementary Figure S8D).
DISCUSSION
Summary and context of findings
Our recent discovery that DNA can catalyze site-specific DNA hydrolysis with 1012 rate enhancement was an important advance, revealing the fundamental ability of DNA to catalyze this challenging chemical reaction (9,14,15). Nevertheless, the key question remained of whether DNA has the catalytic ability to cleave essentially any arbitrarily chosen ssDNA substrate sequence at a specific predetermined site. A priori, it is not obvious that deoxyribozymes can have sufficiently short substrate sequence requirements such that any reasonably sized collection of these catalysts can be general for ssDNA hydrolysis. In the present study, the data demonstrate conclusively that numerous site-specific DNA-hydrolyzing deoxyribozymes can be identified with mere two-nucleotide sequence requirements (Figure 1), thereby establishing the substrate sequence tolerance that was the principal objective of the study.
The four deoxyribozymes 7VH6, 6YR25, 7VK55 and 8VL51 (Figure 3) collectively allow site-specific hydrolysis of any arbitrarily chosen ssDNA substrate sequence that includes an N^G cleavage site (Figure 6). Particularly high rate and yield were observed for 6YR25, whose sequence was optimized by reselection (Figure 6B). These achievements required the development of an in vitro selection approach (Figure 5) that should—with comprehensive follow-up experiments, including systematic reselections of initially identified DNA catalysts—lead to a complete set of deoxyribozymes for ssDNA hydrolysis. This work also provides the first intentional identification of numerous new DNA-hydrolyzing deoxyribozymes from random-sequence pools, whereas in our previous report (9), DNA-catalyzed DNA hydrolysis was a serendipitously identified activity.
It should be noted that cleavage-site sequences other than X^G, although not the explicit objective of the current experiments, were clearly evident in numerous cases, and therefore the observed cleavage activity is not limited to X^G sites. A clear example is the strong T^C cleavage site observed for a large fraction of deoxyribozymes in the uncloned pool for the selection with CG unpaired nucleotides, as shown in Figure 4. Considering all of the selection experiments performed during this study as well as our previously published examples (9,14), DNA-catalyzed DNA hydrolysis has now been validated at N^G, N^T, T^N and C^A sites (in each case for which data is not shown, the evidence is comparable to that shown in Figure 4). Therefore, although the present efforts focused specifically on systematically identifying deoxyribozymes for hydrolysis of X^G sites (Figure 6), finding analogous deoxyribozymes for hydrolysis of any dinucleotide site within ssDNA appears to be feasible.
Future biochemical characterization, and application to double-stranded DNA substrates
Clearly much effort remains to understand the structures and mechanisms of DNA catalysts of all kinds, including those that hydrolyze DNA. Mechanistic studies of deoxyribozymes have been impeded by a lack of functionally relevant structural information (35). Specifically in the case of DNA-catalyzed hydrolysis of ssDNA, we have begun efforts to understand the relationships among practical characteristics such as tolerance of varying pH values and site specificity (14), as well as the functional roles of particular metal ions (15). In ongoing work, we are carefully examining divalent metal ion requirements for DNA-catalyzed DNA hydrolysis, to determine if Zn2+ and Mn2+ together (as used here) constitute the optimal cofactor combination for identifying new catalytic function (V. Dokukin, M. Chandra, and S.K.S., unpublished results). New experiments will investigate mechanistic and structural aspects of many of the new deoxyribozymes identified in the present report.
To achieve DNA-catalyzed cleavage of double-stranded DNA (dsDNA) rather than ssDNA using deoxyribozymes of the kind described in this study (which interact with their substrate using Watson-Crick interactions), separation of the dsDNA substrate into its two ssDNA components would be required. Such separation may be possible via suitable strand invasion strategies, whereupon each of the two single-stranded components would then be cleavable by a corresponding deoxyribozyme. However, such strand invasion is experimentally challenging and often requires chemically sophisticated approaches (36–39). Alternatively, an intact dsDNA substrate can be targeted for cleavage, but achieving this objective will require an entirely different basis for substrate–catalyst interactions that does not rely upon Watson-Crick base pairing of the type shown in Figure 1 (e.g. interactions based on small molecule–DNA contacts, triplex formation, or protein–DNA contacts). Such objectives constitute a longer-term challenge for the development of practical DNA catalysts that hydrolyze DNA.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online: Supplementary Table S1, Supplementary Figures S1–S6.
FUNDING
National Institutes of Health (grant GM065966, to S.K.S.); Defense Threat Reduction Agency (grant BRBAA08-L-2-0001, to S.K.S.); National Science Foundation (grant 0842534, to S.K.S.). The open access publication charge for this paper has been waived by Oxford University Press—NAR Editorial Board members are entitled to one free paper per year in recognition of their work on behalf of the journal.
Conflict of interest statement. None declared.
Supplementary Material
REFERENCES
- 1.Schlosser K, Li Y. Biologically inspired synthetic enzymes made from DNA. Chem. Biol. 2009;16:311–322. doi: 10.1016/j.chembiol.2009.01.008. [DOI] [PubMed] [Google Scholar]
- 2.Silverman SK. DNA as a versatile chemical component for catalysis, encoding, and stereocontrol. Angew. Chem. Int. Ed. 2010;49:7180–7201. doi: 10.1002/anie.200906345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Breaker RR, Joyce GF. A DNA enzyme that cleaves RNA. Chem. Biol. 1994;1:223–229. doi: 10.1016/1074-5521(94)90014-0. [DOI] [PubMed] [Google Scholar]
- 4.Silverman SK. In vitro selection, characterization, and application of deoxyribozymes that cleave RNA. Nucleic Acids Res. 2005;33:6151–6163. doi: 10.1093/nar/gki930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Schlosser K, Li Y. A versatile endoribonuclease mimic made of DNA: characteristics and applications of the 8-17 RNA-cleaving DNAzyme. ChemBioChem. 2010;11:866–879. doi: 10.1002/cbic.200900786. [DOI] [PubMed] [Google Scholar]
- 6.Joyce GF. Directed evolution of nucleic acid enzymes. Annu. Rev. Biochem. 2004;73:791–836. doi: 10.1146/annurev.biochem.73.011303.073717. [DOI] [PubMed] [Google Scholar]
- 7.Joyce GF. Forty years of in vitro evolution. Angew. Chem. Int. Ed. 2007;46:6420–6436. doi: 10.1002/anie.200701369. [DOI] [PubMed] [Google Scholar]
- 8.Silverman SK. Catalytic DNA (deoxyribozymes) for synthetic applications—current abilities and future prospects. Chem. Commun. 2008:3467–3485. doi: 10.1039/b807292m. [DOI] [PubMed] [Google Scholar]
- 9.Chandra M, Sachdeva A, Silverman SK. DNA-catalyzed sequence-specific hydrolysis of DNA. Nat. Chem. Biol. 2009;5:718–720. doi: 10.1038/nchembio.201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fekry MI, Gates KS. DNA-catalyzed hydrolysis of DNA phosphodiesters. Nat. Chem. Biol. 2009;5:710–711. doi: 10.1038/nchembio.224. [DOI] [PubMed] [Google Scholar]
- 11.Carmi N, Shultz LA, Breaker RR. In vitro selection of self-cleaving DNAs. Chem. Biol. 1996;3:1039–1046. doi: 10.1016/s1074-5521(96)90170-2. [DOI] [PubMed] [Google Scholar]
- 12.Carmi N, Balkhi SR, Breaker RR. Cleaving DNA with DNA. Proc. Natl. Acad. Sci. USA. 1998;95:2233–2237. doi: 10.1073/pnas.95.5.2233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Carmi N, Breaker RR. Characterization of a DNA-cleaving deoxyribozyme. Bioorg. Med. Chem. 2001;9:2589–2600. doi: 10.1016/s0968-0896(01)00035-9. [DOI] [PubMed] [Google Scholar]
- 14.Xiao Y, Chandra M, Silverman SK. Functional compromises among pH tolerance, site specificity, and sequence tolerance for a DNA-hydrolyzing deoxyribozyme. Biochemistry. 2010;49:9630–9637. doi: 10.1021/bi1013672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Xiao Y, Allen EC, Silverman SK. Merely two mutations switch a DNA-hydrolyzing deoxyribozyme from heterobimetallic (Zn2+/Mn2+) to monometallic (Zn2+-only) behavior. Chem. Commun. 2011;47:1749–1751. doi: 10.1039/c0cc04575f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Basile LA, Raphael AL, Barton JK. Metal-activated hydrolytic cleavage of DNA. J. Am. Chem. Soc. 1987;109:7550–7551. [Google Scholar]
- 17.Hettich R, Schneider H-J. Cobalt(III) polyamine complexes as catalysts for the hydrolysis of phosphate esters and of DNA. A measurable 10 million-fold rate increase. J. Am. Chem. Soc. 1997;119:5638–5647. [Google Scholar]
- 18.Sreedhara A, Cowan JA. Catalytic hydrolysis of DNA by metal ions and complexes. J. Biol. Inorg. Chem. 2001;6:337–347. doi: 10.1007/s007750100209. [DOI] [PubMed] [Google Scholar]
- 19.Branum ME, Tipton AK, Zhu S, Que L., Jr Double-strand hydrolysis of plasmid DNA by dicerium complexes at 37°C. J. Am. Chem. Soc. 2001;123:1898–1904. doi: 10.1021/ja0010103. [DOI] [PubMed] [Google Scholar]
- 20.Mancin F, Tecilla P. Zinc(II) complexes as hydrolytic catalysts of phosphate diester cleavage: from model substrates to nucleic acids. New J. Chem. 2007;31:800–817. [Google Scholar]
- 21.Shell TA, Mohler DL. Hydrolytic DNA cleavage by non-lanthanide metal complexes. Curr. Org. Chem. 2007;11:1525–1542. [Google Scholar]
- 22.Bof Oliveira MC, Mazera D, Scarpellini M, Cardoso Severino P, Neves A, Terenzi H. Mononuclear Cu(II)-phenolate bioinspired complex is catalytically promiscuous: phosphodiester and peptide amide bond cleavage. Inorg Chem. 2009;48:2711–2713. doi: 10.1021/ic802208v. [DOI] [PubMed] [Google Scholar]
- 23.Ullrich S, Nazir Z, Busing A, Scheffer U, Wirth D, Bats JW, Durner G, Gobel MW. Cleavage of phosphodiesters and of DNA by a bis(guanidinium)naphthol acting as a metal-free anion receptor. ChemBioChem. 2011;12:1223–1229. doi: 10.1002/cbic.201100022. [DOI] [PubMed] [Google Scholar]
- 24.Santoro SW, Joyce GF. A general purpose RNA-cleaving DNA enzyme. Proc. Natl. Acad. Sci. USA. 1997;94:4262–4266. doi: 10.1073/pnas.94.9.4262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cruz RPG, Withers JB, Li Y. Dinucleotide junction cleavage versatility of 8–17 deoxyribozyme. Chem. Biol. 2004;11:57–67. doi: 10.1016/j.chembiol.2003.12.012. [DOI] [PubMed] [Google Scholar]
- 26.Silverman SK, Baum DA. Use of deoxyribozymes in RNA research. Methods Enzymol. 2009;469:95–117. doi: 10.1016/S0076-6879(09)69005-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Schroeder GK, Lad C, Wyman P, Williams NH, Wolfenden R. The time required for water attack at the phosphorus atom of simple phosphodiesters and of DNA. Proc. Natl Acad. Sci. USA. 2006;103:4052–4055. doi: 10.1073/pnas.0510879103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Li Y, Breaker RR. Kinetics of RNA degradation by specific base catalysis of transesterification involving the 2′-hydroxyl group. J. Am. Chem. Soc. 1999;121:5364–5372. [Google Scholar]
- 29.Flynn-Charlebois A, Wang Y, Prior TK, Rashid I, Hoadley KA, Coppins RL, Wolf AC, Silverman SK. Deoxyribozymes with 2′-5′ RNA ligase activity. J. Am. Chem. Soc. 2003;125:2444–2454. doi: 10.1021/ja028774y. [DOI] [PubMed] [Google Scholar]
- 30.Kost DM, Gerdt JP, Pradeepkumar PI, Silverman SK. Controlling regioselectivity and site-selectivity in RNA ligation by Zn2+-dependent deoxyribozymes that use 2’,3’-cyclic phosphate RNA substrates. Org. Biomol. Chem. 2008;6:4391–4398. doi: 10.1039/b813566e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Flynn-Charlebois A, Prior TK, Hoadley KA, Silverman SK. In vitro evolution of an RNA-cleaving DNA enzyme into an RNA ligase switches the selectivity from 3′-5′ to 2′-5′. J. Am. Chem. Soc. 2003;125:5346–5350. doi: 10.1021/ja0340331. [DOI] [PubMed] [Google Scholar]
- 32.Silverman SK. Deoxyribozymes: selection design and serendipity in the development of DNA catalysts. Acc. Chem. Res. 2009;42:1521–1531. doi: 10.1021/ar900052y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sheppard TL, Ordoukhanian P, Joyce GF. A DNA enzyme with N-glycosylase activity. Proc. Natl Acad. Sci. USA. 2000;97:7802–7807. doi: 10.1073/pnas.97.14.7802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nickens DG, Bardiya N, Patterson JT, Burke DH. Template-directed ligation of tethered mononucleotides by T4 DNA ligase for kinase ribozyme selection. PLoS ONE. 2010;5 doi: 10.1371/journal.pone.0012368. e12368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Nowakowski J, Shim PJ, Prasad GS, Stout CD, Joyce GF. Crystal structure of an 82-nucleotide RNA-DNA complex formed by the 10-23 DNA enzyme. Nat. Struct. Biol. 1999;6:151–156. doi: 10.1038/5839. [DOI] [PubMed] [Google Scholar]
- 36.He G, Rapireddy S, Bahal R, Sahu B, Ly DH. Strand invasion of extended, mixed-sequence B-DNA by γPNAs. J. Am. Chem. Soc. 2009;131:12088–12090. doi: 10.1021/ja900228j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ishizuka T, Otani K, Sumaoka J, Komiyama M. Strand invasion of conventional PNA to arbitrary sequence in DNA assisted by single-stranded DNA binding protein. Chem. Commun. 2009:1225–1227. doi: 10.1039/b813975j. [DOI] [PubMed] [Google Scholar]
- 38.Ishizuka T, Tedeschi T, Corradini R, Komiyama M, Sforza S, Marchelli R. SSB-assisted duplex invasion of preorganized PNA into double-stranded DNA. ChemBioChem. 2009;10:2607–2612. doi: 10.1002/cbic.200900381. [DOI] [PubMed] [Google Scholar]
- 39.Lahoud G, Arar K, Hou YM, Gamper H. RecA-mediated strand invasion of DNA by oligonucleotides substituted with 2-aminoadenine and 2-thiothymine. Nucleic Acids Res. 2008;36:6806–6815. doi: 10.1093/nar/gkn755. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.