Abstract
Synthetic scaffolds that permit spatial and temporal organization of enzymes in living cells are a promising post-translational strategy for controlling the flow of information in both metabolic and signaling pathways. Here, we describe the use of plasmid DNA as a stable, robust and configurable scaffold for arranging biosynthetic enzymes in the cytoplasm of Escherichia coli. This involved conversion of individual enzymes into custom DNA-binding proteins by genetic fusion to zinc-finger domains that specifically bind unique DNA sequences. When expressed in cells that carried a rationally designed DNA scaffold comprising corresponding zinc finger binding sites, the titers of diverse metabolic products, including resveratrol, 1,2-propanediol and mevalonate were increased as a function of the scaffold architecture. These results highlight the utility of DNA scaffolds for assembling biosynthetic enzymes into functional metabolic structures. Beyond metabolism, we anticipate that DNA scaffolds may be useful in sequestering different types of enzymes for specifying the output of biological signaling pathways or for coordinating other assembly-line processes such as protein folding, degradation and post-translational modifications.
INTRODUCTION
Metabolic engineering of microbial pathways provides a cost-effective and environmentally benign route for producing numerous valuable compounds, including commodity and specialty chemicals (e.g. biodegradable plastics), biofuels (e.g. ethanol and butanol) and therapeutic molecules (e.g. anticancer drugs and antimicrobial compounds). However, efforts to engineer new functional biosynthetic pathways in well-characterized micro-organisms such as Escherichia coli are still often hampered by issues such as imbalanced pathway flux, formation of side products and accumulation of toxic intermediates that can inhibit host cell growth. One strategy for increasing metabolite production in metabolically engineered microorganisms is the use of directed enzyme organization [for a review see Ref. (1)]. This concept is inspired by natural metabolic systems, for which optimal metabolic pathway performance often arises from the organization of enzymes into specific complexes and, in some cases, enzyme-to-enzyme channeling (a.k.a. metabolic channeling) (1–3).
The most striking naturally occurring examples are enzymes that have evolved three-dimensional structures capable of physically channeling substrates such as tryptophan synthase and carbamoyl phosphate synthase. The crystal structures of these enzymes reveal tunnels that connect catalytic sites and protect reactive intermediates from the bulk solution (4,5). Other notable examples include electrostatic channeling of negatively charged substrates along a positively charged protein surface that leads from one active site to the next (6), direct channeling of substrates via thioester linkages between polyketide synthase enzyme modules (7), compartmentalization of specific enzymes into small volumes within the cell in the form of subcellular organelles (8,9) and dynamic assembly of enzyme complexes, perhaps as a feedback mechanism, to achieve a precise concentration of metabolic product (10,11).
Inspired by these natural systems, several groups have developed methods for artificially assembling enzyme complexes to enhance the performance of biological pathways. For example, direct enzyme fusions have been used to coordinate the expression and localization of two resveratrol biosynthetic enzymes in a manner that increased product titers in yeast and mammalian cells (12). However, fusing more than two enzymes may prove problematic due to misfolding and/or proteolysis of the fusion protein. In a notable departure from fusion proteins, Fierobe and co-workers constructed artificial cellulosomes where selected enzymes were incorporated in specific locations on a protein scaffold (13). Compared to their free enzyme counterparts, the resulting enzyme complexes exhibited enhanced synergistic action on crystalline cellulose. More recently, Dueber et al. (14) expressed scaffolds built from the interaction domains of metazoan signaling proteins to assemble metabolic enzymes that were tagged with their cognate peptide ligands. Significant increases in the production of mevalonate and separately glucaric acid were observed in the presence of several of these scaffolds. Along similar lines, Delebecque et al. (15) created RNA aptamer-based scaffolds to control the spatial organization of two metabolic enzymes involved in biological hydrogen production. Similar to protein scaffolds, RNA-based scaffolds increased the hydrogen output as a function of scaffold architecture.
Here, we describe an alternative method for generating artificial complexes of metabolic pathway enzymes that uses DNA as the scaffold. The choice of DNA for guiding enzyme assembly affords many advantages. First, DNA has a highly predictable local structure. Therefore, scaffolds based on DNA have the potential for arranging enzymes into a predefined order with the caveat that supercoiling of plasmid DNA could affect long-range ordering. For example, the spatial orientation of bound proteins may be tuned by varying the number of nucleotides between the enzyme binding sites. Second, the in vivo stability of DNA scaffolds is largely sequence independent, which means that numerous architectures of virtually any sequence and length can be generated without decreasing the availability of the scaffold. Protein- and RNA-based scaffolds, on the other hand, are subject to issues associated with misfolding, aggregation and susceptibility to degradation (16–19), which may become more pronounced as the scaffold designs become larger and more complex (i.e. more difficult to fold, greatly likelihood of forming off-pathway intermediates and more potential sites for enzymatic degradation). In fact, the folding and stability of protein- and RNA-based scaffolds may change from one design to the next, even for very subtle changes to the RNA or protein sequence. Third, a large number of different DNA-binding proteins exist in nature. Some of them, such as zinc fingers (ZFs), have modular structures that can be engineered to bind unique DNA sequences with nanomolar dissociation constants and discriminate effectively against nonspecific DNA (20,21). As a result of these and other advanced ZF selection methods (22,23), there are already more than 700 experimentally tested ZFs available for use with DNA scaffolds. Relative to the seemingly limitless number of highly active ZF domains and corresponding DNA sequences, there are far fewer characterized protein interaction domains and RNA-binding proteins with ultra-high affinity for their targets. One notable exception is leucine zipper interaction domains, which have picomolar to nanomolar affinities and have been used for some scaffolding applications (24). However, these domains may homodimerize and even aggregate if included in the same polypeptide. Finally, fourth, because of the similar overall fold, different zinc finger domains have comparable in vivo folding and stability profiles compared to the more structurally diverse protein interaction and RNA-binding domains used in earlier systems.
To test the potential of DNA scaffolds, we created chimeras between target biosynthetic enzymes and ZF domains that specifically bind unique DNA sequences. When these modified enzymes were expressed in cells carrying a DNA scaffold comprising corresponding ZF binding domains, significant titer enhancements for three diverse metabolic products including resveratrol, 1,2-propanediol (1,2-PD) and mevalonate were achieved compared to cells expressing unassembled pathway enzymes. These results underscore the potential of DNA scaffolds programmed with distinct protein docking sites as a powerful new tool for assembling biological pathways in a manner that directly impacts their output.
MATERIALS AND METHODS
Plasmid construction for biosynthetic pathways
Chimeric enzymes for the resveratrol biosynthetic pathway were constructed by linking the genes encoding 4CL and STS to the 3′-end of the genes encoding Zif268 and PBSII, respectively. Each construct included a GGSGGGSGGS polypeptide linker separating the enzyme from the ZF domain. 4CL was from Arabidopsis thaliana and STS was from Vitis vinifera and were not codon optimized for E. coli. Genes for Zif268 and PBSII were codon optimized for expression in E. coli and synthesized by GeneArt. PCR products corresponding to the coding regions for enzymes and zinc fingers were fused together by overlap extension PCR. The Zif268-4CL PCR product was restriction digested using XbaI and ApaI and PBSII-STS was digested by ApaI and BamHI. Both fragments were simultaneously ligated in XbaI/BamHI digested pET19b vector to obtain plasmid pET-Res-ZF-Enz. The 4CL-STS fusion protein in plasmid pET28a (pET-ResFusion) (12) was provided by Dr. Oliver Yu (DDPSC).
The genes encoding MgsA, DkgA and GldA were PCR-amplified from E. coli MG1655 genomic DNA. These genes were then cloned into pBAD18 (25) as a polycistron for 1,2-PD synthesis as follows: the mgsA gene was placed between NheI and XbaI, the dkgA gene between XbaI and SphI, and the gldA gene between SphI and HindIII. The same strong ribosomal binding site was placed directly upstream of each gene in the polycistron with an NdeI site at each start codon. To the 3′-end of mgsA, dkgA and gldA, codon-optimized versions of the ZF triplets OZ052 (ZFa), OZ300 (ZFb) and OZ076 (ZFc) (22), were connected, respectively, by a codon optimized L5 polylinker (TSAAA) (18). Each ZF was appended with a C-terminal HA epitope tag. The resulting plasmid was named pBAD-PD-ZF-Enz.
The mevalonate pathway enzymes were tethered to ZF domains directly in the construct pRM178 (14). Here, the linker and ligand at the 3′-end of each gene was excised and replaced exactly with the L5 linker and appropriate ZF domains as above. ZFa was fused to AtoB, ZFb fused to HMGS and ZFc fused to HMGR, with an HA epitope tag introduced on the C-terminus of all proteins. The resulting plasmid was named pTet-Mev-ZF-Enz.
Plasmid construction for DNA scaffolds
Primer pairs encoding Zif268 or PBSII binding sites (Supplementary Table S1) separated by 2-, 4- or 8-bp spacers and flanked by standard Biobrick restriction sites were annealed by 10-min incubation at 95°C and subsequent slow cooling to room temperature. Multiple copies of a DNA scaffold were assembled according to standard Biobrick assembly (26) and cloned into the high copy pSB1K3 vector (http://partsregistry.org/Part:pSB1K3).
For ZFa, ZFb and ZFc binding sites (Supplementary Table S1), pUC19 served as the basis for construction of the DNA scaffold. pUC19 was completely digested with AatII and PvuII and replaced with a polylinker containing the following restriction sites: AatII–SacI–SpeI–XbaI–SphI–ClaI–PvuII. DNA scaffolds were assembled using SpeI and XbaI cohesive ends for ligation. Basic parts were made so that scaffolds would be flanked by SpeI sites on the 5′-end and XbaI sites on the 3′-end. Composite scaffolds were constructed by digesting the backbone with XbaI and ligating an SpeI/XbaI-digested insert at the 3′-end, thus maintaining the SpeI site at the 5′-end and XbaI site at the 3′-end for future ligations. To separate the binding sites, a 4- or 12-bp spacer was employed. For 1,2-PD production, the resulting scaffolds were subcloned into pBAD18, between the β-lactamase and PBAD promoters, directly after the AgeI site. For mevalonate production, the scaffolds were employed directly from the pUC19 plasmid.
Bacterial strains, media and growth conditions
Resveratrol was produced in E. coli Rosetta (DE3) pLysS harboring pET-Res-ZF-Enz encoding the Zif268-4CL and PBSII-STS chimeras or pET-Res-Enz encoding 4CL and STS without the ZF domains in the presence of a DNA scaffold in plasmid pSB1K3. Overnight bacterial cultures were diluted to optical density measured at 600 nm (A600) of 0.2 in 100 ml 2 × YT medium in shake flasks and grown at 30°C and 160 rpm. At A600 of 0.8, 1 mM IPTG and 0.3 mM coumaric acid were added to induce gene expression and provide the substrate for resveratrol production, respectively. Samples were taken for analysis 6 h after induction of the ZF-enzyme chimeras. Escherichia coli strain W3110 harboring the pBAD-PD-ZF-Enz plasmid encoding the ZF-enzyme chimeras and corresponding DNA scaffolds was used for 1,2-PD production. 1,2-PD anaerobic fermentations were followed as described (27) with the following exceptions. l-Arabinose was added to 0.2% (w/v) at the time of inoculation to induce gene expression. All fermentations were run at 37°C, either at 200 or 250 rpm, with tubes held vertically or at a 45° angle. The 10-ml fermentation mixtures were inoculated to an A600 of 0.05 with the overnight culture. Samples were taken for analysis at 9 h post-induction for Western blot analysis and 24 h post-induction for fermentation yields. Mevalonate production was conducted as described (14) but with E. coli DP10 cells harboring the pTet-Mev-ZF-Enz plasmid and a pUC19-based DNA scaffold. An inducer concentration of 250 nM anhydrotetracycline was used for all experiments. Samples were taken for analysis at 25 h post-induction for Western blot analysis and 50 h post-induction for fermentation yields. Antibiotics were provided at the following concentrations: ampicillin, 100 μg/ml; chloramphenicol, 25 μg/ml; and kanamycin, 50 μg/ml.
Product recovery and analysis
At selected time points, resveratrol was extracted from 1 ml of bacterial supernatants by ethyl acetate as described (28). Briefly, supernatants were obtained after removing bacteria from culture by centrifugation at 13 000 rpm. Supernatants were acidified by 1 M HCl (50 µl/1 ml supernatant) and kept at −80°C until extracted twice by equal volumes of ethyl acetate. Ethyl acetate was removed by evaporation in vacuum evaporator and the remaining pellet resuspended in methanol prior to analysis by HPLC (Thermo Finnigan). Separation was performed at 25°C on a stainless-steel column Hypersil ODS C18 (150 × 4.6 mm I.D., particle size: 5 μm, Thermo) protected by Phenomenex HPLC guard cartridge C18 as a precolumn. Adequate separation was achieved in 35 min by a gradient elution and a mobile phase consisting of acetate buffer with pH 5.6 (solvent A) and acetonitrile (solvent B). Gradient elution program applied at flow rate 1.5 ml/min was as follows: 5 min 95% A, 15 min 95–50% A, 3 min 50–5% A, 5 min 5% A, 2 min 5–95% A and 5 min 95% A. Chromatograms were monitored at 303 nm (Supplementary Figure S1a). Quantitative determination of trans-resveratrol was performed using calibration standards (0.1, 0.25, 0.5, 1 and 2 µg/ml) prepared from a stock solution of trans-resveratrol (99%, Sigma Aldrich) in 50% (v/v) methanol in water. The standard solutions were stored at −80°C. The identity of resveratrol was also confirmed by MS analysis (Supplementary Figure S1b). 1,2-PD present in the fermentation media was recovered by removal of cells and quantified as described (27) with the following exceptions. Compounds were measured with a Waters Breeze HPLC system (Supplementary Figure S2a). The mobile phase was a 0.03 N sulfuric acid solution, with a flow rate of 0.45 ml/min, and the column and detector temperatures were 50°C and 40°C, respectively. All samples were filtered through 0.22-μm-pore-size membranes prior to analysis. Mevalonate was recovered by acidifying cell cultures to form mevalonolactone followed by extraction with ethyl acetate exactly as described elsewhere (14). The samples were then run on Agilent Technologies chiral cyclosil-B column (30 m length × 0.25 mm i.d. × 0.25 µm Film) to determine the relative abundance of mevalonolactone as described in detail elsewhere (14) (Supplementary Figure S2b).
Western blot analysis
Escherichia coli Rosetta (DE3) cells co-expressing the Zif268-4CL and PBSII-STS chimeras or expressing the 4CL-STS fusion for 6 h were harvested by centrifugation. Likewise, W3110 cultures expressing the 1,2-PD ZF-enzyme chimeras for 9 h or DP10 cultures expressing the mevalonate ZF-enzyme chimeras for 25 h were harvested by centrifugation. Cell pellets were resuspended in PBS, lysed via sonication and centrifuged at 16 000 rpm for 10 min. The supernatant was retained as the soluble cell lysate. All samples were normalized to the amount of total soluble protein. Immunoblot analysis of soluble lysates was performed with anti-His antibodies (Sigma) to detect Zif268-4CL, PBSII-STS or 4CL-STS chimeras and anti-HA antibodies (Sigma) for detection of 1,2-PD- or mevalonate-related chimeras according to standard procedures. GroEL served as a loading control and was detected with anti-GroEL antibodies (Sigma).
RESULTS
Targeting DNA in vitro and in vivo with ZF domains
We aimed to use plasmid DNA as a scaffold onto which cellular proteins of interest could be docked (Figure 1). This required a method for site-specific targeting of enzymes along the DNA surface. To this end, we focused on five different ZF domains (PBSII, Zif268, ZFa, ZFb and ZFc) that were each comprising three fingers with specificity for unique 9 base-pair DNA sequences (22,29–32) (Supplementary Table S1). The selection criteria for choosing these particular ZF domains was as follows: first, the ZF domain should be non-toxic to the host cells (33); second, the ZF domains should be capable of binding orthogonal sequences with high affinity. Based on our estimation of approximately 127 plasmids per cells (0.2 µM) and approximately 5000 enzyme chimeras/cell (8 µM) (Supplementary Figure S3a), we determined that the zinc fingers should have sub-µM affinity. The five ZF domains tested here all bind DNA with low nanomolar affinity. An additional design goal was to balance fusion protein stability with the number of competitive binding sites in the E. coli genome. We observed that zinc fingers comprising as many as four fingers did not impact the stability or activity of the protein to which they were fused (Supplementary Figure S3b). Of these, we focused our attention on three-finger designs because these were relatively short (84–87 amino acids in length) and minimally cross-reactive with host DNA (only approximately 15 predicted binding sites in the E. coli genome). Importantly, none of the selected ZF domains were predicted to bind functional regions of essential genes in E. coli and thus would be unlikely to hamper bacterial fitness.
Figure 1.
DNA scaffold-assisted assembly of metabolic pathways in E. coli. (a) Schematic of the (1:1)n system developed for resveratrol biosynthesis. Depicted are a representative DNA scaffold (n = 1) (left) and the plasmid pET-Res-ZF-Enz for expression of the ZF-enzyme chimeras (right). E1 and E2 are the enzymes 4CL and STS, respectively, while ZF domains a and b are Zif268 and PBSII, respectively. (b) Schematic of different scaffold arrangements used for the three-enzyme pathways producing either 1,2-PD or mevalonate. E1, E2 and E3 are the 1,2-PD or mevalonate biosynthetic enzymes (see the text for details) and the ZF domains a, b and c are ZFa, ZFb and ZFc, respectively. In all cases where n > 1 (bottom right), the scaffolds were designed such that the first enzyme was always flanked on both sides by the second and third enzyme giving rise to a bidirectional pathway arrangement. Also shown is the plasmid pBAD-PD-ZF-Enz or pTet-Mev-ZF-Env for expressing 1,2-PD or mevalonate ZF-enzyme chimeras, respectively (top right). All enzymes and ZF domains were connected by flexible polypeptide linkers.
As a first test of the system components, we verified DNA binding of candidate ZF domains fused to the N- or C-terminus of different model proteins including fragments of the yellow fluorescent protein (YFP) and E. coli maltose-binding protein (MBP). Following purification from E. coli, all ZF chimeras bound their target DNA sequences when positioned either N- or C-terminally (Supplementary Figure S3c and d). Next, we determined whether ZF domains could bind to neighboring sites on a DNA scaffold. For this, we genetically fused split YFP to the N- and C-termini of the ZF domains PBSII and Zif268, respectively. As expected, reassembly of split YFP did not occur in solution in the absence of a DNA scaffold, or in the presence of a DNA scaffold where the binding sites for the neighboring pairs were scrambled. However, we observed strong fluorescence indicative of YFP reassembly in the presence of a DNA scaffold that contained neighboring binding sites for PBSII and Zif268 separated by only two DNA base pairs (bp) (Supplementary Figure S4a). Binding of these PBSII and Zif268 chimeras to the same DNA scaffold was independently confirmed using surface plasmon resonance (SPR) (Figure S4b). Taken together, these results indicate that (i) the expression and/or activity of different target proteins was not significantly affected when fused with these relatively small ZF domains, (ii) ZF domains retained DNA binding activity when fused to different proteins and (iii) two orthogonal ZF domains can simultaneously bind their target sequences in a DNA scaffold and bring their fused protein domains into close proximity as evidenced by the YFP reassembly.
As a final test, we investigated whether these ZF domains could bind their cognate DNA targets in vivo. To confirm target DNA binding by ZFs in vivo, we generated a simple β-galactosidase (β-gal) screen for ZF activity in E. coli. The assay involved a single, low-copy plasmid encoding a synthetic promoter, PSYN, into which a DNA-binding sequence specific for each ZF domain was inserted (between 35 and 10 sites of the promoter). This promoter was positioned upstream of the lacZ reporter gene, expression of which was controlled by PSYN. The gene encoding the ZF domain was cloned in the same plasmid but under control of the arabinose inducible PBAD promoter. The principle of this screen is that an active ZF domain should bind to its specific target sequence in the PSYN promoter and act as a synthetic repressor, thereby decreasing the basal activity of this promoter and lowering β-gal levels. As expected, induction of each ZF domain resulted in a strong reduction of β-gal activity, whereas β-gal activity was unchanged in controls where the PSYN promoter contained a binding site of an unrelated zinc finger (Supplementary Figure S4c). These results confirm that the ZF domains used in our studies bind specifically to their corresponding DNA target sites in vivo and thus are ideally suited for directing diverse cellular enzymes to specific sites on plasmid DNA.
Enhancing trans-resveratrol biosynthesis in the presence of DNA scaffolds
We next investigated the ability of the ZF domains to assemble the resveratrol (trans-3,5,4′-trihydroxystilbene) biosynthetic enzymes on DNA in the cytoplasm of E. coli. The metabolic pathway for this natural plant product has been reconstituted in microbes (12,28,34). Production of trans-resveratrol from 4-coumaric acid occurs in two steps in which 4-coumaric acid is converted to 4-coumaroyl-CoA by 4-coumarate:CoA ligase (4CL) and trans-resveratrol is formed by condensation of one molecule of 4-coumaroyl-CoA and three molecules of malonyl-CoA by stilbene synthase (STS) (Supplementary Figure 2a). We hypothesized that successful DNA-guided assembly of this simple metabolic pathway would lead to measurable increases in resveratrol titers compared to the unassembled pathway. To test this notion, genes encoding 4CL and STS were fused to the Zif268 and PBSII ZF domains, respectively, in one plasmid while the DNA scaffold was present on a second plasmid. It should be noted that a large number of possible enzyme arrangements on plasmid DNA are possible. The different architectures tested here are described as [E1a:E2b]n for a two-enzyme system, where a and b describe the enzyme stoichiometry within a single scaffold unit [hereafter denoted as (a:b)] and n is the number of times the scaffold unit is repeated in the plasmid (Figure 1a). For resveratrol assembly, we initially focused on a simple (1:1) scaffold unit that was repeated 16 times on the plasmid (n = 16). The rationale for this number of repeats was based on the fact that plasmid DNA copy numbers in E. coli are commonly far below that of overexpressed metabolic enzymes. Thus, we predicted that most simple scaffold units would need to be repeated tens of times on a plasmid to accommodate all of the expressed enzymes. When the plasmids for a (1:1)16 resveratrol system were combined in E. coli, we found that resveratrol production was consistently enhanced by 2- to 3-fold compared to the case where a random scaffold control plasmid was present (Figure 2b).
Figure 2.
Enhancement of trans-resveratrol biosynthesis in the presence of DNA scaffolds. (a) Schematic representation of resveratrol biosynthetic pathway. (b) Comparison of resveratrol titers from E. coli cells expressing the 4CL-STS fusion or Zif268-4CL and PBSII-STS chimeras in the presence of DNA scaffolds (n = 16) with different spacer lengths between ZF binding sites or a random scaffold control plasmid. Cells expressing the ZF-enzyme chimeras in the presence of the random scaffold control served as the control to which all data was normalized. Also shown are data for the separated (1:1)4 scaffold where the spacing between the ZF binding sites was 850 bp. Samples were taken 6-h post-induction. The amount of resveratrol produced in random scaffold control cells was 2.31 ± 0.20 mg/l. Data are the average of three replicate experiments and error bars are the standard error of the mean (SEM). (c) Western blot of enzyme levels in cells expressing the 4CL-STS fusion protein compared to cells co-expressing the Zif268-4CL and PBSII-STS chimeras.
In addition to enzyme stoichiometry, additional degrees of freedom of the DNA scaffold system include the number of repetitive scaffold units and the spacer length between the ZF binding sites. In the case of the (1:1)16 resveratrol system, the largest product enhancement was observed for spacer lengths of 2 and especially 8 bp, while a spacer length of 4 bp showed a smaller yet measureable improvement over the unscaffolded enzymes (Figure 2b). An even larger increase in titer enhancement of nearly 5-fold was observed when the number of scaffold repeats was decreased from 16 to 4 (Figure 2b). These improvements highlight the ability to impact resveratrol production via simple changes in scaffold design that may lead in some cases to optimal arrangements of the enzymes on the DNA. Next, we examined whether the enhanced product titers were dependent upon the close proximity (2–8 bp) of the two pathway enzymes. To test this notion, the ZF binding sites within the (1:1)4 scaffold were separated on the plasmid by either 2 bp or 850 bp. The latter configuration provided the same number of binding sites on the plasmid for both enzymes but prevented the bound enzymes from localizing in close proximity to one another. It is important to note here that no changes were made to either of the chimeric enzymes. As would be expected for a proximity effect, the 5-fold enhancement in resveratrol production observed for the (1:1)4 scaffold was abolished when the binding sites for each enzyme were positioned far apart on the plasmid (Figure 2b).
We also evaluated an alternative strategy for enzyme co-localization using a 4CL-STS fusion protein which was previously reported to increase resveratrol production in yeast up to ∼6-fold (12). In E. coli, however, the (1:1)16 scaffold system produced >50 times more resveratrol than the 4CL-STS fusion (Figure 2b), even though bacterial growth was very similar in both cases and the 4CL-STS fusion protein was expressed at an equal or slightly higher level than both the ZF-enzyme chimeras (Figure 2c). This result may be due to the propensity of multidomain fusion proteins to misfold (and hence be less active) in E. coli (18) and highlights the advantage of the DNA assembly strategy whereby each of the ZF-enzyme fusions fold independently.
Improving the metabolic performance of a three-enzyme pathway for 1,2-PD
To test the generality of our approach, we next focused our attention on a three-enzyme pathway for producing 1,2-PD from dihydroxyacetone phosphate (DHAP) (Figure 3a). We chose this pathway because a biosynthetic route for 1,2-PD in E. coli is well established (27). For targeting the 1,2-PD metabolic pathway enzymes to DNA, we fused methylglyoxal synthase (MgsA), 2,5-diketo-d-gluconic acid reductase (DkgA) and glycerol dehydrogenase (GldA) (all from E. coli) to the N-termini of ZFa, ZFb and ZFc, respectively. For the scaffold design, target DNA sequences corresponding to each of the ZF domains were placed on the same plasmid as the ZF-enzyme chimeras. Given that there are approximately 127 plasmids per cell and approximately 5000 ZF-enzyme chimeras per cell (Figure S3a), DNA scaffolds for the three-enzyme pathway were designed that would provide enough binding sites to accommodate all of the expressed enzymes. Specifically, we constructed and tested scaffolds with enzyme:scaffold ratios in the range of 40:1 to 1:3 [(1:1:1)1 to (1:4:2)32, respectively]. Like the resveratrol results above, E. coli with the (1:1:1)16 1,2-PD system produced ∼3.5 times more 1,2-PD than cells expressing the ZF-enzyme fusions in the presence of no scaffold or a random scaffold control (Figure 3b). Moreover, the growth rate of the cells in all of these cases was nearly identical. Also similar to the resveratrol results was the observation that protein fusions including MgsA-DkgA, DkgA-GldA and MgsA-DkgA-GldA did not improve 1,2-PD titers over the unscaffolded enzymes (data not shown). Interestingly, when the number of scaffold unit repeats, n, was reduced from 16 to 4, 1,2-PD titers increased to approximately 4.5-fold above the unscaffolded controls. Here, only a small drop-off in metabolic performance was observed when the spacing between ZF binding sites was reduced from 12 to 4 bp for the (1:1:1)4 scaffold.
Figure 3.
DNA scaffold-assisted production of 1,2-PD. (a) Schematic representation of 1,2-PD biosynthetic pathway. (b) Comparison of 1,2-PD titers from E. coli cells expressing the MgsA-ZFa, DkgA-ZFb and GldA-ZFc chimeras in the presence of a (1:1:1)n scaffold with n = 4 or 16 and the spacing between ZF binding sites = 4 or 12 bp as indicated. Cells expressing the ZF-enzyme chimeras in the presence of no scaffold served as the control to which all data were normalized. Also shown are data from cells carrying a random scaffold control. The amount of 1,2-PD produced in unscaffolded control cells was 0.13 ± 0.01 g/l. (c) Comparison of enzyme levels and fold improvement of 1,2-PD in cells carrying different (1:2:1)n scaffolds compared to no scaffold and random scaffold controls. Also shown are data for the separated (1:2:1)2 scaffold where the spacing between the ZF binding sites was ∼1000 bp. Data are the average of three replicate experiments and error bars are the standard error of the mean (SEM).
To systematically investigate the relationship between scaffold design variables and product formation, we generated a matrix of additional plasmid-encoded DNA scaffolds where a was always 1, while b and c were varied to give the following stoichiometries: 1:1:1, 1:2:1, 1:2:2, 1:4:1 and 1:4:2. The number of scaffold units, n, was varied to be 1, 2, 4, 8, 16 or 32 and the spacing between ZF binding sites was either 4 or 12 bp. It should also be noted that the first pathway enzyme, MgsA, was flanked on each side by the second and third pathway enzymes, giving rise to a bidirectional pathway arrangement (Figure 1b). To determine the impact of these designs, E. coli cells were transformed with plasmids encoding the ZF-enzyme chimeras and the different scaffolds. Nearly all of the scaffolds with 12-bp spacers between ZF binding sites were observed to enhance 1,2-PD production (Figure S5a). In particular, the [1:1:1]4, [1:1:1]8, [1:2:1]4, [1:2:2]4, and [1:4:2]2 scaffolds each increased 1,2-PD levels by ∼4-5 fold compared to the unscaffolded control. These data also revealed that when only a single scaffold unit was present (n = 1), product titers were largely insensitive to the scaffold stoichiometry. However, as the number of scaffold units was increased, the effect of scaffold stoichiometry on 1,2-PD levels became more varied. We also observed that nearly all scaffolds with 4-bp spacers between ZF binding sites were less effective than their 12-bp counterparts at improving 1,2-PD titers (Figure S5b).
To investigate the factors underlying the observed enhancement of 1,2-PD production, we first measured the cellular expression levels of ZF-enzyme chimeras in the presence of different DNA scaffolds. Regardless of whether the cells carried a (1:2:1)n scaffold, a random scaffold sequence or lacked a scaffold altogether, the expression level of these chimeras were all very similar with the exception of cells carrying the (1:2:1)2 and (1:2:1)4 scaffolds, which appeared to accumulate slightly lower levels of MgsA-ZFa and DkgA-ZFb enzymes (Figure 3c). However, this lower expression on it's own was insufficient to explain the improved 1,2-PD titers conferred by these scaffolds. This is because the (1:2:1)1 and the (1:2:1)8 scaffolds showed similar enhancements in 1,2-PD titers but with enzyme expression levels that were nearly indistinguishable from the unscaffolded controls. Therefore, we conclude that a simple change in cellular enzyme levels is not the cause of the DNA scaffold-guided enhancement of 1,2-PD levels. To test whether enzyme proximity was important for enhanced 1,2-PD titers, the ZF binding sites within the (1:2:1)2 scaffold were separated on the plasmid by ∼1000 bp. As seen above for resveratrol production, the enhancement in 1,2-PD production was abolished when the binding sites for each enzyme were positioned far apart on the plasmid (Figure 3c). Hence, the relative proximity of the enzymes appears to be a key factor underlying the observed titer enhancements.
Extending DNA scaffolds to mevalonate production
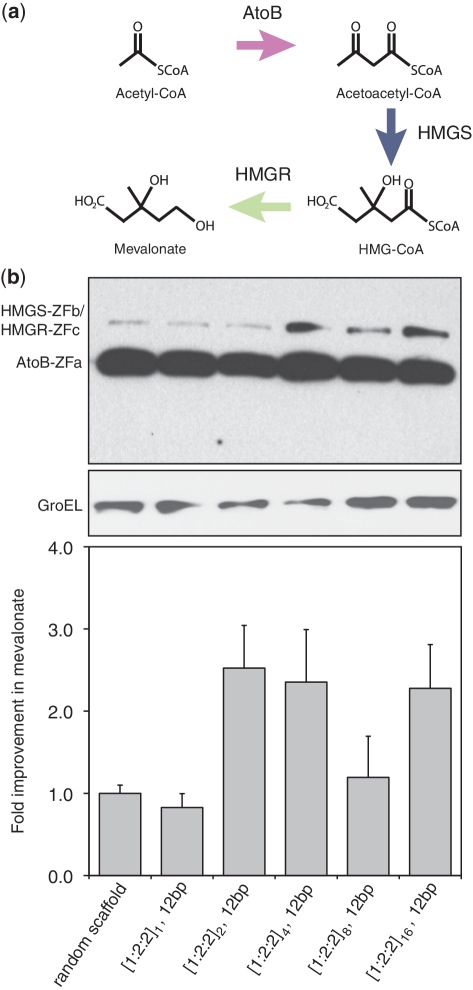
As a final test of the generality of our approach, we attempted to use DNA scaffolds to improve mevalonate production. We chose this pathway because production of mevalonate from acetyl-CoA in E. coli has been described (Figure 4a) (35). Furthermore, as mentioned above, previous studies demonstrated that assembly of the mevalonate biosynthetic enzymes on a protein scaffold caused cells to accrue significantly higher titers of mevalonate (14); therefore, this pathway allows direct comparison between our DNA scaffolds and earlier protein-based scaffolds. We created ZF-enzyme chimeras by fusing the mevalonate biosynthetic enzymes E. coli acetoacetyl-CoA thiolase (AtoB), Saccharomyces cerevisiae hydroxy-methylglutaryl-CoA synthase (HMGS) and S. cerevisiae hydroxy-methylglutaryl-CoA reductase (HMGR) to the N-termini of ZFa, ZFb and ZFc, respectively. For the scaffold design, target DNA sequences corresponding to each of the ZF domains were placed on a separate plasmid. Similar to the resveratrol and 1,2-PD cases above, mevalonate titers were increased 2- to 3-fold in the presence of several different scaffold designs (Figure S6a). While no clear trend was apparent, the greatest titer enhancement—an increase of nearly 3-fold—came from the (1:4:2)16 scaffold. This was followed closely by several of the (1:2:2)n scaffolds (i.e. n = 2, 4 and 16) that each enhanced mevalonate titers by ∼2.5 fold compared to the random scaffold control (Figure 4b). The fact that the best yield enhancement using protein-based scaffolds also came from a 1:2:2 motif (14) suggests that this arrangement may be optimal for balancing pathway flux. Consistent with the results above for 1,2-PD, the smaller 4-bp spacers between the ZF binding sites resulted were less effective than their 12-bp counterparts at improving metabolic performance (Supplementary Figure S6b). In fact, most scaffolds containing 4-bp spacers resulted in little to no enhancement of mevalonate titers compared to unscaffolded enzymes. Finally, while the expression levels of the ZF-enzyme chimeras were largely unaffected by the presence or absence of a specific DNA scaffold, the amount of AtoB-ZFa that accumulated in cells was much greater compared to the HMGS-ZFb/HMGR-ZFc chimeras (Figure 4b). In contrast, the expression levels of the ZF-enzyme chimeras for 1,2-PD were more evenly balanced, which might account for the generally larger fold improvements seen for the production of 1,2-PD versus mevalonate. These data suggest that more balanced expression of the ZF-enzyme chimeras may further increase mevalonate titers in the future.
Figure 4.
DNA scaffold-assisted production of mevalonate. (a) Schematic representation of mevalonate biosynthetic pathway. (b) Comparison of enzyme levels and mevalonate titers from E. coli cells expressing the AtoB-ZFa, HMGS-ZFb and HMGR-ZFc chimeras in the presence of (1:2:2)n scaffolds with n = 1, 2, 4, 8 or 16 and the spacing between ZF binding sites = 12 bp as indicated. Cells expressing the ZF-enzyme chimeras in the presence of the random scaffold control served as the control to which all data were normalized. The amount of mevalonate produced in the random scaffold control cells was 1.7 ± 0.07 g/l. Data are the average of three replicate experiments and error bars are the standard error of the mean (SEM).
DISCUSSION
We have demonstrated DNA scaffold-assisted biosynthesis as a viable strategy for significantly enhancing the titers of three diverse metabolic products. This enhancement appears to arise from the enforced proximity of metabolic enzymes that likely increases the effective concentrations of intermediary metabolites. In every case tested, DNA scaffold-assisted biosynthesis was implemented on an existing microbial metabolic pathway and did not require any a priori knowledge about the structure or function of any of the underlying biosynthetic enzymes, making the implementation of this new approach simple and generalizable to virtually any pathway. This was made possible by the ability to fuse distinct ZF domains to diverse protein targets at will without significant loss of the ZF domains’ DNA binding activity or the target proteins’ enzymatic activity. As a result, this is the first ever report of DNA as an intracellular scaffold for controlling the flow of information in a metabolic or signaling context.
It should be pointed out that scaffolds comprised of comprising expressed proteins and RNAs have recently been reported that have been used for directing new cell signaling behaviors (24,36) and linking together metabolic enzymes to more efficiently synthesize desired chemical products (14,15). Compared to these systems, DNA scaffolds present a number of unique challenges and opportunities for improvement. For example, much larger titer enhancements were observed with both protein- and RNA-based scaffolds (>50-fold) compared to DNA scaffolds (up to ∼5-fold). In the case of protein scaffolds, however, the largest titer enhancements (77-fold) were observed only under conditions where enzyme expression levels were very low. When saturating amounts of the inducer were used, which were on par with the inducer concentration used in our studies (i.e. ∼250 nM anhydrotetracycline), the scaffold-dependent increases in mevalonate titers decreased to levels that were similar to or even below those observed here. Furthermore, when protein scaffolds were applied to a second metabolic system, namely glucaric acid production, the improvement was ∼2–5 fold (14,37), which was consistent with the improvements achieved with DNA scaffolds. In the case of RNA scaffolds, studies were focused on just a single metabolic system, namely biohydrogen production. Thus, whether such a large titer enhancement can be generalized to other pathways remains to be shown. A potential drawback of DNA scaffolds is that the placement of repetitive sequences in plasmid DNA may result in recombination of the plasmid to remove the repeat regions. To date, however, we have performed numerous DNA sequencing and restriction digestion analysis experiments and have never seen evidence of plasmid recombination under any of the conditions tested. Nonetheless, a recA− strain background in which recombination events are minimized could be used as the scaffolding host. Another challenge associated with plasmid DNA is its tendency to become supercoiled in cells. Plasmid supercoiling may restrict the ability to spatially control enzyme orientation especially over long distances. In contrast, the use of RNA permits the assembly of discrete one- and two-dimensional scaffolds (15). However, with the recent development of methods for rationally designing DNA nanostructures with complex secondary structures that assemble in the cytoplasm of E. coli (38), it may be possible in the future to create nanostructured DNA scaffolds in vivo that permit exquisite patterning of target proteins.
Despite some of these challenges, the ultra-stable nature of DNA and its ability to support locally ordered scaffolds, here up to 2.4 kb with over 150 individual ZF binding sites, will enable scalability of DNA scaffolds to large metabolic systems (i.e. comprising more than three enzymes and/or more than one pathway) arranged in virtually any stoichiometry and repeated many times over. Another major advantage of DNA scaffolds is their modularity, which permits a very high degree of freedom with respect to important system variables such as: stoichiometry of enzyme binding sites, number of scaffold units (n), spacing of ZF binding sites, location of binding sites on the plasmid, copy number of the plasmid, and binding affinity of the ZF domain for the DNA target sequence. The number of tunable parameters that can be used to advantageously tailor a metabolic system increases dramatically if one also considers the range of modifications that can be made to the ZF-enzyme fusion (e.g. N- or C-terminal attachment of ZF domain, length and composition of the linker connecting ZF domain to the enzyme, sequence of the ZF domain, etc.). By studying different DNA scaffold architectures, enzyme stoichiometries, and flux balanced or imbalanced scenarios, it should be possible to determine when enzyme co-localization is most beneficial. This, in turn, will be very useful for guiding future design of these systems and in envisioning new applications for enzyme co-localization. It is also worth mentioning that our DNA scaffold approach is highly complementary to many of the existing methods for enzyme, pathway and strain engineering that are already in the cellular engineer's toolkit. Hence, a successful strategy for achieving the production yields, near theoretical maximum, necessary for industrial viability will likely involve a combination of these approaches. Of course, DNA scaffolds could also be used to flexibly control the flow of different classes of biological information that extend beyond metabolic pathways and small-molecule products. For example, DNA scaffolds could be used to rewire intracellular signaling pathways or to coordinate other assembly-line processes such as protein folding, degradation and post-translational modifications. Thus, we anticipate that DNA scaffolds should enable the construction of reliable protein networks to program a range of useful cellular behaviors. Even though the beauty of nature's most elegant compartmentalization strategies such as a protected tunnel (4) or intracellular organelles (8,9) have yet to be recapitulated by engineers, the use of DNA scaffolds is an important early step towards this goal.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online: Supplementary Table S1, Supplementary Figures 1–6, Supplementary Methods and Supplementary References [18,33].
FUNDING
Office of Naval Research Young Investigator Grant Program (N000140610565 and N000140710027 to M.P.D.); Slovenian Research Agency and Centre of Excellence EN-FIST (to R.J.); NSF (CBET-0756801).Funding for open access charge: NSF.
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
We would like to thank Matej Žnidarič Tina Ilc, and Tjaša Stošicki for their help in the iGEM2010 project related to this work. We also thank Dr. Breda Simonovska, Alen Albreht and Mateja Puklavec for assistance with determination of resveratrol, Dr Oliver Yu for providing the construct encoding the 4CL-STS fusion protein, and Katelyn Connell for assistance with determination of mevalonate.
REFERENCES
- 1.Conrado RJ, Varner JD, DeLisa MP. Engineering the spatial organization of metabolic enzymes: mimicking nature's synergy. Curr. Opin. Biotechnol. 2008;19:492–499. doi: 10.1016/j.copbio.2008.07.006. [DOI] [PubMed] [Google Scholar]
- 2.Srere PA. Complexes of sequential metabolic enzymes. Annu. Rev. Biochem. 1987;56:89–124. doi: 10.1146/annurev.bi.56.070187.000513. [DOI] [PubMed] [Google Scholar]
- 3.Miles EW, Rhee S, Davies DR. The molecular basis of substrate channeling. J. Biol. Chem. 1999;274:12193–12196. doi: 10.1074/jbc.274.18.12193. [DOI] [PubMed] [Google Scholar]
- 4.Hyde CC, Ahmed SA, Padlan EA, Miles EW, Davies DR. Three-dimensional structure of the tryptophan synthase alpha 2 beta 2 multienzyme complex from Salmonella typhimurium. J. Biol. Chem. 1988;263:17857–17871. [PubMed] [Google Scholar]
- 5.Thoden JB, Holden HM, Wesenberg G, Raushel FM, Rayment I. Structure of carbamoyl phosphate synthetase: a journey of 96 A from substrate to product. Biochemistry. 1997;36:6305–6316. doi: 10.1021/bi970503q. [DOI] [PubMed] [Google Scholar]
- 6.Stroud RM. An electrostatic highway. Nat. Struct. Biol. 1994;1:131–134. doi: 10.1038/nsb0394-131. [DOI] [PubMed] [Google Scholar]
- 7.Tsuji SY, Cane DE, Khosla C. Selective protein–protein interactions direct channeling of intermediates between polyketide synthase modules. Biochemistry. 2001;40:2326–2331. doi: 10.1021/bi002463n. [DOI] [PubMed] [Google Scholar]
- 8.Bobik TA. Polyhedral organelles compartmenting bacterial metabolic processes. Appl. Microbiol. Biotechnol. 2006;70:517–525. doi: 10.1007/s00253-005-0295-0. [DOI] [PubMed] [Google Scholar]
- 9.Straight PD, Fischbach MA, Walsh CT, Rudner DZ, Kolter R. A singular enzymatic megacomplex from Bacillus subtilis. Proc. Natl Acad. Sci. USA. 2007;104:305–310. doi: 10.1073/pnas.0609073103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Narayanaswamy R, Levy M, Tsechansky M, Stovall GM, O'Connell JD, Mirrielees J, Ellington AD, Marcotte EM. Widespread reorganization of metabolic enzymes into reversible assemblies upon nutrient starvation. Proc. Natl Acad. Sci. USA. 2009;106:10147–10152. doi: 10.1073/pnas.0812771106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.An S, Kumar R, Sheets ED, Benkovic SJ. Reversible compartmentalization of de novo purine biosynthetic complexes in living cells. Science. 2008;320:103–106. doi: 10.1126/science.1152241. [DOI] [PubMed] [Google Scholar]
- 12.Zhang Y, Li SZ, Li J, Pan X, Cahoon RE, Jaworski JG, Wang X, Jez JM, Chen F, Yu O. Using unnatural protein fusions to engineer resveratrol biosynthesis in yeast and mammalian cells. J. Am. Chem. Soc. 2006;128:13030–13031. doi: 10.1021/ja0622094. [DOI] [PubMed] [Google Scholar]
- 13.Fierobe HP, Mechaly A, Tardif C, Belaich A, Lamed R, Shoham Y, Belaich JP, Bayer EA. Design and production of active cellulosome chimeras. Selective incorporation of dockerin-containing enzymes into defined functional complexes. J. Biol. Chem. 2001;276:21257–21261. doi: 10.1074/jbc.M102082200. [DOI] [PubMed] [Google Scholar]
- 14.Dueber JE, Wu GC, Malmirchegini GR, Moon TS, Petzold CJ, Ullal AV, Prather KL, Keasling JD. Synthetic protein scaffolds provide modular control over metabolic flux. Nat. Biotechnol. 2009;27:753–759. doi: 10.1038/nbt.1557. [DOI] [PubMed] [Google Scholar]
- 15.Delebecque CJ, Lindner AB, Silver PA, Aldaye FA. Organization of intracellular reactions with rationally designed RNA Assemblies. Science. 2011;333:470–474. doi: 10.1126/science.1206938. [DOI] [PubMed] [Google Scholar]
- 16.Ponchon L, Dardel F. Recombinant RNA technology: the tRNA scaffold. Nat Methods. 2007;4:571–576. doi: 10.1038/nmeth1058. [DOI] [PubMed] [Google Scholar]
- 17.Baneyx F, Mujacic M. Recombinant protein folding and misfolding in Escherichia coli. Nat. Biotechnol. 2004;22:1399–1408. doi: 10.1038/nbt1029. [DOI] [PubMed] [Google Scholar]
- 18.Chang HC, Kaiser CM, Hartl FU, Barral JM. De novo folding of GFP fusion proteins: high efficiency in eukaryotes but not in bacteria. J. Mol. Biol. 2005;353:397–409. doi: 10.1016/j.jmb.2005.08.052. [DOI] [PubMed] [Google Scholar]
- 19.Netzer WJ, Hartl FU. Recombination of protein domains facilitated by co-translational folding in eukaryotes. Nature. 1997;388:343–349. doi: 10.1038/41024. [DOI] [PubMed] [Google Scholar]
- 20.Greisman HA, Pabo CO. A general strategy for selecting high-affinity zinc finger proteins for diverse DNA target sites. Science. 1997;275:657–661. doi: 10.1126/science.275.5300.657. [DOI] [PubMed] [Google Scholar]
- 21.Rebar EJ, Pabo CO. Zinc finger phage: affinity selection of fingers with new DNA-binding specificities. Science. 1994;263:671–673. doi: 10.1126/science.8303274. [DOI] [PubMed] [Google Scholar]
- 22.Maeder ML, Thibodeau-Beganny S, Osiak A, Wright DA, Anthony RM, Eichtinger M, Jiang T, Foley JE, Winfrey RJ, Townsend JA, et al. Rapid “open-source” engineering of customized zinc-finger nucleases for highly efficient gene modification. Mol. Cell. 2008;31:294–301. doi: 10.1016/j.molcel.2008.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sander JD, Dahlborg EJ, Goodwin MJ, Cade L, Zhang F, Cifuentes D, Curtin SJ, Blackburn JS, Thibodeau-Beganny S, Qi Y, et al. Selection-free zinc-finger-nuclease engineering by context-dependent assembly (CoDA) Nat. Methods. 2011;8:67–69. doi: 10.1038/nmeth.1542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bashor CJ, Helman NC, Yan S, Lim WA. Using engineered scaffold interactions to reshape MAP kinase pathway signaling dynamics. Science. 2008;319:1539–1543. doi: 10.1126/science.1151153. [DOI] [PubMed] [Google Scholar]
- 25.Guzman LM, Belin D, Carson MJ, Beckwith J. Tight regulation, modulation, and high-level expression by vectors containing the arabinose PBAD promoter. J. Bacteriol. 1995;177:4121–4130. doi: 10.1128/jb.177.14.4121-4130.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Shetty RP, Endy D, Knight TF., Jr Engineering BioBrick vectors from BioBrick parts. J. Biol. Eng. 2008;2:5. doi: 10.1186/1754-1611-2-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Altaras NE, Cameron DC. Metabolic engineering of a 1,2-propanediol pathway in Escherichia coli. Appl. Environ. Microbiol. 1999;65:1180–1185. doi: 10.1128/aem.65.3.1180-1185.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Beekwilder J, Wolswinkel R, Jonker H, Hall R, de Vos CH, Bovy A. Production of resveratrol in recombinant microorganisms. Appl. Environ. Microbiol. 2006;72:5670–5672. doi: 10.1128/AEM.00609-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hurt JA, Thibodeau SA, Hirsh AS, Pabo CO, Joung JK. Highly specific zinc finger proteins obtained by directed domain shuffling and cell-based selection. Proc. Natl Acad. Sci. USA. 2003;100:12271–12276. doi: 10.1073/pnas.2135381100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ooi AT, Stains CI, Ghosh I, Segal DJ. Sequence-enabled reassembly of beta-lactamase (SEER-LAC): a sensitive method for the detection of double-stranded DNA. Biochemistry. 2006;45:3620–3625. doi: 10.1021/bi0517032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pavletich NP, Pabo CO. Zinc finger-DNA recognition: crystal structure of a Zif268-DNA complex at 2.1 A. Science. 1991;252:809–817. doi: 10.1126/science.2028256. [DOI] [PubMed] [Google Scholar]
- 32.Stains CI, Porter JR, Ooi AT, Segal DJ, Ghosh I. DNA sequence-enabled reassembly of the green fluorescent protein. J. Am. Chem. Soc. 2005;127:10782–10783. doi: 10.1021/ja051969w. [DOI] [PubMed] [Google Scholar]
- 33.Sander JD, Zaback P, Joung JK, Voytas DF, Dobbs D. An affinity-based scoring scheme for predicting DNA-binding activities of modularly assembled zinc-finger proteins. Nucleic Acids Res. 2009;37:506–515. doi: 10.1093/nar/gkn962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Watts KT, Lee PC, Schmidt-Dannert C. Biosynthesis of plant-specific stilbene polyketides in metabolically engineered Escherichia coli. BMC Biotechnol. 2006;6:22. doi: 10.1186/1472-6750-6-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Martin VJ, Pitera DJ, Withers ST, Newman JD, Keasling JD. Engineering a mevalonate pathway in Escherichia coli for production of terpenoids. Nat. Biotechnol. 2003;21:796–802. doi: 10.1038/nbt833. [DOI] [PubMed] [Google Scholar]
- 36.Park SH, Zarrinpar A, Lim WA. Rewiring MAP kinase pathways using alternative scaffold assembly mechanisms. Science. 2003;299:1061–1064. doi: 10.1126/science.1076979. [DOI] [PubMed] [Google Scholar]
- 37.Moon TS, Dueber JE, Shiue E, Prather KL. Use of modular, synthetic scaffolds for improved production of glucaric acid in engineered E. coli. Metab. Eng. 2010;12:298–305. doi: 10.1016/j.ymben.2010.01.003. [DOI] [PubMed] [Google Scholar]
- 38.Lin C, Rinker S, Wang X, Liu Y, Seeman NC, Yan H. In vivo cloning of artificial DNA nanostructures. Proc. Natl Acad. Sci. USA. 2008;105:17626–17631. doi: 10.1073/pnas.0805416105. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.