Abstract
SINE-VNTR-Alu (SVA) elements are non-autonomous, hominid-specific non-LTR retrotransposons and distinguished by their organization as composite mobile elements. They represent the evolutionarily youngest, currently active family of human non-LTR retrotransposons, and sporadically generate disease-causing insertions. Since preexisting, genomic SVA sequences are characterized by structural hallmarks of Long Interspersed Elements 1 (LINE-1, L1)-mediated retrotransposition, it has been hypothesized for several years that SVA elements are mobilized by the L1 protein machinery in trans. To test this hypothesis, we developed an SVA retrotransposition reporter assay in cell culture using three different human-specific SVA reporter elements. We demonstrate that SVA elements are mobilized in HeLa cells only in the presence of both L1-encoded proteins, ORF1p and ORF2p. SVA trans-mobilization rates exceeded pseudogene formation frequencies by 12- to 300-fold in HeLa-HA cells, indicating that SVA elements represent a preferred substrate for L1 proteins. Acquisition of an AluSp element increased the trans-mobilization frequency of the SVA reporter element by ~25-fold. Deletion of (CCCTCT)n repeats and Alu-like region of a canonical SVA reporter element caused significant attenuation of the SVA trans-mobilization rate. SVA de novo insertions were predominantly full-length, occurred preferentially in G+C-rich regions, and displayed all features of L1-mediated retrotransposition which are also observed in preexisting genomic SVA insertions.
INTRODUCTION
Three different families of non-LTR retrotransposons are actively mobilized in the human genome. These are Long Interspersed Elements 1 (LINE-1, L1), Alu elements (Short Interspersed Elements, SINE) and SVA (SINE-VNTR-Alu) elements. Their success is documented by the fact that non-LTR retrotransposons encompass ~34% of the human genome, making them the most populous group of transposable elements in the human genome (1). L1 elements are the only currently known retrotransposons in the human genome that are coding for the protein machinery required for their own mobilization. Despite the cis-preference (2) of L1 proteins for their own encoding RNA, RNA polymerase III transcripts [Alu, 7SL, U6 and hY sequences (3–6)] mutated full-length L1 RNAs (2), and cellular mRNAs [resulting in processed pseudogene formation (7–9)] were experimentally demonstrated to be trans-mobilized by hijacking the L1-encoded protein machinery
Since genomic preexisting insertions of the hominid-specific non-autonomous non-LTR retrotransposon SVA exhibit the classical hallmarks of L1-mediated retrotransposition (10–12,24), SVA was also assumed to use the L1 protein machinery for its own mobilization (13). These hallmarks are: (i) insertion at a 5′-TTTT/AA-3′ consensus sequence of the L1 endonuclease recognition motif, (ii) 4- to 20-bp target site duplications (TSDs) flanking each SVA insertion, (iii) poly(A) tails of varying lengths at their 3′-ends, (iv) presence of 5′-truncated SVA insertions, (v) internal rearrangements and inversions (13–15) and (vi) 3′-transductions (16,17). SVA is a composite retrotransposon and present in about 2700 copies (12) in the human genome reference sequence. Approximately 30% of them have been integrated after the divergence of humans and chimpanzees (18). The origin of SVA elements can be traced back to the beginnings of hominid primate evolution, ~18–25 Mya. Starting at the 5′-end, a full-length SVA element is composed of a (CCCTCT)n hexamer repeat region; an Alu-like region consisting of two antisense Alu fragments and an intervening unique sequence; a variable number of tandem repeats (VNTR) region, which is made up of copies of a 36- to 42-bp sequence or of a 49- to 51-bp sequence (13), presumably derived from the SVA2 element found in Rhesus macaques and humans (19–22); and a short interspersed element of retroviral origin (SINE-R) region (22). A poly(A) tail is positioned downstream of the predicted conserved polyadenylation signal AATAAA (13). Seven different SVA variants exist in hominid genomes, including 5′- or 3′-transductions, or both 5′- and 3′- transductions (21,23). The in vivo retrotransposition rate of the SVA retrotransposon family was recently estimated to be one in 916 births (25).
Several observations indicate that SVA elements constitute a highly active family of hominid-specific non-LTR retrotransposons whose mobilization rate exceeds processed pseudogene formation frequencies: First, seven SVA insertions were found to be associated with disease [for review see (24)] suggesting that SVA mobilization in vivo is currently more efficient than the formation of high-copy pseudogenes, which have not been found to be associated with any human disease so far. Second, the identified disease-causing SVA insertions are derived from different source elements of the SVA subfamilies D, E, F and F1, and SVAs from these subfamilies are polymorphic in humans (11,12,21). It was estimated that ~40% of the SVA elements in the human genome are polymorphic (11). Also, 14 SVA insertions were recently identified in the HuRef sequence that are not present in the haploid human genome reference sequence from the HGP (25). Lastly, each mRNA pseudogene originates from primarily one source locus, while retrotransposed SVAs are derived from multiple SVA source loci (16,21).
We set out to test the hypothesis that the L1 protein machinery is mobilizing SVA elements in trans by establishing an SVA retrotransposition reporter assay in cell culture. We compared the rate of processed pseudogene formation with the trans-mobilization frequencies of two human-specific SVA elements that were identified as potential source elements. We found that SVA RNAs transcribed from the retrotransposition reporter plasmids are trans-mobilized 12- to 300-fold more efficiently than RNA-Pol II transcripts expressed from a pseudogene formation reporter plasmid in HeLa-HA cells. Furthermore, we demonstrate that the hexameric (CCCTCT)n repeat/Alu-like region at the 5′-end of canonical SVA elements and the 3′-transduced AluSp sequence of an SVA source element have different effects on SVA retrotransposition frequencies in cell culture. Marked SVA de novo insertions were predominantly full-length and exhibited all structural features of L1-mediated retrotransposition that are observed in pre-existing genomic SVA insertions.
MATERIALS AND METHODS
Isolation of genomic SVAE and SVAF1 elements
Based on two recent publications (26,21), we selected two potentially functional human-specific SVA elements as retrotransposition reporter elements. In order to isolate the member of the SVA subfamily E (SVAE) that served as source element of a reported retrotransposition event into the LDLRAP1 gene (26), we performed a BLAT search of the human genome reference sequence (hg17; May 2004), using the partially published sequence of this disease-causing SVA insertion as a query to identify its potential source element. We observed 100% identity between query sequence and genomic SVAE element H19_27 (21). Since the human genome is polymorphic for this SVA insertion, we amplified this SVA element from a BAC clone (RP11-420K14 [AC092364] obtained from the Roswell Park Cancer Institute (Buffalo, NY, USA) via the RZPD-Deutsches Ressourcenzentrum für Genomforschung) by PCR using primers chrom19-gen-FW and chrom19-gen-REV (Supplementary Table S1) which are specific for the genomic sequences flanking SVA H19_27. PCRs were performed using the Expand Long Template PCR System (Roche) and controlled by sequence analyses. The resulting ~2-kb PCR product was subcloned into the pGEM-T Easy vector (Promega). Subsequent sequence analysis of the cloned PCR product revealed that the amplified SVA element harbors only 21 VNTR subunits instead of 33 VNTR subunits specified in the human genome reference sequence. This finding was confirmed by sequence analysis of three independent PCR amplifications of SVA H19_27 located on the BAC clone performed with three different primer pairs. We conclude that the nucleotide sequence of the VNTR region of SVA H19_27 on the BAC clone differs from the corresponding sequence of the human genome reference sequence. To isolate the SVA element H10_1 (21) which is a source element of the SVA subfamily F1 (SVAF1) together with its 3′-flanking AluSp sequence, we amplified the corresponding genomic fragment from the BAC clone RPCIB753F0114Q [AL392107] (ImaGenes) by PCR with primers H10_1 For1 and H10_1 Rev1 (Supplementary Table S1). The resulting 4291-bp PCR product was subcloned into the pTZ57R vector (Fermentas) leading to pTZ H10_1. The subcloned PCR product was verified by sequence analyses.
Retrotransposition reporter constructs and mutant L1 protein donor plasmids
pCEPneo
The mneoI indicator cassette of pJM101/L1.3 (61) was PCR amplified using the primers Neo-Nhe-FW and Neo-BamHI-REV (Supplementary Table S1), and subcloned into pGEM-T Easy. Next, the mneoI cassette was removed by NheI/BamHI digestion and inserted into pCEP4 (Invitrogen) downstream of the CMV promoter (CMVP) and in opposite transcriptional orientation relative to the CMV promoter (Figure 1A).
Figure 1.
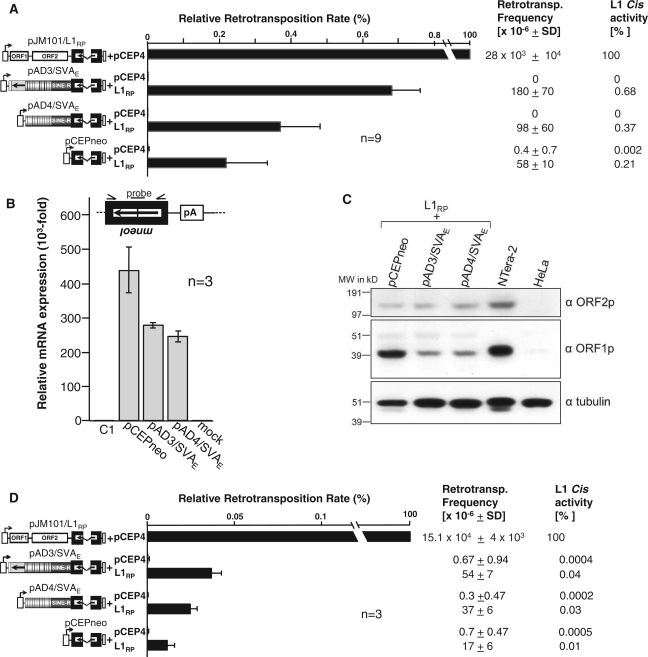
Rationale of the SVA trans-mobilization assay. (A) Schematic representation of SVA retrotransposition reporter plasmids, and pseudogene-formation control construct pCEPneo. SVA reporter elements in pAD3/SVAE, pAD4/SVAE, pSC3/SVAF1 and pSC4/SVAF1 and the processed pseudogene formation cassette in pCEPneo were each tagged with the indicator gene mneoI, and set under transcriptional control of the human CMV promoter (CMVP). Splice donor (SD) and splice acceptor (SA) sites of the oppositely oriented γ-globin intron are indicated. mneoI is flanked by a heterologous promoter (P′) and a polyadenylation signal (A′). Transcripts originating from CMVP driving SVA mneoI or CEP mneoI transcription can splice the intron, but contain an antisense copy of the neo gene. G418 resistant (G418R) colonies arise only if this transcript is reverse transcribed, integrated into chromosomal DNA, and expressed from its own promoter P′. Each SVA reporter element was inserted between CMVP and the mneoI cassette. pAD4/SVAE, differs from pAD3/SVAE in the deletion of the initial 498 nt of the SVA 5′-end covering (CCCTCT)n- and Alu-like region. pSC4/SVAF1 varies from pSC3/SVAF1 in the insertion of the 389-bp AluSp element fused to the A14TTTA26 stretch between SINE-R 3′-end and mneoI cassette. Transcriptional termination signals (ΔAATAAA) at the SINE-R 3′-end and in the sequence flanking the 3′ TSD of AluSp were omitted from the inserted SVA retrotransposition cassettes to ensure transcriptional read-through into the mneoI cassette and polyadenylation at the pCEP4-encoded SV40 polyadenylation signal (pA). pCEPneo is distinguished from the SVA reporter elements by the absence of any SVA sequence. CMVP sequences are highlighted in grey. CMVP major and minor transcription start sites (47) are indicated by arrows. Black rectangles, Alu TSDs; (B) Experimental setup to test for trans-mobilization of mneoI-tagged SVA elements by the L1 protein machinery. Approach 1: Hygromycin selection for the presence of SVA reporter and L1 protein expression plasmid. SVA retrotransposition reporter plasmids or pCEPneo were each cotransfected with L1 protein donor pJM101/L1RPΔneo or pJM101/L1RPΔneoΔORF1 into HeLa cells which were subsequently selected for hygromycin resistance for 14 days. Resulting cell populations were assayed for retrotransposition events by selecting for 9–12 days for G418R HeLa colonies. Approach 2: Transient cotransfection of SVAE reporter plasmids and L1 protein donor plasmid pJM101/L1RPΔneo with subsequent G418R selection. Two days after cotransfection of SVA reporter plasmids or pCEPneo and pJM101/L1RPΔneo, HeLa cells were assayed for L1-mediated trans-mobilization of the respective reporter cassette by selection for G418 resistance.
pAD3/SVAE
The subcloned SVA H19_27 element was reamplified by PCR using primers CT-up-Kpn and DOWN-Δ-pA (Supplementary Table S1). The resulting fragment is devoid of the SINE-R-encoded polyadenylation signal and was inserted between CMVP and the mneoI indicator cassette of pCEPneo via KpnI/NheI. CMVP -driven SVA transcription in pAD3/SVAE reads into the mneo indicator cassette to be terminated by the pCEP4-encoded SV40 polyadenylation signal.
pAD4/SVAE.
pAD4/SVAE differs from pAD3/SVAE in the absence of the initial 499 nt of the 5′-end of SVA H19_27 covering the (CCCTCT)n hexameric repeats and the Alu-like region. To generate pAD4/SVAE, the reamplified SVA H19_27 fragment was digested with AlwNI and NheI. The resulting 1380-bp fragment was cloned between the CMVP and the mneoI indicator cassette of pCEPneo via KpnI/blunt/AlwNI and NheI (Figure 1A).
pSC3/SVAF1
To remove the transcriptional termination signal at the SINE-R 3′-end of SVAF1 source element H10_1 for subsequent cloning steps, the H10_1 sequence was reamplified from pTZ H10_1 using the primer pair H10_1 For2 and H10_1 Rev2 including KpnI and NheI restriction sites, respectively (Supplementary Table S1). The resulting 3527-bp PCR product was inserted into pGEM-T Easy, excised from the resulting plasmid pGEM H10_1 by KpnI/NheI digestion, and inserted between the CMVP and the mneoI indicator cassette of pCEPneo via KpnI/NheI, yielding pSC3/SVAF1.
pSC4/SVAF1
The synthetic oligonucleotide sequence A14TTTA26 (Generi Biotech) was fused as NheI/SpeI fragment into the NheI site of the SINE-R 3′-end of pGEM H10_1, substituting for the AATAAA-containing polyA tail at the 3′-end of the genomic SVA H10_1 element. The resulting plasmid pGEM-H10_1/ΔpA. AluSp was amplified from pTZ H10_1 using oligonucleotides H10_1 AluFor2 and H10_1 AluRev2 including SpeI and SalI restriction sites, respectively (Supplementary Table S1), and subcloned into pGEM-T Easy. The 389-bp SpeI/SalI AluSp fragment was fused with the 3′-end of the A14TTTA26 stretch of pGEM-H10_1/ΔpA via SpeI/SalI resulting in pGEM-H10_1+Alu. The 3940-bp H10_1/A14TTTA26/AluSp-fragment was inserted between CMVP and mneoI cassette of pCEPneo via KpnI and NheI/SalI blunt, yielding pSC4/SVAF1.
pJM101/L1RPΔneoΔORF1 (L1RPΔORF1)
pJM101/L1RPΔneoΔORF1 was generated by introducing a 330-bp in-frame deletion in L1 ORF1 of pJM101/L1RPΔneo (2). The deletion was accomplished by XhoI/SapI-restriction of pJM101/L1RPΔneo and subsequent religation after blunt-ending.
Cell culture, SVA retrotransposition reporter assays and statistical analyses
Cell lines HeLa-JVM and HeLa-HA (28) were cultured in DMEM High Glucose (Biochrom AG, Berlin, Germany) supplemented with 10% FCS (Biowest, Nuaillé, France), 100 µg/ml streptomycin and 100 U/ml penicillin. To perform retrotransposition reporter assays with initial hygromycin selection for the presence of retrotransposition reporter plasmid and L1 protein donor plasmid, 1.8 × 106 cells were plated on 10-cm dishes or T75-flasks. Plated cells were cotransfected with 3 µg of an SVA retrotransposition reporter plasmid or pCEPneo and 3 µg of an L1 protein donor construct (pJM101/L1RPΔneo, pJM101/L1RPΔneoΔORF1) or pCEP4 (negative control) using FUGENE 6 (Roche) according to the manufacturer's instructions. Each cotransfection was performed twice or thrice in quadruplicate: In each case, three cotransfections were used to quantify retrotransposition rates of the SVA reporter elements and pseudogene formation rates of the pCEPneo construct. The fourth cotransfection was used to isolate cell lysates and total RNA in order to analyze expression of retrotransposition reporter cassettes and L1-specific gene products expressed from the L1 donor plasmids. Starting 24 h post-transfection, cells were subjected to hygromycin (200 μg/ml, Invitrogen) selection for 14 days. After trypsinization and reseeding, cells were selected for L1-mediated retrotransposition events in medium containing 400 μg/ml G418 (Invitrogen). After 11–12 days of selection, G418R colonies were either fixed an stained with Giemsa (Merck) to quantify retrotransposition events as described previously (27), or individual G418R colonies were isolated and expanded to characterize SVA de novo retrotransposition events.
To perform retrotransposition reporter assays after transient cotransfection of the respective reporter plasmid and L1 donor plasmid (without hygromycin selection), 2.8 × 106 cells were plated on 15-cm dishes, and transiently cotransfected with 8 µg of the reporter plasmids pAD3/SVAE, pAD4/SVAE or pCEPneo and 8 µg of either pJM101/L1RPΔneo or pCEP4 using FUGENE 6. Each cotransfection was performed four times in parallel. HeLa cells resulting from one transient cotransfection experiment each were used to analyze the expression of ORF1p encoded by pJM101/L1RPΔneo. Cells were trypsinized 2 days after transfection, re-seeded, and cultivated in G418 containing medium for 14 days. The cis-retrotransposition rate which was observed after cotransfection of pJM101/L1RP and pCEP4 served as positive control and was set as 100%. To obtain countable results for retrotransposition in cis, only 1 × 104 cells were plated for G418 selection. Transfection efficiency was determined by cotransfecting 4 µg of pEGFP-N1 (Clontech), 4 µg of pAD3/SVAE, pAD4/SVAE or pCEPneo and 8 µg of the L1 donor plasmid into 2.8 × 106 HeLa-JVM cells using FUGENE 6. EGFP expressing cells were counted 24 h post-transfection by flow cytometry. The percentage of green fluorescent cells was used to determine the transfection efficiency of each sample (2,29). Statistical evaluation was performed by means of an Analysis of Variance (ANOVA). To control the overall type I error α = 0.05, P-values were adjusted according to Dunnett for multiple comparisons. The statistical analysis was performed with SAS®/STAT software, version 9.2 SAS system for Windows.
Quantitative real-time RT-PCR
Fourteen days after cotransfection of HeLa cells with pJM101/L1RPΔneo and either SVA retrotransposition reporter plasmid or pCEPneo, total RNA was extracted from hygromycin-selected HeLa cells using TRIZOL® (Invitrogen) following the manufacturer's instructions. One microgram of total RNA was incubated with 2 U of RNAse-free DNaseI (Invitrogen) for 30 min at room temperature. Using the SuperScript III® First-Strand Synthesis System for RT–PCR (Invitrogen) in combination with an oligo(dt)16–18 primer, first-strand cDNA was synthesized from 0.5 µg of DNaseI-digested, total RNA according to the manufacturer's instructions. To quantify levels of spliced transcripts expressed from the mneo-tagged reporter elements in pAD3/SVAE, pAD4/SVAE, pSC3/SVAF1, pSC4/SCAF1 and pCEPneo, real-time PCR was performed in triplicate applying TaqMan® chemistry (Applied Biosystems) in an Applied Biosystems 7900HT Fast Real-Time PCR System base unit. We used a primer /probe combination (Neofor: 5′-GCTATTCGGCTATGACTGG-3′; Neorev: 5′-GCCACGATAGCCGCGCTGC-3′; probe: 5′-FAM-CCTCGTCCTGAAGCTCATTC-3′) specifically recognizing the spliced mneoI cassette. For normalization, eukaryotic 18srRNA was used as internal control. Cycling conditions were as follows: 95°C for 15 min (initial cycle), 95°C for 15 s and 60°C for 1 min (40 cycles). The software applied to analyze real-time and end point fluorescence was RQ manager 1.2. Relative quantification of RNA expression was carried out using the ΔΔCt method (30).
Immunoblot analysis
To assess L1 ORF1p and L1 ORF2p expression, HeLa cells were cotransfected as described above and harvested 14 days (with hygromycin selection) or 2 days later (without hygromycin selection), respectively. Cells were lysed in RIPA buffer (25 mM Tris, pH 8, 137 mM NaCl, 1% glycerol, 0.5% sodium deoxycholate, 1% Nonidet P-40, 2 mM EDTA, pH 8, 0.1% SDS and protease inhibitors), and lysates were cleared by centrifugation. In the case of transient cotransfection of SVA reporter plasmid and L1 protein donor, 2.8 × 106 cells were plated on 15-cm dishes and transiently cotransfected as described above. Cells of one 15-cm dish carrying the respective plasmids were trypsinized 2 days after cotransfection and lysed with RIPA lysis buffer. Twenty micrograms of each protein lysate were boiled in Laemmli buffer, loaded on 12% polyacrylamide gels, subjected to SDS–PAGE, and electroblotted onto nitrocellulose membranes. After protein transfer, membranes were blocked for 2 h at room temperature in a 10% solution of non-fat milk powder in 1× PBS-T [137 mM NaCl, 3 mM KCl, 16.5 mM Na2HPO4, 1.5 mM KH2PO4, 0.05 % Tween 20 (Sigma)], washed in 1× PBS-T, and incubated overnight with the respective primary antibody at 4°C. To detect L1 ORF1p, the polyclonal rabbit-anti-L1ORF1p antibody #984 (see Supplementary Data) was used in a 1:2000 dilution in 1× PBS-T containing 5% milk powder. L1 ORF2p expression was verified using a rabbit anti-ORF2p-N antibody (31) at a 1:1000 dilution in 1× PBS, 5% milk powder, 0.05 % Tween 20. Membranes were washed thrice in 1× PBS-T and incubated with an HRP-conjugated, secondary anti-rabbit IgG antibody (Amersham Biosciences) at a dilution of 1:30 000 in 1× PBS-T/5% milk powder for 2 h. Subsequently, the membrane was washed three times for 10 min in 1× PBS-T. ß-actin and α-tubulin expression were detected using a monoclonal anti-β-actin antibody (clone AC-74, Sigma-Aldrich) and a polyclonal anti-α-tubulin antibody (ab4074, Abcam) as primary antibodies at dilutions of 1:30 000 and 1:10 000, respectively. Anti-mouse HRP-linked species-specific antibody (from sheep) at a dilution of 1:10 000 and anti-rabbit HRP-linked specific antibody at a dilution of 1:5000 served as secondary antibodies specific for anti-β-actin and anti-α-tubulin antibody, respectively. Immunocomplexes were visualized using lumino-based ECL immunoblot reagent (Amersham Biosciences).
Analysis of SVA de novo insertions
Genomic DNA from expanded G418R -HeLa colonies was isolated applying the Qiagen DNeasy® Tissue Kit according to the manufacturer's protocol. To test for the presence of the spliced mneoI indicator cassette, a diagnostic PCR was performed using the intron-flanking primer pair GS86/GS87 (Figure 4 and Supplementary Table S1). PCR cycling conditions were as follows: 3 min at 96°C, (30 s at 96°C, 15 s at 56°C; 2 min at 72°C) 25 cycles, 7 min at 72°C. To determine genomic pre- and post-integration sites of SVA de novo insertions, we used a modified version (32) of a previously published extension primer tag selection preceding solid-phase ligation-mediated PCR (EPTS/LM-PCR) (33) to isolate 3′ junctions of these insertions. Products of the final PCR were separated in a 1% agarose gel, isolated from the gel using the QIAquick Gel Extraction Kit (Qiagen), and sequenced either directly or after subcloning into pGEM-T Easy. Obtained sequences were mapped to the human genome using the UCSC genome browser at http://genome.ucsc.edu. To characterize 5′-junctions of each SVA de novo insertion, primers specific for the genomic sequence adjacent to the 5′-end of the de novo insertions were designed. The second PCR primer used was GS87, GS88 or, alternatively, HERV-K REV because these primers bind specifically to the retrotransposed SVAE reporter cassette. All oligonucleotides used in this study are listed in Supplementary Table S1. Genomic pre-integration sites and surrounding sequences were characterized using the UCSC genome browser annotation for genes, the Repeatmasker and G+C content tracks. For localization of the integration sites to particular isochores/ G+C-content regions the table published by Costantini et al. (34) was used.
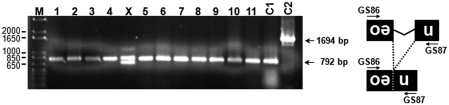
Figure 4.
Diagnostic PCR to test for correct splicing of the intron from the mneoI indicator cassette. Genomic DNA was extracted from 12 G418R HeLa clones that have resulted from cotransfection of pAD3/SVAE (clones 1–8, X) or pAD4/SVAE (clones 9–11) with pJM101/L1RPΔneo and subsequent hygromycin selection. The DNAs were used as template for PCR with primers GS86 and GS87. The PCR allows distinction of the spliced and reverse-transcribed form of the mneoI cassette (792-bp PCR product) from the original unspliced form (1694-bp PCR product) present in the reporter constructs and confirmed integration into the genome via authentic retrotransposition (29). As positive control for an unspliced neoR gene, PCR was performed on pSV2neo (BD Biosciences) mixed with genomic DNA from untransfected HeLa cells (lane C1). PCR performed on pJM101/L1RP DNA that was mixed with genomic HeLa DNA resulted in a fragment specific for the unspliced neoR cassette (lane C2). Lane M, 1-kb Plus DNA ladder (Invitrogen).
Sequence logos
To display patterns of sequence conservation between genomic target sequences of pre-existing SVAE/SVAF, L1-Ta and AluYa5 elements and isolated SVAE de novo retrotransposition events, sequence logos were generated (Figure 6) applying the program WebLogo (35, http://weblogo.berkeley.edu/logo.cgi). We randomly picked 70 genomic target sequences of preexisting members from each of the retrotransposon subfamilies L1Hs-Ta, AluYa5 and SVAE or SVAF. Genomic target sequences of L1Hs-Ta and AluYa5 elements (Supplementary Table S3) were identified from databases (L1_selection.xls; L1_TSD.txt; L1_coord_seq.txt; http://batzerlab.lsu.edu) published recently (36,37). Genomic target sequences of 70 preexisting members of the SVA subfamilies E and F (Supplementary Table S3) were determined from a recently published list of human endogenous SVA insertions (21).
Figure 6.
The nucleotide profile of SVAE de novo insertion sites resembles the consensus target sequence of pre-existing human-non-LTR retrotransposons. Target sequence logos were generated by multiple sequence alignments of genomic integration sites of L1Hs-Ta, AluYa5, SVAA and SVAE insertions using the program WebLogo (35). Logos for the top strand sequence cover four nucleotides of upstream and eight nucleotides of downstream sequences relative to the L1 EN cleavage site (arrow) on the bottom strand. Numbers denote nucleotide positions relative to the nicking site. (A) Comparison of consensus target sequences of L1 and SVAE de novo insertions. Target sequence logos were generated for SVA de novo integration sites (n = 11) and for target sequences of 35 L1 de novo insertions (43). (B) Target sequence logos of 70 preexisting L1-Ta, AluYa5 and SVAE/F insertions. Integration site sequences were identified as described in the ‘Material and Methods’ section.
RESULTS
Identification, isolation and engineering of functional human-specific SVA reporter elements
In order to test the hypothesis that SVA elements are mobilized by the L1-encoded protein machinery, we set out to establish a trans-mobilization assay in which potentially functional marked SVA elements are tested for retrotransposition in the presence of the overexpressed L1 protein machinery. As a first step we identified human-specific, genomic SVA elements that were likely to be retrotransposition-competent (RC). Since sequence and/or structural characteristics of RC SVA source elements were unknown and RC SVA source elements had not been identified when we started our study, we firstly selected SVA insertions reported earlier to be the cause of single cases of genetic disorders. These SVAs were caught red-handed after they were launched from RC source elements and did not have time to accumulate disfiguring mutations in the human genome. We picked the sequence of an SVAE insertion into the LDLRAP1 gene that caused a case of autosomal recessive hypercholesterolemia (ARH) (26), as query, and performed a BLAT search of the human genome reference sequence (hg17; May 2004) to identify the potential source element of the disease-causing insertion. We observed 100% identity between the query sequence and genomic SVAE element H19_27 (Supplementary Figure S1) which displays presence/absence polymorphism in the human genome (21). Given the sequence correlation between the ARH-causing SVA insertion and SVA H19_27, and the polymorphic state of H19_27, we concluded that either H19_27 is the source element of the disease-causing SVA insertion, or both H19_27 and the ARH-causing insertion are derived from the same source element which is not present in the analyzed human genome reference sequence. Therefore, we chose the SVAE element H19_27 as a reporter element for the planned SVA retrotransposition reporter assays. To generate the SVA retrotransposition reporter plasmid pAD3/SVAE, the genomic canonical SVAE element H19_27 was amplified by PCR, tagged at its 3′-end with the mneoI indicator cassette (38), and inserted into the episomal pCEP4 expression vector, where the SVA reporter cassette was set under the control of the human CMV promoter (Figure 1A; see ‘Materials and Methods’ section). To address the question if the (CCCTCT)n repeats and/or the 300-bp Alu-like region at the 5′-end of SVA elements play a role in the efficiency of any potential trans-mobilization of SVA elements, we deleted 498 nt of the 5′-end sequence of the SVA element in pAD3/SVAE to generate pAD4/SVAE (Figures 1 and Supplementary Figure S1).
We chose the genomic SVAF1 element H10_1 as second retrotransposition reporter, because it has been identified recently as source element of at least 13 SVAF1 subfamily members (21,23), including one SVAF1 element that is the progenitor of a disease-associated insertion into the HLA-A gene (39). The human-specific SVAF1 subfamily was generated by the acquisition of a MAST2 sequence via splicing (21,23), includes at least 84 elements and has further evolved by usurping 5′- and 3′-transductions that include Alu sequences. We PCR-amplified SVA H10_1 together with its 3′-flanking functional AluSp element from a BAC clone because it was shown that, due to the weak transcriptional termination signal at the 3′-end of the SINE-R module, transcriptional readthrough into the 3′-flanking genomic AluSp sequence can occur (21) (Figure 1A). Termination of Pol II transcription by a termination signal located downstream of the AluSp TSD caused 3′-transducing H10_1 transcripts that were retrotransposed, leading to at least 13 genomic 3′-transduced SVAF1 insertions (21).
Since there is evidence that both the H10_1 element alone and H10_1 derivatives including the 3′-transduced AluSp sequence have served as source elements for SVA retrotransposition (21,23), we tested both SVAF1 members (Figure 1A; pSC3/SVAF1, pSC4/SVAF1) in our trans-mobilization assay.
For that purpose, we first fused the entire 3.5-kb H10_1 element at its SINE-R 3′-end with the mneoI indicator cassette in opposite orientation, and inserted the marked SVAF1 reporter element into the pCEP4 expression vector, generating pSC3/SVAF1 (Figure 1A). Next, we fused the genomic AluSp element to the 3′-end of SVA H10_1, which resulted in a structure that also constitutes the genomic situation. The fusion was tagged with the mneoI indicator cassette and the entire 6.2-kb retrotransposition reporter cassette was inserted into pCEP4, resulting in pSC4/SVAF1 (Figure 1A). To ensure that the indicator cassette is transcribed as efficiently as the upstream SVA sequences, transcriptional termination signals located at the SINE-R 3′-end and in the sequence downstream of the AluSp 3′ TSD were removed (ΔAATAAA; Figure 1A). Since pSC3/SVAF1 and pSC4/SVAF1 differ exclusively in the presence of an intact AluSp element at the SINE-R 3′-end, we will be able to evaluate if this element plays any role in SVA trans-mobilization efficiency.
L1-encoded proteins facilitate retrotransposition of both SVAE reporter elements
To evaluate if the SVAE reporter elements are trans-mobilized by the L1-encoded protein machinery, we cotransfected pAD3/SVAE and pAD4/SVAE with the L1 protein donor plasmid pJM101/L1RPΔneo or pCEP4 into HeLa-JVM cells (Figure 1B) because it was demonstrated that HeLa cells can efficiently accommodate and express proteins from two different expression vectors (2). Cotransfected cells were subjected to hygromycin selection for the presence of the plasmids (Figure 1B) and subsequently selected for G418 resistance. Each SVA reporter element was tagged with an antisense copy of the selectable marker gene neo encoding neomycin phosphotransferase, a heterologous promoter (P′), and a polyadenylation signal (A′) (Figure 1A). This arrangement ensures that G418-resistant cells (G418R) will only arise if a transcript initiated from the human CMV promoter (CMVP) driving SVAmneoI or CEPmneoI expression is spliced, reverse transcribed, reintegrated into chromosomal DNA, and expressed from promoter P′. G418R foci indicating retrotransposition of an SVA reporter cassette, could be observed only if the L1 expression plasmid was cotransfected suggesting that L1 proteins are required for SVA mobilization (Figures 2A and Supplementary Figure S3).
Figure 2.
Trans-mobilization of mneoI-tagged SVAE reporter elements. (A) SVAE retrotransposition reporter assay after hygromycin selection for the presence of expression plasmids. SVAE reporter plasmids pAD3/SVAE, pAD4/SVAE, or pCEPneo were cotransfected with the L1 protein donor pJM101/L1RPΔneo (L1RP) or the empty vector pCEP4. After hygR selection, G418R selection for retrotransposition events followed and retrotransposition rates were determined by counting G418R HeLa colonies. Each cotransfection experiment and subsequent retrotransposition reporter assay was carried out three times in triplicates. Retrotransposition frequencies per 106 cells are listed and relative retrotransposition rates are indicated as bar diagram. Cis retrotransposition rate of the L1 reporter element pJM101/L1RP was set as 100%. Each bar depicts the arithmetic mean ± SD of the relative retrotransposition rates obtained from nine individual cotransfection experiments (n = 9). (B) qRT–PCR analyses to quantify the relative amounts of spliced transcripts expressed from retrotransposition reporter cassettes. Total RNA was isolated 48 h after cotransfection of pCEPneo, pAD3/SVAE, and pAD4/SVAE with the L1 protein donor plasmid pJM101/L1RPΔneo. The used primer/probe combination (see ‘Materials and Methods’ section) is specific for the spliced mneoI-cassette (black box with arrow). Relative amounts of mRNA expression refer to the signal obtained from total RNA of untransfected HeLa cells which was set as 1 (C1); Total RNA from mock-transfected HeLa cells served as negative control. (C) Immunoblot analysis of L1 protein expression in HeLa cells after cotransfection of the L1 protein donor (L1RP) with retrotransposition reporter plasmids pAD3/SVAE, pAD4/SVAE and pCEPneo. Whole-cell lysates were prepared 14 days after cotransfection upon completion of hygromycin selection and subjected to immunoblot analysis using antibodies against either L1 ORF1p (αORF1p) or L1 ORF2p (αORF2p). An amount of 20 µg of whole-cell extracts were loaded per lane. α-tubulin protein levels (~50 kDa) were analyzed as loading control. Lysates from untransfected HeLa cells and from the germ cell tumor cell line NTera-2 served as negative and positive control for L1 protein detection, respectively. (D) SVAE trans-mobilization assay after transient cotransfection of expression plasmids. pAD3/SVAE, pAD4/SVAE or pCEPneo were transiently cotransfected with pJM101/L1RPΔneo (L1RP) or pCEP4 into HeLa cells. Two days later, cells were G418-selected for de novo retrotransposition events for 14 days. G418R HeLa colonies were Giemsa-stained and counted. Each cotransfection experiment was done in quadruplicate. Subsequent retrotransposition reporter assays were performed in triplicate. Retrotransposition frequencies per 106 cells are listed and relative retrotransposition rates are indicated as bar diagram. Cis retrotransposition rate of L1 reporter element pJM101/L1RP was set as 100%. Each bar depicts the arithmetic mean ± SD of the relative retrotransposition rates obtained from three individual cotransfection experiments (n = 3).
Next, we asked if L1 proteins prefer our reporter SVA mRNA to any other PolII transcript as substrate for trans-mobilization. If this was the case, trans-mobilization frequency of the SVA reporter element would be expected to exceed the frequency of processed pseudogene formation of a regular PolII gene. In order to determine the pseudogene formation rate, we generated the reporter plasmid pCEPneo which differs from pAD3/SVAE exclusively by the absence of the 1876-bp SVA sequence (Figure 1A). Since transcripts expressed from the CMVP of pCEPneo consist exclusively of the mneoI indicator cassette in antisense orientation and do not include any retrotransposon sequences, these transcripts should be trans-mobilized as frequent as random mRNAs encoded by host PolII genes. Overall, trans-mobilization frequencies of the pAD3/SVAE-encoded canonical SVA element exceeded pseudogene formation of the reverse mneoI cassette by 2- to 5-fold whereas the 5′-truncated SVAE cassette encoded by pAD4/SVAE outnumbered the pseudogene formation rate by only 1- to 3-fold (Figures 2A and Supplementary Figure S3). While the differences in trans-mobilization rates between pAD3/SVAE and pCEPneo were statistically significant (p1 = 0.0001), those between pAD4/SVAE and pCEPneo were not (p2 = 0.2212). To verify that the observed differences in trans-mobilization rates between pAD3/SVAE and pAD4/SVAE are not resulting from discrepancies in mRNA production or stability between the different SVA reporter constructs, we tested for the presence of similar amounts of spliced, tagged SVA mRNAs by quantitative real-time RT–PCR (qRT–PCR) (Figure 2B). Using primer/probe combinations specifically recognizing the spliced mneoI reporter cassette, we quantified the relative amounts of spliced mRNA expressed from the reporter plasmids after 14 days of hygromycin selection (Figure 2B). Clearly, the observed differences in the amounts of spliced SVA reporter mRNA of 1.1-fold between pAD3/SVAE- and pAD4/SVAE-tranfected cells (Figure 2B) is only minor compared to the 2- to 5-fold differences in trans-mobilization rates (Figure 2A). Therefore, we draw the conclusion that the 498-nt fragment deleted from the 5′-end of the canonical SVA element in pAD3/SVAE makes the 5′-truncated element in pAD4/SVAE a somewhat less attractive substrate for trans-mobilization. Interestingly, although the amount of spliced pCEPneo transcripts exceeds those derived from transcription of the SVAE reporter cassettes, trans-mobilization of SVAE transcripts is still more efficient, emphasizing that SVAE RNA is a preferred substrate for the L1 protein machinery. In order to evaluate if the observed differences in trans-mobilization rates can be attributed to varying L1 protein levels, we assessed L1 ORF1p and ORF2p expression in a parallel set of cotransfected hygR -selected HeLa cell cultures (Figure 2C). Immunoblot analysis of cell extracts isolated the day before the onset of G418 selection of the remaining cotransfected cultures, shows that comparable amounts of ORF2p are expressed in the differently cotransfected cells. Although L1 ORF1p levels are elevated in HeLa cells cotransfected with pCEPneo relative to pAD3/SVAE- and pAD4/SVAE-cotransfected cells, the observed trans-mobilization rate is the highest in cells cotransfected with pAD3/SVAE. This indicates that the observed differences between pseudogene formation rate and SVAE trans-mobilization frequencies did not result from diverse L1 protein levels.
Next, we wanted to evaluate if we can confirm these results on trans-mobilization of the SVAE reporter elements in an experimental setup, in which we transiently cotransfect HeLa cells with retrotransposition reporter and L1 protein donor plasmid, and select for retrotransposition events 2 days later (Figure 1B, approach 2). In compliance with the results obtained in the case of approach 1, we observed in this transient-cotransfection setup trans-mobilization rates of the SVA reporter cassettes in pAD3/SVAE and pAD4/SVAE which exceeded pseudogene formation rates by 2- to 5-fold (arithmetic mean: 3.5-fold) and 1- to 3-fold (arithmetic mean: 2.4-fold), respectively (Figure 2D). Again, the difference in trans-mobilization rates between pAD3/SVAE and pCEPneo was statistically significant (p3 = 0.0021), whereas differences between pAD4/SVAE and pCEPneo were not (p2 = 0.0354). To test for comparable expression levels of the L1 protein machinery, we assessed L1 ORF1p expression in a parallel set of transiently cotransfected HeLa cultures. Immunoblot analysis of cell lysates harvested three days after cotransfection uncovered that there were no detectable differences in L1 ORF1p expression levels between the differently cotransfected cells (Supplementary Figure S5). This confirms that also in the transient-cotransfection setting, the observed discrepancies in trans-mobilization rates (Figure 2D and Supplementary Figure S4) are not a consequence of varying expression levels of the L1 protein machinery.
The 3′-transduced AluSp element increases the trans-mobilization rate of the SVAF1 source element by ~25-fold
In order to evaluate if the canonical SVAE reporter element differs in its trans-mobilization rate from SVAF elements that were reported to be source elements of the highly successful human-specific SVAF1 subfamily, we tested pAD3/SVAE, pSC3/SVAF1 and pSC4/SVAF1 (Figure 1) in our trans-mobilization assay in parallel. Cotransfection of pAD3/SVAE, pSC3/SVAF1 and pCEPneo with the L1 protein donor plasmid pJM101/L1RPΔneo into HeLa-HA cells uncovered that SVAE reporter and SVAF1 source element H10_1 are trans-mobilized by ~18 and ~12-fold relative to pseudogene formation rate, respectively (Figure 3A and B). Surprisingly, the mobilization rate of the pSC4/SVAF1-encoded SVAF1 source element that includes the 3′-transduced AluSp sequence was exceeding pseudogene formation rate by ~300-fold. This corresponds with a relative retrotransposition rate of ~11% of L1 cis activity (Figure 3A and B). Data suggest that the acquired AluSp sequence makes the SVA element a significantly more attractive substrate for the L1 protein machinery resulting in a mobilization rate that corresponds to AluY reporter elements (3).
Figure 3.
L1 ORF1p is required for trans-mobilization of SVA reporter elements. (A) Trans-mobilization of SVAE and SVAF1 reporter elements in presence/absence of the entire L1 protein machinery. SVA reporter plasmids pAD3/SVAE, pSC3/SVAF1 and pSC4/SVAF1 were each cotransfected with intact (L1RP) and mutant (ΔORF1) L1 protein donor plasmid and pCEP4. pCEPneo was cotransfected with pJM101/L1RPΔneo (L1RP) or the empty vector pCEP4. After hygR selection, cotransfected HeLa cells were G418-selected for retrotransposition events and retrotransposition rates were determined. Each cotransfection experiment and subsequent retrotransposition reporter assay was carried out in triplicates twice (n = 6). Retrotransposition rates per 106 cells including standard deviations (±SD) are listed. Arithmetic means of mobilization rates relative to the cis-activity of pJM101/L1RP (L1 cis activity) are specified and depicted as bar diagram. Error bars, ± SD; Primary data of trans-mobilization assays are summarized in Supplementary Figure S6. (B) The 3′ terminal AluSp sequence of SVAF1 element H10_1 increases its trans-mobilization rate by ~25-fold. For reasons of clarity, a subset of the information presented in Figure 3A is displayed. Relative retrotransposition rates (L1 cis activity [%]) and retrotransposition frequencies that resulted from cotransfections with the L1 protein donor pJM101/L1RP (L1RP) are compared. (C) Immunoblot analysis of L1 protein expression after cotransfection of L1 protein donors with SVA retrotransposition reporter plasmids or pCEPneo. Whole-cell lysates were prepared 14 days after cotransfection upon completion of hygromycin selection and subjected to immunoblot analysis using antibodies against either L1 ORF1p (αORF1p) or L1 ORF2p (αORF2p). An amount of 70 µg of whole-cell lysates were loaded per lane. α-tubulin protein levels (~50 kDa) were analyzed as loading control. Lysates from untransfected HeLa cells and from the germ cell tumor cell line 2102Ep served as negative and positive control for L1 protein detection, respectively. Shorter (exp.1) and longer exposures (exp.2) of the αORF1p immunoblot are presented to demonstrate expression of endogenous L1 ORF1p. (D) qRT–PCR analyses to quantify relative amounts of spliced transcripts encoded by the diverse retrotransposition reporter cassettes. Total RNA was isolated after 14 days of hygromycin selection following cotransfection of the reporter constructs pSC3/SVAF1, pSC4/SVAF1 and pCEPneo with pJM101/L1RPΔneo. The used primer/probe combination is specific for the spliced mneoI-cassette (Figure 2B). Relative amounts of mRNA expression refer to the signal obtained from total RNA of untransfected HeLa cells which was set as 1 (C1); Total RNA from mock-transfected HeLa cells served as negative control.
SVA trans-mobilization requires both L1 ORF1p and L1 ORF2p
While it is obvious that the generation of G418R foci after expression of the L1 protein machinery requires ORF2-encoded RT activity, it was unclear if L1 ORF1p is essential for SVA trans-mobilization. To uncover, if L1 ORF1p is required for trans-mobilization of SVA elements, we generated a second L1 protein donor plasmid, termed pJM101/L1RPΔneoΔORF1, which differs from pJM101/L1RPΔneo exclusively in a 330-bp in-frame deletion in L1 ORF1 (Figure 1B). The in-frame deletion of pJM101/L1RPΔneoΔORF1 ensures that an ORF1p mutant which lacks amino acid positions 99–208 is expressed, and that initiation of ORF2 translation within the bicistronic RNA is not perturbed. The deletion comprises the C-terminal half of the coiled coil (cc) domain and the N-terminal half of the RNA-recognition motif (RRM) domain of ORF1p (40). We confirmed by immunoblot analysis that both L1 protein donor plasmids facilitated expression of similar amounts of ORF2p after cotransfection with each of the three SVA reporter plasmids (Figures 3C). Coexpression of ORF1p deletion mutant and functional ORF2p from pJM101/L1RPΔneoΔORF1 did not result in trans-mobilization of the SVA reporter elements encoded by pAD3/SVAE and pSC3/SVAF1 indicating that an intact ORF1p is essential for SVA retrotransposition. Surprisingly, in the absence of both overexpressed L1 proteins or ORF1p alone, trans-mobilization of the SVAF1 reporter element of pSC4/SVAF1 was still exceeding pseudogene formation by 2- to 3-fold (Figure 3A). Since the 3′-transduced AluSp sequence obviously makes the SVA RNA a preferred substrate for L1 proteins, mobilization in the absence of overexpressed L1 proteins could be explained by trans-mobilization of the pSC4/SVAF1 reporter cassette beyond pseudogene formation by the moderately expressed endogenous L1 protein machinery in HeLa cells (41,42). Quantification of spliced SVA reporter RNAs by qRT–PCR (Figure 3D) and of overexpressed L1 proteins by immunoblot analysis (Figure 3C) show that the observed differences in G418R colonies are neither a consequence of differences in the amounts of spliced retrotransposition reporter transcripts nor the result of varying amounts of overexpressed L1 proteins.
Structural features of SVA de novo integrants
To verify that the G418R HeLa colonies that resulted from SVA trans-mobilization assays were a consequence of marked SVA de novo retrotransposition events, genomic DNA was extracted from 12 randomly chosen single expanded G418R HeLa cell colonies which had resulted from cotransfection of the SVAE reporter plasmids with pJM101/L1RPΔneo (Figure 2A). First, we carried out a diagnostic PCR on genomic DNA with primers spanning the intron of mneoI (Figure 4). The generation of a 792-bp PCR product shows that transcription, splicing, and L1-mediated reverse transcription of the SVA reporter elements as well as integration of the SVAE-mneoI cDNA into the genome has occurred. A 1694-bp PCR product that would be specific for the unspliced indicator cassette encoded by the SVAE retrotransposition reporter plasmids could not be detected (Figure 4). Since PCR products obtained from HeLa clone X differed from the expected pattern, this clone was excluded from further analyses. Next, we examined the structures of the genomic mneo-tagged SVAE de novo insertions for attributes of L1-mediated retrotransposition. Analysis of pre- and post-integration sites of eight pAD3/SVAE-derived and three pAD4/SVAE-derived de novo insertions isolated from G418R HeLa colonies (Figure 5) revealed that 5/11 insertions occurred into genes (Supplementary Table S2) and each insertion is indeed distinguished by structural hallmarks of L1-mediated retrotransposition such as a 30- to 78-nt poly(A) tail. The nucleotide profile of the target sites of SVAE de novo insertions resembles the L1 EN consensus target sequence 5′-TTTT/AA-3′ of L1-Ta de novo insertions and preexisting L1-Ta, AluYa5 and SVAE/F insertions (Figure 6). This indicates that the sequence specificity of L1 EN determines SVA integration site properties at the local level. With one exception, all characterized insertions are flanked by TSDs ranging from 8 to 19 nt (Figure 5). SVA de novo insertion 9 led to the formation of an 11-bp target site deletion, and the deleted sequence was replaced by a 5′-truncated mneoI-tagged SVA retrotransposition event lacking TSDs. The formation of similar genomic target site deletions ranging from 1 bp to >11 kb associated with 5′-truncated de novo insertions has been reported for EN-dependent L1 retrotransposition events earlier (43–45). Target site deletions associated with L1-mediated retrotransposition events are believed to arise as a consequence of a second-strand cleavage event that occurred upstream of the initial first-strand cleavage (43,46).
Figure 5.
Genomic SVAE de novo insertions display hallmarks of L1-mediated retrotransposition. Both pre- and post-integration sites are presented. Insertions 1–8 and 9–11 are derived from SVAE reporter elements encoded by pAD3/SVAE and pAD4/SVAE, respectively. Nucleotide positions at the 5′-end of each SVA insertion refer to the reference sequence of SVA H19_27 [(21); Supplementary Figure S1]. Marked full-length insertions cover ~3.3 kb (pAD3/SVAE-derived) and ~2.8 kb (pAD4/SVAE-derived), respectively. Identified 5′-truncated insertions comprise 1580 bp (insertion 6) and 1620 bp (insertion 9), respectively. The L1EN cleavage site on the bottom strand is indicated in blue. Extra deoxyguanylates at the 5′-ends of de novo insertions are indicated in green. CMVP-derived sequences are highlighted in yellow. Nucleotides representing patches of microcomplementarity are underlined. Black bars, Alu TSD sequences. Red lettering, SVA TSD sequences; neo, neomycin-phosphotransferase gene.
Only 2 out of the 11 characterized SVA de novo insertions were characterized by 5′-truncations encompassing 1716 and 680 bp (insertions 6 and 9; Figure 5), respectively. The remaining nine insertions are full-length and cover 3347 (pAD3/SVAE-derived) and 2850 nt (pAD4/SVAE derived). The 5′-ends of SVA full length de novo insertions derived from both SVA reporter elements coincide with positions 3 or 4 downstream of the CMV promoter transcription initiation site (47) indicating that transcription of the SVA reporter elements is controlled by CMVP and not by any potential SVA-specific internal promoter. Full-length SVA de novo insertions include the same 44–45 nt of non-SVA sequences at their 5′-ends which result from transcriptional initiation within the CMVP region (Figure 1A).
In three cases we observed an untemplated G nucleotide between the 3′-end of the 5′ TSD and the 5′-end of full-length SVA insertions. (Figure 5A; insertions 2, 5 and 8). We identified short patches of microcomplementarity of 1–2 nt at the 5′-genomic DNA/SVA junction of 5/11 SVA de novo insertions which is consistent with previous studies analyzing preexisting 5′-genomic DNA/L1 junctions (48,36) and 5′-junctions of L1 de novo insertions (44,45). Taken together, each of the described structural features of SVA de novo insertions have been reported for L1 de novo insertions before. This is indicating that the analyzed SVA de novo insertions are a consequence of the trans-activity of the L1 protein machinery acting on SVA transcripts.
Target site preferences of de novo SVA integrants
SVA de novo insertions showed a clear integration preference for TSDs or sequences flanking endogenous non-LTR retrotransposons. Five out of the 11 characterized SVA insertions occurred into Alu-TSDs or adjacent sequences (Figure 5). SVA copies inserted into an L1 3′-end (insertion 10) and within the first 100 nt of the 3′-flanking region of an L1 element (insertion 5), respectively.
We calculated the average G+C content of the genomic sequences flanking de novo SVA insertion within 5-kb windows and 30-kb windows, and found that it amounted to 44% and 44.2%, respectively (Table 1). This was significantly higher than the genome average of 41% (p5kb< 0.025; p30kb< 0.005), and is in accordance with the overall distribution of preexisting members of the SVA subfamilies E and F which were reported to have accumulated in G+C-rich regions of the human genome (12). Several scenarios have been proposed for the apparent enrichment of SVAE and SVAF elements in G+C-rich regions (12). Sequence analyses of the 5-kb and 30-kb windows also indicate that SVA de novo insertions occurred into genomic regions with an increased Alu density and a relatively poor L1 density compared to the overall genomic situation (Table 1). Distribution of SVA de novo insertions to different isochores/ G+C-content domains of the genome is consistent with the recently described situation of genomic SVA copies (49). Eight integrants are found in isochore H1, while the remainder insertions are located in isochore L2 (two insertions) and H2 (one insertion).
Table 1.
G+C- and retrotransposon content of genomic sequences flanking SVA de novo insertions
| SVA insertion | G+C 5 kb (%) | Alu 5 kb (%) | L1 5 kb (%) | G+C 30 kb (%) | Alu 30 kb (%) | L1 30 kb (%) |
|---|---|---|---|---|---|---|
| 1 | 46.44 | 35.8 | 1.52 | 43.91 | 45.51 | 7.04 |
| 2 | 42.40 | 44.54 | – | 42.32 | 31.01 | 9.42 |
| 3 | 40.57 | 40.86 | 3.58 | 40.54 | 36.62 | 2.74 |
| 4 | 48.39 | 39.88 | – | 45.98 | 44.36 | 7.88 |
| 5 | 38.93 | 13.54 | 22.28 | 42.30 | 16.52 | 6.11 |
| 6 | 40.03 | 17.34 | – | 38.28 | 11.81 | – |
| 7 | 39.42 | 36.42 | 11.62 | 42.41 | 19.89 | 1.94 |
| 8 | 45.43 | 39.92 | 2.66 | 47.02 | 36.46 | 7.30 |
| 9 | 50.85 | 13.28 | – | 49.17 | 12.26 | 7.28 |
| 10 | 44.81 | 29.96 | 24.1 | 48.75 | 31.78 | 5.75 |
| 11 | 47.00 | 32.98 | 0.48 | 45.42 | 30.25 | 4.19 |
| Arith. Mean 1–11 | 44.02 | 31.32 | 5.21 | 44.19 | 28.77 | 5.97 |
| Genome average | 40.91 | 10.60 | 16.89 | 40.91 | 10.60 | 16.89 |
Analyzed sequences encompass 5-kb and 30-kb windows; G+C 5 kb/G+C 30 kb, G+C content (in percent) of 5-kb or 30-kb genomic sequence windows flanking the SVA de novo insertions; Alu 5 kb/Alu 30 kb (L1 5 kb/L1 30 kb), fraction of Alu (L1) sequences (in percent) covering 5-kb or 30-kb genomic sequence windows flanking the SVA de novo insertion.
DISCUSSION
L1-mediated trans-mobilization of SVA elements requires both L1 encoded proteins
In order to gain insight into the molecular mechanisms of the mobilization of the non-autonomous SVA elements, we tested the hypothesis that SVA elements are trans-mobilized by the protein machinery encoded by functional L1 elements. We established an SVA retrotransposition reporter assay which enabled us to experimentally confirm that SVA RNA recruits the L1 protein machinery for its own mobilization. Trans-mobilization frequencies of the tested human-specific SVA reporter elements exceeded the processed pseudogene formation rate of cellular mRNAs by 2- to 5-fold in HeLa-JVM cells and by 12- to 300-fold in HeLa-HA cells. During the revision of this manuscript, Hancks and coworkers published in rough accordance with our results that, in their hands, L1-mediated SVA retrotransposition exceeds processed pseudogene formation in HeLa-HA cells by 1- to 54-fold (50). We observed a strict requirement of L1 ORF1p and L1 ORF2p for the mobilization of the three tested SVA reporter elements in HeLa cells. While ORF1p was also shown to be essential for processed pseudogene formation (2,7), it is dispensable for Alu retrotransposition (3). The stern need for ORF1p for the mobilization of both versions of the SVAF1 source element on chromosome 10 which differ from each other exclusively in the presence/absence of the ~390 bp AluSp 3′ transduction was confirmed by Hancks et al. (50). However, while we also observed the same requirement of ORF1p for the mobilization of the canonical SVAE reporter element H19_27, the Kazazian laboratory obtained conflicting results when they analyzed the role of ORF1p in the mobilization of a canonical SVAD source element (50): Consistent with our findings, they reported that expression of an L1 driver with a double mutation in the RRM domain of ORF1p blocked trans-mobilization of the canonical SVAD source element almost entirely in HeLa-HA cells, and that coexpression of ORF2p alone with the EGFP-marked SVAD element led to barely any detectable retrotransposition events. In contrast, coexpression of ORF2p with the mneoI-marked SVAD element produced more G418R foci than transfection with the intact full-length L1 driver plasmid suggesting that ORF2p alone is essential for trans-mobilization (50). It is unlikely that the ~600-bp difference in the extension of the VNTR region between the ~1.9-kb SVAE element and the ~2.5-kb SVAD element played any role in the observed differences in ORF1p requirement.
Structural features of SVA elements affecting trans-mobilization rates
The observation that the trans-mobilization rates of the canonical SVAE reporter element in pAD3/SVAE exceeds retropseudogene formation by 2- to 5-fold in HeLa-JVM cells and by ~18-fold in HeLa-HA cells, raises the question if potential SVA-specific structural features might qualify SVA transcripts as preferred substrates for the L1 protein machinery. First, as indicated by Alus, tRNA-derived SINEs (51), and tailless tRNAs, the ability of an RNA to localize to the ribosome determines its retrotranspositional success. After transcription, the SVA RNA needs to come in contact with L1 ORF2p, and out-compete the L1 RNA for the attention of ORF2p. L1 and Alu RNA competition for ORF2p presumably takes place at the ribosome (52,53). It has been hypothesized that the SVA-encoded Alu-like domain localizes SVA RNA to the ribosome by annealing with Alu RNAs which were suggested to be docked on ribosomes via the SRP9/14 complex (52,54). This hypothesis is consistent with our observation that the retrotransposition rate of the SVA reporter element in pAD4/SVAE which is devoid of hexameric (CCCTCT)n repeats and Alu-like domain, is reduced in average by 32–46% (Figure 2A and D) relative to the full-length SVA reporter element in pAD3/SVAE. A potential relevance of the Alu-like region for trans-mobilization is also supported by the finding that our pSC3/SVAF1 encoded SVA source element which also lacks the (CCCTCT)n repeats and almost the entire Alu-like domain exceeds pseudogene formation rate by only ~12-fold while the canonical SVAE element is trans-mobilized ~18-fold more efficiently than the pseudogene in HeLa-HA cells (Figure 3A and B). Second, the structures of preexisting SVA insertions imply that several modules of a canonical full-length SVA element are dispensable and not essential for successive rounds of retrotransposition. The existence of SVA2 elements which consist exclusively of VNTRs fused to short non-SVA sequences and display hallmarks of L1-mediated retrotransposition (20,21,55), suggests that the VNTR region is essential for SVA mobilization. This is supported by the fact that the VNTR region which can vary in length significantly among different SVA insertions, is the only module all mobilized SVA RNAs have in common. It was suggested that the VNTR alone or within the context of SVA may increase RNA stability (24).
We show that the 3′ transduction-mediated acquisition of the AluSp sequence by the SVAF1 source element H10_1 (pSC4/SVAF1) led to a 3′-transduced SVAF1 RNA which is mobilized by the L1 protein machinery ~25-fold more efficiently than the same RNA lacking the AluSp sequence (pSC3/SVAF1) (Figure 3A and B). The relatively high trans-mobilization rate of the 3′-transduced SVAF1 element H10_1 is obviously based on the observed preference of L1 proteins for SVA transcripts carrying intact AluSp sequences at their 3′-ends and is consistent with the presence of numerous genomic SVAF1 copies characterized by the 3′-transduced AluSp sequences (21,23,39). The same mechanism hypothesized recently to be responsible for the preferential trans-mobilization of Alu elements by the L1-encoded protein machinery (3,52) could explain the favored mobilization of SVA RNAs harbouring intact Alu elements at their 3′-terminus (21). In this model, the Alu sequence is docked on ribosomes via the SRP9/14 complex and captures the L1 ORF2 protein as it is translated from an active L1 element mRNA (52). Provided that the 3′-terminal AluSp elements in SVA transcripts allow formation of the three-dimensional structure required for SRP9/14 interaction (56), Alu sequences could mediate docking of the respective SVA RNA to the ribosome via SRP9/14 and thus facilitate efficient capture of ORF2 proteins (3,52,54). Interestingly, the AluSc element in the 5′-region of the SVAF1 source element (Figure 1A) does not seem to be beneficial for trans-mobilization, because the canonical SVAE reporter element in pAD3/SVAE which is devoid of AluSc, is mobilized at a similar frequency (~18-fold) as the SVAF1 source element in pSC3/SVAF1 (~12-fold) (Figure 3B).
Comparison of trans-mobilization rates of SVA and Alu elements
The trans-mobilization frequency of our canonical SVAE reporter element in HeLa cells after hygromycin selection equates to a relative retrotransposition rate of 0.4–0.9% as to the cis retrotransposition frequency of the pJM101/L1RP-encoded functional L1RP element which was set as 100% (Figures 2A and 3B). Trans-mobilization of the full-length SVAE reporter element exceeded pseudogene formation rates of 0.01–0.04% in HeLa-JVM and HeLa-HA cells by 2- to 5-fold and ~18-fold, respectively (Figures 2D and 3B) and coincides well with the processed pseudogene formation frequencies of 0.01–0.05% reported previously by Moran et al. (2). Differences in retrotransposition activity between human cell lines have been reported earlier (50) and are also known to exist between different HeLa cell lines (Moran and Deininger, personal communication). The ~25-fold increase of the trans-mobilization rate of the SVAF1 source element H10_1 after the addition of the intact AluSp sequence (Figure 3) equates to a relative retrotransposition rate of ~11%. This corresponds approximately to the relative trans-mobilization frequency (~10%) of the AluYa5a2 NF1 element (3) which is one of the most active Alus described to date (53). Trans-mobilization rates of the canonical SVAE reporter element (pAD3/SVAE, 0.68%) and the SVAF1 source element that lacks AluSp (pSC4/SVAF1, 0.43%) are close to the activity of a subset of polymorphic AluY elements which were found to reach only 10% of the AluYa5a2 NF1 retrotransposition rate (53). The relative retrotransposition rates of L1, Alu and SVA elements determined in cell culture assays do not reflect their recently estimated in vivo retrotransposition rates of one in 212, 21 and 916 births, respectively (25). One possible explanation for discrepancies, for example, in the case of SVA and L1 elements, could be an excessive upregulation of SVA source element transcription in the germ line or during early stages of embryonal development relative to L1 transcription.
Hancks et al. (50) referred only a ≤2-fold increase in trans-mobilization rate of the SVAF1 source element after acquisition of the 3′-transduced AluSp sequence and that AluY is trans-mobilized 30-fold more efficiently than the AluSp-including SVAF1 element. There are several differences in the design of SVA reporter and L1 donor plasmids between the two reports which make a comparison of the presented results rather complicated. First, the 3′-transduced sequence including the AluSp element in the SVAF1 reporter used by Hancks and coworkers (50) differed from the genomic sequence in several nucleotide substitutions which were reported to affect trans-mobilization rates significantly. In contrast, nucleotide sequences of all SVA reporter elements tested in our study match genomic sequences. Second, unlike Hancks et al., we removed transcriptional termination signals at the SINE-R 3′-ends and in the 3′-flanking sequence of the AluSp 3′ TSD to ensure transcriptional read-through into the mneoI cassettes and consistent polyadenylation at the pCEP4-encoded SV40 polyadenylation signal (Figure 1A). Third, we fused the mneoI indicator cassette always with the 3′-end of the respective SVA reporter element, while Hancks and coworkers inserted this cassette between SINE-R and AluSp sequence of the SVAF1 element. As a result, the transcriptional start site of the mneoI cassette is located ~430-bp upstream of the 3′-end of the SVAF1 RNA, while the transcription start site of the same cassette in our analogous construct pSC3/SVAF1 is located at the RNA 3′-end. Since 5′-truncations occur during SVA trans-mobilization, it could well be that the location of the indicator cassette upstream of the SVA 3′-end leads to the formation of less G418R foci or EGFP expressing cells in cell culture assays.
SVA de novo insertions bear the hallmarks of mobilization by the L1 protein machinery
Analysis of pre- and post-integration sites of 11 SVA de novo insertions uncovered the hallmarks of L1-mediated retrotransposition. We found variable TSDs of 8–19 bp in length which correspond well to the TSD lengths of preexisting genomic L1 insertions ranging from 9 to 27 bp (15). The nucleotide profile of the target sites of the 11 analyzed SVAE de novo insertions resembles the L1 consensus target sequence 5′-TTTT/AA-3′ (Figure 6). SVA insertions into or next to Alu TSDs can be explained by the fact that their AT-rich TSDs represent recognition sequences for the L1 endonuclease in an otherwise AT-poor environment (Figure 5).
Poly(A) tail lengths of SVA de novo insertions (30–78 nt) exceeded those of pre-existing SVAs (2–72 nt) (21) significantly. Similar differences have been reported for L1 de novo insertions (3–150 nt) and preexisting L1s (approximately 13 adenosines) (43,57). One reason for the differences in polyA tail lengths between preexisting and de novo insertions might be the fact that each de novo insertion derived from an SVAE reporter cassette was polyadenylated at the sole SV40pA site present at the 3′-end of the reporter construct (Figure 1A), while RNAs derived from genomic SVA insertions are polyadenylated at sites encoded by the SINE-R region.
We found 9/11 (~82%) SVA de novo insertions to be full-length covering 3.35 and 2.85 kb, respectively (Figure 5 and Supplementary Table S2). Full-length insertions included the entire spliced transcript expressed from the SVA retrotransposition reporter cassette, starting with position +3 or +4 relative to the transcriptional initiation site of the CMV promoter. The fact that the percentage of 63% of preexisting full-length SVA insertions differs from the proportion of de novo full-length insertions could be a consequence of the relative small number of analyzed de novo insertions relative to the statistically more significant pool of ~2760 preexisting SVAs analyzed by Wang et al. (12). Since it was reported that the length of de novo insertions generated by L1 proteins in trans depends on the activity of this particular L1 element (58), the observed difference might alternatively be attributed to the use of L1RP, one of the most active human L1 elements identified to date (59), as L1 protein donor in our reporter assays. In vivo trans-mobilization of genomic SVA elements, however, is probably mediated by a multitude of different functional L1 elements with most of them being less active than L1RP (59). Therefore, lower frequencies of endogenous full-length SVA retrotransposition events would be expected. In the case of L1, only ~6% of all de novo retrotransposition events (43–45) and ~5% of preexisting copies (15) are full-length. Clearly, one reason for the significantly larger fraction of full-length SVA elements compared to L1 is the fact that L1 RT can process more of the comparatively short full-length SVA-encoded mRNAs which are only 700–4000 bp in length (12,23) than 6-kb transcripts that are encoded by full-length L1s. Since the average size of recovered L1RP de novo insertions is ~3165 bp (44), and our de novo SVA full-length insertions are 3303 or 2805 bp in length (Supplementary Table S2), one would expect an increased rate of full-length SVA insertions. The fact that L1 RT successfully reverse transcribes the GC-rich VNTR region of the SVA reporter elements militates against the hypothesis that L1 5′-truncations are a consequence of an extenuated L1 RT activity. Hancks et al. reported that mobilization of an SVAE source element that was not under transcriptional control of an external promoter generated only 5′-truncated de novo insertions (50). This result suggests that SVA elements per se do not include strong promoter sequences that could facilitate full-length transcription, and that mneoI insertions derived from the promoterless SVAE reporter might have been produced from a cryptic promoter located upstream of the SVA sequence on the reporter plasmid. Alternatively, these SVAE insertions might not represent 5′-truncations at all and SVA transcription started within the VNTR region as numerous transcription start sites exist throughout the SVA sequence (23,50).
We found untemplated G nucleotides at the 5′-end of three full-length SVA de novo insertions. They have been described originally for L1 insertions (60) and were attributed to reverse transcription of the 7-methyl guanosine cap (44). Endogenous SVAs are very likely transcribed by RNA polymerase II. 5′ capping of SVA RNAs was suggested in earlier reports because of the presence of guanosine residues at the 5′-end of about 33% of pre-existing SVA insertions (12). The 5/11 insertions are characterized by short patches of microcomplementarity of 1–2 nt at the junctions between SVA 5′-end and TSD. Such short patches of microhomology were reported earlier for L1 5′ junctions (36) and indicate the involvement of double strand break repair by error-prone non homologous endjoining (NHEJ) in the attachment of the SVA 5′-end to the chromosomal DNA.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online: Supplementary Tables 1–3, Supplementary Figures 1–6, Supplementary Methods, and Supplementary References [61–63].
FUNDING
Deutsche Forschungsgemeinschaft (DA 545/2-1 to A.D. and G.G.S.). Funding for open access charge: Paul-Ehrlich-Institut and Deutsche Forschungsgemeinschaft.
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
The authors thank John Moran for providing plasmids pJM101/L1.3, pJM101/L1RP, and pJM101/L1RPΔneo and the cell lines HeLa-HA and HeLa-JVM. The authors are grateful to John Goodier and Haig Kazazian who provided us with the reliable αL1ORF2p-N antibody. The authors want to thank Mark Batzer for giving us access to his genomic AluYa5 database. The authors are indebted to Kay-Martin Hanschmann for statistical analyses.
REFERENCES
- 1.Lander ES, Linton LM, Birren B, Nusbaum C, Zody MC, Baldwin J, Devon K, Dewar K, Doyle M, FitzHugh W, et al. Initial sequencing and analysis of the human genome. Nature. 2001;409:860–921. doi: 10.1038/35057062. [DOI] [PubMed] [Google Scholar]
- 2.Wei W, Gilbert N, Ooi SL, Lawler JF, Ostertag EM, Kazazian HH, Boeke JD, Moran JV. Human L1 retrotransposition: cis preference versus trans complementation. Mol. Cell Biol. 2001;21:1429–1439. doi: 10.1128/MCB.21.4.1429-1439.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Dewannieux M, Esnault C, Heidmann T. LINE-mediated retrotransposition of marked Alu sequences. Nat. Genet. 2003;35:41–48. doi: 10.1038/ng1223. [DOI] [PubMed] [Google Scholar]
- 4.Kroutter EN, Belancio VP, Wagstaff BJ, Roy-Engel AM. The RNA polymerase dictates ORF1 requirement and timing of LINE and SINE retrotransposition. PLoS. Genet. 2009;5:e1000458. doi: 10.1371/journal.pgen.1000458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Buzdin A, Ustyugova S, Gogvadze E, Vinogradova T, Lebedev Y, Sverdlov E. A new family of chimeric retrotranscripts formed by a full copy of U6 small nuclear RNA fused to the 3' terminus of l1. Genomics. 2002;80:402–406. doi: 10.1006/geno.2002.6843. [DOI] [PubMed] [Google Scholar]
- 6.Garcia-Perez JL, Doucet AJ, Bucheton A, Moran JV, Gilbert N. Distinct mechanisms for trans-mediated mobilization of cellular RNAs by the LINE-1 reverse transcriptase. Genome Res. 2007;17:602–611. doi: 10.1101/gr.5870107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Esnault C, Maestre J, Heidmann T. Human LINE retrotransposons generate processed pseudogenes. Nat. Genet. 2000;24:363–367. doi: 10.1038/74184. [DOI] [PubMed] [Google Scholar]
- 8.Pavlicek A, Paces J, Elleder D, Hejnar J. Processed pseudogenes of human endogenous retroviruses generated by LINEs: their integration, stability, and distribution. Genome Res. 2002;12:391–399. doi: 10.1101/gr.216902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhang Z, Harrison P, Gerstein M. Identification and analysis of over 2000 ribosomal protein pseudogenes in the human genome. Genome Res. 2002;12:1466–1482. doi: 10.1101/gr.331902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Shen L, Wu LC, Sanlioglu S, Chen R, Mendoza AR, Dangel AW, Carroll MC, Zipf WB, Yu CY. Structure and genetics of the partially duplicated gene RP located immediately upstream of the complement C4A and the C4B genes in the HLA class III region. Molecular cloning, exon-intron structure, composite retroposon, and breakpoint of gene duplication. J. Biol. Chem. 1994;269:8466–8476. [PubMed] [Google Scholar]
- 11.Bennett EA, Coleman LE, Tsui C, Pittard WS, Devine SE. Natural genetic variation caused by transposable elements in humans. Genetics. 2004;168:933–951. doi: 10.1534/genetics.104.031757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wang H, Xing J, Grover D, Hedges DJ, Han K, Walker JA, Batzer MA. SVA elements: a hominid-specific retroposon family. J. Mol. Biol. 2005;354:994–1007. doi: 10.1016/j.jmb.2005.09.085. [DOI] [PubMed] [Google Scholar]
- 13.Ostertag EM, Goodier JL, Zhang Y, Kazazian HH., Jr SVA elements are nonautonomous retrotransposons that cause disease in humans. Am. J. Hum. Genet. 2003;73:1444–1451. doi: 10.1086/380207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ostertag EM, Kazazian HH., Jr Twin priming: a proposed mechanism for the creation of inversions in L1 retrotransposition. Genome Res. 2001;11:2059–2065. doi: 10.1101/gr.205701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Szak ST, Pickeral OK, Makalowski W, Boguski MS, Landsman D, Boeke JD. Molecular archeology of L1 insertions in the human genome. Genome Biol. 2002;3:research0052. doi: 10.1186/gb-2002-3-10-research0052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Xing J, Wang H, Belancio VP, Cordaux R, Deininger PL, Batzer MA. Emergence of primate genes by retrotransposon-mediated sequence transduction. Proc. Natl Acad. Sci. USA. 2006;103:17608–17613. doi: 10.1073/pnas.0603224103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Moran JV, DeBerardinis RJ, Kazazian HH., Jr Exon shuffling by L1 retrotransposition. Science. 1999;283:1530–1534. doi: 10.1126/science.283.5407.1530. [DOI] [PubMed] [Google Scholar]
- 18.Mills RE, Bennett EA, Iskow RC, Luttig CT, Tsui C, Pittard WS, Devine SE. Recently mobilized transposons in the human and chimpanzee genomes. Am. J. Hum. Genet. 2006;78:671–679. doi: 10.1086/501028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jurka J, Kapitonov VV, Pavlicek A, Klonowski P, Kohany O, Walichiewicz J. Repbase Update, a database of eukaryotic repetitive elements. Cytogenet. Genome Res. 2005;110:462–467. doi: 10.1159/000084979. [DOI] [PubMed] [Google Scholar]
- 20.Han K, Konkel MK, Xing J, Wang H, Lee J, Meyer TJ, Huang CT, Sandifer E, Hebert K, Barnes EW, et al. Mobile DNA in Old World monkeys: a glimpse through the rhesus macaque genome. Science. 2007;316:238–240. doi: 10.1126/science.1139462. [DOI] [PubMed] [Google Scholar]
- 21.Damert A, Raiz J, Horn AV, Lower J, Wang H, Xing J, Batzer MA, Lower R, Schumann GG. 5'-Transducing SVA retrotransposon groups spread efficiently throughout the human genome. Genome Res. 2009;19:1992–2008. doi: 10.1101/gr.093435.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ono M, Kawakami M, Takezawa T. A novel human nonviral retroposon derived from an endogenous retrovirus. Nucleic Acids Res. 1987;15:8725–8737. doi: 10.1093/nar/15.21.8725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hancks DC, Ewing AD, Chen JE, Tokunaga K, Kazazian HH., Jr Exon-trapping mediated by the human retrotransposon SVA. Genome Res. 2009;19:1983–1991. doi: 10.1101/gr.093153.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hancks DC, Kazazian HH., Jr SVA retrotransposons: Evolution and genetic instability. Semin. Cancer Biol. 2010;20:234–245. doi: 10.1016/j.semcancer.2010.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Xing J, Zhang Y, Han K, Salem AH, Sen SK, Huff CD, Zhou Q, Kirkness EF, Levy S, Batzer MA, et al. Mobile elements create structural variation: analysis of a complete human genome. Genome Res. 2009;19:1516–1526. doi: 10.1101/gr.091827.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wilund KR, Yi M, Campagna F, Arca M, Zuliani G, Fellin R, Ho YK, Garcia JV, Hobbs HH, Cohen JC. Molecular mechanisms of autosomal recessive hypercholesterolemia. Hum. Mol. Genet. 2002;11:3019–3030. doi: 10.1093/hmg/11.24.3019. [DOI] [PubMed] [Google Scholar]
- 27.Moran JV, Holmes SE, Naas TP, DeBerardinis RJ, Boeke JD, Kazazian HH., Jr High frequency retrotransposition in cultured mammalian cells. Cell. 1996;87:917–927. doi: 10.1016/s0092-8674(00)81998-4. [DOI] [PubMed] [Google Scholar]
- 28.Hulme AE, Bogerd HP, Cullen BR, Moran JV. Selective inhibition of Alu retrotransposition by APOBEC3G. Gene. 2007;390:199–205. doi: 10.1016/j.gene.2006.08.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ostertag EM, Prak ET, DeBerardinis RJ, Moran JV, Kazazian HH., Jr Determination of L1 retrotransposition kinetics in cultured cells. Nucleic Acids Res. 2000;28:1418–1423. doi: 10.1093/nar/28.6.1418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Δ Δ C(T)) Method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 31.Goodier JL, Ostertag EM, Engleka KA, Seleme MC, Kazazian HH., Jr A potential role for the nucleolus in L1 retrotransposition. Hum. Mol. Genet. 2004;13:1041–1048. doi: 10.1093/hmg/ddh118. [DOI] [PubMed] [Google Scholar]
- 32.Kirilyuk A, Tolstonog GV, Damert A, Held U, Hahn S, Lower R, Buschmann C, Horn AV, Traub P, Schumann GG. Functional endogenous LINE-1 retrotransposons are expressed and mobilized in rat chloroleukemia cells. Nucleic Acids Res. 2008;36:648–665. doi: 10.1093/nar/gkm1045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Schmidt M, Hoffmann G, Wissler M, Lemke N, Mussig A, Glimm H, Williams DA, Ragg S, Hesemann CU, von KC. Detection and direct genomic sequencing of multiple rare unknown flanking DNA in highly complex samples. Hum. Gene Ther. 2001;12:743–749. doi: 10.1089/104303401750148649. [DOI] [PubMed] [Google Scholar]
- 34.Costantini M, Clay O, Auletta F, Bernardi G. An isochore map of human chromosomes. Genome Res. 2006;16:536–541. doi: 10.1101/gr.4910606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Crooks GE, Hon G, Chandonia J-M, Brenner SE. WebLogo: A sequence logo generator. Genome Res. 2004;14:1188–1190. doi: 10.1101/gr.849004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zingler N, Willhoeft U, Brose HP, Schoder V, Jahns T, Hanschmann KM, Morrish TA, Lower J, Schumann GG. Analysis of 5' junctions of human LINE-1 and Alu retrotransposons suggests an alternative model for 5'-end attachment requiring microhomology-mediated end-joining. Genome Res. 2005;15:780–789. doi: 10.1101/gr.3421505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Otieno AC, Carter AB, Hedges DJ, Walker JA, Ray DA, Garber RK, Anders BA, Stoilova N, Laborde ME, Fowlkes JD, et al. Analysis of the human Alu Ya-lineage. J. Mol. Biol. 2004;342:109–118. doi: 10.1016/j.jmb.2004.07.016. [DOI] [PubMed] [Google Scholar]
- 38.Freeman JD, Goodchild NL, Mager DL. A modified indicator gene for selection of retrotransposition events in mammalian cells. Biotechniques. 1994;17:46, 48–49, 52. [PubMed] [Google Scholar]
- 39.Takasu M, Hayashi R, Maruya E, Ota M, Imura K, Kougo K, Kobayashi C, Saji H, Ishikawa Y, Asai T, et al. Deletion of entire HLA-A gene accompanied by an insertion of a retrotransposon. Tissue Antigens. 2007;70:144–150. doi: 10.1111/j.1399-0039.2007.00870.x. [DOI] [PubMed] [Google Scholar]
- 40.Khazina E, Weichenrieder O. Non-LTR retrotransposons encode noncanonical RRM domains in their first open reading frame. Proc. Natl Acad. Sci. USA. 2009;106:731–736. doi: 10.1073/pnas.0809964106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Belancio VP, Roy-Engel AM, Pochampally RR, Deininger P. Somatic expression of LINE-1 elements in human tissues. Nucleic Acids Res. 2010;38:3909–3922. doi: 10.1093/nar/gkq132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Comeaux MS, Roy-Engel AM, Hedges DJ, Deininger PL. Diverse cis factors controlling Alu retrotransposition: What causes Alu elements to die? Genome Res. 2009;19:545–555. doi: 10.1101/gr.089789.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Gilbert N, Lutz-Prigge S, Moran JV. Genomic deletions created upon LINE-1 retrotransposition. Cell. 2002;110:315–325. doi: 10.1016/s0092-8674(02)00828-0. [DOI] [PubMed] [Google Scholar]
- 44.Gilbert N, Lutz S, Morrish TA, Moran JV. Multiple fates of L1 retrotransposition intermediates in cultured human cells. Mol. Cell Biol. 2005;25:7780–7795. doi: 10.1128/MCB.25.17.7780-7795.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Symer DE, Connelly C, Szak ST, Caputo EM, Cost GJ, Parmigiani G, Boeke JD. Human l1 retrotransposition is associated with genetic instability in vivo. Cell. 2002;110:327–338. doi: 10.1016/s0092-8674(02)00839-5. [DOI] [PubMed] [Google Scholar]
- 46.Ichiyanagi K, Okada N. Mobility pathways for vertebrate L1, L2, CR1, and RTE clade retrotransposons. Mol. Biol. Evol. 2008;25:1148–1157. doi: 10.1093/molbev/msn061. [DOI] [PubMed] [Google Scholar]
- 47.Isomura H, Stinski MF, Kudoh A, Nakayama S, Murata T, Sato Y, Iwahori S, Tsurumi T. A cis element between the TATA Box and the transcription start site of the major immediate-early promoter of human cytomegalovirus determines efficiency of viral replication. J. Virol. 2008;82:849–858. doi: 10.1128/JVI.01593-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Martin SL, Li WL, Furano AV, Boissinot S. The structures of mouse and human L1 elements reflect their insertion mechanism. Cytogenet. Genome Res. 2005;110:223–228. doi: 10.1159/000084956. [DOI] [PubMed] [Google Scholar]
- 49.Pozzoli U, Menozzi G, Fumagalli M, Cereda M, Comi GP, Cagliani R, Bresolin N, Sironi M. Both selective and neutral processes drive GC content evolution in the human genome. BMC. Evol. Biol. 2008;8:99. doi: 10.1186/1471-2148-8-99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Hancks DC, Goodier JL, Mandal PK, Cheung LE, Kazazian HH., Jr Retrotransposition of marked SVA elements by human L1s in cultured cells. Hum.Mol.Genet. 2011;20:3386–3400. doi: 10.1093/hmg/ddr245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Okada N, Hamada M, Ogiwara I, Ohshima K. SINEs and LINEs share common 3' sequences: a review. Gene. 1997;205:229–243. doi: 10.1016/s0378-1119(97)00409-5. [DOI] [PubMed] [Google Scholar]
- 52.Boeke JD. LINEs and Alus–the polyA connection. Nat. Genet. 1997;16:6–7. doi: 10.1038/ng0597-6. [DOI] [PubMed] [Google Scholar]
- 53.Bennett EA, Keller H, Mills RE, Schmidt S, Moran JV, Weichenrieder O, Devine SE. Active Alu retrotransposons in the human genome. Genome Res. 2008;18:1875–1883. doi: 10.1101/gr.081737.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Mills RE, Bennett EA, Iskow RC, Devine SE. Which transposable elements are active in the human genome? Trends Genet. 2007;23:183–191.. doi: 10.1016/j.tig.2007.02.006. [DOI] [PubMed] [Google Scholar]
- 55.Jurka J. Repbase update: a database and an electronic journal of repetitive elements. Trends Genet. 2000;16:418–420. doi: 10.1016/s0168-9525(00)02093-x. [DOI] [PubMed] [Google Scholar]
- 56.Weichenrieder O, Wild K, Strub K, Cusack S. Structure and assembly of the Alu domain of the mammalian signal recognition particle. Nature. 2000;408:167–173. doi: 10.1038/35041507. [DOI] [PubMed] [Google Scholar]
- 57.Ovchinnikov I, Troxel AB, Swergold GD. Genomic characterization of recent human LINE-1 insertions: evidence supporting random insertion. Genome Res. 2001;11:2050–2058. doi: 10.1101/gr.194701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Farley AH, Luning Prak ET, Kazazian HH., Jr More active human L1 retrotransposons produce longer insertions. Nucleic Acids Res. 2004;32:502–510. doi: 10.1093/nar/gkh202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Brouha B, Schustak J, Badge RM, Lutz-Prigge S, Farley AH, Moran JV, Kazazian HH., Jr Hot L1s account for the bulk of retrotransposition in the human population. Proc. Natl Acad. Sci. USA. 2003;100:5280–5285. doi: 10.1073/pnas.0831042100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Lavie L, Maldener E, Brouha B, Meese EU, Mayer J. The human L1 promoter: variable transcription initiation sites and a major impact of upstream flanking sequence on promoter activity. Genome Res. 2004;14:2253–2260. doi: 10.1101/gr.2745804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Sassaman DM, Dombroski BA, Moran JV, Kimberland ML, Naas TP, DeBerardinis RJ, Gabriel A, Swergold GD, Kazazian HH., Jr Many human L1 elements are capable of retrotransposition. Nat. Genet. 1997;16:37–43. doi: 10.1038/ng0597-37. [DOI] [PubMed] [Google Scholar]
- 62.Waterborg JH, Matthews HR. The electrophoretic elution of proteins from polyacrylamide gels. Methods Mol. Biol. 1994;32:169–175. doi: 10.1385/0-89603-268-X:169. [DOI] [PubMed] [Google Scholar]
- 63.Kimberland ML, Divoky V, Prchal J, Schwahn U, Berger W, Kazazian HH., Jr Full-length human L1 insertions retain the capacity for high frequency retrotransposition in cultured cells. Hum. Mol. Genet. 1999;8:1557–1560. doi: 10.1093/hmg/8.8.1557. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.