Abstract
Both kinetochore function and sister chromatid cohesion can depend upon pericentromere chromatin structure, and factors associated with heterochromatin have been proposed to have general, conserved roles in distinguishing centromeres and pericentromeres and in conferring pericentromere-intrinsic functions. We applied genome-wide sequencing approaches to quantify RNA expression, DNA methylation and histone modification distributions in maize (Zea mays), focusing on two maize chromosomes with nearly fully sequenced centromeres and pericentromeres. Aside from the presence of the Histone H3 variant common to all centromeres, Centromeric Histone H3 (CENH3), we found no RNA expression or chromatin modifications that clearly differentiate pericentromeres from either centromeres or from chromosome arms, nor did we identify an epigenetic signature that accurately predicts CENH3 location. RNA expression and chromatin modification frequencies were broadly associated with distance from centromeres, gradually peaking or dipping toward arms. When interpreted in the context of experimental data from other systems, our results suggest that centromeres may confer essential functions (such as cohesion retention) to flanking sequence regardless of the local heterochromatin profile.
INTRODUCTION
The sequence of steps in chromosome movement during cell division depends on a unique chromatin environment in centromeres. Many factors can contribute to the critical attributes of centromere chromatin, but the histone variant CENH3 (also referred to as CENP-A) is viewed as a defining feature of an active centromere [reviewed in (1,2)]. In organisms where centromeres exist within a single domain on each chromosome, the adjacent domains are generally referred to as pericentromeres. Pericentromeres also have essential roles in chromosome movement during cell division as they provide the framework for keeping sister chromatids oriented correctly until anaphase. Between prophase and anaphase, sister chromatids remain attached along their pericentromeres specifically, as cohesin is removed everywhere else [reviewed in (3)]. Various factors associated with heterochromatin during interphase have been proposed to set the stage for the retention of cohesin at pericentromeres, including Heterochromatin Protein 1 (HP1), trimethylation of lysine 9 on histone H3 (H3K9me3) and RNAi (4,5). Transient deposition of histone modifications during early mitosis has also been proposed to function in this regard, such as phosphorylation of serines 10 and 28 on histone H3 and phosphorylation of serine 121 on histone H2A (6,7).
Extensive investigation in the fission yeast Schizosaccharomyces pombe pericentromere chromatin has led to the discovery that specific chromatin factors and an RNAi-related mechanism are key not only for proper cohesin dynamics but also for proper placement of CENH3 (8,9). The DNA composition itself contributes to pericentromere function, as repetitive sequences in the pericentromere are transcribed and give rise to siRNAs that recruit heterochromatin factors (5,10,11). Despite the comparatively simple composition and small size of S. pombe centromeres and pericentromeres, similarities have been identified in the much more complex centromeric regions of diverse plants and animals. In addition to heterochromatin factors in pericentromeres, histone modifications associated with euchromatin have been observed in centromeres of both S. pombe and animals (12,13). Schizosaccharomyces pombe is different from many other eukaryotes in that it lacks DNA methylation, which is an important component of heterochromatin in organisms which are capable of it. Hence extrapolation from S. pombe suggests that DNA methylation could contribute to pericentromere heterochromatin function (though not actually in S. pombe itself).
The similarities between S. pombe and other organisms have led to much conjecture about a conserved, functional connection between pericentromere heterochromatin and both centromere function and sister chromatin cohesin (14,15). However, the extent to which the S. pombe model applies to other organisms, with its reliance on control elements intrinsic to pericentromeres, is unclear [see discussions in (6,16,17) and references therein]. These arguments against broad applicability include: (i) a general lack of mitotic centromere or cohesin defects in RNAi and chromatin modification mutants in plants and animals; (ii) Profound differences in both DNA structure and siRNA expression patterns between S. pombe and other organisms; (iii) Dispersed rather than pericentromere-concentrated heterochromatin marks in plants with large genomes; (iv) Existence of functional centromeres that lack canonical pericentromere sequences or even heterochromatin domains (18,19); and (v) Lack of a clear link between HP1 and centromere function in animals (20–22) and an established euchromatic function for the HP1 homolog in plants (23–25). These observations support the notion that the predominant controlling factors are derived from within the centromere itself, both for internal control (CENH3-containing chromatin) and external (pericentromere cohesion) (17).
Pericentromere-intrinsic control models derived from S. pombe predict that the pericentromere chromatin should be functionally and visibly distinct both from the centromere chromatin and the arm chromatin. The presence of CENH3 within the centromere specifically is one such distinction; however, centromeres also contain measurable amounts of cannonical H3 and of course H2A, H2B and H4 [reviewed in (1,2)]. Within the scope of histone modifications, DNA methylation, and RNA expression patterns, one would expect to see a pericentromere chromatin signature that could be connected to its functional role. Such a signature is clearly evident in S. pombe and others (12). Maize presents a useful organism for direct testing of this expectation because of its large, sequenced centromeres and high-resolution maps of CENH3 occupancy (26,27) and because of its rich history of genetic and cytological analysis of chromosome structure. Previous cytological analyses in maize have revealed that its heterochromatin is dispersed along the length of its chromosomes rather than being highly concentrated just at pericentromeres, and the euchromatin histone modification H3K4me2 that is enriched in S. pombe and animal centromeres shows no enrichment in maize centromeres (28–30).
In order to more fully characterize RNA expression and chromatin patterns in maize centromere regions and to search for evidence of a pericentromere chromatin signature, we examined siRNA and poly(A) RNA expression, DNA methylation, and four euchromatic histone modifications patterns—H3K4me3, H3K36me3, H3K27me3 and acetylation of H3K9—across chromosomes in maize using both our own and publicly available deep sequence data. We found no evidence for anything peculiar to pericentromeres, but instead found that the centromere itself was the predominant feature in terms of large-scale patterns. The general trend manifest from these analyses was the centromere rather than pericentromere providing the most visible mark along the chromosome, representing either the top or bottom of a large curve with opposite extremes toward the arms.
MATERIALS AND METHODS
Derivation of small RNA sequencing libraries
Wild-type maize seeds, B73 stock, were germinated in vermiculite at 30°C for 3 days and the last 2–3 mm of the primary root tips cut off with a razor blade and frozen on dry ice. After grinding to a fine powder in liquid nitrogen, small RNA was extracted using the mirVana™ miRNA Isolation Kit (Ambion). A Solexa/Illumina library was then prepared using oligos as described in (31) and a ligation scheme derived from (32) and (33), which selectively captures Dicer products and other small RNAs with both a 3′-OH and a 5′-monophosphate. In order to distinguish this sample from others also included in the same sequencing run, we included a barcode CGT on the 5′-adapter. Samples were sequenced with the Illumina Genome Analyzer II. The 5′-barcode and 3′-adapter sequence were trimmed off as described previously except that the 3′-adapter was identified and trimmed based on a perfect match to the first 8 nt of the adapter (CTGTAGGC) (31). The raw, untrimmed reads are available at the NCBI Sequence Read Archive, accession SRR218319.1.
Derivation of CENH3-chromatin immunoprecipitation bisulfite sequencing libraries
Chromatin immunoprecipitation (ChIP) was carried out as previously described (34) using anti-CENH3 antibodies (35). The 14-day-old B73 seedlings were used as the source tissue. The ChIPed DNA was treated with sodium bisulfite and prepared for Illumina sequencing using a method similar to that of Lister et al. (36). Reads are available at the NCBI Sequence Read Archive, accession SRR218318.1.
Read processing and alignments
After removal of barcodes and adapters from the small RNA reads, known microRNAs were filtered out by Blasting against a set of maize mature miRNA sequences obtained from miRBase (release 16) [www.mirbase.org; (37)]. We used blastall with default parameters except the expectation value (E) was set to 1 e–6. The resulting miRNA-filtered reads, which we refer to as siRNA reads, were sorted by read length to produce individual fasta files corresponding to each length from 20 to 26 nt. Each file was then aligned to the complete maize genome version 2, obtained from www.maizesequence.org. Alignments were carried out with bowtie software (38). For analysis of uniquely-aligning reads, i.e. those with a single best alignment, the following parameters were used: ‘-n 0 -l 20 -m 1 –best’. For analysis of total aligning reads, the parameters were: ‘-n 0 -l 20 -M 1 –best’. Alignment of the 454-sequenced CENH3 reads (27) to the reference genome to mark the centromeres in figures was carried out with bowtie similarly to siRNA reads except that only uniquely-aligning reads were analyzed and a longer seed length was used ‘-n 0 -l 100 -m 1 –best’.
Control reads were derived from a sample of randomly-fragmented B73 genomic DNA sequenced with Illlumina paired-end technology (39). We combined the reads from both ends into a single set, which we treated as single-end, trimmed to the appropriate length (20, 26, or 36 nt), and processed identically to the siRNA reads (except without removal of miRNA matches). This control was intended primarily as an indicator for domain-level effects from errors in the physical map, particularly omissions, as a large proportion of centromere repeats have not yet been placed on the physical map. However, any biases inherent to Illumina sequencing may also be reflected in this sample.
Bisulfite reads were aligned to the genome and to individual repeat sequences using BS Seeker (40). Only reads that could be unambiguously assigned to a single locus with no mismatches were used for quantifying methylation status, using default parameters except ‘-t N -e 36 -m 0’. Control, non-bisulfite-treated reads from Tenaillon et al. (39) were processed exactly the same. The low error reads were derived by extracting only the first 36 nt; the high error reads were from nucleotides 49 to 84 of the reads.
Poly(A) enriched, ChIP reads (35 nt) and the control reads (low error reads; see above) were processed similarly to the siRNA reads, but without removal of miRNA matches.
RESULTS
siRNA expression patterns
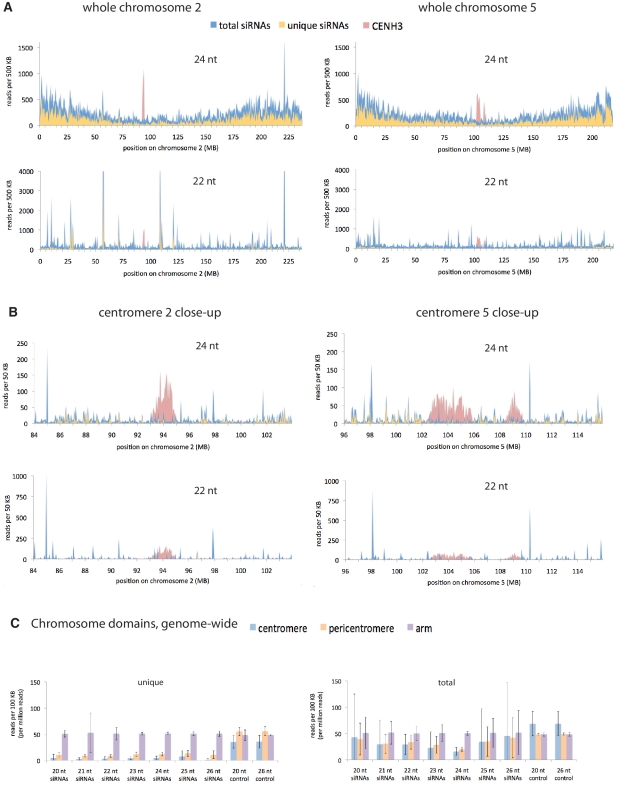
Given the importance of siRNAs in directing histone modifications in S. pombe pericentromeres and in directing DNA methylation of repetitive elements in plants, we first surveyed siRNA expression patterns genome-wide for evidence of a pericentromeric chromatin signature. We prepared Illumina small RNA sequencing libraries from RNA extracted from maize root tips (which contain a large fraction of dividing cells) and aligned the resulting siRNA reads onto the maize genome. A complication with aligning short reads to a highly repetitive genome is that in many cases, it is impossible to determine which of the multiple identical loci is the source of a read. Two common solutions are to disregard all but unique reads (those that can unambiguously be assigned to a single locus), or to include all reads and to assign a locus at random from the possible options. One method loses information, especially when dealing with highly repetitive centromeres; the other entails a higher level of misinformation, since it will assign a portion of repetitive reads to the wrong loci. We opted to use both methods, as displayed in Figure 1 and Supplementary Figure S1. Since one of the distinguishing characteristics of functionally distinct types of siRNAs is length, we split the siRNA reads into sets based on length, from 20 to 26 nt. The most abundant, and potentially most interesting because of their connection to heterochromatin, are the 22-nt and 24-nt types (41,42)
Figure 1.
Comparison of siRNA abundance across chromosome domains. (A and B) Chromosome-wide views and centromere close-ups of 24- and 22-nt siRNAs. (A) Whole chromosome views of chromosomes 2 and 5, and (B) close up views of the centromeres plus 5 Mb of sequence on either side. For small RNAs, the Y-axes show read counts, normalized per million total genomic reads for each length at each 500-kb or 50-kb interval. Unique siRNAs are those corresponding to a single locus whereas total siRNAs include those which could be derived from multiple loci (one of which was chosen at random). Note that some peaks extend off the chart. The CENH3 reads mark unique loci within centromeres and are derived from CENH3 ChIP [described in (27)]. CENH3 read count was normalized per hundred thousand unique genomic reads. See also Supplementary Figure S1. (C) Average siRNA count for each chromosome domain. Centromeres are the domains occupied by CENH3, as defined by the CENH3 ChIP reads. For the purposes of this study, pericentromeres extend 5 MB on either side of the centromeres, and arms are the rest of the chromosomes beyond the pericentromeres. Error bars are standard deviations arising from differences in the values from the 10 chromosomes. The control reads are derived from an Illumina-sequenced, randomly-fragmented genomic DNA sample (39) and are included to provide an estimate of the bias introduced in each domain due to inaccuracies in the physical map and biases arising from Illumina sequencing.
Centromeres were defined to include all the chromatin within the outer bounds of the region of the chromosomes enriched for CENH3 [defined by ChIP with CENH3 antibodies (27)]. In all but two chromosomes (chromosomes five and seven) a single, MB-scale domain of CENH3 was designated as the centromere in a prior study (27). These regions are also referred to as centromere cores, designating that they are defined precisely by CENH3 occupancy. Although centromeric sequences are highly repetitive, there is still enough variation in the DNA content to align even short reads uniquely, especially in centromeres two and five, which have been traversed and sequenced (27). For the 22-nt siRNAs, 11% of the putative centromere matching reads were unique. Small RNAs have also been successfully mapped to rice centromere 3 (43). We note that in some rice centromere cores, CENH3 domains are interspersed with extended regions of H3-containing chromatin (44). This is probably true in maize as well, but at this stage we lack the resolution to determine where such regions may lie.
The 22-nt siRNAs were observed at a consistently low baseline level across chromosome domains, with large spikes in specific regions (these are not known microRNAs). In contrast, the chromosomal distribution of 24-nt siRNAs revealed a pattern of higher density toward the arms and a low point within the centromere, reminiscent of both recombination frequency and gene density (26,45,46). On chromosome 5 there was a particularly low density of 24-nt siRNAs over the region associated with CENH3, however a similar dip was not observed on chromosome 2. We also examined the chromosomal distribution of other lower abundance siRNAs between 20 and 26 nt on chromosome 2; however, none showed anything unusual in the pericentromeric region (Supplementary Figure S1A and B). These high-resolution analyses of siRNAs within centromere core regions and surrounding areas provide additional evidence that centromeric domains are compatible with both transcription and siRNA production, but only at a low level in both centromere and pericentromere relative to chromosome arms in root tip cells.
DNA methylation patterns
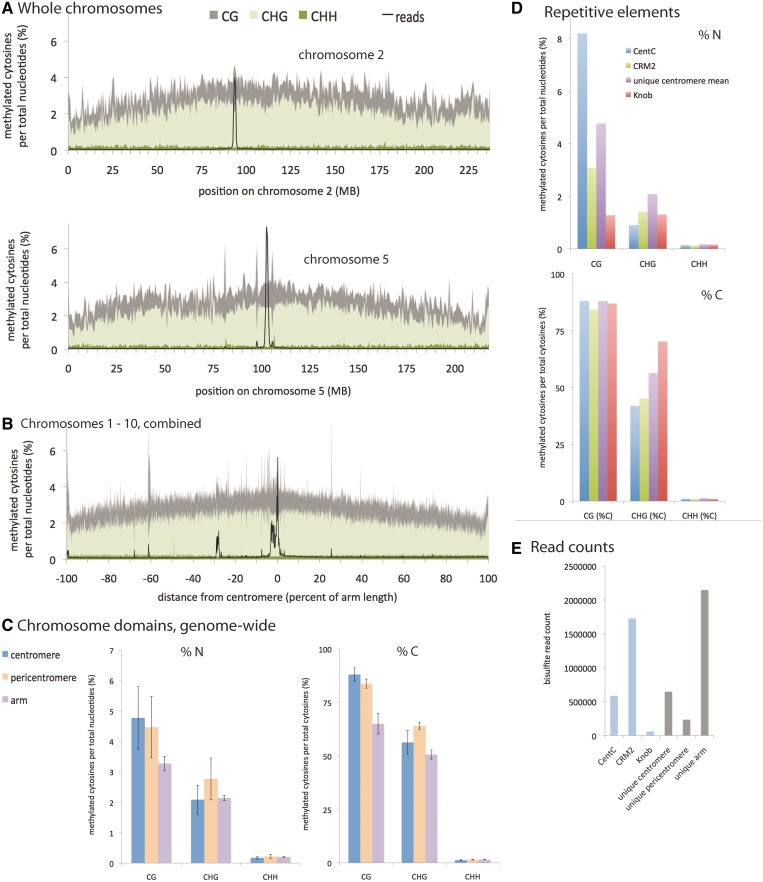
The lack of a pericentromeric 24-nt siRNA signature suggested that associated DNA methylation might also lack a distinguishing pericentromeric pattern. DNA methylation in plants exists in three forms: cytosine methylation in CG, CHG and CHH contexts, where H is anything but G. The CHH type is most heavily dependent upon 24-nt siRNAs [reviewed in (47)]. Our intent was to focus on the centromere core regions, which had been shown in some cases to display hypomethylation in prior immunostaining studies using methyl-cytosine-specific antibodies (48–51). To this end, we first performed ChIP using maize CENH3 antibodies to enrich for centromeric sequences. We then treated the sample with bisulfite and sequenced it with Illumina technology. The experiment conducted in this way clearly delineated the boundaries of the centromere cores while retaining sufficient read depth to cover the rest of the maize genome (Figure 2 and Supplementary Figure S2A and B). The analysis revealed gentle, chromosome-level trends of CG and CHG methylation increasing toward centromeres and decreasing toward arms, as has been observed in many other species (52,53). In contrast, CHH methylation was generally similar across different domains of the chromosomes as has been observed previously in rice (52). Although overall methylation was similar in centromeres and pericentromeres, within the centromere core we noted a subtle increase in CG methylation and decrease in CHG methylation, with notable variation among centromeres (Figures 2, Supplementary Figure S2A and B). Such variation may be attributable to varying sequence coverage and/or the relative sizes of the CentC arrays in different centromeres, which range from <300 kb to several megabases (27,54).
Figure 2.
Comparison of DNA methylation across chromosome domains. (A) Chromosome-wide view of DNA methylation in each sequence context for unique loci on chromosomes 2 and 5. Each chromosome was divided into 500-kb intervals, and the methylation percentage was calculated for each interval. Also displayed (as a black line) is the number of unique bisulfite reads at each position, making the enrichment for centromeres in this ChIP sample clearly evident. To make the scale appropriate for the figure, the total read count was divided by 10 000. See also Supplementary Figure S2. (B) Chromosome-wide view of DNA methylation in each sequence context for unique loci for all 10 chromosomes combined. All the chromosomes were aligned at the center of their centromeres and their arm lengths normalized such that positions were converted to a percentage of total distance from the centromere (0% to 100% of arm length, X-axis). The arms were divided into 1000 intervals, the length of which depended on the particular chromosome. The methylation percent (Y-axis) is a cumulative value calculated from all reads on all chromosomes within a particular interval. Note that several maize centromeres contain mainly the CentC repeat and have been poorly sequenced, thus, the combined view (also for C below) could be biased toward the better sequenced centromeres. Several chromosomes have errors in their physical maps leading to substantial CENH3 signals on arms (black line). (C) Average DNA methylation in unique loci in each chromosome domain. Included are both percent of methylated cytosines per total nucleotides, as in (A), and percent of methylated cytosines per total cytosines in each context. Centromeres are the domains occupied by CENH3, as defined by the CENH3 ChIP reads. For the purposes of this study, pericentromeres extend five MB on either side of the centromeres, and arms are the rest of the chromosomes beyond the pericentromeres. Error bars are standard deviations arising from differences in the values from the 10 chromosomes. (D) DNA methylation in repetitive loci. CentC is the tandem repeat of maize centromeres, CRM2 is the dominant centromere retrotransposon, and Knob is a tandem repeat found on arms. Shown are both percent of methylated cytosines per total nucleotides, as in (A), and percent of methylated cytosines per total cytosines in each context. (E) Abundance of bisulfite reads. Raw counts are depicted for each chromosome domain (uniquely aligning reads in gray) and for repeats (blue). Due to enrichment for CENH3 domains by ChIP prior to bisulfite treatment and sequencing, a substantial number of uniquely aligning reads come from centromeres, despite the high density of repeats in centromere repeats.
This genome-wide analysis was initially limited to unique loci in order to ensure high accuracy in quantification of methylation frequency (Supplementary Figure S2C). To assess the level of methylation within repetitive areas, which are under-presented in the genome assembly, we analyzed reads that match the tandem repeat (satellite) CentC and the retrotransposon CRM2 independently of map position and copy number (Figure 2D). These two elements are the most abundant of the repetitive elements in maize centromeres (26,27). We found that CHH methylation was present at very low levels for CentC and CRM2, similar to CHH methylation at unique centromere loci (∼1% of cytosines in a CHH context were methylated in each case) The average CHG methylation values for both CentC and CRM2 as a whole were lower than the CHG values in unique centromeric loci (42% for CentC and 45% for CRM2 as compared with 65% for unique centromeric loci). These results may partially explain the prior observation that active maize centromeres stain poorly with methyl-cytosine-specific antibodies in maize and other plants (44,46). In contrast, CG methylation in centromere repeats was not notably different from either single copy pericentromeric or single copy centromeric regions. The data reveal that 88% of the CG dinucleotides within the CentC repeats are methylated on average (Figure 2D); however it is likely that the levels of CG methylation vary within long arrays (44–47).
mRNA and euchromatin histone modification patterns
Transcription has been observed at both centromeres and pericentromeres in many other organisms, as evidenced by sequence identity with known centromere- or pericentromere-specific repeats. In S. pombe, transcription driven by RNA polymerase II occurs at pericentromeres but not centromeres (11,12). Similarly, in rice, genes exist in centromere core regions but they tend to lie within subdomains of normal H3 chromatin and not the subdomains identified by CENH3 (44). Nevertheless, a direct comparison is difficult because repetitive sequences in plants can be transcribed via a complicated interaction of three RNA polymerases, II, IV and V, of which only II exists in S. pombe [reviewed in (55)].
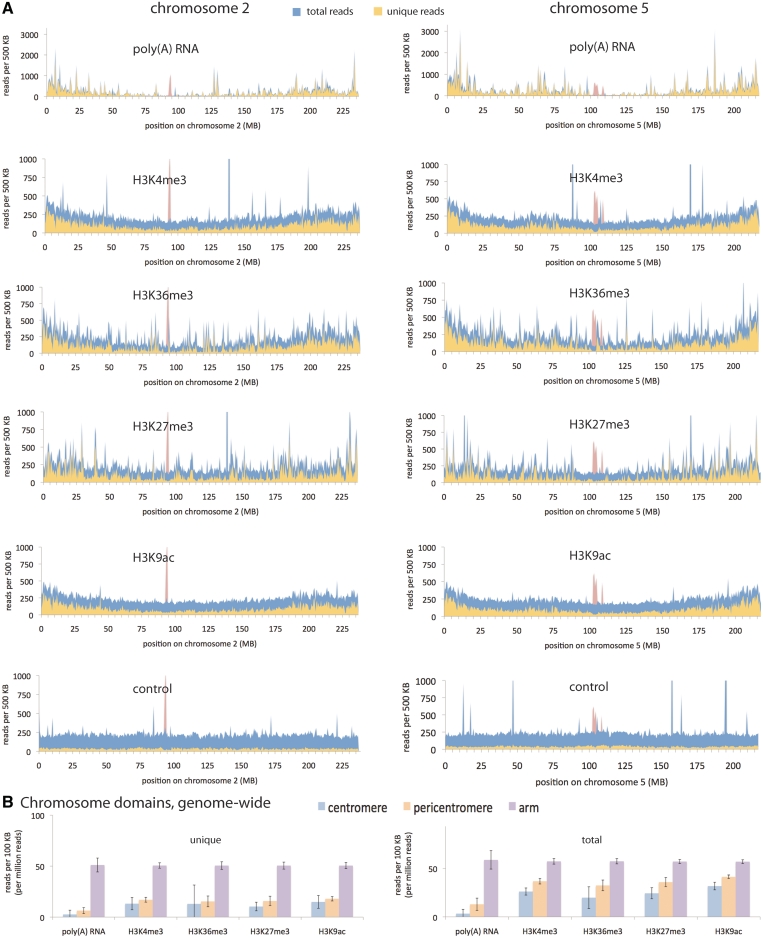
Maize has the distinction of being the first organism for which centromere RNA was found to associate directly with CENH3 chromatin (56) but the most detailed analyses have been done in mammals, where transcription has been associated with both chromatin modifications and kinetochore integrity [reviewed in (57)]. In order to quantify RNA expression over the entire centromere and pericentromere in maize, we examined a previously published set of poly(A)-enriched RNA fragments (58). We took the raw, unfiltered data from this study, and produced genome-wide alignments to measure steady-state RNA levels (Figure 3). The clear image that emerges from this is a general depletion of poly(A) RNA toward the centromere and enrichment in chromosome arms, as expected based on density of protein-coding genes. However, low but detectable expression was also present in centromeres.
Figure 3.
Poly(A) RNA and euchromatin histone modification patterns. (A) Chromosome-wide view of reads from poly(A) RNA fragments and from ChIP with antibodies against H3K4me3, H3K36me3, H3K27me3 and H3K9ac [derived from (58)]. Also included are a set of control, genomic DNA fragments (39). All counts are normalized per million reads. Note that some peaks extend off the chart. The CENH3 reads mark unique loci within centromeres and are derived from CENH3 ChIP [described in (27)]. See also Supplementary Figure S3. (B) Average read count for each chromosome domain. Centromeres are the domains occupied by CENH3, as defined by the CENH3 ChIP reads. For the purposes of this study, pericentromeres extend 5 MB on either side of the centromeres, and arms are the rest of the chromosomes beyond the pericentromeres. In order to lessen biases from errors in the physical map and Illumina sequencing, all values were normalized by the same genomic DNA fragment control reads as displayed in Figure 1C. In this case, the control reads were trimmed to 36 nt to match the read length of the poly(A)-enriched RNA and histone modification reads. For the unique alignments, this normalization also removed the bias against repetitive regions, since the control reads were subject to the same unique alignment criteria. Error bars are standard deviations arising from differences in the values from the 10 chromosomes.
To measure transcript abundance for the tandem repeat CentC and the retrotransposon CRM2, we counted the reads that matched to either element. Of the 26 071 222 genome-matching, poly(A)-enriched reads, 4 matched CentC and 318 matched CRM2, and only a handful of these mapped uniquely. An analysis of the non-unique CRM2 alignments on centromere 2 suggested that most of the transcripts were derived from the centromere core areas (Supplementary Figure S3), as expected based on the distribution of CRM2 elements (27). We also observed a substantial number of siRNAs derived from CentC and CRM2: In a sample of 1 550 399 total genome-matching siRNA reads, 127 matched CentC and 545 matched CRM2 (Supplementary Figure S1C and D). These siRNAs are likely derived from non-polyadenylated or otherwise unstable templates. CentC transcripts of unusual sizes have also been detected by RNA blotting and in sequence libraries from non-poly(A)-enriched RNA (56,59,60). Based on the centromeric location of CentC DNA (27), and the physical association of CentC transcripts with CENH3 chromatin (56), these data provide compelling evidence for transcription from centromeres in maize, though not in a form that is readily captured and sequenced by poly(A) enrichment. In contrast, evidence for S. pombe-type transcription from pericentromeres is lacking in maize.
Euchromatin histone modifications are generally thought to be depleted from pericentromeres, and in both S. pombe and multiple animal species, certain euchromatin modifications (H3K4me2 and H3K36me3) have been reported to be enriched with the centromere itself relative to pericentromeres (12,13,61,62). Such an enrichment is not observed by immunostaining in maize (28,29), nor in the filamentous fungus Neurospora crassa, where H3K4me2 is absent in centromeres but enriched in flanking areas (19). The Wang et al., study from which we derived our analysis of poly(A) RNA fragments also included a large-scale ChiP with antibodies for four euchromatin histone modifications. We used the reads from this study to ask whether an enrichment could be seen in centromeres, or whether any clear difference between centromeres and pericentromeres could be detected (Figure 3 and Supplementary Figure S3). While some reduction in H3 modifications is expected in centromeres by virtue of CENH3 occupancy, all four of the modifications examined—H3K4me3, H3K36me3, H3K27me3 and H3K9ac—were generally depleted within both centromeres and pericentromeres, similar to 24-nt siRNAs, poly(A) RNA, density of protein coding genes, and crossover suppression, and opposite to that of CG and CHG methylation. The chromosome landscape visible from these views is consistently the centromere at the summit of the hill or bottom of the valley, and the pericentromere occupying the slopes on either side. The landscapes are notably gentle, though highly variable, and if not for the presence of CENH3, we would find it difficult to pinpoint the locations of centromeres by standard epigenetic profiling.
DISCUSSION
Using genome-wide epigenetic profiling, we found that maize pericentromeres differed little from centromeres (with the key exception that CENH3 replaces canonical of H3 in the kinetochore domain). There are many caveats to this study relating to the repetitive nature of centromeres and the fact that only two maize centromeres are fully assembled, and none are fully sequenced base for base. The results apply most firmly to centromeres two and five where nearly complete sequences are available. In the most conservative sense, our data can be viewed as pertaining to regions that are unique by a 36-nt or less sequence tag. However, despite the short size of the reads, a large proportion can be aligned to regions within the centromere core and flanking pericentromeric areas (e.g., Figure 2E and 1C).
If arms are viewed as one extreme and centromeres as the other, pericentromeres represent an intermediate state but are much more similar to centromeres than to arms. This general trend was true for siRNAs, DNA methylation, poly(A)-enriched RNA, and for four histone modifications—H3K4me3, H3K36me3, H3K27me3 and H3K9ac. These results are similar to what has been described in Caenorhabditis elegans, Neurospora crassa, Candida albicans, Saccharomyces cerevisiae and many other species in that they lack defined pericentromeric heterochromatin regions. It is of course possible that other marks or specific combinations of marks delineate the pericentromere in these species, but such an unidentified code would not generally be classified as heterochromatin as we understand it.
Our data showing that maize heterochromatin gradually decreased in relation to the distance from the centromere differs substantially from what has been shown in several widely studied species that have an enrichment of heterochromatin directly flanking centromeres, including S. pombe, Drosophila melanogaster, A. thaliana, and humans. However, a close look at these systems suggest that they are dissimilar in fundamental ways [reviewed in (16,17)]. For example, HP1 performs a critical function of ensuring cohesion at S. pombe pericentromeres but appears to be dispensable for cohesion in humans (20–22) and in A. thaliana (23–25). Furthermore, centromeres are orders of magnitude smaller in S. pombe than in these other species. These data suggest that while heterochromatin may frequently be found in close proximity to centromeres, its role in chromosome segregation varies among species. S. pombe may provide an extreme example with its dependence on heterochromatin for de novo deposition of CENH3 and for proper cohesion dynamics (4,5,8,9)
In contrast to pericentromeric heterochromatin, CENH3 is always found at active centromeres, making it a prime candidate as regulator of its own deposition, although perhaps indirectly [reviewed in (1,2)]. The most compelling arguments in favor of the view that centromeres and pericentromeres can function independently comes from cases where centromeres are known to have moved to new locations. In the yeast S. cerevisiae, centromeres that have been empirically moved to chromosome arms are sufficient for apparently normal cohesion behavior flanking the new centromere (63). Furthermore, it is well established that human neocentromeres can form on chromosome arms and in the absence of both canonical pericentromeric DNA sequence and of visible flanking heterochromatin. Conversely, when a centromere has been epigenetically deactivated on a dicentric maize chromosome, pericentromeric phosphorylation of H3 serine 10 and cohesion are lost, demonstrating a reliance on the centromere (64). We emphasize, however, that CENH3 must interact with H3 at a local level since they are in close proximity and H3 is present in centromere cores. For example, multiple experiments with human artificial chromosomes have revealed interactions between CENH3 and the local chromatin environment (61,65–67).
Our findings reveal that the centromere is the single most prominent feature of the entire centromere-pericentromeric region in maize, and suggest that CENH3 alone may be responsible for functions often ascribed to pericentromeres (summarized in Figure 4). Cohesion may be controlled indirectly by kinetochores, and the vast domain of recombination suppression flanking centromeres may also be an indirect outcome of the total recombination block within centromere cores (68) and other mechanical constraints on recombination events that lie in close proximity to centromeres at meiosis [reviewed in (17)]. We emphasize, however, that our data do not demonstrate a mechanistic connection between the centromere and whole-chromosome patterns of RNA expression and chromatin marks. Furthermore, our data leave open the possibility that specific epigenetic marks such as phosphorylation or other transient modifications can be associated with pericentromeric areas during mitosis (6,7). What seems clear is that pericentromere structures vary dramatically across species but pericentromere function is conserved. Our data, combined with multiple studies pointing to similar conclusions, suggest that new models are needed to explain how and why kinetochores are invariably flanked by reduced recombination and regulated retention of cohesion.
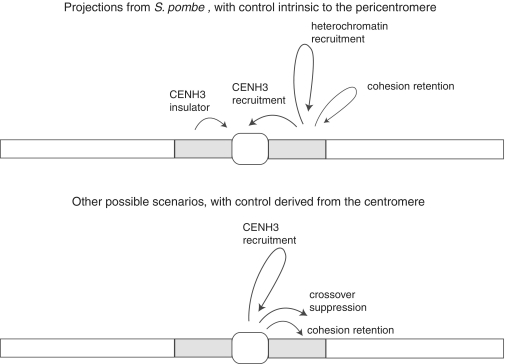
Figure 4.
Models for centromere/pericentromere interactions. The arrows in each diagram depict proposed directions of causality, either arising from centromere (center circle) or pericentromere (flanking rectangles).
ACCESSION NUMBER
NCBI Sequence Read Archive accession SRR218319.1.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online: Supplementary Figures 1–3.
FUNDING
National Science Foundation (0421671, 0607123, 0922703); National Institutes of Health (NIGMS F32GM095223). Funding for open access charge: National Science Foundation.
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
The authors thank Xiaoyu Zhang, Richard M. Gell, Gernot G. Presting, Jeffrey Ross-Ibarra, and Nathanael A. Ellis for expert advice and helpful discussions; Yeching Huang, Tao Zhang and Yufeng Wu for technical help with bioinformatic software; and Sarah Wolf, Rashin Ghaffari and Kathryn S. Hodges for logistical support.
REFERENCES
- 1.Black BE, Cleveland DW. Epigenetic centromere propagation and the nature of CENP-a nucleosomes. Cell. 2011;144:471–479. doi: 10.1016/j.cell.2011.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Henikoff S, Furuyama T. Epigenetic Inheritance of Centromeres. Cold Spring Harb. Symp. Quant. Biol. 2010;75:51–60. doi: 10.1101/sqb.2010.75.001. [DOI] [PubMed] [Google Scholar]
- 3.Nasmyth K, Haering CH. Cohesin: its roles and mechanisms. Annu. Rev. Genet. 2009;43:525–558. doi: 10.1146/annurev-genet-102108-134233. [DOI] [PubMed] [Google Scholar]
- 4.Bernard P, Maure JF, Partridge JF, Genier S, Javerzat JP, Allshire RC. Requirement of heterochromatin for cohesion at centromeres. Science. 2001;294:2539–2542. doi: 10.1126/science.1064027. [DOI] [PubMed] [Google Scholar]
- 5.Volpe T, Kidner C, Hall I, Teng G, Grewal S, Martienssen R. Regulation of heterochromatic silencing and histone H3 lysine-9 methylation by RNAi. Science. 2002;297:1833–1837. doi: 10.1126/science.1074973. [DOI] [PubMed] [Google Scholar]
- 6.Zhang X, Li X, Marshall JB, Zhong CX, Dawe RK. Phosphoserines on maize CENTROMERIC HISTONE H3 and histone H3 demarcate the centromere and pericentromere during chromosome segregation. Plant Cell. 2005;17:572–583. doi: 10.1105/tpc.104.028522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kawashima SA, Yamagishi Y, Honda T, Ishiguro K, Watanabe Y. Phosphorylation of H2A by Bub1 prevents chromosomal instability through localizing shugoshin. Science. 2010;327:172–177. doi: 10.1126/science.1180189. [DOI] [PubMed] [Google Scholar]
- 8.Folco H, Pidoux A, Urano T, Allshire R. Heterochromatin and RNAi are required to establish CENP-A chromatin at centromeres. Science. 2008;319:94–97. doi: 10.1126/science.1150944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kagansky A, Folco H, Almeida R, Pidoux A, Boukaba A, Simmer F, Urano T, Hamilton G, Allshire R. Synthetic heterochromatin bypasses RNAi and centromeric repeats to establish functional centromeres. Science. 2009;324:1716–1719. doi: 10.1126/science.1172026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chen ES, Zhang K, Nicolas E, Cam HP, Zofall M, Grewal SI. Cell cycle control of centromeric repeat transcription and heterochromatin assembly. Nature. 2008;451:734–737. doi: 10.1038/nature06561. [DOI] [PubMed] [Google Scholar]
- 11.Djupedal I, Portoso M, Spåhr H, Bonilla C, Gustafsson C, Allshire R, Ekwall K. RNA Pol II subunit Rpb7 promotes centromeric transcription and RNAi-directed chromatin silencing. Genes Dev. 2005;19:2301–2306. doi: 10.1101/gad.344205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cam HP, Sugiyama T, Chen ES, Chen X, FitzGerald PC, Grewal SI. Comprehensive analysis of heterochromatin- and RNAi-mediated epigenetic control of the fission yeast genome. Nat. Genet. 2005;37:809–819. doi: 10.1038/ng1602. [DOI] [PubMed] [Google Scholar]
- 13.Sullivan BA, Karpen GH. Centromeric chromatin exhibits a histone modification pattern that is distinct from both euchromatin and heterochromatin. Nat. Struct. Mol. Biol. 2004;11:1076–1083. doi: 10.1038/nsmb845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Allis DC, Jenuwein T, Reinberg D, Caparros M-L, editors. Epigenetics. Cold Spring Harbor, New York: Cold Spring Harbor Laboratory Press; 2007. [Google Scholar]
- 15.Ekwall K. Epigenetic control of centromere behavior. Annu. Rev. Genet. 2007;41:63–81. doi: 10.1146/annurev.genet.41.110306.130127. [DOI] [PubMed] [Google Scholar]
- 16.Lu J, Gilbert DM. Cell cycle regulated transcription of heterochromatin in mammals vs.. fission yeast: functional conservation or coincidence? Cell Cycle. 2008;7:1907–1910. doi: 10.4161/cc.7.13.6206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Topp CN, Dawe RK. Reinterpreting pericentromeric heterochromatin. Curr. Opin. Plant Biol. 2006;9:647–653. doi: 10.1016/j.pbi.2006.09.008. [DOI] [PubMed] [Google Scholar]
- 18.Alonso A, Hasson D, Cheung F, Warburton PE. A paucity of heterochromatin at functional human neocentromeres. Epigenetics Chromatin. 2010;3:6. doi: 10.1186/1756-8935-3-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Smith KM, Phatale PA, Sullivan CM, Pomraning KR, Freitag M. Heterochromatin is required for normal distribution of Neurospora CenH3. Mol. Cell. Biol. 2011;31:2528–2542. doi: 10.1128/MCB.01285-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kang J, Chaudhary J, Dong H, Kim S, Brautigam CA, Yu H. Mitotic centromeric targeting of HP1 and its binding to Sgo1 are dispensable for sister-chromatid cohesion in human cells. Mol. Biol. Cell. 2011;22:1181–1190. doi: 10.1091/mbc.E11-01-0009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Koch B, Kueng S, Ruckenbauer C, Wendt KS, Peters JM. The Suv39h-HP1 histone methylation pathway is dispensable for enrichment and protection of cohesin at centromeres in mammalian cells. Chromosoma. 2008;117:199–210. doi: 10.1007/s00412-007-0139-z. [DOI] [PubMed] [Google Scholar]
- 22.Serrano A, Rodríguez-Corsino M, Losada A. Heterochromatin protein 1 (HP1) proteins do not drive pericentromeric cohesin enrichment in human cells. PLoS One. 2009;4:e5118. doi: 10.1371/journal.pone.0005118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Turck F, Roudier F, Farrona S, Martin-Magniette ML, Guillaume E, Buisine N, Gagnot S, Martienssen RA, Coupland G, Colot V. Arabidopsis TFL2/LHP1 specifically associates with genes marked by trimethylation of histone H3 lysine 27. PLoS Genet. 2007;3:e86. doi: 10.1371/journal.pgen.0030086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Xu L, Shen WH. Polycomb silencing of KNOX genes confines shoot stem cell niches in Arabidopsis. Curr. Biol. 2008;18:1966–1971. doi: 10.1016/j.cub.2008.11.019. [DOI] [PubMed] [Google Scholar]
- 25.Zhang X, Germann S, Blus BJ, Khorasanizadeh S, Gaudin V, Jacobsen SE. The Arabidopsis LHP1 protein colocalizes with histone H3 Lys27 trimethylation. Nat. Struct. Mol. Biol. 2007;14:869–871. doi: 10.1038/nsmb1283. [DOI] [PubMed] [Google Scholar]
- 26.Schnable P, Ware D, Fulton R, Stein J, Wei F, Pasternak S, Liang C, Zhang J, Fulton L, Graves T, et al. The B73 maize genome: complexity, diversity, and dynamics. Science. 2009;326:1112–1115. doi: 10.1126/science.1178534. [DOI] [PubMed] [Google Scholar]
- 27.Wolfgruber T, Sharma A, Schneider K, Albert P, Koo D, Shi J, Gao Z, Han F, Lee H, Xu R, et al. Maize centromere structure and evolution: sequence analysis of centromeres 2 and 5 reveals dynamic Loci shaped primarily by retrotransposons. PLoS Genet. 2009;5:e1000743. doi: 10.1371/journal.pgen.1000743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jin W, Lamb JC, Zhang W, Kolano B, Birchler JA, Jiang J. Histone modifications associated with both A and B chromosomes of maize. Chromosome Res. 2008;16:1203–1214. doi: 10.1007/s10577-008-1269-8. [DOI] [PubMed] [Google Scholar]
- 29.Shi J, Dawe RK. Partitioning of the maize epigenome by the number of methyl groups on histone H3 lysines 9 and 27. Genetics. 2006;173:1571–1583. doi: 10.1534/genetics.106.056853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Houben A, Demidov D, Gernand D, Meister A, Leach CR, Schubert I. Methylation of histone H3 in euchromatin of plant chromosomes depends on basic nuclear DNA content. Plant J. 2003;33:967–973. doi: 10.1046/j.1365-313x.2003.01681.x. [DOI] [PubMed] [Google Scholar]
- 31.Gent JI, Schvarzstein M, Villeneuve AM, Gu SG, Jantsch V, Fire AZ, Baudrimont A. A Caenorhabditis elegans RNA-directed RNA polymerase in sperm development and endogenous RNA interference. Genetics. 2009;183:1297–1314. doi: 10.1534/genetics.109.109686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lau NC, Lim LP, Weinstein EG, Bartel DP. An abundant class of tiny RNAs with probable regulatory roles in Caenorhabditis elegans. Science. 2001;294:858–862. doi: 10.1126/science.1065062. [DOI] [PubMed] [Google Scholar]
- 33.Pfeffer S, Lagos-Quintana M, Tuschl T. Cloning of small RNA molecules. Curr. Protoc. Mol. Biol. 2005;Chapter 26 doi: 10.1002/0471142727.mb2604s72. Unit 26.24. [DOI] [PubMed] [Google Scholar]
- 34.Nagaki K, Talbert P, Zhong C, Dawe R, Henikoff S, Jiang J. Chromatin immunoprecipitation reveals that the 180-bp satellite repeat is the key functional DNA element of Arabidopsis thaliana centromeres. Genetics. 2003;163:1221–1225. doi: 10.1093/genetics/163.3.1221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhong C, Marshall J, Topp C, Mroczek R, Kato A, Nagaki K, Birchler J, Jiang J, Dawe R. Centromeric retroelements and satellites interact with maize kinetochore protein CENH3. Plant Cell. 2002;14:2825–2836. doi: 10.1105/tpc.006106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lister R, Pelizzola M, Dowen RH, Hawkins RD, Hon G, Tonti-Filippini J, Nery JR, Lee L, Ye Z, Ngo QM, et al. Human DNA methylomes at base resolution show widespread epigenomic differences. Nature. 2009;462:315–322. doi: 10.1038/nature08514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Griffiths-Jones S. The microRNA Registry. Nucleic Acids Res. 2004;32:D109–D111. doi: 10.1093/nar/gkh023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Langmead B, Trapnell C, Pop M, Salzberg SL. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 2009;10:R25. doi: 10.1186/gb-2009-10-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tenaillon MI, Hufford MB, Gaut BS, Ross-Ibarra J. Genome size and transposable element content as determined by high-throughput sequencing in maize and Zea luxurians. Genome Biol. Evol. 2011;3:219–229. doi: 10.1093/gbe/evr008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Chen PY, Cokus SJ, Pellegrini M. BS Seeker: precise mapping for bisulfite sequencing. BMC Bioinformatics. 2010;11:203. doi: 10.1186/1471-2105-11-203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Jia Y, Lisch D, Ohtsu K, Scanlon M, Nettleton D, Schnable P. Loss of RNA-dependent RNA polymerase 2 (RDR2) function causes widespread and unexpected changes in the expression of transposons, genes, and 24-nt small RNAs. PLoS Genet. 2009;5:e1000737. doi: 10.1371/journal.pgen.1000737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Nobuta K, Lu C, Shrivastava R, Pillay M, De Paoli E, Accerbi M, Arteaga-Vazquez M, Sidorenko L, Jeong DH, Yen Y, et al. Distinct size distribution of endogeneous siRNAs in maize: Evidence from deep sequencing in the mop1-1 mutant. Proc. Natl Acad. Sci. USA. 2008;105:14958–14963. doi: 10.1073/pnas.0808066105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yan H, Ito H, Nobuta K, Ouyang S, Jin W, Tian S, Lu C, Venu RC, Wang GL, Green PJ, et al. Genomic and genetic characterization of rice Cen3 reveals extensive transcription and evolutionary implications of a complex centromere. Plant Cell. 2006;18:2123–2133. doi: 10.1105/tpc.106.043794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yan H, Talbert P, Lee H, Jett J, Henikoff S, Chen F, Jiang J. Intergenic locations of rice centromeric chromatin. PLoS Biol. 2008;6:e286. doi: 10.1371/journal.pbio.0060286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Gore MA, Chia JM, Elshire RJ, Sun Q, Ersoz ES, Hurwitz BL, Peiffer JA, McMullen MD, Grills GS, Ross-Ibarra J, et al. A first-generation haplotype map of maize. Science. 2009;326:1115–1117. doi: 10.1126/science.1177837. [DOI] [PubMed] [Google Scholar]
- 46.Liu S, Yeh C, Ji T, Ying K, Wu H, Tang H, Fu Y, Nettleton D, Schnable P. Mu transposon insertion sites and meiotic recombination events co-localize with epigenetic marks for open chromatin across the maize genome. PLoS Genet. 2009;5:e1000733. doi: 10.1371/journal.pgen.1000733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Law JA, Jacobsen SE. Establishing, maintaining and modifying DNA methylation patterns in plants and animals. Nat. Rev. Genet. 2010;11:204–220. doi: 10.1038/nrg2719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zhang W, Lee H, Koo D, Jiang J. Epigenetic modification of centromeric chromatin: hypomethylation of DNA sequences in the CENH3-associated chromatin in Arabidopsis thaliana and maize. Plant Cell. 2008;20:25–34. doi: 10.1105/tpc.107.057083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Yan H, Kikuchi S, Neumann P, Zhang W, Wu Y, Chen F, Jiang J. Genome-wide mapping of cytosine methylation revealed dynamic DNA methylation patterns associated with genes and centromeres in rice. Plant J. 2010;636:353–365. doi: 10.1111/j.1365-313X.2010.04246.x. [DOI] [PubMed] [Google Scholar]
- 50.Koo DH, Han F, Birchler JA, Jiang J. Distinct DNA methylation patterns associated with active and inactive centromeres of the maize B chromosome. Genome Res. 2011;21:908–914. doi: 10.1101/gr.116202.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Koo DH, Jiang J. Super-stretched pachytene chromosomes for fluorescence in situ hybridization mapping and immunodetection of DNA methylation. Plant J. 2009;59:509–516. doi: 10.1111/j.1365-313X.2009.03881.x. [DOI] [PubMed] [Google Scholar]
- 52.Feng S, Cokus SJ, Zhang X, Chen PY, Bostick M, Goll MG, Hetzel J, Jain J, Strauss SH, Halpern ME, et al. Conservation and divergence of methylation patterning in plants and animals. Proc. Natl Acad. Sci. USA. 2010;107:8689–8694. doi: 10.1073/pnas.1002720107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zemach A, McDaniel IE, Silva P, Zilberman D. Genome-wide evolutionary analysis of eukaryotic DNA methylation. Science. 2010;328:916–919. doi: 10.1126/science.1186366. [DOI] [PubMed] [Google Scholar]
- 54.Jin W, Melo JR, Nagaki K, Talbert PB, Henikoff S, Dawe RK, Jiang J. Maize centromeres: organization and functional adaptation in the genetic background of oat. Plant Cell. 2004;16:571–581. doi: 10.1105/tpc.018937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Matzke M, Kanno T, Daxinger L, Huettel B, Matzke AJ. RNA-mediated chromatin-based silencing in plants. Curr. Opin. Cell Biol. 2009;21:367–376. doi: 10.1016/j.ceb.2009.01.025. [DOI] [PubMed] [Google Scholar]
- 56.Topp C, Zhong C, Dawe R. Centromere-encoded RNAs are integral components of the maize kinetochore. Proc. Natl Acad. Sci. USA. 2004;101:15986–15991. doi: 10.1073/pnas.0407154101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Vourc'h C, Biamonti G. Transcription of Satellite DNAs in Mammals. Prog. Mol. Subcell. Biol. 2011;51:95–118. doi: 10.1007/978-3-642-16502-3_5. [DOI] [PubMed] [Google Scholar]
- 58.Wang X, Elling AA, Li X, Li N, Peng Z, He G, Sun H, Qi Y, Liu XS, Deng XW. Genome-wide and organ-specific landscapes of epigenetic modifications and their relationships to mRNA and small RNA transcriptomes in maize. Plant Cell. 2009;21:1053–1069. doi: 10.1105/tpc.109.065714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Du Y, Topp C, Dawe R. DNA binding of centromere protein C (CENPC) is stabilized by single-stranded RNA. PLoS Genet. 2010;6:e1000835. doi: 10.1371/journal.pgen.1000835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Singh M, Goel S, Meeley RB, Dantec C, Parrinello H, Michaud C, Leblanc O, Grimanelli D. Production of viable gametes without meiosis in maize deficient for an ARGONAUTE protein. Plant Cell. 2011;23:443–458. doi: 10.1105/tpc.110.079020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Bergmann JH, Rodríguez MG, Martins NM, Kimura H, Kelly DA, Masumoto H, Larionov V, Jansen LE, Earnshaw WC. Epigenetic engineering shows H3K4me2 is required for HJURP targeting and CENP-A assembly on a synthetic human kinetochore. EMBO J. 2010;30:328–340. doi: 10.1038/emboj.2010.329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Ribeiro SA, Vagnarelli P, Dong Y, Hori T, McEwen BF, Fukagawa T, Flors C, Earnshaw WC. A super-resolution map of the vertebrate kinetochore. Proc. Natl Acad. Sci. USA. 2010;107:10484–10489. doi: 10.1073/pnas.1002325107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Weber SA, Gerton JL, Polancic JE, DeRisi JL, Koshland D, Megee PC. The kinetochore is an enhancer of pericentric cohesin binding. PLoS Biol. 2004;2:E260. doi: 10.1371/journal.pbio.0020260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Han F, Lamb JC, Birchler JA. High frequency of centromere inactivation resulting in stable dicentric chromosomes of maize. Proc. Natl Acad. Sci. USA. 2006;103:3238–3243. doi: 10.1073/pnas.0509650103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Nakashima H, Nakano M, Ohnishi R, Hiraoka Y, Kaneda Y, Sugino A, Masumoto H. Assembly of additional heterochromatin distinct from centromere-kinetochore chromatin is required for de novo formation of human artificial chromosome. J. Cell Sci. 2005;118:5885–5898. doi: 10.1242/jcs.02702. [DOI] [PubMed] [Google Scholar]
- 66.Lam AL, Boivin CD, Bonney CF, Rudd MK, Sullivan BA. Human centromeric chromatin is a dynamic chromosomal domain that can spread over noncentromeric DNA. Proc. Natl Acad. Sci. USA. 2006;103:4186–4191. doi: 10.1073/pnas.0507947103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Okamoto Y, Nakano M, Ohzeki J, Larionov V, Masumoto H. A minimal CENP-A core is required for nucleation and maintenance of a functional human centromere. EMBO J. 2007;26:1279–1291. doi: 10.1038/sj.emboj.7601584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Shi J, Wolf SE, Burke JM, Presting GG, Ross-Ibarra J, Dawe RK. Widespread gene conversion in centromere cores. PLoS Biol. 2010;8:e1000327. doi: 10.1371/journal.pbio.1000327. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.