Abstract
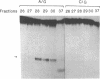
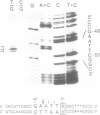
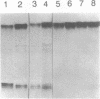
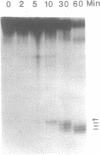
An endonuclease activity (called MS-nicking) for all possible base mismatches has been detected in the extracts of yeast, Saccharomyces cerevisiae. DNAs with twelve possible base mismatches at one defined position are cleaved at different efficiencies. DNA fragments with A/G, G/A, T/G, G/T, G/G, or A/A mismatches are nicked with greater efficiencies than C/T, T/C, C/A, and C/C. DNA with an A/C or T/T mismatch is nicked with an intermediate efficiency. The MS-nicking is only on one particular DNA strand, and this strand disparity is not controlled by methylation, strand break, or nature of the mismatch. The nicks have been mapped at 2-3 places at second, third, and fourth phosphodiester bonds 5' to the mispaired base; from the time course study, the fourth phosphodiester bond probably is the primary incision site. This activity may be involved in mismatch repair during genetic recombination.
Full text
PDF





Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Au K. G., Cabrera M., Miller J. H., Modrich P. Escherichia coli mutY gene product is required for specific A-G----C.G mismatch correction. Proc Natl Acad Sci U S A. 1988 Dec;85(23):9163–9166. doi: 10.1073/pnas.85.23.9163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Au K. G., Clark S., Miller J. H., Modrich P. Escherichia coli mutY gene encodes an adenine glycosylase active on G-A mispairs. Proc Natl Acad Sci U S A. 1989 Nov;86(22):8877–8881. doi: 10.1073/pnas.86.22.8877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhagwat A. S., Sohail A., Lieb M. A new gene involved in mismatch correction in Escherichia coli. Gene. 1988 Dec 25;74(1):155–156. doi: 10.1016/0378-1119(88)90274-0. [DOI] [PubMed] [Google Scholar]
- Bishop D. K., Andersen J., Kolodner R. D. Specificity of mismatch repair following transformation of Saccharomyces cerevisiae with heteroduplex plasmid DNA. Proc Natl Acad Sci U S A. 1989 May;86(10):3713–3717. doi: 10.1073/pnas.86.10.3713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bishop D. K., Kolodner R. D. Repair of heteroduplex plasmid DNA after transformation into Saccharomyces cerevisiae. Mol Cell Biol. 1986 Oct;6(10):3401–3409. doi: 10.1128/mcb.6.10.3401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bishop D. K., Williamson M. S., Fogel S., Kolodner R. D. The role of heteroduplex correction in gene conversion in Saccharomyces cerevisiae. Nature. 1987 Jul 23;328(6128):362–364. doi: 10.1038/328362a0. [DOI] [PubMed] [Google Scholar]
- Brooks P., Dohet C., Almouzni G., Méchali M., Radman M. Mismatch repair involving localized DNA synthesis in extracts of Xenopus eggs. Proc Natl Acad Sci U S A. 1989 Jun;86(12):4425–4429. doi: 10.1073/pnas.86.12.4425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown T. C., Jiricny J. A specific mismatch repair event protects mammalian cells from loss of 5-methylcytosine. Cell. 1987 Sep 11;50(6):945–950. doi: 10.1016/0092-8674(87)90521-6. [DOI] [PubMed] [Google Scholar]
- Brown T. C., Jiricny J. Different base/base mispairs are corrected with different efficiencies and specificities in monkey kidney cells. Cell. 1988 Aug 26;54(5):705–711. doi: 10.1016/s0092-8674(88)80015-1. [DOI] [PubMed] [Google Scholar]
- Claverys J. P., Lacks S. A. Heteroduplex deoxyribonucleic acid base mismatch repair in bacteria. Microbiol Rev. 1986 Jun;50(2):133–165. doi: 10.1128/mr.50.2.133-165.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox E. C. Bacterial mutator genes and the control of spontaneous mutation. Annu Rev Genet. 1976;10:135–156. doi: 10.1146/annurev.ge.10.120176.001031. [DOI] [PubMed] [Google Scholar]
- Detloff P., Sieber J., Petes T. D. Repair of specific base pair mismatches formed during meiotic recombination in the yeast Saccharomyces cerevisiae. Mol Cell Biol. 1991 Feb;11(2):737–745. doi: 10.1128/mcb.11.2.737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujii H., Shimada T. Isolation and characterization of cDNA clones derived from the divergently transcribed gene in the region upstream from the human dihydrofolate reductase gene. J Biol Chem. 1989 Jun 15;264(17):10057–10064. [PubMed] [Google Scholar]
- Hare J. T., Taylor J. H. Hemi-methylation dictates strand selection in repair of G/T and A/C mismatches in SV40. Gene. 1988 Dec 25;74(1):159–161. doi: 10.1016/0378-1119(88)90276-4. [DOI] [PubMed] [Google Scholar]
- Hare J. T., Taylor J. H. One role for DNA methylation in vertebrate cells is strand discrimination in mismatch repair. Proc Natl Acad Sci U S A. 1985 Nov;82(21):7350–7354. doi: 10.1073/pnas.82.21.7350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holliday R. Molecular aspects of genetic exchange and gene conversion. Genetics. 1974 Sep;78(1):273–287. doi: 10.1093/genetics/78.1.273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holmes J., Jr, Clark S., Modrich P. Strand-specific mismatch correction in nuclear extracts of human and Drosophila melanogaster cell lines. Proc Natl Acad Sci U S A. 1990 Aug;87(15):5837–5841. doi: 10.1073/pnas.87.15.5837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiricny J., Hughes M., Corman N., Rudkin B. B. A human 200-kDa protein binds selectively to DNA fragments containing G.T mismatches. Proc Natl Acad Sci U S A. 1988 Dec;85(23):8860–8864. doi: 10.1073/pnas.85.23.8860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones M., Wagner R., Radman M. Mismatch repair and recombination in E. coli. Cell. 1987 Aug 14;50(4):621–626. doi: 10.1016/0092-8674(87)90035-3. [DOI] [PubMed] [Google Scholar]
- Kramer B., Kramer W., Williamson M. S., Fogel S. Heteroduplex DNA correction in Saccharomyces cerevisiae is mismatch specific and requires functional PMS genes. Mol Cell Biol. 1989 Oct;9(10):4432–4440. doi: 10.1128/mcb.9.10.4432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kramer W., Kramer B., Williamson M. S., Fogel S. Cloning and nucleotide sequence of DNA mismatch repair gene PMS1 from Saccharomyces cerevisiae: homology of PMS1 to procaryotic MutL and HexB. J Bacteriol. 1989 Oct;171(10):5339–5346. doi: 10.1128/jb.171.10.5339-5346.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lieb M., Allen E., Read D. Very short patch mismatch repair in phage lambda: repair sites and length of repair tracts. Genetics. 1986 Dec;114(4):1041–1060. doi: 10.1093/genetics/114.4.1041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lieb M. Specific mismatch correction in bacteriophage lambda crosses by very short patch repair. Mol Gen Genet. 1983;191(1):118–125. doi: 10.1007/BF00330898. [DOI] [PubMed] [Google Scholar]
- Linton J. P., Yen J. Y., Selby E., Chen Z., Chinsky J. M., Liu K., Kellems R. E., Crouse G. F. Dual bidirectional promoters at the mouse dhfr locus: cloning and characterization of two mRNA classes of the divergently transcribed Rep-1 gene. Mol Cell Biol. 1989 Jul;9(7):3058–3072. doi: 10.1128/mcb.9.7.3058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu A. L., Chang D. Y. A novel nucleotide excision repair for the conversion of an A/G mismatch to C/G base pair in E. coli. Cell. 1988 Sep 9;54(6):805–812. doi: 10.1016/s0092-8674(88)91109-9. [DOI] [PubMed] [Google Scholar]
- Lu A. L., Chang D. Y. Repair of single base-pair transversion mismatches of Escherichia coli in vitro: correction of certain A/G mismatches is independent of dam methylation and host mutHLS gene functions. Genetics. 1988 Apr;118(4):593–600. doi: 10.1093/genetics/118.4.593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mankovich J. A., McIntyre C. A., Walker G. C. Nucleotide sequence of the Salmonella typhimurium mutL gene required for mismatch repair: homology of MutL to HexB of Streptococcus pneumoniae and to PMS1 of the yeast Saccharomyces cerevisiae. J Bacteriol. 1989 Oct;171(10):5325–5331. doi: 10.1128/jb.171.10.5325-5331.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maxam A. M., Gilbert W. Sequencing end-labeled DNA with base-specific chemical cleavages. Methods Enzymol. 1980;65(1):499–560. doi: 10.1016/s0076-6879(80)65059-9. [DOI] [PubMed] [Google Scholar]
- Modrich P. DNA mismatch correction. Annu Rev Biochem. 1987;56:435–466. doi: 10.1146/annurev.bi.56.070187.002251. [DOI] [PubMed] [Google Scholar]
- Muster-Nassal C., Kolodner R. Mismatch correction catalyzed by cell-free extracts of Saccharomyces cerevisiae. Proc Natl Acad Sci U S A. 1986 Oct;83(20):7618–7622. doi: 10.1073/pnas.83.20.7618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prudhomme M., Martin B., Mejean V., Claverys J. P. Nucleotide sequence of the Streptococcus pneumoniae hexB mismatch repair gene: homology of HexB to MutL of Salmonella typhimurium and to PMS1 of Saccharomyces cerevisiae. J Bacteriol. 1989 Oct;171(10):5332–5338. doi: 10.1128/jb.171.10.5332-5338.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Radicella J. P., Clark E. A., Fox M. S. Some mismatch repair activities in Escherichia coli. Proc Natl Acad Sci U S A. 1988 Dec;85(24):9674–9678. doi: 10.1073/pnas.85.24.9674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Radman M. Mismatch repair and the fidelity of genetic recombination. Genome. 1989;31(1):68–73. doi: 10.1139/g89-014. [DOI] [PubMed] [Google Scholar]
- Radman M., Wagner R. Mismatch repair in Escherichia coli. Annu Rev Genet. 1986;20:523–538. doi: 10.1146/annurev.ge.20.120186.002515. [DOI] [PubMed] [Google Scholar]
- Raposa S., Fox M. S. Some features of base pair mismatch and heterology repair in Escherichia coli. Genetics. 1987 Nov;117(3):381–390. doi: 10.1093/genetics/117.3.381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rayssiguier C., Thaler D. S., Radman M. The barrier to recombination between Escherichia coli and Salmonella typhimurium is disrupted in mismatch-repair mutants. Nature. 1989 Nov 23;342(6248):396–401. doi: 10.1038/342396a0. [DOI] [PubMed] [Google Scholar]
- Stephenson C., Karran P. Selective binding to DNA base pair mismatches by proteins from human cells. J Biol Chem. 1989 Dec 15;264(35):21177–21182. [PubMed] [Google Scholar]
- Su S. S., Lahue R. S., Au K. G., Modrich P. Mispair specificity of methyl-directed DNA mismatch correction in vitro. J Biol Chem. 1988 May 15;263(14):6829–6835. [PubMed] [Google Scholar]
- Su S. S., Modrich P. Escherichia coli mutS-encoded protein binds to mismatched DNA base pairs. Proc Natl Acad Sci U S A. 1986 Jul;83(14):5057–5061. doi: 10.1073/pnas.83.14.5057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas D. C., Roberts J. D., Kunkel T. A. Heteroduplex repair in extracts of human HeLa cells. J Biol Chem. 1991 Feb 25;266(6):3744–3751. [PubMed] [Google Scholar]
- Tsai-Wu J. J., Radicella J. P., Lu A. L. Nucleotide sequence of the Escherichia coli micA gene required for A/G-specific mismatch repair: identity of micA and mutY. J Bacteriol. 1991 Mar;173(6):1902–1910. doi: 10.1128/jb.173.6.1902-1910.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiebauer K., Jiricny J. In vitro correction of G.T mispairs to G.C pairs in nuclear extracts from human cells. Nature. 1989 May 18;339(6221):234–236. doi: 10.1038/339234a0. [DOI] [PubMed] [Google Scholar]
- Wiebauer K., Jiricny J. Mismatch-specific thymine DNA glycosylase and DNA polymerase beta mediate the correction of G.T mispairs in nuclear extracts from human cells. Proc Natl Acad Sci U S A. 1990 Aug;87(15):5842–5845. doi: 10.1073/pnas.87.15.5842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williamson M. S., Game J. C., Fogel S. Meiotic gene conversion mutants in Saccharomyces cerevisiae. I. Isolation and characterization of pms1-1 and pms1-2. Genetics. 1985 Aug;110(4):609–646. doi: 10.1093/genetics/110.4.609. [DOI] [PMC free article] [PubMed] [Google Scholar]