Abstract
Bacterial Hfq is a protein that plays an important role in the regulation of genes in cooperation with sRNAs. Escherichia coli Hfq (EcHfq) has two or more sites that bind RNA(s) including U-rich and/or the poly(A) tail of mRNA. However, functional and structural information about Bacillus subtilis Hfq (BsHfq) including the RNA sequences that specifically bind to it remain unknown. Here, we describe RNA aptamers including fragment (AG)3A that are recognized by BsHfq and crystal structures of the BsHfq–(AG)3A complex at 2.2 Å resolution. Mutational and structural studies revealed that the RNA fragment binds to the distal site, one of the two binding sites on Hfq, and identified amino acid residues that are critical for sequence-specific interactions between BsHfq and (AG)3A. In particular, R32 appears to interact with G bases in (AG)3A. Poly(A) also binds to the distal site of EcHfq, but the overall RNA structure and protein–RNA interaction patterns engaged in the R32 residues of BsHfq–(AG)3A differ from those of EcHfq–poly(A). These findings provide novel insight into how the Hfq homologue recognizes RNA.
INTRODUCTION
Gene expression is controlled in bacteria through responses to environmental changes that affect growth. Hfq is an important component in the regulation of gene expression in cooperation with sRNAs. Hfq (also known as HF-I) was originally identified as an Escherichia coli host factor required for the RNA replication of phage Qβ (1) and further studies revealed that Hfq in Gram-negative bacteria functions as a post-transcriptional regulator by interacting with various RNAs. Escherichia coli Hfq (EcHfq), which is the most studied among this family of proteins, promotes interactions between small untranslated RNA regulatory molecules (such as OxyS, DsrA, Spot42, RyhB and SgrS sRNAs) and their target mRNAs (2–6). Escherichia coli Hfq interacts with many target mRNAs as an RNA chaperone (7,8) and stimulates polyadenylation that is catalyzed by poly(A) polymerase I, by binding to the poly(A) tail (9–11). Furthermore, EcHfq is involved in RNA stability control by protecting RNA from degradation by the 3′–5′ exoribonucleases, RNase II and PNPase (12,13).
About half of all sequenced Gram-negative (such as E. coli spp., Salmonella enterica, Pseudomonas aeruginosa, Neisseria meningitidis and Vibrio cholerae) and Gram-positive (e.g. Staphylococcus aureus, Listeria monocytogenes and Bacillus subtilis) bacteria express Hfq [reviewed in Ref. (14)]. Growth rates of Hfq deletion strains of E. coli K-12 and P. aeruginosa O1 are reduced (15,16). In addition, growth of the Hfq deletion strain of N. meningitidis is impaired in nutrient-rich media and proteomics analysis has revealed that the expression of 28 genes is affected in this strain (17). Moreover, Hfq is required for intestinal colonization by V. cholerae (18). On the other hand, little is known about the function of Hfq in Gram-positive bacteria. The growth of S. aureus and L. monocytogenes Hfq deletion strains is not affected (19,20). Although Hfq in S. aureus (SaHfq) does not play important role in the stress response, RNA stability or exoprotein expression (19), L. monocytogenes Hfq controls the expression of numerous stress- and virulence-associated genes and binds to sRNAs (20,21).
The deletion of B. subtilis Hfq (BsHfq), which is encoded by the ymaH gene, does not affect either the growth or sporulation (22). BsHfq binds to SR1 sRNA and to ahrC mRNA and the interaction of SR1 sRNA with its primary target ahrC mRNA causes the inhibition of ahrC mRNA translation. However, BsHfq neither stabilizes SR1 sRNA, nor promotes complex formation between SR1 sRNA and ahrC mRNA (23,24). Alternatively, BsHfq is required to activate ahrC mRNA translation (24).
Crystal structures for several bacterial Hfqs and archaeal Hfq-like proteins show that Hfq forms a homohexameric ring consisting of Sm-like folds and two RNA-binding sites at proximal/distal locations in both sides of the ring (25–30). A co-crystal structure of SaHfq in complex with AU5G has revealed that the RNA is recognized by residues at its proximal site (two loops between β2–β3 and β4–β5), although evidence that RNA binds to the distal site of SaHfq has not been uncovered (25). Site-directed mutagenesis of EcHfq has revealed that the two binding sites recognize their respective RNAs, namely poly(A) and U-rich RNA; the poly(A) tail of the target mRNA interacts with residues (Y25 and I30) on the distal site of EcHfq and short U-rich RNA (AU5G) recognizes residues (Y55, K56 and H57) on the proximal site (31). Moreover, a co-crystal structure of EcHfq in complex with poly(A) RNA has recently been described. The structure shows that the poly(A) RNA binds to the distal site (on loop between β1–β2, β4 and β5) and the poly(A)-binding site comprises an adenosine specificity site, a purine nucleotide selectivity site, and a sequence-non-discriminating RNA entrance/exit site (30).
In this article, we present the RNA sequence motif that is recognized by BsHfq and the crystal structure of BsHfq–RNA complex. We first performed a systematic evolution of ligands by exponential enrichment (SELEX) experiment and identified a single stranded AG repeat sequence in specific RNA sequences that are recognized by BsHfq. Structures of BsHfq in complex with the RNA fragment (AG)3A were determined at a resolution of 2.2 Å by X-ray crystallography using molecular replacement. The overall structure of BsHfq is a hexameric ring comprising a Sm-like motif, and the RNA fragment (AG)3A is bound to the distal site of BsHfq. This binding mode was confirmed by gel electrophoresis mobility shift assays (EMSA) with site-directed mutagenesis. The present results provide novel insight into the recognition pattern of RNA with AG repeats located at the distal site of Hfq.
MATERIALS AND METHODS
Plasmid construction
We constructed pQE60Hfq for C-terminal 6× His fusion BsHfq (BsHfq–His) overexpression by inserting a PCR-amplified ymaH gene into the NcoI–BglII digested pQE60 expression vector (Qiagen). The PCR product was obtained using ymaH-NcoI and ymaH-BglII primers (Supplementary Table S1). Site-directed mutagenesis was performed using a QuikChange site-directed mutagenesis kit (Stratagene). Single specific mutations were introduced into BsHfq–His protein using pQE60Hfq as a template DNA together with mutagenic primers (see primer list in Supplementary Table S1). All constructs were verified by DNA sequencing.
Protein overexpression and purification
The expression of BsHfq–His in E. coli M15/pREP4 was induced for 5 h in the presence of 2 mM IPTG. The cells were harvested by centrifugation at 4000 g at 4°C for 15 min. Wet cells were suspended in lysis buffer (10 mM Tris–HCl, 100 mM NaH2PO4, 8 M Urea, pH 8.0), lysed during incubation at room temperature for 30 min and centrifuged at 15 000 g at room temperature for 15 min. The supernatant was incubated with His-select™ Nickel Affinity gel (Sigma) at room temperature for 1 h, and eluted with elution buffer A (10 mM Tris–HCl, 100 mM NaH2PO4, 8 M Urea, 200 mM Imidazole, pH 6.3). The purity of the BsHfq–His protein was analyzed by 15% SDS–PAGE. Eluted samples were dialyzed against 20 mM Tris–HCl buffer (pH 7.5) containing 500 mM NaCl and 50% glycerol. The present study assesses hexameric conformations of purified single specific BsHfq mutations by gel filtration chromatography on Superdex™ 200 10/300 GL columns (GE Healthcare) equilibrated with binding buffer A (10 mM Tris–HCl, pH 7.5, 50 mM NaCl, 50 mM KCl, 1 mM MgCl2). Mutant BsHfqs (48 µg) with lysozyme (50 µg) were eluted with the same buffer. We purified GST-tagged BsHfq and removed the GST-tag using PreScission protease for crystal structure analysis and EMSA experiments with short RNAs. The purified BsHfq contained an extra GPLGS sequence at the N-terminus as described previously (32).
SELEX experiment
RNA selection experiments proceeded as described (33) using the template 5′-TAATACGACTCACTATAGGGACACAATGGACG – N30 – TAACGGCCGACATGAGAG-3′ where N30 represents 30 random nucleotide positions (T7 RNA polymerase promoter is underlined) and the primer sequence, 5′-CTCTCATGTCGGCCGTTA-3′. We enzymatically synthesized RNA pools using AmpliScribe™ T7 High Yield Transcription kit (Epicentre Biotechnologies). After the transcribed RNAs were incubated with DNase I at 37°C for 15 min, RNAs were separated by 6% PAGE under denaturing condition with 8 M urea and then purified from the gel.
Aptamers that bind to BsHfq–His were obtained after nine rounds of selection. The first six rounds comprised filtering the Hfq–RNA aptamer complex through 0.22 µm nitrocellulose filter (Millipore). The samples were mixed in binding buffer A. The filters were washed with the binding buffer A and then bound RNA aptamers were eluted from the filter using elution buffer B (0.3 M NaOAc, 0.1% SDS). The RNA aptamers were reverse transcribed into cDNA, amplified by PCR and then RNA was synthesized from the synthesized cDNA at the next round of selection. The next two rounds of selection comprised purification using the MagneHis Protein Purification System (Promega). The final round of selection consisted of filtration through nitrocellulose filter. The yielded ligand DNA was cloned into the pGEM-T vector (Promega) and individual clones were sequenced using the ABI PRISM 310 genetic Analyzer with the Big-Dye Terminator v3.1 cycle sequencing kit (Applied Biosystems).
Preparation of RNA samples
Samples of short RNAs (≤18 nt) were chemically synthesized using a DNA/RNA synthesizer (Expedite 8909, Perseptive). Long RNA samples (≥45 nt) were enzymatically synthesized by in vitro transcription using the AmpliScribe™ T7 High Yield Transcription kit. Template DNA plasmids for transcription were constructed by inserting the DNA fragment into the pGEM-3zf(+) vector (Promega) digested with EcoRI–HindIII and cleaved by HindIII digestion to enable run-off transcription with T7 RNA polymerase. The DNA fragments were prepared by annealing primer pairs (see the primer list in Supplementary Table S1). RNA samples were purified by resolution on PAGE under denaturing conditions with 8 M urea and concentrated by ethanol precipitation.
EMSA experiments
Long and short RNAs were mixed in binding buffer A containing 1 mM DTT, 0.3% ribonucleoside vanadyl complex and 5% glycerol and in binding buffer B (10 mM Tris–HCl, pH 7.5 and 10 mM NaCl), respectively, before EMSA with PAGE. The mixtures were reacted for 30 min at 37 and 4°C for the long and short RNAs, respectively. Incubation temperatures and buffer conditions were optimized to establish appropriate binding conditions and reproducibility between long or short RNAs and BsHfqs. Protein-bound long and short RNAs were separated from free RNA by 6% or 8% PAGE using acrylamide:bisacrylamide (40:1) in 1 × and 0.5 × TEB (Tris–borate–EDTA) buffer, respectively. Non-denaturing gel electrophoresis proceeded for 60–70 min at 100 V/cm on 100 × 100 mm plates at an ambient temperature of 4°C. Bands containing RNA on the gels were visualized by staining with ethidium bromide or Toluidinblau O (Chroma Gesellschaft Schmidt & Co.).
Footprinting assay
We prepared 5′- or 3′- 32P-labeled RNAs (0.07 or 0.3 pmol) in RNA structure buffer (Ambion) containing 1 µg of yeast tRNA in the absence or presence of 0.58 pmol BsHfq–His. The samples were incubated at 37°C for 30 min before digestion with RNases in a reaction volume of 10 µl. Each sample was then adjusted with either 2 µl of 0.003 U/ml RNase T1 (Ambion), 2 µl of 0.0005 U/ml RNase V1 (Ambion) or 2 µl of 5 U/ml RNase S1 (Ambion) at 37°C for 5 min. The reaction samples were quenched by adding 20 µl of inactivation/precipitation buffer (Ambion) at −20°C for 1 h, separated by centrifugation at 20 000 g at 4°C for 20 min, and then pellets were washed with 70% ethanol. Dried pellets were dissolved in gel loading buffer II (Ambion) and then the digests were separated by 10% PAGE under denaturing conditions with 8 M Urea, and detected by autoradiography (Fujifilm).
UV cross-link
Samples of 32P-RNA and BsHfq–His were incubated at 37°C for 30 min in binding buffer A containing 1 mM DTT and 5% glycerol. Concentrations of the samples in the binding buffer A were adjusted to 50 000 cpm/µl for the labeled RNA and 0.8 µM for BsHfq–His. The mixtures were exposed for 15 min to five 8 W germicidal lamps (254 nm wavelength) using UV Stratalinker 1800 (Stratagene) and then incubated with RNase A at 37°C for 10 min. Cross-linked samples were analyzed by 15% SDS–PAGE and visualized using the Bio-imaging Analyzer System (Fujifilm).
X-ray crystallography
GST-tagged protein was expressed and purified for crystal structure analysis and BsHfq–RNA complexes were crystallized as described previously (32). The crystal structures of BsHfq–RNA were resolved at BsHfq:RNA molar ratios of 1:1 and 1:2 by molecular replacement with a model based on the structure of S. aureus Hfq (1KQ2) using the program MOLREP (34) in the CCP4 package (35) and refined with the program REFMAC in the CCP4 package and CNS 1.2 (36). Atomic models were fitted into electron density maps using the graphics program XtalView/Xfit (37) and COOT (38). The quality of both crystal structures, assessed using PROCHECK (39), were as predicted or better for a structure at this resolution.
RESULTS
In vitro selection of RNA bound to BsHfq
To define a specific RNA sequence that BsHfq recognizes, RNA aptamers were obtained by nine SELEX cycles using nitrocellulose filter and magnetic beads. Table 1 summarizes the concentrations of BsHfq and RNAs for each cycle. Sequencing the 47 clones isolated from the RNA pool after nine selection cycles showed that the 22 RNA aptamers possessed AG repeats (AG)n where n ≥ 2. The other 25 RNA aptamers did not contain AG repeats.
Table 1.
Summary of in vitro selection of RNA aptamers
| Cycle | RNA (µM) | BsHfq (µM) | tRNA (µM) |
|---|---|---|---|
| 1 | 5.0 | 0.17 | |
| 2 | 2.5 | 0.08 | |
| 3 | 2.0 | 0.03 | |
| 4 | 2.0 | 0.03 | |
| 5 | 2.0 | 0.02 | 10 |
| 6 | 2.0 | 0.02 | 20 |
| 7 | 2.5 | 0.33 | |
| 8 | 2.0 | 0.33 | 20 |
| 9 | 2.0 | 0.02 | 20 |
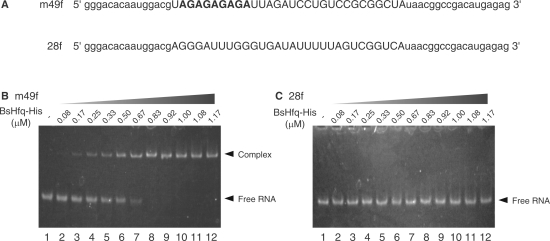
To elucidate how BsHfq recognizes AG repeats in RNA aptamers, we prepared 25 RNA aptamers with or without AG repeats by in vitro transcription using T7 RNA polymerase and investigated their ability to bind BsHfq using an EMSA (Supplementary Figure S1). Bands corresponding to RNA–protein complexes of aptamers containing AG repeats shifted, except for aptamer m36f that contained a (AG)2A sequence but did not bind BsHfq. In contrast, bands for aptamers without AG repeats did not shift (m4f, m18f and m33f in Supplementary Figure S1). These findings indicated that the AG repeat plays a key role in BsHfq binding. Figure 1 shows the sequences of RNA aptamers with high binding affinity for BsHfq in EMSA. Analysis of the aligned sequences indicated that all A residues in the sequence of the AG repeat were strictly conserved, whereas G residues could be replaced with A or U residues. To reconfirm the relationship between AG repeats and BsHfq binding ability using EMSA, we used aptamer m49f of which the binding affinity of the (AG)4A sequence was the highest among the aptamers and 28f isolated from the SELEX round 6 pool that contained no AG repeats (Figure 2A). The amount of the m49f RNA–protein complex obviously increased with increasing amounts of BsHfq (Figure 2B). The band corresponding to free RNA (m49f) disappeared at a protein:RNA molar ratio of 1:1 (Figure 2B, lane 10), which agreed with the experimental finding that bacterial Hfqs form hexamers (2,4,25,30,40). The apparent dissociation constant for the complex of m49f and BsHfq estimated by the ImageJ program (National Institutes of Health, Bethesda, MD) was approximately 10 nM. Gel bands corresponding to free RNA in the case of 28f were not shifted despite the presence of an equimolar amount of BsHfq (Figure 2C), indicating the absence of affinity for binding BsHfq. These results indicate that an AG repeat sequence is necessary for the specific binding of BsHfq to its target RNA.
Figure 1.
Random sequence regions of selected aptamers with high binding affinity for BsHfq determined by EMSA. Names of individual clones are left of sequences. Shared sequence motif (AG repeats) is shown in bold. Multiple sequences were aligned using ClustalW and manually edited.
Figure 2.
Affinity and specificity of BsHfq-binding RNA. (A) RNA sequences isolated by SELEX: m49f (round 9) and 28f (round 6). Nucleotides comprising AG repeats highlighted in bold. Nucleotides in lower-case letters indicate constant flanking sequences. EMSA of m49f (B) and 28f (C) (1 µM each) titrated with increasing amounts of BsHfq–His to determine binding to BsHfq.
Enzymatic footprinting assays
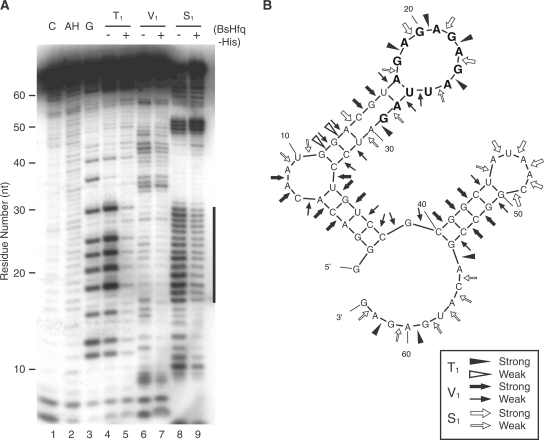
The results of the SELEX experiment suggested (AG)n (n ≥ 2) as a candidate sequence for BsHfq binding. To identify protein interaction sites in RNA aptamers, RNA secondary structure models for several RNA aptamers were predicted using the MFOLD program package (41). All predicted secondary structure models indicated that the AG repeat sequence adopts a loop structure (Supplementary Figure S2). We then performed a ribonuclease probing experiment using 32P-5′- or 3′- m49f digested with RNase T1, RNase V1 and RNase S1 (Figure 3). The RNase digestion data showed that the AG repeat region forms a single-stranded structure (Figure 3A, lanes 4 and 8). Figure 3B shows the most probable secondary structure of m49f resulting from the enzymatic cleavage determined by probing analysis and secondary structure prediction. Footprinting analysis of 32P-5′-m49f with BsHfq using RNase T1 and S1 located protected residues in a region containing the AG repeat sequence [Figure 3A (lanes 5 and 9) and B]. Residues in this region were also protected by BsHfq in the experiment using 32P-3′-m49f (data not shown). Consequently, BsHfq binds to the loop structure formed by the AG repeat sequence.
Figure 3.
Identification of BsHfq-binding site on RNA aptamer m49f. (A) Enzymatic footprinting of BsHfq bound to m49f. Cleavage products of 32P-5′- m49f modified by RNase T1, V1 and S1 with or without BsHfq-His were analyzed by denaturing gel electrophoresis. Lanes C, AH and G represent untreated RNA, alkaline hydrolysis ladder and G ladder, respectively. The vertical line on the right of the gel indicates protected residues that are resistant to RNase digestion in the presence of BsHfq-His. (B) Summarized results of enzymatic probing analysis of RNA aptamer m49f. Secondary structure was obtained by combining experimental results with MFOLD predictions (41). Major and minor cuts are indicated as symbols (see box). Residues corresponding to vertical line in (A) are highlighted in bold.
Binding affinity of mutant RNAs for BsHfq
We constructed mutant RNAs based on the sequence of aptamer m49f (hereafter referred to as 49R, Figure 4A) to determine the importance of residues in AG repeat sequences for BsHfq binding. We initially examined the BsHfq-binding affinities of three mutants in which GA residues were replaced with CC residues (named 49R-mt1, 49R-mt2 and 49R-mt3; Figure 4A). Mutant RNAs were labeled with 32P, exposed to short-wavelength UV light in the presence of BsHfq and then analyzed by SDS–PAGE (Figure 4B). The intensity of bands corresponding to three 32P-labeled mutants (49R-mt1, 49R-mt2 and 49R-mt3) was apparently reduced, compared with that of 49 R that contains an (AG)4A sequence (Figure 4B, lanes 1–4). The EMSA results showed that the mutants cannot form stable complexes with BsHfq even in the case of 49R-mt1 (data not shown). These findings indicated that reducing the number of AG residue repeats decreased their protein binding affinity.
Figure 4.
Effects of RNA mutations on RNA binding to BsHfq. (A) Mutant RNA sequences (bold type) constructed for UV cross-linking analysis. Tag sequence was added to 3′- and 5′-ends of RNA (italic) for in vitro transcription. AG repeat sequence is underlined. (B) UV cross-linking analysis of mutant RNA binding to BsHfq. 32P-RNA (50 000 cpm) was incubated with 8.3 pmol of BsHfq-His and then loaded on gels. Lanes 1 and 5, BsHfq–His and 49R; lanes 2–4, BsHfq–His and mutants with GA(s) in AG repeat sequence replaced by CC(s) (49R-mt1, 49R-mt2 and 49R-mt3); lanes 6–9, BsHfq–His with mutants in which G in AG repeat sequence was replaced by U/A/C/GG (49R-U, 49R-A, 49R-C and 49R-GG).
The sequence of aptamers obtained from the SELEX experiment also indicated that G residues in the AG repeat sequence can be replaced by either A or U residues (Figure 1). The G residue at the sixth position of the AG repeat sequence is in fact, frequently substituted in this manner. To determine the nucleotide sequence specificity for the binding of these aptamers by BsHfq, we generated four more mutants based on the 49 R sequence (bottom of Figure 4A); the G residue at the sixth position of the AG repeat sequence of 49 R was replaced with U, A, C or GG residues and the mutants were named 49R-U, 49R-A, 49R-C and 49R-GG, respectively. The results of UV cross-linking indicated that mutant 49R-A and 49 R formed similarly stable complexes (Figure 4B, lanes 5 and 7). However, the 32P signal intensity of 49R-U, 49R-C and 49 R-GG was reduced, indicating that these mutants do not easily form complexes (Figure 4B, lanes 6, 8 and 9). The results with 49 R and mutant proteins in EMSA were the same (data not shown). Thus, dinucleotide repeat motifs (AG) play an important role in the efficiency of RNA aptamer binding to BsHfq.
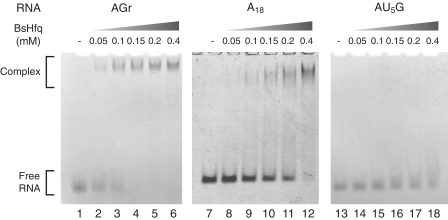
We constructed a series of RNA fragments by deleting residues from the 3′-end of the 9-mer 5′-(AG)4A-3′ and examined interactions between the fragments and BsHfq using EMSA to analyze crystal structures. The protein binding ability of fragments that were shorter than 7-mer (AG)3A, was reduced compared with that of 7-, 8- and 9-mer fragments (data not shown). Therefore, the minimal RNA fragment suitable for structure analysis was defined as a 7-mer (AG)3A (hereafter referred to as AGr). In practice, EMSA with the AGr fragment showed that the intensity of the shifted bands increased with increasing protein concentrations (Figure 5, lanes 2–6). Furthermore, adding equimolar or excess amounts of BsHfq led to significantly weakened bands for free RNA. The apparent dissociation constant for the binding of AGr to BsHfq estimated by the ImageJ software was ~1 μM. The same sequence (AGAGAGA) was present in both AGr and 49R-mt1, although 49R-mt1 contains an additional sequence outside the AG repeat sequence. However, 49R-mt1 had lower BsHfq-binding affinity than AGr (data not shown). This finding suggests that the additional sequence in 49R-mt1 inhibits the binding of 49R-mt1 to BsHfq.
Figure 5.
Affinity of BsHfq for RNAs with sequences AGr, A18 and AU5G. RNA (0.1 mM) was titrated with increasing amounts of BsHfq.
Escherichia coli Hfq and SaHfq reportedly bind to A18 and AU5G, respectively (25,31,42). We performed EMSAs to determine whether BsHfq can bind to these short RNAs (Figure 5). The band corresponding to free RNA disappeared when the concentration of BsHfq was 4-fold higher than that of A18 (Figure 5, lane 12). This indicates that BsHfq can interact with A18; however, the dissociation constant for the A18 binding to BsHfq was ~10 μM, indicating that BsHfq has weaker binding affinity for A18 than for AGr. Moreover, a dissociation constant of 10 nM has been established between EcHfq and A18 (42), indicating stronger binding than that between BsHfq and A18. In contrast, EMSA showed that AU5G did not bind BsHfq (Figure 5, lanes 14–18). Taken together, these findings demonstrated that the AGr fragment possesses significant BsHfq-binding affinity.
Structure of BsHfq–RNA complex
We experimentally verified that AGr has higher binding affinity than A18 for BsHfq, as described above. To analyze the molecular recognition pattern of BsHfq for AG repeats, we determined the structures of the BsHfq–AGr complex by X-ray crystallography. Crystals of BsHfq–AGr were obtained under molar ratios of 1:1 (co-crystal 1) and 1:2 (co-crystal 2) (32). The space groups of these crystals were I422 and F222 for co-crystals 1 and 2, respectively. The asymmetric unit of co-crystal 1 contained six monomers of BsHfq and one molecule of RNA, that of co-crystal 2 contained three monomers of BsHfq and one molecule of RNA (32). Crystal structures of co-crystals 1 and 2 were determined at 2.2 Å resolution by molecular replacement using a polyalanine model of SaHfq (PDB code: 1KQ2) (25), and refined to R/Rfree values of 21.4/22.8% and 25.9/26.0%, respectively (Table 2).
Table 2.
Diffraction data and refinement statistics for BsHfq–AGr complexes
| Data collection statistics | Co-crystal 1 | Co-crystal 2 |
|---|---|---|
| Date collection | ||
| Space group | I422 | F222 |
| Unit cell parameters (Å) | a = b = 123.70, c = 119.13 | a = 91.92, b = 92.50, c = 114.92 |
| Wavelength (Å) | 1.0000 | 1.0000 |
| Resolution (Å) | 25.00–2.20 (2.28–2.20) | 30.00–2.20 (2.28–2.20) |
| Multiplicity | 14.8 (14.9) | 7.4 (7.4) |
| Number of observations | 350 287 | 92 082 |
| Number of unique reflections | 23 732 | 12 626 |
| Completeness (%) | 100 (100) | 100 (100) |
| Rmarge (%)a | 6.3 (32.5) | 3.3 (30.7) |
| Average I/ σ(I) | 38.02 (7.07) | 45.75 (4.84) |
| Matthews coefficient (Å3 Da−1) | 2.34 | 2.51 |
| Solvent content (%) | 48 | 51 |
| Refinement | ||
| Data range (Å) | 25.00–2.20 | 30.00–2.20 |
| R-factor (%) | 21.4 | 25.9 |
| Rfree (%) | 22.8 | 26.0 |
| Number of protein atoms | 3333 | 1632 |
| Number of RNA atoms | 154 | 132 |
| Number of solvent atoms | 138 | 25 |
| RMSD of bond lengths (Å) | 0.022 | 0.035 |
| RMSD of bond angles (°) | 2.004 | 2.957 |
| Ramachandran Plot | ||
| Most favored regions | 93.1 | 88.7 |
| Additional allowed regions | 6.6 | 10.2 |
| Generously allowed regions | 0.3 | 1.1 |
| Disallowed regions | 0.0 | 0.0 |
aRmerge = Σhkl Σi | Ii(hkl) − <I(hkl)> | / Σhkl ΣiIi(hkl). Values in parentheses are for the highest resolution shell.
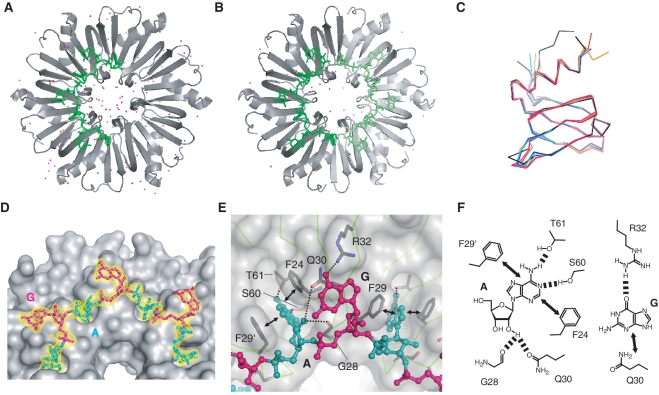
The quaternary structure of BsHfq in these complexes was a homohexameric ring and each subunit of homohexameric BsHfq contained an Sm-like domain fold consisting of an N-terminal α-helix followed by a five-stranded antiparallel β-barrel (Figure 6A, B and C), as found in other bacterial and archaeal Hfqs (25–30). The superposition of these monomer structures between co-crystals 1 and 2 yielded a low RMSD value of 0.27 Å for backbone (N, Cα, C) atoms of secondary structure elements. The AGr bound to the distal site of BsHfq with a circular conformation (Figure 6), although electron densities in a pocket surrounded by F41 and its neighboring residues at the proximal site could not be interpreted. A full-length AGr (7-mer) was identified in co-crystal 1. On the other hand, a 3′-end A residue of AGr was not found in co-crystal 2; instead, a 5′-end A residue from neighboring AGr molecule was found in that location. In this manner, we found that although the observed length of AGr differed between co-crystals 1 (7-mer) and 2 (6-mer), the nucleotide binding patterns were the same for both crystals except for the 3′-end A residue. All riboses in AGr exhibited the C2′-endo form. The A and G residues in AGr adopted anti and syn glycosidic bond conformations, respectively (Figures 6A and B), although one G residue in co-crystal 2 adopted the anti conformation because of intermolecular crystal contact with Y20 from a neighboring molecule. Both structures revealed that A bases in the anti form were inserted into the binding pockets formed by two aromatic rings of F24 and F29, and that BsHfq was recognized by π–π stacking interactions and four hydrogen bonding interactions (A N1 – S60 γOH, A N6 (NH2) – T61 γO, A OH2′ – G28 CO and A OH2′ – Q30 εO1) (Figure 6D, E and F). All three G bases in the syn form in co-crystal 1 formed a stacking interaction with the side chain of Q30 and two of them were involved in a hydrogen bonding interaction between O6 of G base and ηNH2 of R32 (Figure 6E and F). The G base in the anti form in co-crystal 2 mentioned above displayed neither the stacking interaction with Q30 nor the hydrogen bonding interaction with R32. The remaining two G bases in the syn form in co-crystal 2 revealed stacking interactions with the side chain of Q30, but only one of them was engaged in hydrogen bonding interaction with the side chain of R32. Notably, the atomic coordinates of the side chain of R32 were not clearly determined due to an ambiguous electron density. As a result, except for the hydrogen bonding interaction of the G base, co-crystals 1 and 2 represented a common interaction mode between dinucleotide repeat motifs (AG) and BsHfq, namely two stacking and four hydrogen bonding interactions for the A base and one stacking interaction for the G base.
Figure 6.
Overall structures of BsHfq and the RNA aptamer, AGr. (A) Quaternary structure of BsHfq–AGr in co-crystal 1. One RNA molecule binds to BsHfq hexamer. Water molecules are shown as magenta spheres. (B) Quaternary structure of BsHfq–AGr in co-crystal 2. Symmetry-related molecule in BsHfq–AGr is shown in light gray and light green. Two RNA molecules bind to one BsHfq hexamer. Water molecules are shown as magenta and gray spheres. (C) Superposition of Cα trace of subunits from several bacterial Hfqs. Co-crystals 1 and 2 are shown in black and gray, respectively. SaHfq (PDB ID: 1KQ1) is shown in magenta, SaHfq–AU5G (PDB ID: 1KQ2) in red, EcHfq (PDB ID: 1HK9) in cyan, EcHfq–poly(A) (PDB ID: 3GIB) in blue and P. aeruginosa Hfq (PDB ID: 1U1S) in orange. Pairwise root-mean-square deviations of backbone (N, Cα, C) atoms for secondary structure elements in these subunits from co-crystal 1 structure were <0.36 Å. (D) BsHfq and AGr are shown as gray surface with cyan (A residue) or magenta (G residue) sticks, respectively. Part of 2Fo–Fc electron density map (yellow) contoured at 1.5 σ is shown for AGr. (E) Close-up view of BsHfq–AGr interface. BsHfq is shown as semi-transparent molecular surface (light gray). Polypeptide backbone is drawn as line representation (light green), while AGr is shown as ball-and-stick representations and colored as in (D). Side chains involved in RNA-binding are labeled and are shown as gray bonds colored according to atom type. Hydrogen bonding and stacking interactions between RNA and Hfq are shown as dashed lines and arrows, respectively. (F) Schematic representations of interactions between BsHfq and A/G residues in AGr. Dashed lines and arrows indicate hydrogen bonding and stacking interactions, respectively.
Effects of point mutations in BsHfq on binding to the RNA aptamer
The crystal structures of BsHfq–AGr showed that AGr binds to the distal site of BsHfq. On the other hand, the electron density was ambiguous in the proximal site of BsHfq, rendering feasible the notion that the AG repeat sequence also binds to the proximal site. Therefore, we examined whether AG repeats (49 R) specifically interact with the distal or proximal site of BsHfq. Based on the information gained from RNA–protein interaction networks in the distal site of BsHfq and in the proximal site of SaHfq (25), we constructed mutant BsHfqs containing a single amino acid substitution in the distal (F24A, F29A, Q30A and R32A) and proximal (N40A, F41A, K56A and H57A) sites (Figure 7A). We then analyzed all of the mutant proteins and 49 R RNA using EMSA (Figure 7B). Shifted bands corresponding to RNA–protein complexes were not evident for mutant proteins with substituted residues on the distal site (F24A, F29A, Q30A and R32A) (Figure 7B, lanes 3–6), indicating that the four mutant proteins lose the ability to bind 49 R RNA. This observation suggests that hydrogen bonding interaction between atoms O6 of G base and ηNH2 of R32 is needed for AGr recognition by BsHfq (Figure 6E and F). Notably, the four residues are highly conserved among Gram-positive bacteria (Supplementary Figure S3). On the other hand, shifted bands for mutant proteins N40A and F41A in which residues on the proximal site were substituted, were similar to those of the wild-type (Figure 7B, lanes 2, 7 and 8). Electrophoretic bands of free RNA were smeared due to the dissociation of complexes with mutant proteins K56A and H57A in which residues on the proximal site were substituted (Figure 7B, lanes 1, 9 and 10). Gel filtration chromatography and SDS–PAGE analysis revealed that the ability of K56A and H57A to form hexameric structure was reduced to ~60 and 30% of that of the wild-type, respectively, and that mutant proteins were more sensitive to denaturation by SDS than the wild-type (peak $ in Figure 7C and D, lanes 2, 4 and 5). Consequently, the dissociation of complexes with K56A and H57A was caused by disruption of the hexameric formation of BsHfq. On the other hand, wild-type and mutant proteins, except for K56A and H57A, adopted a hexameric structure in solution since their ability to form the hexameric structure was maintained and the proteins were insensitive to denaturation by SDS (peak $ in Figure 7C and D, lanes 2 and 3). Taken together, only the distal site of BsHfq possesses an AG repeat-containing RNA-binding site.
Figure 7.
Point mutational analysis of distal and proximal sites of BsHfq. (A) Surface representations of distal (left panel) or proximal (right panel) sites of BsHfq. Mutated residues are labeled and their side chains are shown in cyan. These figures were generated using PyMOL software. (B) EMSA of wild-type and mutant BsHfq binding to RNA (49R). RNA (1 µM) was incubated in the absence (lane 1) or presence of 1.5 µM wild-type (WT, lane 2) or mutant (lanes 3–10) BsHfqs. (C) Gel filtration profiles of wild-type (black), F29A (magenta), K56A (cyan) and H57A (green) mutants. Peaks corresponding to BsHfq and lysozyme are indicated as $ and #, respectively. (D) A coomassie-stained SDS acrylamide gel shows wild-type and mutant BsHfqs. Samples collected from peak $ of gel filtration were resolved by SDS–PAGE. Plus and minus symbols on the top of the gel indicate samples boiled or not in SDS buffer for 10 min at 95°C, respectively. Apparent molecular weights of monomeric and hexameric BsHfqs are ~9.6 and 58 kDa, respectively.
DISCUSSION
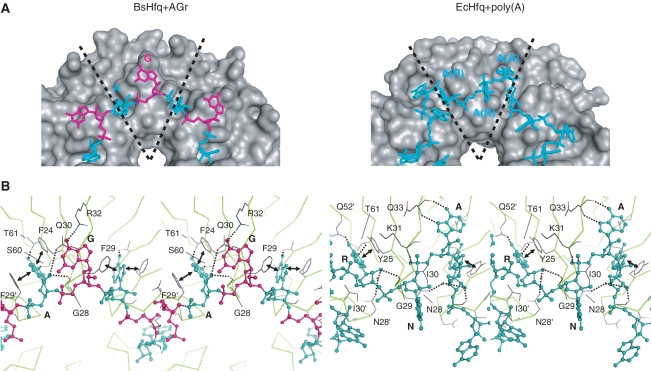
Mutagenesis studies of EcHfq and structure analysis of the EcHfq–poly(A) complex have revealed that the distal site of EcHfq is used to bind to poly(A) (30,31,42). The superposition of the hexameric Hfqs of EcHfq–poly(A) and BsHfq–AGr results in the RMSD of 0.44 Å for backbone (N, Cα, C) atoms of secondary structure elements. Moreover, studies of the electrostatic surface potentials of EcHfq (30) and BsHfq have revealed that the distal and proximal sites of these Hfqs are all positively charged (Supplementary Figure S4). However, the sequences and structures differ between RNA-binding sites at the distal site of EcHfq–poly(A) and BsHfq–AGr, and they produce distinct binding patterns, as shown below. Firstly, Hfq-bound RNAs in the BsHfq–AGr conformationally differ from those in EcHfq–poly(A). At least both the R and A sites (where R is a purine nucleotide) are necessary for poly(A) recognition in EcHfq (30), and a residue at an N site (where N is any nucleotide) is therefore likely to be essential to connect to nearby residues at R and A sites. Hence, EcHfq might recognize A-R-N trinucleotide as the smallest unit of poly(A) (right panel in Figure 8A). On the other hand, we demonstrated that residues at A (F24 and F29) and G (Q30 and R32) sites are absolutely required for specific RNA binding at the distal site in BsHfq (Figure 7). Thus, BsHfq interacts with AG dinucleotide as the smallest unit of the AG repeat motif (left panel in Figure 8A). As a consequence, both RNA shapes adopted a circular conformation, whereas poly(A) in EcHfq assumed a waving conformation, and then the N base (the base in the N site) in EcHfq is exposed to the solvent (Figure 8). The RNA–protein binding patterns differed between BsHfq–AGr and EcHfq–poly(A). The G base at the G site of BsHfq formed a hydrogen bonding interaction with the side chain of R32 (left panel in Figure 8B). Mutational analysis also revealed that R32 is critical for interaction with the AG repeat motif (Figure 7). The residue corresponding to R32 is Q33 in EcHfq (right panel in Figure 8B), and the side chain of Q33 does not interact with the N base in EcHfq (30). Instead, the interaction of Q33 of EcHfq with the A base (the base in the A site) is important for poly(A) binding (30), although this binding pattern does not exist in BsHfq (Figure 8B). Notably, R32 of BsHfq and Q33 of EcHfq are highly conserved among Gram-positive and Gram-negative bacteria, respectively (Supplementary Figure S3). These findings indicate that the binding pattern of residues at G site to target RNA is specific to BsHfq and not EcHfq. Lastly, both the A base of BsHfq and the R base (the base in a R site) of EcHfq are inserted into the pocket formed by residues corresponding to F24 and F29 of BsHfq. However, distinct binding pattern of hydrogen bonding interactions in BsHfq and EcHfq directly influence specific structural features inside the pocket. The A base of BsHfq forms hydrogen bonding interactions with S60 and T61 (Figures 6E, and F and 8B), whereas the R base of EcHfq does so with Q52 and T61 (30). In particular, the S60 residue of BsHfq locates near the bottom of the pocket, whereas the Q52 residue locates outside the pocket (Figure 8B). Thus, only the binding pattern of S60 in BsHfq allows insertion of the A base deeply into the pocket without steric clashes. These results show that the distal site of BsHfq can selectively bind to AGr in a sequence- and/or structure-specific manner.
Figure 8.
Molecular recognition of RNA by distal site of Hfq. (A) Left panel: surface representation of hexameric BsHfq (gray) with stick representation of A (cyan) and G (magenta) residues of AGr. Right panel: surface representation of hexameric EcHfq (gray) with stick representation of poly(A) (cyan). Subunit boundaries are indicated by dashed lines. (B) Stereo view of BsHfq–AGr and EcHfq–poly(A) (left and right panel, respectively). Molecules are shown using same representation and color scheme as shown in Figure 6E. R and N indicate purine nucleotide and any nucleotide, respectively. These figures were generated using PyMOL software.
Co-crystals 1 and 2 had some ambiguous, but non-protein electron density at the proximal site, especially in a pocket surrounded by F41 and its neighboring residues (Supplementary Figure S5). The electron densities cannot be interpreted as AGr, because our mutational analysis revealed that AGr does not interact with the proximal site of BsHfq (Figure 7). Moreover, we confirmed that the BsHfq used in this study contained no residual nucleic acid after purification by the Warburg–Christian method (43). A positively charged region in the proximal site of SaHfq appears to interact with small U-rich RNA (AU5G) (25) (Supplementary Figure S4). Moreover, AU5G-bound amino acid residues in the proximal site were identified by the structure of SaHfq–AU5G complex (25). Structure-based sequence alignment revealed that AU5G-bound amino acid residues and the conformations of their side chains in SaHfq are also conserved in BsHfq (Supplementary Figures S3 and S5). Thus, BsHfq is likely to bind to AU5G, but in fact, AU5G did not bind to BsHfq (Figure 5). On the other hand, the crystal structure of Salmonella typhimurium Hfq (StHfq) in complex with U6 RNA has recently been determined, in which U6 RNA interacts with the proximal site of Hfq (44), and a free 3′ hydroxyl group of RNA is crucial for high-affinity binding to StHfq. In particular, the 3′-terminal guanosine in RNA obviously reduced StHfq binding. Thus, we speculate that the absence of interactions between AU5G and BsHfq could be due to the guanosine residue at the 3′-end of AU5G. Therefore, these findings raise the notion that BsHfq still associates with some RNA(s) through the positively charged proximal site, as found in SaHfq or EcHfq (25,31) (Supplementary Figure S4). However, whether or how the proximal site of BsHfq recognizes RNA(s) remains unclear.
Hfq in Gram-negative bacteria functions as a post-transcriptional regulator that acts by mediating interactions between many sRNAs and their mRNA targets. On the other hand, BsHfq and SaHfq that are both from Gram-positive bacteria interacted with different RNA sequences (mRNA and/or sRNA) via structurally distinct RNA-binding surfaces (distal or proximal site). BsHfq-binding sequences in SR1 sRNA and ahrC mRNA have been elucidated (24), and they are involved in short AG repeats GAAUAAGAGA (SR1 sRNA) or AAAUAGAG (ahrC mRNA). Therefore, these results when combined with the findings of the SELEX experiments indicated that the target sequence for BsHfq certainly comprises AG repeats. Taken together with these data, the results presented herein suggest that SR1 and ahrC mRNA recognize the same distal site of BsHfq. We further examined whether the intergenic regions of B. subtilis 168 genome include the AGr sequence using the SubtiList database (http://genolist.pasteur.fr/SubtiList/). We identified the sequence in 110 intergenic regions and in 215 protein coding regions, indicating that the AGr sequence does in fact exists in the B. subtilis genome.
Amino acid residues on the surface of the distal site, including those that bind to AGr in BsHfq, are highly conserved in Gram-positive bacteria such as S. aureus and L. monocytogenes (Supplementary Figure S3). Moreover, lmo0850 mRNA, which binds to L. monocytogenes Hfq, also possesses AG repeats (45). Therefore, we speculate that several other Hfqs might function through interaction with the AG repeat motif among Gram-positive bacteria. To date, various RNA target sequences have been identified and they can discriminate the RNA-binding surface(s) of hexameric Hfq at the atomic level. Furthermore, multiple functional roles of Hfq have been identified in several bacterial species, probably as a consequence of this. Hence, bacterial Hfqs might have species-specific functions among bacterial phyla. To understand Hfq activities, recognition patterns between RNA(s) and Hfq across various biological species should be further elucidated in future studies from both structural and functional perspectives.
ACCESSION NUMBERS
Coordinates and structure factors for the co-crystal 1 and co-crystal 2 of BsHfq–AGr have been deposited with the Protein Data Back under accession codes 3HSB and 3AHU.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR online: Supplementary Table 1, Supplementary Figures 1–5.
FUNDING
Funding for open access charge: University of Tsukuba.
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Ms S. Tamura, S. Suzuki, and A. Shino for excellent technical assistance and the staff of SPring-8 BL38B1 (2008B1506 and 2011A2043) for help with collecting the X-ray data. We also thank Ms N. Foster for critical reading of the manuscript.
REFERENCES
- 1.Franze de Fernandez MT, Hayward WS, August JT. Bacterial proteins required for replication of phage Qβ ribonucleic acid. Purification and properties of host factor I, a ribonucleic acid-binding protein. J. Biol. Chem. 1972;247:824–831. [PubMed] [Google Scholar]
- 2.Zhang A, Wassarman KM, Ortega J, Steven AC, Storz G. The Sm-like Hfq protein increases OxyS RNA interaction with target mRNAs. Mol. Cell. 2002;9:11–22. doi: 10.1016/s1097-2765(01)00437-3. [DOI] [PubMed] [Google Scholar]
- 3.Sledjeski DD, Whitman C, Zhang A. Hfq is necessary for regulation by the untranslated RNA DsrA. J. Bacteriol. 2001;148:1997–2005. doi: 10.1128/JB.183.6.1997-2005.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Møller T, Franch T, Højrup P, Keene DR, Bächinger HP, Brennan RG, Valentin-Hansen P. Hfq: a bacterial Sm-like protein that mediates RNA-RNA interaction. Mol. Cell. 2002;9:23–30. doi: 10.1016/s1097-2765(01)00436-1. [DOI] [PubMed] [Google Scholar]
- 5.Geissmann TA, Touati D. Hfq, a new chaperoning role: binding to messenger RNA determines access for small RNA regulator. EMBO J. 2004;23:396–405. doi: 10.1038/sj.emboj.7600058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Morita T, Maki K, Aiba H. RNase E-based ribonucleoprotein complexes: mechanical basis of mRNA destabilization mediated by bacterial noncoding RNAs. Genes Dev. 2005;19:2176–2186. doi: 10.1101/gad.1330405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Moll I, Leitsch D, Steinhauser T, Bläsi U. RNA chaperone activity of the Sm-like Hfq protein. EMBO Rep. 2003;4:284–289. doi: 10.1038/sj.embor.embor772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Večerek B, Moll I, Bläsi U. Translational autocontrol of the Escherichia coli hfq RNA chaperone gene. RNA. 2005;11:976–984. doi: 10.1261/rna.2360205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hajnsdorf E, Régnier P. Host factor Hfq of Escherichia coli stimulates elongation of poly(A) tails by poly(A) polymerase I. Proc. Natl Acad. Sci. USA. 2000;97:1501–1505. doi: 10.1073/pnas.040549897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mohanty BK, Maples VF, Kushner SR. The Sm-like protein Hfq regulates polyadenylation dependent mRNA decay in Escherichia coli. Mol. Microbiol. 2004;54:905–920. doi: 10.1111/j.1365-2958.2004.04337.x. [DOI] [PubMed] [Google Scholar]
- 11.Folichon M, Allemand F, Régnier P, Hajnsdorf E. Stimulation of poly(A) synthesis by Escherichia coli poly(A) polymerase I is correlated with Hfq binding to poly(A) tails. FEBS J. 2005;272:454–463. doi: 10.1111/j.1742-4658.2004.04485.x. [DOI] [PubMed] [Google Scholar]
- 12.Folichon M, Arluison V, Pellegrini O, Huntzinger E, Régnier P, Hajnsdorf E. The poly(A) binding protein Hfq protects RNA from RNase E and exoribonucleolytic degradation. Nucleic Acids Res. 2003;31:7302–7310. doi: 10.1093/nar/gkg915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Afonyushkin T, Večerek B, Moll I, Bläsi U, Kaberdin VR. Both RNase E and RNase III control the stability of sodB mRNA upon translational inhibition by the small regulatory RNA RyhB. Nucleic Acids Res. 2005;33:1678–1689. doi: 10.1093/nar/gki313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chao Y, Vogel J. The role of Hfq in bacterial pathogens. Curr. Opin. Microbiol. 2010;13:24–33. doi: 10.1016/j.mib.2010.01.001. [DOI] [PubMed] [Google Scholar]
- 15.Tsui H-CT, Leung HCE, Winkler ME. Characterization of broadly pleiotropic phenotypes caused by an hfq insertion mutation in Escherichia coli K-12. Mol. Microbiol. 1994;13:35–49. doi: 10.1111/j.1365-2958.1994.tb00400.x. [DOI] [PubMed] [Google Scholar]
- 16.Sonnleitner E, Hagens S, Rosenau F, Wilhelm S, Habel A, Jäger KE, Bläsi U. Reduced virulence of a hfq mutant of Pseudomonas aeruginosa O1. Microb. Pathog. 2003;35:217–228. doi: 10.1016/s0882-4010(03)00149-9. [DOI] [PubMed] [Google Scholar]
- 17.Pannekoek Y, Huis in't Veld R, Hopman CTh P, Langerak AAJ, Speijer D, van der Ende A. Molecular characterization and identification of proteins regulated by Hfq in Neisseria meningitidis. FEMS Microbiol. Lett. 2009;294:216–224. doi: 10.1111/j.1574-6968.2009.01568.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ding Y, Davis BM, Waldor MK. Hfq is essential for Vibrio cholerae virulence and downregulates sigma expression. Mol. Microbiol. 2004;53:345–354. doi: 10.1111/j.1365-2958.2004.04142.x. [DOI] [PubMed] [Google Scholar]
- 19.Bohn C, Rigoulay C, Bouloc P. No detectable effect of RNA-binding protein Hfq absence in Staphylococcus aureus. BMC Microbiol. 2007;7:10. doi: 10.1186/1471-2180-7-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Christiansen JK, Larsen MH, Ingmer H, Søgaard-Andersen L, Kallipolitis BH. The RNA-binding protein Hfq of Listeria monocytogenes: Role in stress tolerance and virulence. J. Bacteriol. 2004;186:3355–3362. doi: 10.1128/JB.186.11.3355-3362.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Christiansen JK, Nielsen JS, Ebersbach T, Valentin-Hansen P, Søgaard-Andersen L, Kallipolitis BH. Identification of small Hfq-binding RNAs in Listeria monocytogenes. RNA. 2006;12:1383–1396. doi: 10.1261/rna.49706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Silvaggi JM, Perkins JB, Losick R. Small Untranslated RNA antitoxin in Bacillus subtilis. J. Bacteriol. 2005;187:6641–6650. doi: 10.1128/JB.187.19.6641-6650.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Heidrich N, Chinali A, Gerth U, Brantl S. The small untranslated RNA SR1 from the Bacillus subtilis genome is involved in the regulation of arginine catabolism. Mol. Microbiol. 2006;62:520–536. doi: 10.1111/j.1365-2958.2006.05384.x. [DOI] [PubMed] [Google Scholar]
- 24.Heidrich N, Moll I, Brantl S. In vitro analysis of the interaction between the small RNA SR1 and its primary target ahrC mRNA. Nucleic Acids Res. 2007;35:4331–4346. doi: 10.1093/nar/gkm439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Schumacher MA, Pearson RF, Møller T, Valentin-Hansen P, Brennan RG. Structures of the pleiotropic translational regulator Hfq and an Hfq-RNA complex: a bacterial Sm-like protein. EMBO J. 2002;21:3546–3556. doi: 10.1093/emboj/cdf322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sauter C, Basquin J, Suck D. Sm-like proteins in Eubacteria: the crystal structure of the Hfq protein from Escherichia coli. Nucleic Acids Res. 2003;31:4091–4098. doi: 10.1093/nar/gkg480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nikulin A, Stolboushkina E, Perederina A, Vassilieva I, Blaesi U, Moll I, Kachalova G, Yokoyama S, Vassylyev D, Garber M, et al. Structure of Pseudomonas aeruginosa Hfq protein. Acta. Crystallogr. D Biol. Crystallogr. 2005;61:141–146. doi: 10.1107/S0907444904030008. [DOI] [PubMed] [Google Scholar]
- 28.Nielsen JS, Bøggild A, Andersen CB, Nielsen G, Boysen A, Brodersen DE, Valentin-Hansen P. An Hfq-like protein in archaea: crystal structure and functional characterization of the Sm protein from Methanococcus jannaschii. RNA. 2007;13:2213–2223. doi: 10.1261/rna.689007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Boggild A, Overgaard M, Valentin-Hansen P, Brodersen DE. Cyanobacteria contain a structural homologue of the Hfq protein with altered RNA-binding properties. FEBS J. 2009;276:3904–3915. doi: 10.1111/j.1742-4658.2009.07104.x. [DOI] [PubMed] [Google Scholar]
- 30.Link TM, Valentin-Hansen P, Brennan RG. Structure of Escherichia coli Hfq bound to polyriboadenylate RNA. Proc. Natl Acad. Sci. USA. 2009;106:19292–19297. doi: 10.1073/pnas.0908744106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mikulecky PJ, Kaw MK, Brescia CC, Takach JC, Sledjeski DD, Feig AL. Escherichia coli Hfq has distinct interaction surfaces for DsrA, rpoS and poly(A) RNAs. Nat. Struct. Mol. Biol. 2004;11:1206–1214. doi: 10.1038/nsmb858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Baba S, Someya T, Kawai G, Nakamura K, Kumasaka T. Expression, crystallization and preliminary crystallographic analysis of RNA-binding protein Hfq (YmaH) from Bacillus subtilis in complex with an RNA aptamer. Acta. Crystallogr. Sect. F Struct. Biol. Cryst. Commun. 2010;66:563–566. doi: 10.1107/S1744309110009942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Conrad RC, Giver L, Tian Y, Ellington AD. In vitro selection of Nucleic Acid Aptamer that bind proteins. Methods Enzymol. 1996;267:336–367. doi: 10.1016/s0076-6879(96)67022-0. [DOI] [PubMed] [Google Scholar]
- 34.Vagin A, Teplyakov A. MOLREP: an automated program for molecular replacement. J. Appl. Cryst. 1997;30:1022–1025. [Google Scholar]
- 35.Collaborative Computational Project, Number 4. The CCP4 suite: programs for protein crystallography. Acta. Crystallogr. D Biol. Crystallogr. 1994;50:760–763. doi: 10.1107/S0907444994003112. [DOI] [PubMed] [Google Scholar]
- 36.Brünger AT, Adams PD, Clore GM, DeLano WL, Gros P, Grosse-Kunstleve RW, Jiang JS, Kuszewski J, Nilges M, Pannu NS, et al. Crystallography & NMR system: A new software suite for macromolecular structure determination. Acta. Crystallogr. D Biol. Crystallogr. 1998;54:905–921. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
- 37.McRee DE. XtalView/Xfit—A versatile program for manipulating atomic coordinates and electron density. J. Struct. Biol. 1999;125:156–165. doi: 10.1006/jsbi.1999.4094. [DOI] [PubMed] [Google Scholar]
- 38.Emsley P, Cowtan K. Coot: Model-building tools for molecular graphics. Acta. Crystallogr. D Biol. Crystallogr. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 39.Laskowski RA, Macarthur MW, Moss DS, Thornton JM. Procheck – a program to check the stereochemical quality of protein structures. J. Appl. Crystallogr. 1993;26:283–291. [Google Scholar]
- 40.Fender A, Elf J, Hampel K, Zimmermann B, Wagner EG. RNAs actively cycle on the Sm-like protein Hfq. Genes Dev. 2010;24:2621–2626. doi: 10.1101/gad.591310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zuker M. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 2003;31:3406–3415. doi: 10.1093/nar/gkg595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sun X, Wartell PM. Escherichia coli Hfq binds A18 and DsrA domain II with similar 2:1 Hfq6/RNA stoichiometry using different surface site. Biochemistry. 2006;45:4875–4887. doi: 10.1021/bi0523613. [DOI] [PubMed] [Google Scholar]
- 43.Stoscheck CM. Quantitation of protein. Methods Enzymol. 1990;182:50–68. doi: 10.1016/0076-6879(90)82008-p. [DOI] [PubMed] [Google Scholar]
- 44.Sauer E, Weichenrieder O. Structural basis for RNA 3′-end recognition by Hfq. Proc. Natl Acad. Sci. USA. 2011;108:13065–13070. doi: 10.1073/pnas.1103420108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Nielsen JS, Lei LK, Ebersbach T, Olsen AS, Klitgaard JK, Valentin-Hansen P, Kallipolitis BH. Defining a role for Hfq in Gram-positive bacteria: evidence for Hfq-dependent antisense regulation in Listeria monocytogenes. Nucleic Acids Res. 2010;38:907–919. doi: 10.1093/nar/gkp1081. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.