Abstract
Solution structures of DNA duplexes containing oxanine (Oxa, O) opposite a cytosine (O:C duplex) and opposite a thymine (O:T duplex) have been solved by the combined use of 1H NMR and restrained molecular dynamics calculation. One mismatch pair was introduced into the center of the 11-mer duplex of [d(GTGACO6CACTG)/d(CAGTGX17GTCAC), X = C or T]. 1H NMR chemical shifts and nuclear Overhauser enhancement (NOE) intensities indicate that both the duplexes adopt an overall right-handed B-type conformation. Exchangeable resonances of C17 4-amino proton of the O:C duplex and of T17 imino proton of O:T duplex showed unusual chemical shifts, and disappeared with temperature increasing up to 30°C, although the melting temperatures were >50°C. The O:C mismatch takes a wobble geometry with positive shear parameter where the Oxa ring shifted toward the major groove and the paired C17 toward the minor groove, while, in the O:T mismatch pair with the negative shear, the Oxa ring slightly shifted toward the minor groove and the paired T17 toward the major groove. The Oxa mismatch pairs can be wobbled largely because of no hydrogen bond to the O1 position of the Oxa base, and may occupy positions in the strands that optimize the stacking with adjacent bases.
INTRODUCTION
DNA polymerase makes errors by misincorporating natural DNA bases and base analogs. Because of the wide variety of possible mismatches and the varying efficiency with which they are repaired, structural studies are necessary to understand in detail how these mismatch pairs differ and can be distinguished from standard Watson–Crick base pairs. Duplex structures with various mismatch pairs of natural bases, such as A:C (1–4), G:T (5–7), have been reported and effects of the mismatch on the structures and biological implications have been extensively studied (8). And also, various unusual mismatch pairs have been studied by X-ray crystallography and NMR to explain their mutagenic properties.
Oxanosine (5-amino-3-β-D-ribofuranosyl-3H-imidazo[4,5-d]oxazin-7-one) was originally isolated from Streptomyces capreolus MG265-CF3 and has antibiotic properties in both its ribo and 2′-deoxyribo forms (9). Oxanine (Oxa, O) is a unique deaminated base in which an endocyclic nitrogen atom of guanine is substituted by an oxygen atom. It was reported that 2′-deoxyoxanosine (dOxa, Figure 1A) was produced by the reaction of 2′-deoxyguanosine with nitric oxide (NO)- or nitrous acid (HNO2)-induced nitrosative oxidation (10). Since Oxa, which is a DNA lesion of guanine, could be produced in the cellular system by NO, HNO2 or other nitrosating agent, its genotoxic properties including deglycosylation susceptibility, base pairing stability and base incorporation patterns have been analyzed (11,12). Cytosine (Cyt) and thymine (Thy) were incorporated by Escherichia coli Klenow Fragment to pair with Oxa in a DNA template with similar efficiency. If Oxa is formed in DNA strands, dOxa can exist for a sufficient time since dOxa moiety is not easily hydrolyzed due to its stable N-glycosidic bond between base and sugar moieties. Therefore, the Oxa generated in cellular genomes may induce the misincorporation of incorrect nucleotides causing G:C to A:T transition.
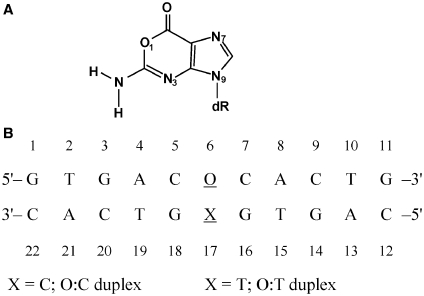
Figure 1.
Chemical structures analyzed in the study. (A) Structures of 2′-deoxyoxanosine and (B) DNA sequence contexts of the O:C duplex.
On the basis of the chemical structure, Oxa was expected to form two hydrogen bonds with either Cyt or Thy (11). By measuring melting temperatures (Tm) and thermodynamic parameters, the base pair of O:C showed relatively high stability in DNA duplexes compared with other base combinations. The orders were O:C > O:T > O:A > O:G (13). In terms of the most stable mismatch of O:C, two models have been proposed for this interaction: a Watson–Crick geometry involving two hydrogen bonds and a wobble geometry. A wide range of theoretical methods have been used to analyze the mutagenic properties of Oxa (14). The theoretical study have focused on the stability of the Oxa mismatch pairs in DNA: Molecular dynamics (MD) simulation showed that the d(G:C) → d(O:C) mutation increases the flexibility of the hydrogen bonded dimer, which fluctuates between the co-existence of different binding modes calculated in the gas phase (14). By examining the cleavage of oligodeoxyribonucleotides containing Oxa by bacterial endonuclease V, hypothetic models of Oxa-containing base pairs and deaminated base recognition mechanism were also proposed (15). However, there has not been as of yet consensus on its structure, because structural studies on DNA oligomers containing the O:C mismatch have not been carried out by NMR or X-ray crystallography so far on account of difficulties in preparing a dOxa amidite monomer. Since the monomer has been successfully synthesized and employed to the automatic DNA synthesizer (13,16), it has become possible to investigate how the mismatch of Oxa base pair affects the structure and properties of the Oxa duplexes at an atomic resolution. Recently, biotechnological utilization of the dOxa oligonucleotides has been reported. For example, T4 DNA ligase-based mismatch detection methods have been proposed as useful strategies for single nucleotide polymorphism (SNP) analyses, and biomolecular response of oxanine in DNA strands to T4 polynucleotide kinase, T4 DNA ligase and restriction enzymes (17,18). In order to make further development of these biotechnological and biomedical utilizations, it has been more important to elucidate the mismatch structures in detail for the oligonucleotides containing Oxa.
In the present study, we have focused on the effects of the O:C and O:T mismatches on the structure and stability of DNA duplex, and examined the theoretical data obtained for the base pairing of the O:C and O:T (14). The mismatch pairs were introduced into the center of the same 11-mer DNA duplex. The DNA sequence was designed and chosen to avoid the signal overlapping and to obtain structural information around the mismatch pair. In order to obtain information about the base pairing of O:C, interstrand nuclear Overhauser enhancement (NOE) from the cytosine 4-amino group of the mismatch to exchangeable protons in the adjacent base pairs of O:C is significant because the 1H–1H distances between those protons are short and sensitive to the DNA conformations (19). For the O:T duplex, the structure has been mainly compared with the G:T duplex. Based on the base pairing geometry and hydrogen bond of the O:C and O:T base pair, biological implication will be discussed.
MATERIALS AND METHODS
Nomenclature
In order to avoid confusing the number of the oxanine ring atoms with those corresponding guanine, the number of the oxanine ring in the present study was changed to be the same as that of the guanine ring. The numbering system for the [d(GTGACOCACTG)/d(CAGTGXGTCAC)] is shown in Figure 1B, where O indicates an oxanosine moiety, and X is C (O:C duplex) or T (O:T duplex). The mismatch pair (O6:X17) is underlined. And the same numbering system was applied to two reference DNA duplexes, [d(GTGACGCACTG)/d(CAGTGCGTCAC)] (G:C duplex) and [d(GTGACGCACTG)/d(CAGTGTGTCAC)] (G:T duplex).
Sample preparation
The DNA strands were synthesized on an automatic synthesizer by the phosphoramidite method and purified by gel filtration and reverse phase HPLC as previously reported (13). The concentration of the NMR sample was estimated using the extinction coefficients at 260 nm which were calculated by the nearest neighbor method (13). Each duplex was dissolved at a concentration of 2 mM in a buffer containing 10 mM sodium phosphate, 100 mM NaCl and 0.2 mM ethylenediaminetetraacetic acid. The pH of the solution was adjusted to 7.0, unless otherwise stated, by the addition of HCl and NaOH. The solution was heated at 80°C for 10 min and was gradually cooled down to room temperature immediately prior to the NMR measurements.
NMR spectroscopy
NMR experiments were carried out on a Bruker ARX-500 spectrometer (500.13 MHz for 1H) and JEOL JNM-ECAseries-920 (920 MHz for 1H). A set of 2D NMR spectroscopy experiments [DQF-COSY (20), TOCSY (21), E-COSY (22), NOESY (23)] was obtained for the duplexes dissolved in the deuterated buffer. NOESY spectra obtained with mixing times of 50, 100 and 200 ms were measured with a 6.5 s relaxation delay. Taking the duplex stability and peak separation into account, several temperatures used for the NMR measurements were employed between 5–25°C for the DNA duplexes. The jump-and-return NOESY (24) by the 500 MHz NMR machine and water gated NOESY (25) by the 920 MHz NMR machine were measured for the samples dissolved in the 90% H2O/10% D2O buffer at both pH 6.0 and 7.0 to assign labile protons and evaluate NOE cross-peak intensities for the O:T and G:T duplexes, because exchange rates of the imino proton with water is the slowest at pH 6 and that of amino protons with water at pH 7 (19). 1H chemical shifts were referred to internal sodium 3-(trimethylsilyl)propionate-2,2,3,3-d4. Acquired data were processed using the program UXNMR (Bruker) and NMRpipe (LA systems, Tokyo, Japan). The NMR sample conditions for the Tm measurements were identical to those for the 2D measurements. The 1D 1H-NMR spectra were collected after the attainment of the equilibration at each temperature. The chemical shift change of well-isolated peaks was plotted as a function of temperature, and the collected data points were fitted to a sigmoidal curve by a non-linear least squares refinement procedure of ORIGIN (Microcal Software Inc.). All the Tm values were reproducible from the curves with temperature increasing and decreasing, and the standard deviations were ±0.5–1.0°C on repetitions of the experiments.
Restrained MD
The NOESY cross-peak intensities were obtained using a measure-volume tool of NMRpipe. The volumes were normalized against C H5-H6 and H2′-H2′′ NOEs, which correspond to fixed distances. All the NOE restraints between the non-exchangeable protons were derived from 50-ms NOESY spectra with a long delay (6.5 s) in D2O. The upper and lower bound corrections of the NOE restraints (a force constant of 20.0 kcal mol/Å2) were set to be 15 and 10% of the obtained interproton distances, respectively. The sequential NOEs regarding exchangeable protons were converted to medium (3.0–4.2 Å) and weak (4.0–5.0 Å) restraints. The numbers of the interproton distances were 231 and 225 for O:C and O:T duplex, respectively. The initial A- and B-structures were generated using Maestro (Schrödinger, MA, USA). The structure calculations were done by XPLOR3.8 (26). Geometrical parameters and the atomic charges of the oxanosine residue were built by using those of the guanosine residue, and the no charge was employed on the O1 atom to avoid the effect of the force field parameters. These parameters were added to the database that was referred to during the structure calculation. Restrained molecular dynamics (rMD) and energy minimization calculations included 102 backbone torsion angle restraints with a force constant of 5.0 kcal/mol/rad2, and six distance restraints per Watson–Crick base pair (a force constant of 50.0 kcal/mol/Å2) to maintain Watson–Crick base pairing (27) with bounds of ±0.1 Å. For the mismatch pair, one hydrogen bond was enforced using canonical distances with bounds of ±0.1 Å between O6 NH2 and C17 N3 atom for the O:C duplex, and between O6 NH2 and T17 O2 atom for the O:T duplex. The reasons of those hydrogen bonds in the mismatch pairs will be described in the ‘Results’ section. The backbone angle restraints, keeping the range between the A- and B-form, were defined as previously described (28,29). The rMD calculations of 20 000 steps at 300 K were carried out with a simulated annealing protocol provided by Grünger (26) to search conformational space for structures consistent with the NMR restraints. The resulting structures were submitted to a final energy minimization, giving rise to the final structures. Seventeen structures of the O:C duplex and 15 structures of the O:T duplex were selected from 100 calculations on the basis of the lowest energy values. There were no violated distance constraints by >0.25 Å and no violated dihedral angle constraints by >5°. The helical parameters were analyzed using CURVES 5.3 (30,31).
RESULTS
Resonance assignment
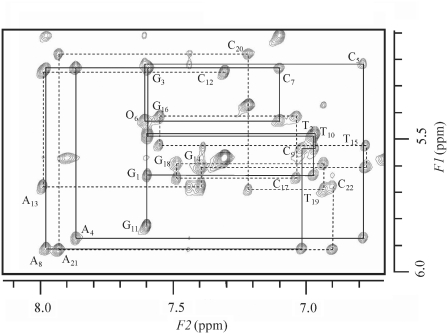
Proton resonances were assigned for all the duplexes (O:C, O:T, G:T and G:C) from a complete set of homonuclear 2D NMR data using established NMR techniques for nucleic acid analyses (19). The assignment of non-exchangeable DNA protons was achieved by analyzing all regions of the NOESY spectra obtained with various mixing times in combination with DQF-COSY and TOCSY data, and H5′/5′′ proton assignments were not assigned because of signal overlapping. For example, the NOESY spectrum of the O:C duplex is shown in Figure 2. The 1H resonance assignments are summarized in Supplementary Table S1. All the non-exchangeable protons were sharp, indicating that no chemical exchange occurs between conformers in an NMR time scale. Intra-residue and sequential base to H1′, H2′, H2′′ and H3′ cross-peaks characteristic of right-handed DNA duplex were observed for all the duplexes. These connectivities were seen throughout the duplexes, including the lesion site and surrounding residues. The intra-residue NOE intensities of the O6 H8-H1′ and paired C17 H6-H1′ cross-peaks were weak, and of similar intensity as the other intraresidue aromatic-H1′ cross-peaks, suggesting that those mismatch residues are anti, which excludes unusual geometries such as those found in Hoogsteen or reverse-Hoogsteen base pairs. For the O:T duplex, the mismatch residues of O6 and T17 also showed an anti conformation. The NOESY spectra of the O:C and O:T duplexes in D2O showed intranucleotide sugar–base NOE patterns of B-form DNA [H2′(i)-H8/H6(i) >> H1′(i)-H8/H6(i) > H3′(i)-H8/H6(i)]. Well-digitized E-COSY spectra were used to measure the coupling constants. The obtained J coupling data (3J1′2′ and 3J1′2′) were in the range of 5.4–8.0 Hz for both of the O:C and O:T duplexes. The sugar pucker and glycosidic bond conformations probably lie in the O4′-endo to C2′-endo and anti ranges, respectively. All the duplexes take an overall B-form DNA.
Figure 2.
1H 2D-water gated NOESY spectrum (mixing time 200 ms) of the DNA duplex containing the O:C mismatch collected at 22°C. Sequential NOE connectivities for a strand of d(GTGACOCACTG) are indicated by a solid line, and those for d(CAGTGCGTCAC) are indicated by a broken line. Intraresidue NOE connectivities between H1′ and H8/H6 are indicated by residue names.
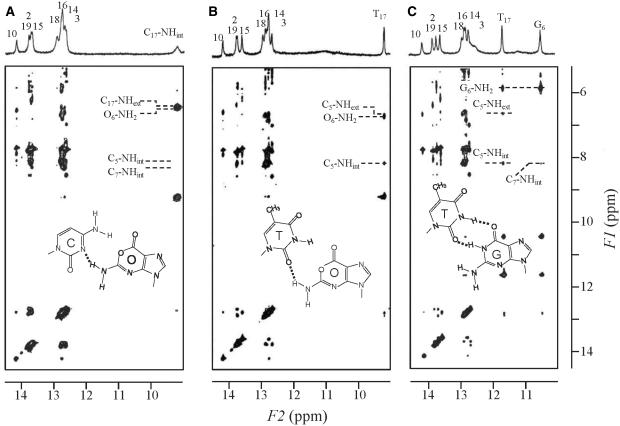
Exchangeable imino and amino proton resonances for each duplex were also assigned by measuring the samples in H2O and by observing NOE cross-peaks between the exchangeable and non-exchangeable protons (Figure 3). The imino and amino proton cross-peaks due to interactions with adjacent bases establish base pair formation and normal base stacking throughout the duplex. For the G:C duplex, the inter- and intrastrand imino–imino NOEs were observed through the duplex, except for the terminal imino protons. On the other hand, for the O:C duplex, the similar NOE cross-peaks were observed between the imino protons in the regions of T2-G3-T19-G18 and G16-T15-G14-T10 because of the absence of the imino proton in the central O:C mismatch pair. Two internal and external amino proton resonances of C17 (C17-NHint and C17-NHext in Figure 3A) were separately observed at 6.43 and 9.21 ppm, respectively. The downfield resonance at 9.21 ppm should be an internal amino proton (C17-NHint). The C17-NHint resonance exhibited weak NOE cross-peaks with the G16- and G18-NH resonances, and also a strong and broad NOE cross-peak with the amino protons of O6 (O6-NH2) at 6.47 ppm, which was overlapped with the intraresidue NOE between the C17-NHint and C17-NHext. For the O:T duplex, the imino proton resonance of T17 (T17-NH) was observed at 9.21 ppm (Figure 3B), and the G:T duplex exhibited two imino proton resonances of G6 and T17 at 10.45 and 11.62 ppm, respectively (Figure 3C). The T17-NH of the O:T duplex was shifted to the more upfield by 2.4 ppm than that of the G:T duplex which is hydrogen bonded to the opposite guanine. NOE cross-peak pattern of the G6:T17 wobble pair of the G:T duplex coincides with that reported for self-complementary DNA duplex with the G:T mismatch pair (5,32,33): The G6-NH2 protons, observed at 5.82 ppm showed a strong NOE cross-peak with G6-NH at 10.45 ppm, and a medium NOE cross-peak with that of T17-NH at 11.62 ppm. Weak sequential NOEs were observed for the G:T duplex between the imino protons of G18-T17-G16 and G18-G6-G16. For the O:T duplex, weak sequential NOEs were also observed between imino protons of the intrastrand, G18-T17-G16. And the T17-NH proton showed a medium NOE cross-peak with the O6-NH2 resonance. The O:T and G:T duplexes resemble each other in the NOE cross-peak pattern, although the chemical shift of the T17-NH proton of the O:T duplex was different from that of the G:T duplex.
Figure 3.
1H jump-and return 2D-NOESY spectra (mixing time 200 ms) of the DNA duplexes containing (A) O:C, (B) O:T and (C) G:T mismatches collected at 5°C. The spectrum of the O:C duplex was measured at pH 7.0, and those of the O:T and G:T at pH 6.0. There was no significant difference in the chemical shift of the non-exchangeable protons at pH 6.0 and 7.0.
Comparison of chemical shift values between the duplexes
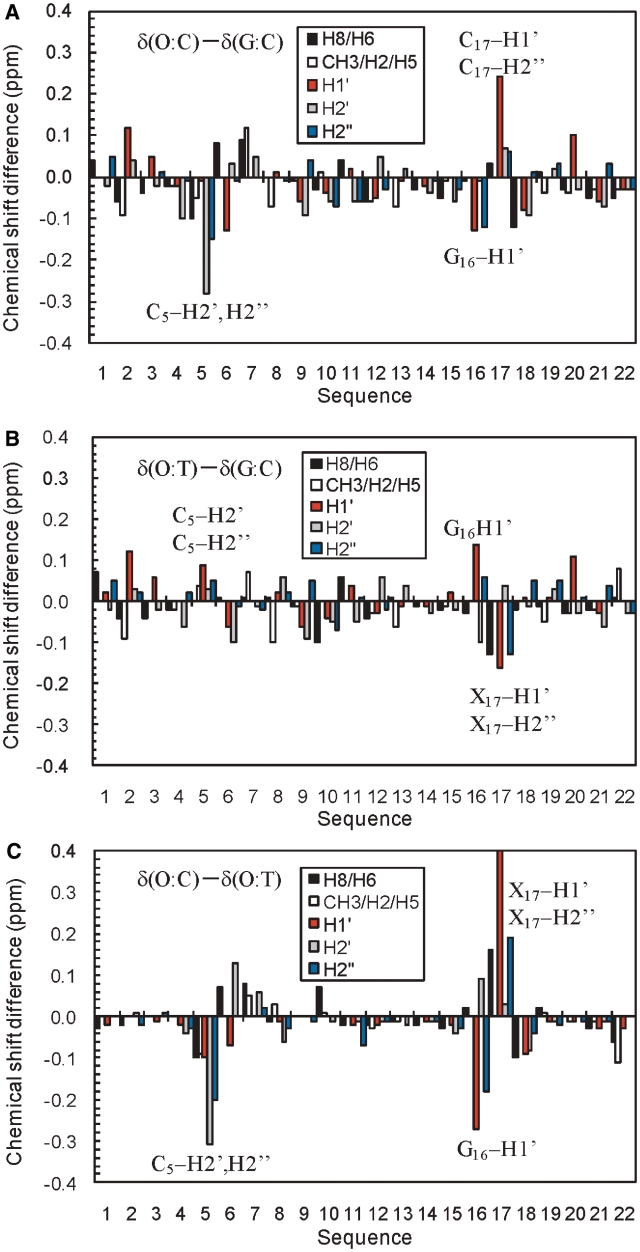
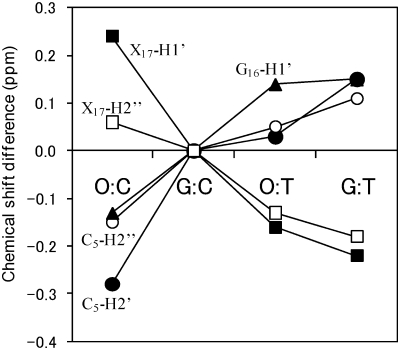
Chemical shift difference (Δδ) provides a first approximation of the structure change. In order to examine the effect of the O:C and O:T mismatch on the chemical shifts, the chemical shift differences were compared with the G:C duplex (Figure 4) or to the G:T duplex (Supplementary Figure S1) that contains the wobble G:T pair. As for the non-exchangeable protons, almost all proton resonances exhibited only a small difference (|Δδ| < 0.1 ppm). Significant chemical shift differences were observed for the protons of the positions of C5-O6-C7, and G18-X17-G16, indicating that the structure change caused by the O:C or O:T mismatch is restricted to its neighboring Watson–Crick base pairs. The comparison of the chemical shift between the duplexes showed that large differences in the chemical shift (|Δδ| > 0.2 ppm) were observed for C5-H2′/H2′′, G16-H1′ and X17-H1′/H2′′ (X = C or T). The non-exchangeable proton resonances of O6 in the O:C and O:T duplexes showed almost identical chemical shifts to those corresponding of G6 in the G:C duplex (Figure 4A and B). It is of interest that the largest differences were observed for the neighboring residue (C5) and residues of G16 and C17 in the counter strand, but not for O6 itself. This indicates that the introduction of Oxa influences not only base pairing with the base at the position of 17, but also the duplex structure of the adjacent residues, possibly resulting in base stacking. For the largest shifted protons, their deviations of the O:C, O:T and G:T duplexes from those of the G:C duplex are shown in Figure 5. It should be noted that the deviations of the O:C duplex from the G:C duplex is opposite to those of the O:T and G:T duplexes for the largest shifted protons. For example, C5-H2′ showed a large upfield shift for O:C, but a downfield shift for O:T, while C17-H1′ showed a downfield shift for O:C, but an upfield shift for O:T. Thus, the chemical shift differences between O:C and O:T (or G:T) were large (Figures 4C and 5), and the chemical shifts of O:T agreed more closely with those of G:T than those of O:C (Supplementary Figure S1B). Taking it into account that the G:C and G:T duplexes take a Watson–Crick and wobble geometry, respectively, the geometry of the O:T pair including its adjacent base pairs may take an intermediate geometry between them, while the O:C pair takes a different geometry.
Figure 4.
1H chemical shift differences between the DNA duplexes for H8/H6 (black), T CH3/A H2/C H5 (white), H1′ (red), H2′ (gray), H2′′ (blue): (A) δ(O:C duplex) – δ(G:C duplex), (B) δ(O:T duplex) – δ(G:C duplex), (C) δ(O:C duplex) – δ(O:T duplex). The protons which showed the large differences between the O:C and O:T duplexes are indicated by the names and sequential numbers.
Figure 5.
1H chemical shift deviations of the largely shifted protons of the mismatched DNA duplexes (O:C, O:T and G:T) from the G:C duplex. Those protons are indicated in the Figure 4: C5-H2′ (closed circle), C5-H2′′ (open circle), G16-H1′ (closed triangle), X17-H1′ (closed square) and X17-H2′′ (open square) (X = C for O:C and X = T for O:T and G:T).
Among exchangeable proton resonances, the C17-NHint proton resonance of the O:C duplex (Figure 3A) and the T17-NH proton resonance of the O:T duplex (Figure 3B) showed outstanding differences between the duplexes. The C17-NHint proton resonance (9.21 ppm) of the O:C duplex was shifted to the downfield as compared with single-stranded cytosine amino protons (7.0–8.0 ppm) and was even downfield relative to the hydrogen-bonded cytosine amino protons in standard Watson–Crick base pairs (8.0–8.6 ppm, in Supplementary Table S1). The corresponding C17-NHint proton of the G:C duplex were observed at 8.28 ppm (data not shown). Therefore, the C17-NHint proton resonance of the O:C duplex shifted to the downfield by 1 ppm, as compared with that of the G:C duplex. This downfield shift of the C17-NHint proton resonance involved in the mismatched base pair suggests that the base pairing geometry is different from the canonical Watson–Crick of G:C. On the other hand, the imino proton of T17 of the O:T duplex was shifted to the upfield by 2.5 ppm, as compared with that of the G:T duplex. Thus, between the O:T and G:T duplexes, the large difference was only observed for T17-NH although there are small differences for all the non-exchangeable protons, indicating that the hydrogen bond pattern of O:T base pair is different from the reported wobble pattern of G:T, while the duplex structures resemble each other.
Melting profile of the O:C and O:T duplex
In order to examine whether the stability of the duplexes could correlate with the structural differences, thermal melting profile was investigated for all the duplexes by 1H NMR spectra. The duplex stability is also considered to be important for the estimation of mismatch DNA. Based on the temperature dependence of chemical shift change of isolated non-exchangeable proton resonances (A4 H8, T15 H6), these isolated protons were cooperatively shifted with the temperature change, and the Tm values were determined. The Tm values for the O:C and O:T duplexes were 58 and 52°C, respectively, while those for the G:C and G:T duplexes were 63 and 57°C, respectively. The O:C duplex was less stable than the G:C duplex by 5°C, and as stable as the G:T duplex. The O:T duplex was less stable than the G:T duplex. The determined orders of Tm were G:C > O:C = G:T > O:T, which coincides with the previous report by using UV melting profile of Oxa mismatch DNAs (13). The base pair of O:C shows relatively high stability in DNA duplex compared with other base combinations. These results demonstrate that the introduction of the Oxa mismatch causes rather small change in the Tm values as previously reported (13).
We also observed the temperature dependence of the exchangeable proton resonances to obtain the information about the duplex stability and fluctuation. The imino protons at the terminal and penultimate base pairs disappeared ∼10°C, and all the imino proton resonances disappeared <40°C, much lower than the Tm values, determined by the chemical shift change of the non-exchangeable protons, because the line broadening of the imino protons would depend on the fast exchange between the imino proton and water or the fluctuation of the base pairs. For the mismatch DNA duplexes (O:C, O:T, and G:T), unique temperature dependences of the exchangeable protons were observed in the temperature range between 5 and 30°C. The C17-NHint resonance of the O:C duplex was broader than those of C5, C7, C9 and C20, which are involved in the internal of the duplex. With elevating temperature, the C17-NHint resonance unchanged below 15°C, but with a further increase of the temperature, disappeared like the imino proton resonances of T2 and T10 located in the peripheral of the duplex (Supplementary Figure S2). This exchange rate was considerably faster than those of the other G:C pairs, except for the terminal G:C pairs. Since the non-exchangeable protons involved in the mismatch pair did not show any broadening in this temperature range, the broadening of the C17-NHint resonance would be caused by the exchange with bulk water. NMR spectrum of the O:T duplex at 5°C showed that the T17-NH resonance at 9.21 ppm was as sharp as those of T15 and T19 involved in the center of the duplex. With temperature increasing from 5 to 25°C, the T17-NH resonance became broad and completely disappeared ∼30°C as well as the imino protons of T2 and T10 located in the peripheral of the duplex. The exchange rate was faster than those of T15 and T19 involved in the A:T pairs (Supplementary Figure S2). The broadening of the T17-NH proton would also be caused by the exchange with bulk water. The same phenomenon was observed for the imino protons of G6 and T17 in the G:T duplex: The T17-NH resonance of the G:T duplex was broadened and disappeared ∼30°C, indicating that relatively rapid exchange of the imino protons with the solvent also occurs for the wobble pair of the G:T mismatch as well.
Characterization of the mismatch pair of O:C duplex
The chemical shift values (6.43 and 9.21 ppm) and its separation (2.78 ppm) of the C17-NH2 protons provide indications that the base pair of O6:C17 is formed and stacked in the duplex. However, the downfield shift and broadness of the C17-NH2 protons suggest that the base pair does not take a Watson–Crick geometry, and a relatively rapid exchange occurs with water molecules. Similar results were reported for a 2-aminopruine(AP):cytosine mismatch in DNA duplex (34,35). The internal 4-amino proton in the AP:C mismatch of CAPC/GCG resonated at 8.94 ppm, and the resonance was broadened into the base line at 25°C but sharpen with decreasing temperature (34). They pointed that the C-NHint proton resonance in the AP:C mismatch may shift to the further downfield than usual, due to an aromatic ring current shift, because the C-NHint proton is likely to be 1.7–2.1 Å from the edge of the 2-AP base, and in a G:C pair, the C-NHint proton is ∼2.6 Å from the edge of the guanine base, leading to a smaller ring current shift. The AP:C mismatch pair was considered to take a wobble pair: The C-NHint proton of AP:C is hydrogen bonded to the AP N1 atom, and the AP 2-amino group to the C N3 atom. The unusual downfield shift of the C17-NHint proton resonance may be caused by the short distance from the C17-NHint proton to the Oxa aromatic ring in the same wobble pair as the AP:C mismatch. According to previous theoretical calculation, oxanine and guanine show very similar electrostatic potential distribution on the aromatic plane (14), so the short distance to the aromatic ring could result in the more downfield shift. Strong NOE cross-peak of C17-NHint to O6-NH2 supports that the O:C base pair takes the wobble geometry similar to the AP:C mismatch, where the O6-NH2 proton resonance probably forms a hydrogen bond to the C17 N3 atom, and the C17-NHint and O6-NH2 protons are adjoining each other. The broadness of the O6-NH2 resonance also implies the hydrogen bond of O6-NH2 proton to the C17 N3 atom. In general, guanine and adenine amino groups often rotate at or near intermediate exchange on the NMR time scale, causing severe broadening of the amino proton resonances and making assignment of these protons difficult. The O6-NH2 resonance was broad, which were more likely to behave similarly to guanine and adenine amino protons involved in the hydrogen bond. The broad line shape of the C17-NHint proton resonance at 5°C and its disappearance with elevating the temperature suggest that the C17-NHint proton is likely located in the center of the wobble geometry, but no or very weak hydrogen bond is formed with the O6 O1 atom. The pH dependence of the exchangeable protons of the O:C duplex showed the broadening of the C17-NHint proton resonance below pH 6 (data not shown). No resonance originated from the protonated cytosine hydrogen bonded to the Oxa moiety was observed at slightly acidic pH. The pH dependence of NMR spectra of the O:C duplex indicate that protonation of the C17 N3 position does not occur in the mismatch pair. These results of the pH titration also support the wobble geometry of the O:C mismatch.
In order to determine the geometry of the mismatch region, we focused on NOE cross-peaks from C17-NHint to G16- and G18-imino protons and to C5- and C7-NHint protons. These NOE cross-peaks provide us with significant information for base pairing of O6:C17 and its mutual position to the neighbor base pairs of C5:G18 and C7:G16, although the NOEs are influenced by spin-diffusion effect and rapid exchange with water. The NOE cross-peaks from C17-NHint to G16- and G18-NH protons were observed, indicating the base pair formation and stacking of O6:C17 in the duplex. However, no NOE cross-peak from C17-NHint to C5-NHint or C7-NHint proton was observed in the NOESY spectra even with a long mixing time (Figure 3A). Assuming that three consecutive base pairs of C5O6C7/G16C17G18 take a Watson–Crick geometry in regular B- or A-type DNA conformation, the NOE from C17-NHint to C5-NHint should be observable: in the regular B-type DNA, the distance from C17-NHint to C5-NHint is 2.9 Å, and that from C17-NHint to C7-NHint is >5 Å. On the other hand, in the regular A-type DNA conformation, the distances from C17-NHint to C5-NHint and to C7-NHint are comparable (3.5 Å). Judging from no NOE cross-peak between C17-NHint and C5-NHint, the base pair of O6:C17 does not take a typical Watson–Crick geometry in the regular B-type DNA. Shortly afterward it will be discussed in detail on the basis of the determined structures.
Characterization of the mismatch pair of O:T duplex
There are two possibilities in the geometry of the O:T mismatch pair: a Watson–Crick geometry like G:C and a wobble geometry like G:T. In order to determine the geometry, NOE cross-peaks related to the imino proton of T17 (T17-NH) were analyzed by NOESY spectra and compared between the O:T and G:T duplexes. The T17-NH proton resonance of the O:T duplex was as sharp as that of the G:T duplex at 5°C, and resonated at 9.2 ppm. As compared to the chemical shift value of 11.62 ppm of the G:T duplex, the upfield shift by >2 ppm indicates that the T17-NH proton of the O:T duplex is not involved in the hydrogen bond of the wobble geometry like G:T. However, in terms of the NOE cross-peaks and their intensities involved in T17-NH, the NOE pattern of the O:T duplex was almost identical to those of the G:T duplex, although those NOEs contain some spin diffusion effects because of the long mixing time (200 ms). In particular, the NOE intensity between O6-NH2 and T17-NH of the O:T duplex was almost equal to that between G6-NH2 and T17-NH of the G:T duplex. Both the NOEs were much weaker than that between G6-NH2 and G6-NH of the G:T duplex. These NOEs indicate that, in the O:T mismatch pair, O6-NH2 and T17-NH protons are not adjoining each other, denying the Watson–Crick geometry. The NOE cross-peaks from T17-NH to G16-NH, G18-NH and to C5-NHint of the O:T and G:T duplexes (Figure 3B and C) exhibited the almost identical pattern with some spin diffusion effects. The conclusion that the O:T duplex takes an intermediate structure between the G:C and G:T duplexes is supported by following observation of the line width of the O6-NH2. The O6-NH2 resonance of the O:T duplex was much sharper than that of the O:C duplex, and was almost identical to that of the G:T duplex. As mentioned before, the line width of the G-NH2 proton resonance involved in the hydrogen bond of the Watson–Crick pair is broad, because of its rotation at or near intermediate exchange on the NMR time scale. The G-NH2 resonance of the G:T mismatch pair is known to be sharp, meaning the fast rotation of the amino group, because the G:T mismatch pair usually has two hydrogen bonds (N–H…O=C) in the wobble base pair, where the G-NH2 is not involved in the hydrogen bonds (6). Therefore, the sharpness of the O6-NH2 of the O:T duplex also suggests that the O6-NH2 protons are not involved in the strong hydrogen bond of the Watson–Crick pairing. Consequently, the O:T mismatch likely takes an intermediate base pairing between G:T (wobble) and G:C (Watson–Crick), and a hydrogen bond may exist between O6-NH2 and T17 O2 atom. The chemical shift deviations of the mismatch DNAs from the G:C duplex (Figure 5) agree with the intermediate structure of the O:T duplex.
Structure calculation and helical parameters
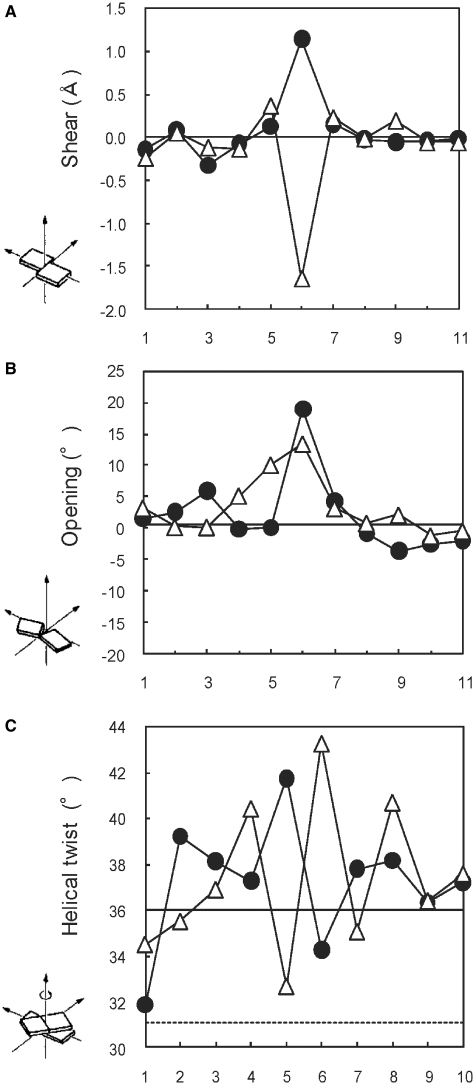
The investigation on the helical parameters provides us with the information about the local conformation of the O:C and O:T mismatches in detail. The rMD calculations were performed with the distance restraints derived from well-resolved NOE cross-peak intensities, and good convergence has been reached: The 17 final structures of the O:C duplex exhibited pairwise root-mean-squared deviation (RMSD) values of 0.81 ± 0.14 Å for all heavy atoms, and the O:T duplex exhibited the pairwise RMSD values of 0.85 ± 0.10 Å for all heavy atoms of the 15 final structures (Supplementary data). Energy terms of the final structures and the number of the NOE and dihedral violations were obtained and summarized in Table 1. These results indicate that all the distances were consistent with the final structures. The obtained structures of the O:C and O:T duplexes (Figure 6) were in the conformational range expected for B-form. The structural differences between the duplexes were small, and the prominent differences were observed around the mismatch pairs (Figure 7). Most of the conformational parameters calculated by Curves 5.3 are interrelated as shown previously (30,31). Selected helical parameters (shear, opening and helical twist) are given in Figure 8, and the complete list is given as Supplementary Figure S3. The shear parameter at the O6:C17 pair reached a large positive value for the O:C duplex and a large negative value for the O:T duplex (Figure 8A), indicating that the bases in the base pair of O6:C17 and the O6:T17 moved oppositely. The helical twist showed symmetrical pattern at the base steps of 5 and 6 for the two duplexes (Figure 8C). The helical twists in the steps 5–6 and 6–7 were compensated each other. The common feature of the helical parameter of the two duplexes is large positive opening parameter (Figure 8B). The both base pairs of O6:C17 and the O6:T17 are opened toward major groove, as compared with typical Watson–Crick geometry.
Table 1.
NMR constraints and structural statistics for the O:C and O:T duplexes
| O:C duplex | O:T duplex | |
|---|---|---|
| NMR constraints | ||
| Distance constraints | 231 | 225 |
| Intraresidue | 141 | 150 |
| Interresidue | 90 | 75 |
| Dihedral angles | 102 | 102 |
| Hydrogen bonds | 62 | 62 |
| Structural statistics for final structures (kcal/mol) | ||
| Etotal | 297 ± 11 | 281 ± 13 |
| Ebond | 36 ± 1 | 36 ± 1 |
| Eangle | 370 ± 3 | 342 ± 2 |
| Eimproper | 60 ± 1 | 60 ± 1 |
| Evdw | −309 ± 3 | −311 ± 3 |
| Enoe | 17 ± 1 | 12 ± 1 |
| Ecdih | 0 ± 0 | 0 ± 0 |
| RMSD from idealized geometry | ||
| Bond length (Å) | 0.007 ± 0 | 0.007 ± 0 |
| Bond angle (°) | 1.34 ± 0.004 | 1.29 ± 0.003 |
| Impropers (°) | 0.9 ± 0.003 | 0.9 ± 0.004 |
| Number of NOE violations | ||
| d > 0.25 Å | 0 | 0 |
| 0.25 > d > 0.15 Å | 7 | 4 |
| RMSD of violation (Å) | 0.05 ± 0.001 | 0.04 ± 0.002 |
| Dihedral violation | ||
| Number of violation (>5°) | 0 | 0 |
| Pairwise RMSD for all heavy atoms (Å) | 0.81 ± 0.14 | 0.85 ± 0.10 |
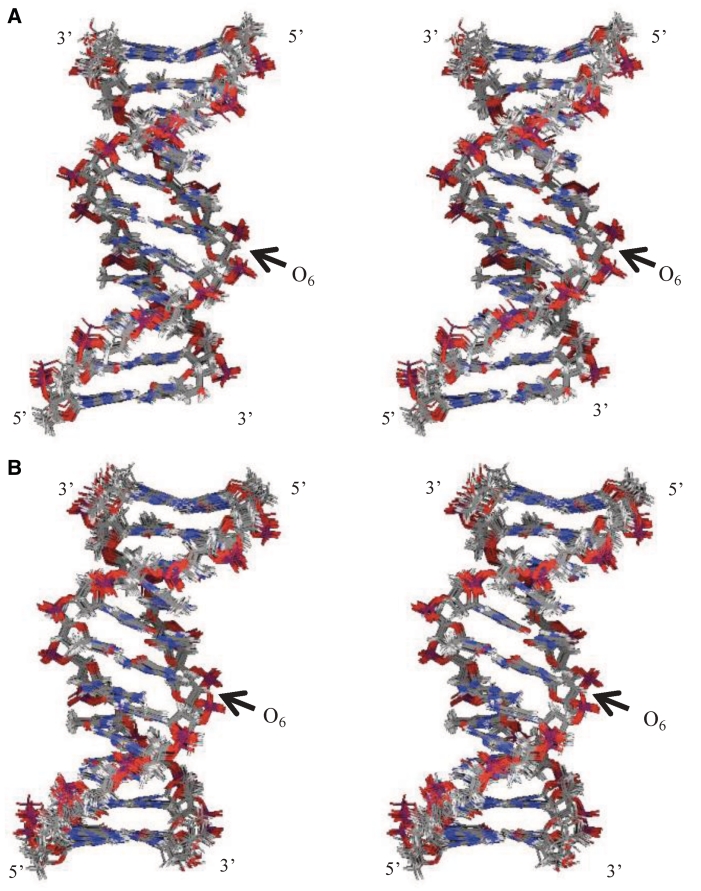
Figure 6.
Superposition of overlapped final structures of (A) O:C duplex and (B) O:T duplex. View of the major groove of the superposition of the heavy atoms of the duplexes. The O6 position is indicated by an arrow.
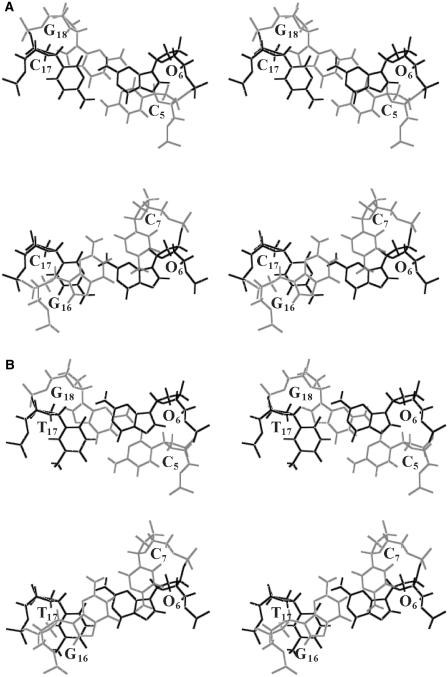
Figure 7.
Base stacking interaction of (A) C5O6/C17G18 and O6C7/G16C17 in the O:C duplex, and (B) C5O6/T17G18 and O6C7/G16T17 in the O:T duplex. The mismatch pairs of O6:C17 and O6:T17 are shown in dark, and the canonical G:C pairs in light.
Figure 8.
Helical parameters (A) shear, (B) opening and (C) helical twist of the O:C and O:T duplexes. Open triangles and closed circles indicate the O:C and O:T duplex, respectively.
The obtained structures of the O:T duplex showed that the distance between T17-NH and O6 O6 atom was 2.21 ± 0.13 Å (Figure 9), suggesting that a weak hydrogen bond exists between them. On the other hand, for the O:C duplex, the distance between C17-NHint and O6 O6 atoms was 3.39 ± 0.19 Å, indicating that there is no direct hydrogen bond between them. The obtained structures of the O:C duplex showed rather short distance (d < 3.0 Å) between C17-NHint and C5-NHint which did not show any NOE cross-peak (Figure 3A). It was reported for C:T mismatch pairs that exchangeable protons involved in the hydrogen bond via water molecule did not show any observable NOEs (36,37). Since the line width of C17-NHint was broad and it disappeared with elevating the temperature as above mentioned, no NOE cross-peak between C17-NHint and C5-NHint is probably caused by the rapid exchange of the amino protons with water. Taking account of the chemical shift values (6.43 and 9.21 ppm), its separation (2.78 ppm), the broad line width of the C17-NH2 protons, and rather high melting temperature of the O:C duplex, there may be a hydrogen bond from the C17-NHint to O6 O6 atom (d = 3.4 Å) via water molecule (Figure 9). Since such a water molecule involved in the hydrogen bond is known to make the exchangeable NH proton, such as imino and amino proton, exchange faster with water (36–38), the proximity of C17- and C5-NHint to the water molecule hydrogen bonded to the O6 O6 atom may contribute to diminishing the NOE cross-peak.
Figure 9.
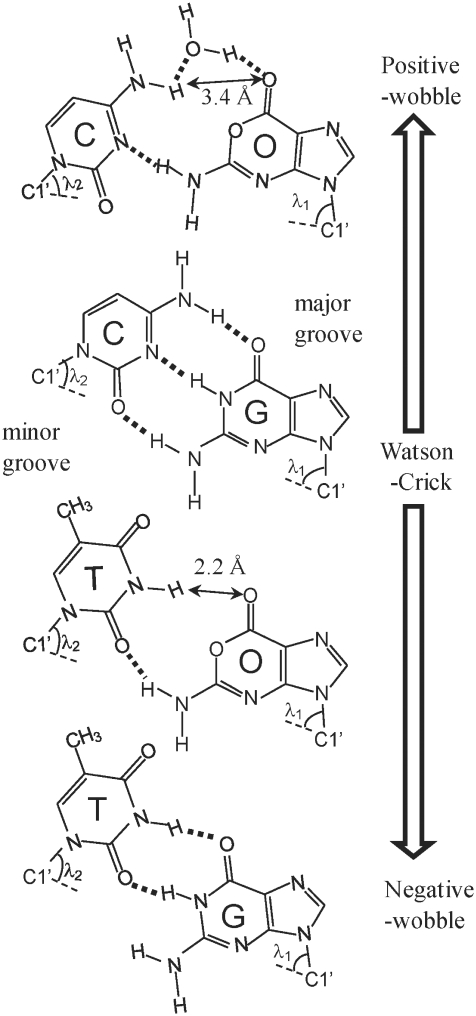
Possible base pairings of the O:C, O:T, G:C and G:T pairs with expected hydrogen bonds.
The angle formed by the purine N9 or pyrimidine N1 and C1′-C1′ intrabase pair vector defines λ. Each base pair may be characterized by two λ values, relating to each strand of the duplex (5,8). The values of λ1 and λ2 (the subscript 1 corresponds to residues 1–11 and 2 designates residues 12–22) were determined for the O:C and O:T duplexes. For all the base pairs except the O:C and O:T mismatch pairs, a high degree of symmetry was observed in the λ1 and λ2 values in the range of 51–57°. However, the O:C mismatch pair showed asymmetry of the λ1 (67.4° ± 1.1°) and λ2 (55.4° ± 1.6°) values, and for the O:T mismatch pair, the values of λ1 and λ2 were 50.0 ± 1.0° and 68.8 ± 0.6°, respectively. In B-DNA containing a G:T mismatched base pair, the values of λ1 and λ2 were reported to be ∼40° and 70°, respectively (5,8). The λ2 value of the O:T mismatch pair is as large as that of the reported G:T wobble pair, although its λ1 value is in the range expected for the Watson–Crick pair. On the other hand, the λ1 and λ2 values of the O:C duplex are reversed; the large λ1 like λ2 of G:T, and the moderate λ2 in the range expected for the Watson–Crick pair.
DISCUSSION
We have presented that the DNA duplexes containing one dOxa residue take two kinds of wobble geometries, depending on the opposite bases (Figure 9). The two wobble geometries are characterized by the mutual position of the Oxa moiety to the pyrimidine base (C or T) in the counter strand. The O:C duplex exhibits that the Oxa moiety shifted toward the major groove and the paired cytosine of C17 shifted toward the minor groove, where the O6-NH2 is hydrogen bonded to the C17-N3 atom. On the other hand, the O:T duplex exhibits that the Oxa moiety shifted toward the minor groove and the paired T17 shifted toward the major groove, where the internal proton of O6-NH2 is partially hydrogen bonded to the T17-O2 atom. The obtained geometries of the O:C and O:T mismatches fairly coincide with those derived from theoretical MD calculations of the duplex DNA with O:C and O:T pairs (14). The relative position and the direction of the displacement of the pyrimidine:purine pair in the present study agreed with those in the theoretical calculation. In terms of the helical parameters, the most prominent change exists in the value of shear parameter. The shear value is around zero in Watson–Crick geometry, where imino protons of G and T are hydrogen bonded to C N3 and A N1 atoms, respectively. The shear values of the O:C and O:T duplexes were positive and negative, respectively, so hereafter, we call the wobble geometry with the positive shear value, ‘positive-wobble’ geometry, and that with the negative shear, ‘negative-wobble’ geometry. The negative-wobble geometries have been reported for many mismatch pairs, such as G:T and A:C (or A+:C). In particular, the G:T mismatch has been studied extensively and shown to take the negative-wobble geometry (7,39). Previous studies of A:C mismatch pair in DNA suggested that the A:C mismatch also existed in the negative-wobble geometry, in which the exocyclic amino group of mispaired Ade hydrogen bonded with Cyt N3 imino nitrogen (1–4). In the negative-wobble geometries of the G:T and C:A mismatch pairs, the purine base projects in the minor groove and pyrimidine into the major groove. The characteristics of the negative-wobble geometry are a rather short distance between the C1′ protons of the mismatch pair and a large difference between the bond angles (λ1 and λ2) of the pyrimidine and purine residues (8). NMR spectra of the negative-wobble geometry of typical G:T in the B-DNA duplex are characterized by two hydrogen bonds where the G-NH resonates at 10–11 ppm and T-NH at 11–12 ppm (5,32,33). In the case of partially distorted G:T mismatch where the G-NH and G-NH2 are hydrogen bonded to T O2 atom, the T-NH proton resonance was shifted to the upfield in the range of 9–10 ppm (40). According to the chemical shift value (9.2 ppm) of the T17-NH proton of the O:T duplex, the O:T mismatch pair takes an intermediate geometry between the negative-wobble and Watson–Crick geometries. Recently, non-hydrogen bonding guanine–difluorotoluene (G:F) pair was reported to take also an intermediate geometry between the negative-wobble of G:T and the Watson–Crick geometries (41). The G:F mismatch pair stacks relatively well into the helix without any hydrogen bond between the mismatch pair, suggesting that the intermediate geometry is likely stabilized by the base stacking.
On the other hand, the O:C mismatch pair takes the positive-wobble geometry differing in the mutual geometry of the purine:pyrimidine pair from the G:T and A:C mismatch pairs. Not many positive-wobble geometries of mismatch DNA have been reported, and it is remarkable that in the AP:C mismatch in DNA duplex, the purine 2-amino group is hydrogen bonded to the pyrmidine N3 (34,35). In the positive-wobble geometry of the O:C mismatch, the O6-NH2 group is hydrogen bonded to the C17 N3 atom. Addition to the chemical shift values and the relatively rapid exchange of the C17-NHint protons of the O:C duplex, its unusual NOE pattern around the C17-NHint proton indicate that the O:C mismatch pair takes the positive-wobble geometry but not Watson–Crick geometry, and that the expected hydrogen bond between O6 O1 and C17-NHint should be weak or could not be formed. It is possibly because the O1 atom of Oxa has an sp3 hybrid orbital and the lone pairs of electrons exist out of the plane. Detailed NOE analyses revealed that there are some differences in the positive-wobble geometries between the O:C and AP:C duplexes. In the AP:C wobble pair of C4AP5C6/G13C14G15, the C14-NHint proton was hydrogen bonded to the AP5 N1 atom, and showed NOE cross-peaks with the C4-NH2 protons (34). As mentioned in the ‘Results’ section, the NOE pattern in the region of C5O6C7/G16C17G18 was different from that of the typical B-DNA and the base pair dynamics of the O:C was also changed. The O:C mismatch pair takes an intermediate geometry between positive-wobble (AP:C) and Watson–Crick (G:C) geometries, and the C17-NHint proton is likely hydrogen bonded to O6 O6 atom of oxanine moiety via water molecule. As U:C mismatch pair which form only a single base–base hydrogen bond are stabilized by a water molecule, which bridges between the ring nitrogens (42), the hydrogen bond via the water molecule probably contributes to the high thermodynamic stability of the O:C duplex. The mismatch pair of the O:C duplex should be more locally fluctuated, as compared with that of the AP:C duplex. Consequently, the difference between the two mismatch duplexes is not only the geometry but the base pair dynamics. It was reported for the AP:C mismatch pair that protonation of the mismatched AP N1 or C N3 positions allowed formation of a second hydrogen bond and stabilization of the AP:C pair, and that the stability of the AP:C mismatch was dependent upon pH (34,35). The protonation of C17 in the O:C mismatch was not observed, possibly because the hydrogen bond of protonated cytosine to the oxanine O1 does not stabilize the duplex structure due to the sp3 hybrid orbital of the O1 atom.
G:T mismatch pair is among the most commonly observed mismatches in genomic DNA (43). It might be expected that the more stable the mismatch, the less efficient is its repair. However, the stable G:T mismatch is among the most efficiently repaired mismatches in DNA, which may suggest that its repair is based on recognition of structure (44–46). While thermodynamics may play a major role in the frequency of the occurrences of different mismatches, it is more likely that the enzymatic recognition and repair of mismatches are influenced by the geometry and 3D structure of the mismatch (7,8,47,48). Repair enzymes that recognize and excise G:T mismatches may recognize subtle backbone perturbations such as in the torsion angle perturbations or the base pair parameters (5). It has been proposed that mismatch repair enzymes may directly recognize the base pair parameters λ1 and λ2, which are approximately the same in canonical G:C and A:T pairs but are highly asymmetric for G:T mismatches (λ1 = 40° and λ2 = 70°) (5,8). In the present study, the positive-wobble geometry of the O:C mismatch showed the large λ1 value (67.4°) of the O6 purine base, indicating that the O:C wobble geometry is reversely asymmetric, as compared with the G:T mismatch. The negative-wobble geometry of the O:T mismatch showed the large λ2 value (68.8°) of the T17 pyrimidine base, but the moderate λ1 value (50.0°) of the O6 base. Similar tendency of the λ values was reported for the non-hydrogen bonding G:F pair (λ1 = 53.5° and λ2 = 70.0°) (41), indicating that the Oxa mismatch pairs with no hydrogen bond to the O1 position of the Oxa base can be wobbled widely, and may occupy some positions in the double strands that optimize the stacking with adjacent bases. The asymmetric patterns of the λ1 and λ2 values of the O:C and O:T duplexes may be one of the reasons for not recognized by the G:T mismatch repairing enzyme.
The determined structures exhibit that the wobble geometries of the O:C and O:T mismatches make some influence on only neighboring base pairs in the helical parameters, such as helical twist. Aside from the lesion site, the duplex structure, including the flanking base pair, are not highly perturbed by the presence of the lesion. In the previous studies of the mismatches of G:T, G:A and A:C, the perturbations caused by the mismatches also extend only to its neighboring Watson–Crick base pair, thus providing a structural basis for the applicability of the nearest-neighbor model to the thermodynamics of internal mismatches (5,49,50). The flanking wobble base pairs of O:C and O:T seem quite stable in terms of the thermodynamic property, and do not perturb the cooperative feature of the duplex stability, so that the nearest-neighbor model could also be applicable to the mismatch containing Oxa. The large increases in the exchange rate of the exchangeable protons in the mismatch pairs with water indicate that the local base pair fluctuations exist in the mismatches of the O:C and O:T, and the fluctuations may be as large as those at the terminus of the duplex. Previous theoretical calculation (14) indicates that the G:C → O:C substitution reduces the stability of the duplex, but probably less than would be expected from the existence of repulsive interactions in the O:C dimer between oxanine O1 and cytosine N3 atoms, and that the G:C → O:C mutation was expected to increase the flexibility of the hydrogen bonded dimer, which fluctuates between binding modes. Oxa forms relatively stable base pairing with C compared with other matches with T, G and A. The O:C mismatch would maintain thermodynamically stable interaction without severe influence on the local structure of DNA duplex, but from a dynamic point of view, the mismatch pair has been largely changed by the substitution of Oxa with G. Similar local fluctuations in the base pairing were reported for DNA duplex containing 6-thioguanine (51,52). They pointed that the fast exchange of the imino proton with water may be caused by a faster apparent opening rate. The base pairing geometry of Oxa to C or T could largely change between the positive- and negative-wobble geometries, and the formed mismatched base pairs themselves are fluctuating. In the case of the base excision repair (BER) system, bacterial AlkA (3-methyladenine DNA glycosylase II) and endonuclease VIII was found to possess repair activities on Oxa in oligodeoxynucleotides (53). Cao and co-workers (15,54) also reported that bacterial endonuclease V and human AAG alkyladenine glycosylase shows BER activity on Oxa. The uniqueness of the Oxa base paring may be one of the reasons for the difficulties in finding the specific repairing enzyme for Oxa. In addition, Oxa mediates a novel genotoxic mechanism related to the formation of DNA–protein cross-link (DPC) (55). It was demonstrated that in the case of Oxa-related genotoxicity (Oxa-mediated DPC formation and its relevant events), the nucleotide excision and recombination repair system would play a more efficient role than BER system (56). The structures solved in this work indicate that the presence of the O:C or O:T mismatches causes the localized distortion and fluctuation of the DNA duplexes. The fluctuation of the O:C or O:T base pair may possibly cause the reaction of DPC, leading to the cytotoxity. The static structure and local fluctuation of the O:C and O:T base pair would be related to the recognition and repair of Oxa mismatch sites in DNA.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online: Supplementary Tables 1–4, Supplementary Figures 1–3.
FUNDING
Grant-in-Aid for Scientific Research from the Ministry of Education, Science, Sports, and Culture (17570092 to K.K.); Basic Science Research Program through the National Research Foundation of Korea (NRF) funded by the Ministry of Education, Science and Technology (2010-0010110 to S.P.P.). This work was partly performed in Nanotechnology Support Project in Central Japan (Institute for Molecular Science), financially supported by Nanotechnology Network of the Ministry of Education, Culture, Sports, Science and Technology (MEXT), Japan. Funding for open access charge: Grant-in-Aid for Scientific Research from the Ministry of Education, Science, Sports, and Culture.
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Ms Sachiko Dairaku for her assistance with measuring NMR spectra.
REFERENCES
- 1.Hunter WN, Brown T, Anand NN, Kennard O. Structure of an adenine-cytosine base pair in DNA and its implications for mismatch repair. Nature. 1986;320:552–555. doi: 10.1038/320552a0. [DOI] [PubMed] [Google Scholar]
- 2.Gao XL, Patel DJ. NMR studies of A·C mismatches in DNA dodecanucleotides at acidic pH. Wobble A(anti).C(anti) pair formation. J. Biol. Chem. 1987;262:16973–16984. [PubMed] [Google Scholar]
- 3.Patel DJ, Kozlowski SA, Ikuta S, Itakura K. Deoxyadenosine·deoxycytidine pairing in the d(C-G-C-G-A-A-T-T-C-A-C-G) duplex: conformation and dynamics at and adjacent to the dA·dC mismatch site. Biochemistry. 1984;23:3218–3226. doi: 10.1021/bi00309a016. [DOI] [PubMed] [Google Scholar]
- 4.Sarma MH, Gupta G, Sarma RH, Bald R, Engelke U, Oei SL, Gessner R, Erdmann VA. DNA structure in which an adenine-cytosine mismatch pair forms an integral part of the double helix. Biochemistry. 1987;26:7707–7715. doi: 10.1021/bi00398a026. [DOI] [PubMed] [Google Scholar]
- 5.Allawi HT, SantaLucia J., Jr NMR solution structure of a DNA dodecamer containing single G·T mismatches. Nucleic Acids Res. 1998;26:4925–4934. doi: 10.1093/nar/26.21.4925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Allawi HT, SantaLucia J., Jr Thermodynamics and NMR of internal G·T mismatches in DNA. Biochemistry. 1997;36:10581–10594. doi: 10.1021/bi962590c. [DOI] [PubMed] [Google Scholar]
- 7.Hunter WN, Brown T, Kneale G, Anand NN, Rabinovich D, Kennard O. The structure of guanosine-thymidine mismatches in B-DNA at 2.5-Å resolution. J. Biol. Chem. 1987;262:9962–9970. doi: 10.2210/pdb113d/pdb. [DOI] [PubMed] [Google Scholar]
- 8.Goodman MF. Hydrogen bonding revisited: geometric selection as a principal determinant of DNA replication fidelity. Proc. Natl Acad. Sci. USA. 1997;94:10493–10495. doi: 10.1073/pnas.94.20.10493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Shimada N, Yagisawa N, Naganawa H, Takita T, Hamada M, Takeuchi T, Umezawa H. Oxanosine, a novel nucleoside from actinomycetes. J. Antibiot. 1981;34:1216–1218. doi: 10.7164/antibiotics.34.1216. [DOI] [PubMed] [Google Scholar]
- 10.Suzuki T, Yamaoka R, Nishi M, Ide H, Makino K. Isolation and characterization of a novel product, 2′-deoxyoxanosine, from 2′-deoxyguanosine, oligonucleotide, and calf thymus DNA treated by nitrous acid and nitric oxide. J. Am. Chem. Soc. 1996;118:2515–2516. [Google Scholar]
- 11.Suzuki T, Matsumura Y, Ide H, Kanaori K, Tajima K, Makino K. Deglycosylation susceptibility and base-pairing stability of 2′-deoxyoxanosine in oligodeoxynucleotide. Biochemistry. 1997;36:8013–8019. doi: 10.1021/bi970166l. [DOI] [PubMed] [Google Scholar]
- 12.Suzuki T, Ide H, Yamada M, Endo N, Kanaori K, Tajima K, Morii T, Makino K. Formation of 2′-deoxyoxanosine from 2′-deoxyguanosine and nitrous acid: mechanism and intermediates. Nucleic Acids Res. 2000;28:544–551. doi: 10.1093/nar/28.2.544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pack SP, Nonogawa M, Kodaki T, Makino K. Chemical synthesis and thermodynamic characterization of oxanine-containing oligodeoxynucleotides. Nucleic Acids Res. 2005;33:5771–5780. doi: 10.1093/nar/gki865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hernández B, Soliva R, Luque FJ, Orozco M. Misincorporation of 2′-deoxyoxanosine into DNA: a molecular basis for NO-induced mutagenesis derived from theoretical calculations. Nucleic Acids Res. 2000;28:4873–4883. doi: 10.1093/nar/28.24.4873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hitchcock TM, Gao H, Cao W. Cleavage of deoxyoxanosine-containing oligodeoxyribonucleotides by bacterial endonuclease V. Nucleic Acids Res. 2004;32:4071–4080. doi: 10.1093/nar/gkh747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pack SP, Makino K. Synthesis of 2′-deoxyoxanosine from 2′-deoxyguanosine, conversion to its phosphoramidite, and incorporation into oxanine-containing oligodeoxynucleotides. Curr. Protoc. Nucleic Acid Chem. 2010 doi: 10.1002/0471142700.nc0439s41. Chapter 4, Unit 4 39. [DOI] [PubMed] [Google Scholar]
- 17.Pack SP, Doi A, Choi YS, Kim HB, Makino K. Accurate guanine:cytosine discrimination in T4 DNA ligase-based single nucleotide polymorphism analysis using an oxanine-containing ligation fragment. Anal. Biochem. 2010;398:257–259. doi: 10.1016/j.ab.2009.11.014. [DOI] [PubMed] [Google Scholar]
- 18.Pack SP, Doi A, Choi YS, Kodaki T, Makino K. Biomolecular response of oxanine in DNA strands to T4 polynucleotide kinase, T4 DNA ligase, and restriction enzymes. Biochem. Biophys. Res. Commun. 2010;391:118–122. doi: 10.1016/j.bbrc.2009.11.013. [DOI] [PubMed] [Google Scholar]
- 19.Wüthrich K. NMR of Proteins and Nucleic Acids. New York: Wiley-Interscience; 1986. [Google Scholar]
- 20.Piantini U, Sørensen OW, Ernst RR. Multiple quantum filters for elucidating NMR coupling networks. J. Am. Chem. Soc. 1982;104:6800–6801. [Google Scholar]
- 21.Davis DG, Bax A. Assignment of complex proton NMR spectra via two-dimensional homonuclear Hartmann-Hahn spectroscopy. J. Am. Chem. Soc. 1985;107:2820–2821. [Google Scholar]
- 22.Bax A, Davis DG. MLEV-17-based two-dimensional homonuclear magnetization transfer spectroscopy. J. Magn. Reson. 1985;65:355–360. [Google Scholar]
- 23.Jeener J, Meier BH, Bachmann P, Ernst RR. Investigation of exchange processes by two-dimensional NMR spectroscopy. J. Chem. Phys. 1979;71:4546–4553. [Google Scholar]
- 24.Williamson JR, Boxer SG. Multinuclear NMR studies of DNA hairpins. 1. Structure and dynamics of d(CGCGTTGTTCGCG) Biochemistry. 1989;28:2819–2831. doi: 10.1021/bi00433a012. [DOI] [PubMed] [Google Scholar]
- 25.Piotto M, Saudek V, Sklenar V. Gradient-tailored excitation for single-quantum NMR spectroscopy of aqueous solutions. J. Biomol. NMR. 1992;2:661–665. doi: 10.1007/BF02192855. [DOI] [PubMed] [Google Scholar]
- 26.Brünger AT. X-PLOR Version 3.1: A System for X-ray Crystallography and NMR. New Haven: Yale University Press; 1993. [Google Scholar]
- 27.Kuszewski J, Schwieters C, Clore GM. Improving the accuracy of NMR structures of DNA by means of a database potential of mean force describing base-base positional interactions. J. Am. Chem. Soc. 2001;123:3903–3918. doi: 10.1021/ja010033u. [DOI] [PubMed] [Google Scholar]
- 28.Mujeeb A, Kerwin SM, Kenyon GL, James TL. Solution structure of a conserved DNA sequence from the HIV-1 genome: Restrained molecular dynamics simulation with distance and torsion angle restraints derived from two-dimensional NMR spectra. Biochemistry. 1993;32:13419–13431. doi: 10.1021/bi00212a007. [DOI] [PubMed] [Google Scholar]
- 29.Kanaori K, Tamura Y, Wada T, Nishi M, Kanehara H, Morii T, Tajima K, Makino K. Structure and stability of the consecutive stereoregulated chiral phosphorothioate DNA duplex. Biochemistry. 1999;38:16058–16066. doi: 10.1021/bi9909344. [DOI] [PubMed] [Google Scholar]
- 30.Lavery R, Sklenar H. Defining the structure of irregular nucleic acids: conventions and principles. J. Biomol. Struct. Dyn. 1989;6:655–667. doi: 10.1080/07391102.1989.10507728. [DOI] [PubMed] [Google Scholar]
- 31.Lavery R, Sklenar H. The definition of generalized helicoidal parameters and of axis curvature for irregular nucleic acids. J. Biomol. Struct. Dyn. 1988;6:63–91. doi: 10.1080/07391102.1988.10506483. [DOI] [PubMed] [Google Scholar]
- 32.Kalnik MW, Kouchankdjian M, Li BF, Swann PF, Patel DJ. Base pair mismatches and carcinogen-modified bases in DNA: an NMR study of G·T and G·O4meT pairing in dodecanucleotide duplexes. Biochemistry. 1988;27:108–115. doi: 10.1021/bi00401a018. [DOI] [PubMed] [Google Scholar]
- 33.Hare D, Shapiro L, Patel DJ. Wobble dG·dT pairing in right-handed DNA: solution conformation of the d(C-G-T-G-A-A-T-T-C-G-C-G) duplex deduced from distance geometry analysis of nuclear Overhauser effect spectra. Biochemistry. 1986;25:7445–7456. doi: 10.1021/bi00371a029. [DOI] [PubMed] [Google Scholar]
- 34.Fagan PA, Fabrega C, Eritja R, Goodman MF, Wemmer DE. NMR study of the conformation of the 2-aminopurine:cytosine mismatch in DNA. Biochemistry. 1996;35:4026–4033. doi: 10.1021/bi952657g. [DOI] [PubMed] [Google Scholar]
- 35.Sowers LC, Boulard Y, Fazakerley GV. Multiple structures for the 2-aminopurine-cytosine mispair. Biochemistry. 2000;39:7613–7620. doi: 10.1021/bi992388k. [DOI] [PubMed] [Google Scholar]
- 36.Boulard Y, Cognet JA, Fazakerley GV. Solution structure as a function of pH of two central mismatches, C. T and C. C, in the 29 to 39 K-ras gene sequence, by nuclear magnetic resonance and molecular dynamics. J. Mol. Biol. 1997;268:331–347. doi: 10.1006/jmbi.1997.0975. [DOI] [PubMed] [Google Scholar]
- 37.Allawi HT, SantaLucia J., Jr Thermodynamics of internal C·T mismatches in DNA. Nucleic Acids Res. 1998;26:2694–2701. doi: 10.1093/nar/26.11.2694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bhaumik SR, Chary KV, Govil G, Liu K, Miles HT. Homopurine and homopyrimidine strands complementary in parallel orientation form an antiparallel duplex at neutral pH with A-C, G-T, and T-C mismatched base pairs. Biopolymers. 1997;41:773–784. doi: 10.1002/(SICI)1097-0282(199706)41:7<773::AID-BIP6>3.0.CO;2-M. [DOI] [PubMed] [Google Scholar]
- 39.Allawi TH, SantaLucia JJ. NMR solution structure of a DNA dodecamer containing single G-T mismatches. Nucleic Acids Res. 1998;26:4925–4934. doi: 10.1093/nar/26.21.4925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Chin KH, Chen FM, Chou SH. Solution structure of the ActD-5'-CCGTT3GTGG-3' complex: drug interaction with tandem G·T mismatches and hairpin loop backbone. Nucleic Acids Res. 2003;31:2622–2629. doi: 10.1093/nar/gkg353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Pfaff DA, Clarke KM, Parr TA, Cole JM, Geierstanger BH, Tahmassebi DC, Dwyer TJ. Solution structure of a DNA duplex containing a guanine-difluorotoluene pair: A wobble pair without hydrogen bonding? J. Am. Chem. Soc. 2008;130:4869–4878. doi: 10.1021/ja7103608. [DOI] [PubMed] [Google Scholar]
- 42.Holbrook SR, Cheong C, Tinoco IJ, Kim SH. Crystal structure of an RNA double helix incorporating a track of non-Watson-Crick base pairs. Nature. 1991;353:579–581. doi: 10.1038/353579a0. [DOI] [PubMed] [Google Scholar]
- 43.Sloane DL, Goodman MF, Echols H. The fidelity of base selection by the polymerase subunit of DNA polymerase III holoenzyme. Nucleic Acid Res. 1998;16:6465–6475. doi: 10.1093/nar/16.14.6465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Modrich P. DNA mismatch correction. Annu. Rev. Biochem. 1987;56:435–466. doi: 10.1146/annurev.bi.56.070187.002251. [DOI] [PubMed] [Google Scholar]
- 45.Kramer B, Kramer W, Fritz H-J. Different base/base mismatches are corrected with different efficiencies by the methyl-directed DNA mismatch-repair system of E. coli. Cell. 1984;38:879–887. doi: 10.1016/0092-8674(84)90283-6. [DOI] [PubMed] [Google Scholar]
- 46.Dohet C, Wagner R, Radman M. Repair of defined single base-pair mismatches in Escherichia coli. Proc. Natl Acad. Sci. USA. 1985;82:503–505. doi: 10.1073/pnas.82.2.503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Moran S, Ren RX, Sheils CJ, Rumney S, Kool ET. Non-hydrogen bonding ‘terminator’ nucleosides increase the 3′-end homogeneity of enzymatic RNA and DNA synthesis. Nucleic Acid Res. 1996;24:2044–2052. doi: 10.1093/nar/24.11.2044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hunter WN. Crystallographic studies of DNA containing mismatches, modified and unpaired bases. Methods Enzymol. 1992;211:221–231. doi: 10.1016/0076-6879(92)11014-a. [DOI] [PubMed] [Google Scholar]
- 49.Allawi HT, SantaLucia J., Jr Nearest neighbor thermodynamic parameters for internal G·A mismatches in DNA. Biochemistry. 1998;37:2170–2179. doi: 10.1021/bi9724873. [DOI] [PubMed] [Google Scholar]
- 50.Allawi HT, SantaLucia J., Jr Nearest-neighbor thermodynamics of internal A·C mismatches in DNA: sequence dependence and pH effects. Biochemistry. 1998;37:9435–9444. doi: 10.1021/bi9803729. [DOI] [PubMed] [Google Scholar]
- 51.Somerville L, Krynetski EY, Krynetskaia NF, Beger RD, Zhang W, Marhefka CA, Evans WE, Kriwacki RW. Structure and dynamics of thioguanine-modified duplex DNA. J. Biol. Chem. 2003;278:1005–1011. doi: 10.1074/jbc.M204243200. [DOI] [PubMed] [Google Scholar]
- 52.Bohon J, de los Santos CR. Structural effect of the anticancer agent 6-thioguanine on duplex DNA. Nucleic Acids Res. 2003;31:1331–1338. doi: 10.1093/nar/gkg203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Terato H, Masaoka A, Asagoshi K, Honsho A, Ohyama Y, Suzuki T, Yamada M, Makino K, Yamamoto K, Ide H. Novel repair activities of AlkA (3-methyladenine DNA glycosylase II) and endonuclease VIII for xanthine and oxanine, guanine lesions induced by nitric oxide and nitrous acid. Nucleic Acids Res. 2002;30:4975–4984. doi: 10.1093/nar/gkf630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Hitchcock TM, Dong L, Connor EE, Meira LB, Samson LD, Wyatt MD, Cao W. Oxanine DNA glycosylase activity from Mammalian alkyladenine glycosylase. J. Biol. Chem. 2004;279:38177–38183. doi: 10.1074/jbc.M405882200. [DOI] [PubMed] [Google Scholar]
- 55.Nakano T, Terato H, Asagoshi K, Masaoka A, Mukuta M, Ohyama Y, Suzuki T, Makino K, Ide H. DNA-protein cross-link formation mediated by oxanine. A novel genotoxic mechanism of nitric oxide-induced DNA damage. J. Biol. Chem. 2003;278:25264–25272. doi: 10.1074/jbc.M212847200. [DOI] [PubMed] [Google Scholar]
- 56.Nakano T, Katafuchi A, Shimizu R, Terato H, Suzuki T, Tauchi H, Makino K, Skorvaga M, Van Houten B, Ide H. Repair activity of base and nucleotide excision repair enzymes for guanine lesions induced by nitrosative stress. Nucleic Acids Res. 2005;33:2181–2191. doi: 10.1093/nar/gki513. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.