Abstract
Strategies to regulate gene function frequently use small interfering RNAs (siRNAs) that can be made from their shRNA precursors via Dicer. However, when the duplex components of these siRNA effectors are expressed from their respective coding genes, the RNA interference (RNAi) activity is much reduced. Here, we explored the mechanisms of action of shRNA and siRNA and found the expressed siRNA, in contrast to short hairpin RNA (shRNA), exhibits strong strand antagonism, with the sense RNA negatively and unexpectedly regulating RNAi. Therefore, we altered the relative levels of strands of siRNA duplexes during their expression, increasing the level of the antisense component, reducing the level of the sense component, or both and, in this way we were able to enhance the potency of the siRNA. Such vector-delivered siRNA attacked its target effectively. These findings provide new insight into RNAi and, in particular, they demonstrate that strand antagonism is responsible for making siRNA far less potent than shRNA.
INTRODUCTION
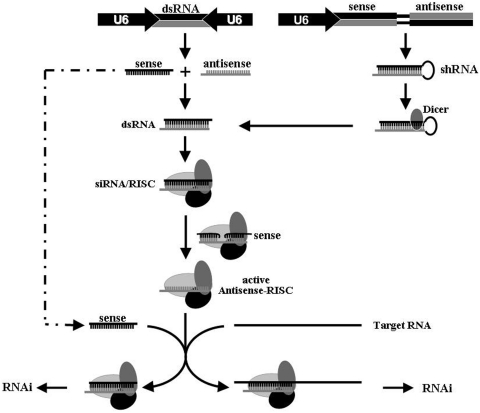
RNA interference (RNAi) is an RNA-dependent gene-silencing phenomenon and is initiated by ∼20-bp double-stranded RNA in the cytoplasm (1–3). The trigger for RNAi can be exogenous, for example, a synthetic small interfering RNA (siRNA), or endogenous, for example, a pre-micro RNA (miRNA)-like shRNA or siRNA that is expressed from its respective gene within the cell (4–9). Endogenous short hairpin RNA (shRNA) and siRNA are transcribed in the nucleus and must be exported to the cytoplasm, where the characteristic stem–loop of shRNA is cleaved by Dicer (10,11). The pathways involving siRNA and its shRNA precursor converge at RNA-induced silencing complex (RISC), with subsequent removal of the sense strand from the siRNA duplex, presumably cleaved first and then degraded, so that the antisense strand within RISC can recognize sequence-homologous RNAs and mediate target specificity (Figure 1). The recruited RNA is cleaved and degraded without any effect on the antisense strand, such that the antisense-programmed RISC is able to identify and degrade multiple copies of the target RNA (1,12).
Figure 1.
Schematic representation of RNAi and of the parameters that influence the difference between the potencies of siRNA and shRNA generated within cells. The ability of the integral sense RNA to the active RISC (dotted line) in the presence of the true RNA target renders siRNA far less potent than shRNA.
The selective and robust effect of RNAi on gene expression both in cell culture and in living organisms makes it a valuable research tool (10). Compared to exogenous siRNA, endogenous shRNA or siRNA, expressed via a plasmid or a viral vector, has enormous advantages in terms both of costless regeneration and long-term gene silencing (4–7,13–15). siRNA is generated via concomitant transcription of the integral sense and antisense strand from the respective coding sequences, which can be either a tandem or a convergent cassette (Figure 1); the latter system holds further promise for the construction of siRNA libraries for reasons both of convenience and of template stability (6–8). shRNA is transcribed from its unique template (16–19), a long and inverted-repeat DNA sequence (palindrome), and then it is translocated to the cytoplasm, where the hairpin loop is deleted to generate siRNA.
The broad applicability of shRNA renders it very attractive as a strong and versatile silencer of various genes both via transient transfection and via stable transduction (4,5,13–15). By contrast, expressed siRNA, even though it is the actual trigger and, theoretically, should be more efficient, is far less potent than its shRNA precursor (6–9). To date and to our knowledge, expressed siRNA delivered by viral vectors has never been shown to silence any target gene with useful efficiency. This problem remains a challenge to those researchers in the RNAi field and, especially to those who are using lentiviral siRNA and corresponding libraries for as potential biological and therapeutic tools (8).
The sequences of expressed siRNA and its shRNA precursor in this study are identical except a loop that links the sense and antisense duplexes in the latter case (Figure 1). Many potential factors, either related or unrelated to this loop, might reduce the potency of siRNA, such as poor transcription, due to potential interference between opposing promoters; poor nucleo-cytoplamic translocation; and/or poor incorporation into RISC due to the absence of the loop. All these possibilities remain to be explored. Recently, Berkhout group has tested the impact of different hairpin loop sequences, varying in size and structure, and found that the nature of a shRNA has a rather major impact on the shRNA activity (20). In this report, we provide evidence to suggest yet another possibility, namely, that sense strand-mediated strand antagonism attenuates the potency of expressed siRNA. Moreover, we show that reduction of this negative effect via alterations in the relative levels of strands of siRNA duplexes during their generation significantly enhances the potency of siRNA, in particular, in cases of stable transduction. Our findings provide new insight into the pathway of RNAi and should help us to develop strategies for exploiting expressed siRNA as a more efficient tool in the future.
MATERIALS AND METHODS
Construction of various shRNA- and siRNA-expression cassettes
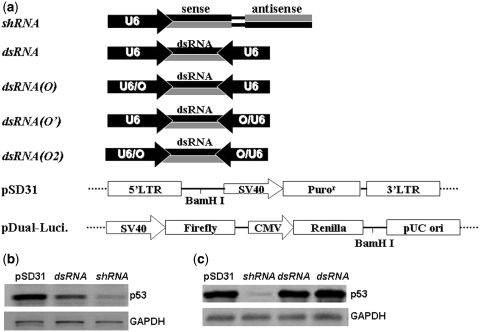
We constructed shRNA and siRNA, two p53-targeting siRNA-expression cassettes, by PCR ligation as described previously (21,22). We also constructed shRNA(O), siRNA(O), siRNA(O′) and siRNA(O2), four shRNA- or siRNA-expression cassettes with one or two inducible U6 promoters (Figure 2a), by the same method. Additional four inducible siRNA expression cassettes targeting firefly luciferase were also constructed with the siRNA sequence as gtgcgctgctggtgccaaccc. We utilized a similar approach for construction of siRNA-antisense, siRNA-sense, shRNA-sense and shRNA-antisense, four siRNA- or shRNA-expression cassettes that allowed an additional antisense or sense strand to be generated, and hU6/U6-siRNA-mU6/U6, hU6/U6/O-siRNA-mU6/U6, hU6-siRNA-mU6/U6, hU6/O-siRNA-mU6/U6 and hU6-siRNA-O/mU6/U6, five siRNA-expression cassettes with one or two chimeric promoters. pSD31 was a lentivector derived from pHIV-7 that was used for transfection and transduction of siRNA, as reported previously (22).
Figure 2.
(a) Schematic representation of shRNA-and siRNA-expression cassettes with the U6 or an inducible U6 promoter. pSD31 is a DNA vector derived from pHIV-7 and pDual-Luci is a dual luciferase reporter system for simultaneous expression of firefly and renilla luciferase. (b) Western blotting analysis of p53 knockdown in 293FT cells transfected with pSD31-shRNA and pSD31-siRNA. (c) Western blotting analysis of p53 knockdown in 293FT cells transduced with pSD31-shRNA and pSD31-siRNA.
Cell culture, transient transfection, stable transduction and packaging and titration of the lentivector
The mammalian cells used for most of this study were 293FT cells, cultured in Dulbecco's modified Eagle's medium supplemented with 10% fetal calf serum. The experimental protocols for expression of siRNA via transient transfection and stable transduction have been described previously (8,22). Experiments involving lentiviral packaging were performed by standard protocols (Invitrogen, San Diego, CA, USA). In brief, sub-confluent 293FT packaging cells were co-transfected with 20 µg of a recombinant lenti-plasmid, 15 µg of pCMV-DR8.91 and 5 µg of pMD2G-VSVG by calcium phosphate precipitation. Viral vectors were harvested 3 days after transfection. After filtration, the titer of the suspension of vector was determined as described by ‘Invitrogen's’ protocol.
Western blotting analysis
Cells were washed with phosphate-buffered saline (PBS) buffer, lysed in lysis buffer for 5 min and passed through a 27-gauge needle. Lysates were cleared by centrifugation at 12 000g for 1 min, and the concentration of protein was determined with a protein assay kit from Bio-Rad with bovine serum albumin (BSA) as standard. Equal amounts of protein were fractionated by sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS-PAGE; 4–20% polyacrylamide) before transfer to nitrocellulose membranes. Membranes were blocked with 3% BSA in TBST (10 mM Tris–HCl, pH 7.5, 150 mM NaCl, 0.1% Tween 20) for 1 h at room temperature. Primary and secondary antibodies were used according to the manufacturer's instructions, with detection with an enhanced chemiluminescence kit (Amersham Biosciences, Piscataway, NJ, USA).
Quantitation of mRNA, expressed siRNA duplexes and shRNA by real-time reverse transcriptase-polymerase chain reaction (RT-PCR)
Total RNA was prepared from 293FT cells in TRIzol® reagents (Invitrogen, San Diego, CA, USA). Nuclear and cytoplasmic RNA were isolated as described previously (23). Standard quantitative real-time reverse transcriptase-polymerase chain reaction (RT-PCR) was performed to determine levels of expression of p53 mRNA, as described previously (22,24). Quantitative analysis of expressed siRNA duplexes and shRNA in transfected or transduced cells by real-time RT-PCR was based on methods developed by others groups (25–27). In brief, 1 µg of total RNA was polyadenylated by poly(A) polymerase plus adenosine triphosphate (ATP) at 37°C for 1 h in a 20 -µl reaction mixture. After phenol–chloroform extraction and ethanol precipitation, the RNA was dissolved in diethylpyrocarbonate (DEPC)-treated water and reverse-transcribed with 200 U of SuperScript™ II reverse transcriptase (Invitrogen, San Diego, CA, USA) and 0.5 µg of poly(T) adapter according to the protocol from Invitrogen. For each real-time PCR, 1 µl of template cDNA, equivalent to ∼100 pg of total RNA, was mixed with 12.5 µl of 2 × SYBR Green PCR master mix and 5 pmol each of the forward and reverse primers in a final volume of 25 µl. Programs for amplification by PCR were the same as those described elsewhere (25–27) and all reactions were run in triplicate. The variation among triplicate results did not exceed 15%.
Mapping of expressed siRNA and shRNA generated from various expression cassettes
In order to verify siRNA effectors generated from various expression cassettes were identical, the RNA transcripts were captured by reverse transcription and amplified by PCR. In brief, 1 µl of cDNA template from the reverse transcription of the total RNA, 47 µl of PCR SuperMix (Invitrogen, San Diego, CA, USA) and 1 µl of each forward and reverse primer with 200 nM as final concentration were mixed and then carried out PCR according to the manufacturer's instructions. The PCR products were purified by 2% agarose gel, cloned into TA vector and then harvested from TOP10 cells for sequencing.
Testing the quality of the nuclear-cytosol fractionation
The nuclear-cytosol RNA fractionation was carried out using Norgen's Cytoplasmic & Nuclear RNA Purification Kit (Norgen, Ontario, Canada), which has been proved as a rapid and efficient method for the isolation of cytoplasmic and nuclear RNA from cultured animal cells and small tissue samples. According to the manufacturer's instructions, a small nuclear RNA, U2 snRNA, and a house-keeping transcript in the cytoplasm, s14 RNA, were utilized as indicators via RT-PCR to demonstrate the quality of nuclear-cytosol fractionation. The specific primers for U2 snRNA were 5′-CATCGCTTCTCGGCCTTTTG-3′ (forward) and 5′-TGGAGGTACT GCAATACCAGG-3′ (reverse). The primers for amplification of S14 were 5′-GGCA GACCGAGATGAATCCTC-3′ (forward) and 5′-CAGGTCCAGGGGTCTTGGTCC-3′ (reverse). In addition, ribosomal RNA 28S and 18S were also utilized as indicators to test the integrity and quality of the isolated RNA.
RESULTS AND DISCUSSION
Expressed siRNA was far less potent than the corresponding precursor shRNA
Unlike shRNA, siRNA expressed intracellularly has not been extensively exploited for inactivation of gene expression by either transient transfection or stable transduction. We chose the well-characterized p53 siRNA for our analysis (4,22) and cloned two forms of its DNA coding sequence (shRNA versus siRNA) into the vector pSD31 (Figure 2a). Then we compared their potency in the inactivation of the expression of the p53 gene. Transient transfection with pSD31-shRNA and pSD31-siRNA, in parallel, of 293FT cells indicated that expressed siRNA was far less potent than shRNA, when analyzed by western blotting: almost all expression of p53 was prevented by the shRNA, while the expressed siRNA was only moderately effective (Figure 2b). In stable transduction experiments with the lentiviral vector pSD31, siRNA was inactive, while shRNA was a strong silencer of its target gene (Figure 2c). The lower potency of expressed siRNA as compared to shRNA has been reported similarly by other groups who studied a variety of genes (6–8). To date siRNA delivered via a viral vector has not been shown to substantially disrupt the expression of any target gene. However, we do not know why expressed siRNA, the actual trigger of RNAi, is so much less effective than its shRNA precursor.
Comparisons of intracellular expression of siRNA versus shRNA
Many factors might be expected to affect the potency of siRNA. Transcription might be impaired as a consequence of opposing promoters, which might act in conflict with one another during convergent expression of the sense and antisense strands, with generation of fewer transcripts. In order to examine this possibility, we quantified the levels of siRNA and shRNA transcripts in transfected cells. Northern blotting analysis indicated that DIG-labeled probes were not sensitive enough to detect short RNA (Supplementary Figure 1). Moreover, even when detection was possible, quantitation of the tiny amounts of RNA and the variations in levels was very difficult, and the data were often unreproducible (Supplementary Figure 1).
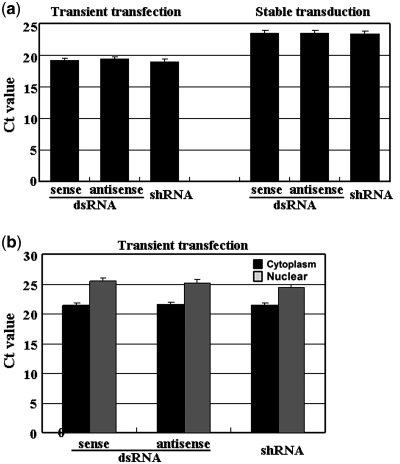
To overcome this problem, we used a previously reported real-time RT-PCR technique (25–27) to quantify expressed siRNA and shRNA (Supplementary Figure 2). The empty vector pSD31 was used as a negative control for comparisons of the relative levels of siRNA duplexes and shRNA generated in transfected and transduced cells. As shown in Figure 3a, we found that the sense and antisense strands of siRNA gave almost the same cycle threshold (Ct) values, namely 19.5 versus 19.4, indicating that the strands were generated with equal efficiency. After normalization (Supplementary Figure 2), the copy numbers of the sense and antisense strands of siRNA in each well of a 6-well plate (∼0.25 × 106 cells transfected with 2 µg of DNA with an efficiency of ∼90%, as determined from a control with the gene for green fluorescent protein) were 7.1 × 107 and 7.3 × 107, respectively. In addition, shRNA generated in a parallel experiment had a Ct value of 19.2, which was very similar to that of siRNA (Figure 3a), indicating that the number of copies of shRNA in transfected cells was the same as that of siRNA (Figure 3a). On the basis of these results, we were able to predict that each transfected cell contained an average of ∼300 copies of siRNA or shRNA transcripts. These results excluded the possibility of poor generation of siRNA, at least the concentration of siRNA detected, which is the sum of synthesis and degradation, was almost similar as that of shRNA. Those data also suggest that the opposing promoters within the siRNA cassette did not interfere with one another and transcription was efficient as from a single promoter.
Figure 3.
(a) Levels of the expression of siRNA duplexes and shRNA in transfected and transduced 293FT cells, as determined by real-time RT–PCR. (b) The distribution of siRNA duplexes and shRNA in the cytoplasm and the nucleus. All data shown reflect results from experiments conducted in triplicate for each sample.
siRNA transcripts were able to export from the nucleus into the cytoplasm
Export from the nucleus might also affect the potency of expressed siRNA. Both expressed siRNA and shRNA are generated in the nucleus but function in the cytoplasm and, thus, must be exported across the nuclear envelope (1,3,11). It has been demonstrated that shRNA, a structural homolog of pre-miRNA, exploits essentially the same nucleocytoplasmic shuttle protein, Exportin-5, as pre-miRNA. Exportin-5 recognizes the 3′ two-nucleotide overhang of the pre-miRNA hairpin for transportation of this RNA into the cytoplasm (28–31). The absence of a hairpin loop might impair the translocation of expressed siRNA, causing lower levels of siRNA in the cytoplasm, which is where RNAi is triggered.
To examine this possibility, we isolated nuclear and cytoplasm RNAs from transfected cells and quantified the respective levels of expressed siRNA and shRNA. We first tested the integrity and quality of the isolated RNA. Analysis of U2 snRNA by RT-PCR indicated that an intense product was observed when the nuclear fraction rather than the cytoplasmic fraction was used as the template (Supplementary Figure 3a). In contrast, the majority of the PCR product for S14 was from the cytoplasmic fraction rather than the nuclear as the template (Supplementary Figure 3b). In addition, the majority of ribosomal RNA (28S and 18S) was in cytoplasm rather than in nuclear as shown in Figure 3c, also indicating effective separation of cytoplasmic and nuclear RNA. Combination of these data indicated that the cytoplasmic and nuclear RNA were effectively separated.
As shown in Figure 3b, siRNA duplexes were detected in the cytoplasm and nucleus with Ct values of 21.4 versus 25.5 for the sense strand and 21.5 and 25.2 for the antisense strand, respectively. We also detected shRNA in both the cytoplasm and nucleus with Ct values of 21.4 and 24.4, respectively. These data indicate that the siRNA duplex, despite the absence of the hairpin loop, was able to transport into the cytoplasm just like shRNA. It remains unclear how siRNA, either as separate individual strands or as paired strands, is exported, but it was clearly not a difference in translocation that caused the difference in potency between siRNA and shRNA.
Expressed siRNA exhibited strand antagonism in triggering RNAi
In a previous study, we found that the lentiviral packaging system was an ideal model for elucidating the roles of individual strands of siRNA in triggering RNAi since the siRNA-delivering viral vector itself is an inherent target (8). We found, previously, that blocking transcription of the antisense strand inactivated RNAi completely. By contrast, blocking transcription of the sense strand had almost no effect on the potency of expressed siRNA (8). These observations let us to ask a fundamental question: is the sense strand really necessary for RNAi? In addition, we wondered whether ‘extra’ integral sense or antisense RNA might enhance the potency of siRNA. To answer these questions, we designed a series of experiments to detect the effects of changes in relative levels of the strands of siRNA duplexes on RNAi.
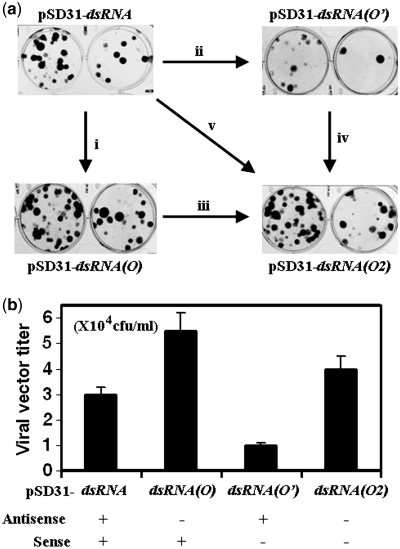
As reported previously (8,21,22), an inducible U6 promoter with a tetracycline operator (TetO or O) reduced transcription of shRNA by ∼20%. Therefore, we replaced one of the two U6 promoters within the siRNA-expression cassette with the inducible promoter (Figure 2a), which reduced the expression of the antisense or sense RNA to the same extent (20%) as reported previously (8,22). Map mapping data indicated that the sequences of siRNA duplexes generated from various siRNA-expression cassettes were identical (Supplementary Figure 4). Such a reduction in the level of the antisense RNA [pSD31-siRNA(O)] increased the titer of lentivector almost 2-fold (Figure 4a and b), suggesting the potency of RNAi decreased slightly since the siRNA-carrying viral RNA itself is an inevitable target of RNAi (8,22). This result was consistent with the hypothesis that the antisense RNA is the predominant effective inducer of RNAi (32–35). By contrast, reduction in the level of the sense RNA [pSD31-siRNA(O’)] tended to increase RNAi with a lower titer of the lentivector being produced (Figure 4a and b), hinting that the sense component might act as an inhibitory factor. Consistently, the increase in potency caused by reduction in the level of the sense component vanished with a similar simultaneous reduction in the level of the antisense component [pSD31-siRNA(O2)]. Together, these observations suggested that equivalent levels of the sense and antisense components rendered the siRNA less potent.
Figure 4.
(a) Production of the viral vector in 293FT cells transfected with pSD31-siRNA, pSD31-siRNA(O), pSD31-siRNA(O’) and pSD31-siRNA(O2) respectively, namely, the response to unilateral reduction in the level of antisense RNA (i), in response to that of sense RNA (iii), in response to reversal these parameter (ii and iv), and in response to simultaneous reductions in levels of both sense and antisense RNA (v). The vector stocks were diluted 1000- and 3000-fold for tittering. (b) The effect of regular expression (+) and an ∼20% reduction (−) in the level of the sense or the antisense RNA or both on production of the viral vector. The vector titers shown are derived from results of an experiment that was conducted in triplicate for each sample.
Confirmation of strand antagonism in RNAi
To confirm that the sense strand really reduced the potency of the siRNA, we expressed ‘extra’ sense RNA by ligating a sense-expression cassette to the siRNA cassette (Figure 5a). As expected, this cassette engineering (siRNA-sense) eliminated the equivalence of the sense versus the antisense component, with the level of the former being close to twice that of the latter (Figure 5b). Generation of ‘extra’ sense RNA really did lower the potency of siRNA since knockdown of p53 was less extensive (Figure 5c). In a reciprocal experiment, ligating an antisense-expression cassette to the siRNA cassette (siRNA-antisense), we found that the potency of the siRNA was significantly enhanced, with the level of antisense RNA being twice that of the sense RNA (Figure 5b and c). We also mapped the siRNA effectors generated from various siRNA and shRNA vectors via RT-PCR to demonstrate their sequence identity. As shown in Supplementary Figure 4, the sequence of the sense RNA and antisense RNA generated from each respective cassette were identical. Combination of their expression profiling with the mapping data supported that the siRNA effectors made from siRNA versus shRNA vectors were identical. Clearly, unilateral increase in the level of the sense RNA reduced the potency of the siRNA while an elevated level of the antisense RNA enhanced such potency.
Figure 5.
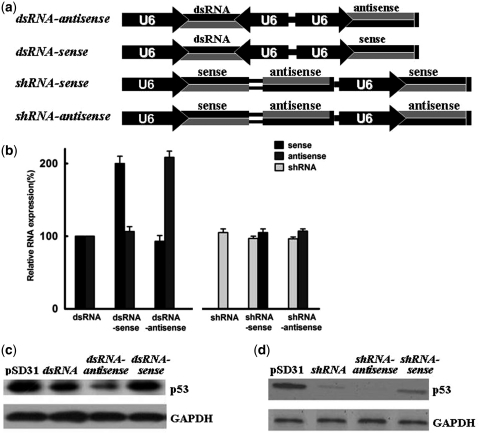
(a) Schematic representation of siRNA- and shRNA-expression cassettes ligated with sense- or antisense-expression cassettes, as indicated, for confirmation of strand antagonism. (b) Relative levels of siRNA duplexes and shRNA in 293FT cells transfected with siRNA-sense, siRNA-antisense, shRNA-sense and shRNA-antisense vectors in parallel. The data shown are relative to levels of siRNA duplexes in 293FT cells transfected with pSD31-siRNA. All samples were analyzed in triplicate. (c) Western blotting analyses of the effects of generation of ‘extra’ sense or antisense RNA on siRNA-mediated knockdown of expression of p53. (d) The effects of generation of ‘extra’ sense and antisense RNA on shRNA-mediated knockdown of expression of p53, as determined by western blotting.
In order to evaluate whether the integral sense component also regulates the potency of shRNA, we ligated the shRNA cassette with the sense-expression cassette (shRNA-sense). As expected, the potency of shRNA fell markedly upon concomitant expression of the sense RNA (Figure 5b and d). In a reciprocal experiment, ligating the shRNA cassette with an antisense-expression cassette (shRNA-antisense), we found that expression of the antisense strand enhanced the potency of shRNA. All these observations together indicate that siRNA duplexes exhibit strand antagonism and the sense component negatively interferes with RNAi, presumably behaving as a substrate for RISC.
We have also developed the Tet-regulated siRNA system, including four TetO-tethered siRNA-expression cassettes targeting firefly luciferase (Figure 2a) and the Dual-Luciferase Reporter, to test strand antagonism. Renilla luciferase reporter was used as an internal control to reduce the significant reading variations existing in luciferase assay. We found siRNA(O’), in which the sense RNA was driven by an inducible promoter, displayed further potency increase following initial dox titration (5, 10, 25 nM), but such enhancement trend reversed following continuous dox titration (Supplementary Figure 5). It supposed that the inducible level of RNA was dependent on the dox concentration, i.e. low concentration marginally induced RNA expression while high concentration restored RNA expression as we proved previously (22). This was in contrast to siRNA(O) and siRNA(O2) which displayed potency increase with dox titration from low concentration to high concentration (Supplementary Figure 5). These data also supported existence of sense RNA negative effect in siRNA-mediated gene knockdown.
Disruption of molar equivalence rendered siRNA functional in stably transducted cell
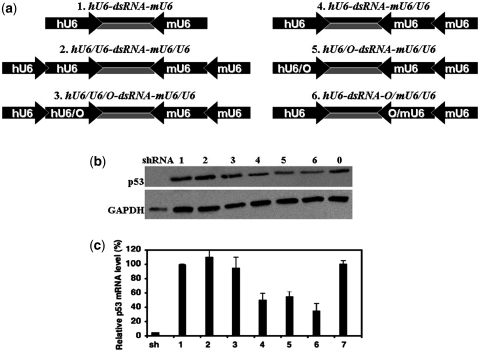
Our group and others have demonstrated that opposing-promoter cassettes are very useful for the generation of siRNA libraries (6–8). When such a cassette is functional in stable transduction, it should have broad application in many contexts. Our discovery of strand antagonism suggested a potential strategy for enhancing siRNA potency via adjustment of the relative levels of the antisense and sense components during their generation. As reported previously, the combination of Pol II with a pol II promoter (36–38) or of Pol II with a pol III promoter (39) has a significant positive effect on RNA transcription. Therefore, we ligated a TATA box-deleted U6 promoter to an intact U6 promoter to generate a pol III/pol III chimeric promoter (Supplementary Figure 6a) and examined whether this combination enhanced the transcription of siRNA. Deletion of the TATA box from the upstream U6 promoter was designed to prevent transcription of RNA other than siRNA duplexes (Supplementary Figure 6a). We constructed a siRNA-expression cassette with chimeric promoters, hU6/U6-siRNA-mU6/U6, with a human U6 chimeric promoter (hU6/U6) and a mouse U6 chimeric promoter (mU6/U6) to avoid long sequence repetition that destabilizes DNA (Figure 6a).
Figure 6.
(a) Schematic representation of symmetric and asymmetric siRNA-expression cassettes, with the expression of either the sense or the antisense RNA or both being driven by a chimeric promoter(s) for disruption of their equivalent molar levels of production. (b) Western blotting analysis of knockdown of p53 expression in 293FT cells transduced with shRNA and with various siRNA-expression cassettes via lentivector pSD31. (c) Levels of expression of p53 mRNA in transduced 293FT cells, as determined by real-time RT-PCR. The data shown are relative to levels of p53 mRNA in pSD31-transduced 293FT cells. Data shown are derived from an experiment that was conducted in triplicate for each sample.
Analysis after transfection of 293T cells with chimeric hU6/U6-siRNA-mU6/U6 in parallel with regular hU6-siRNA-mU6 via the vector pSD31 indicated that the former was more potent than the latter in knockdown of p53 expression (Supplementary Figure 6b), with ∼50% more siRNA transcripts generated in the former case than in the latter (Supplementary Figure 6c). We then used the chimeric promoter to construct a variety of siRNA-expression cassettes, including symmetric hU6/U6-siRNA-mU6/U6 and hU6/U6-siRNA-O/mU6/U6 and asymmetric hU6-siRNA-O/mU6/U6 and hU6/O-siRNA-mU6/U6 (Figure 6a). In the various cassettes, the mouse U6 promoter (mU6) and the corresponding chimeric promoter (mU6/U6) drove transcription of the antisense strand, while the human promoter (hU6) and the corresponding chimeric promoter (hU6/U6) drove transcription of the sense strand. As shown in Figure 6b and c, stable transduction with the vector that harbored the symmetric hU6/U6-siRNA-mU6/U6 and hU6/U6-siRNA-O/mU6/U6 has almost no effect on the expression of p53, as was the case with hU6-siRNA-mU6, indicating that simultaneous increases in levels of both strands of siRNA duplexes had no significant effect on RNAi, at least in stable transduction experiments. By contrast, stable transduction with asymmetric hU6-siRNA-mU6/U6, hU6/O-siRNA-mU6/U6 and hU6-siRNA-O/mU6/U6, such that the chimeric promoter generated more antisense strand than sense strand (Supplementary Figure 6d), resulted in obvious knockdown of p53 at the level of both the protein (>50%; Figure 6b) and its mRNA (>70%; Figure 6c). To our knowledge, this is the first report of siRNA, delivered by a lentivector, having substantial ability to disrupt expression of a target gene, even though its potency remained much lower than that of shRNA.
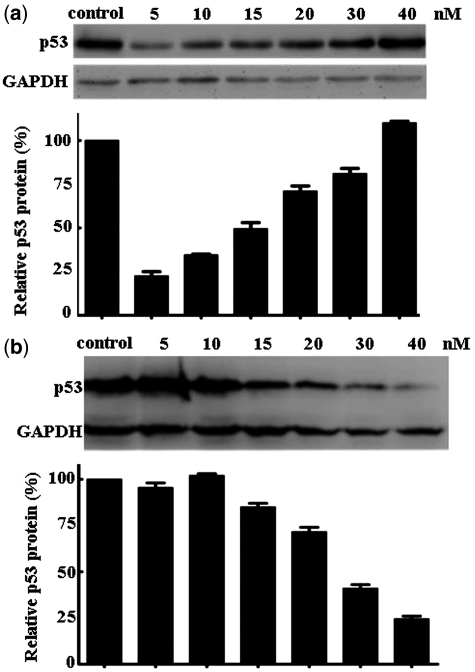
Using asymmetric hU6/O-siRNA-mU6/U6 and hU6-siRNA-O/mU6/U6, we found that the former cassette, despite generation of more antisense RNA and less sense RNA than the latter, did not further enhance the potency of the siRNA (Figure 6b and c). This observation hints that the potential reward might not extend indefinitely with further alterations in the proportions of two strands. Prior to a given ratio of levels, there is enhancement of RNAi; beyond that point, the opposite occurs. This conclusion is consistent with the previous observation that the complete absence of the sense strand does not enhance RNAi (8). In an ongoing experiment aimed at further proving less sense RNA is beneficial, we co-transfected the antisense oligo at fixed concentration (15 nM) with the sense oligo varying at concentration from 5 to 40 nM. We found RNAi with the sense RNA at lower concentration than 15 nM was much potent that at higher concentration (Figure 7a and b). Combination of these data also supported the sense RNA and antisense RNA exerts strand antagonistic effect during RNA interference and less sense RNA is beneficial.
Figure 7.
Confirmation of strand antagonism based on transfection of synthetic sense and antisense oligomers at a variety of molar ratios. (a) The level of the antisense oligomer was fixed at 15 nM and the sense oligomer was varied from 5 to 40 nM. (b) The level of the sense oligomer was fixed at 15 nM and the level of the antisense oligomer was varied from 5 to 40 nM.
Mechanistic differences between the siRNA and shRNA in the induction of RNAi
The events that occur during RNAi, from incorporation of the siRNA duplex to degradation of the target mRNA, have been well studied (Figure 1). The key complex of antisense–RISC, after one round of activity, can be recycled, with multiple turnovers, to destroy all complementary RNAs (1,11). In the case of siRNA duplexes generated within cells, it is very possible that only part of the nascent sense RNA forms base pairs with the antisense RNA because of the cellular environment wherein various macromolecules interfere with one another. Unpaired sense RNAs might be diverted directly to the antisense–RISC complex and might, thus, compete with the real target (Figure 1). The strand antagonism observed in the present study strongly supports the possibility that sense RNAs do, indeed, reduce the levels of effectors of RNAi that are available for the real target, reducing the extent of RNAi. Our observations also help to clarify the fate of sense RNA in the initiation of RNAi. They appear to act as the first turned-over substrate, after cleavage within the loaded RISC complex (Figure 1). Then subsequent turnover of the true RNA substrates is triggered.
In the case of shRNA, the loop sequence linking the sense and antisense strand together lowers the kinetic barrier for duplex formation significantly (Figure 1). More importantly, the hairpin structure renders shRNA itself and other RNAs harboring shRNA structure poor targets for RNAi, as demonstrated previously (8,40–43). This scenario stands in significant in contrast to that involving siRNA since the free sense RNA is an inevitable substrate of RNAi and, once recruited to the active antisense–RISC complex, it reduces the potency of RNAi. Thus, the present study provides new insight into RNAi, and in particular, into the way in which the sense RNA reduces the potency of siRNA.
The negative effect of the integral sense RNA is intrinsic and, even after further optimization, must always exist, preventing siRNA from being as effective as shRNA. However, by adjusting the molar ratio of antisense to sense RNA, namely, by over-expressing the antisense strand of siRNA relative to the sense strand, we were able to enhance the activity of siRNA in cells to a significant extent. As a result, we were able, for the first time, to suppress expression of a target gene using siRNA-expression vectors in transduced cells. Our strategy, the over-expression of the antisense strand, might also be useful for enhancing the activity of widely used shRNA-expression vectors.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online: Supplementary Figures 1–6.
FUNDING
Funding for open access charge: The National Basic Research Program of China (973 Program; grant no. 2010CB12300); and the National Natural Science Foundation of China (grant nos. 20932001 and 91029711).
Conflict of interest statement. None declared.
Supplementary Material
REFERENCES
- 1.Sontheimer EJ, Carthew RW. Argonaute journeys into the heart of RISC. Science. 2004;305:140–141. doi: 10.1126/science.1103076. [DOI] [PubMed] [Google Scholar]
- 2.Liu J, Carmell MA, Rivas FV, Marsden CG, Thomson M, Song J, Hammond SM, Joshua-Tor L, Hannon GJ. Argonaute2 is the catalytic engine of mammalian RNAi. Science. 2004;305:1437–1441. doi: 10.1126/science.1102513. [DOI] [PubMed] [Google Scholar]
- 3.Meister G, Tuschl T. Mechanisms of gene silencing by double-stranded RNA. Nature. 2004;431:343–349. doi: 10.1038/nature02873. [DOI] [PubMed] [Google Scholar]
- 4.Brummelkamp TP, Bernards R, Agami R. A system for stable expression of short interfering RNAs in mammalian cells. Science. 2002;296:550–553. doi: 10.1126/science.1068999. [DOI] [PubMed] [Google Scholar]
- 5.Sui G, Sooboo C, Affar EB, Gay F, Forrester WC, Shi Y. A DNA vector-based RNAi technology to suppress gene expression in mammalian cells. Proc. Natl Acad. Sci. USA. 2002;99:5515–5520. doi: 10.1073/pnas.082117599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zheng L, Liu J, Batalov S, Zhou D, Orth A, Ding S, Schultz PG. An approach to genomewide screens of expressedsmall interfering RNAs in mammalian cells. Proc. Natl Acad. Sci. USA. 2004;101:135–140. doi: 10.1073/pnas.2136685100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tran N, Cairns MJ, Dawes IW, Arndt GM. Expressing functional siRNAs in mammalian cells using convergent transcription. BMC Biotechnol. 2003;3:21. doi: 10.1186/1472-6750-3-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhou D, Zhang J, Wang CY, He QS, Joshua R, Zhang L, Wong-Staal F. A method for detecting and preventing negative RNA interference in preparation of lentiviral vectors for siRNA delivery. RNA. 2009;15:732–740. doi: 10.1261/rna.985209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Miyagishi M, Taira K. U6 promoter-driven siRNAs with four uridine 3' overhangs efficiently suppress targeted gene expression in mammalian cells. Nat. Biotechnol. 2002;20:497–500. doi: 10.1038/nbt0502-497. [DOI] [PubMed] [Google Scholar]
- 10.Hannon GJ, Rossi JJ. Unlocking the potential of the human genome with RNA interference. Nature. 2004;431:371–378. doi: 10.1038/nature02870. [DOI] [PubMed] [Google Scholar]
- 11.Mello CC, Conte DJ. Revealing the world of RNA interference. Nature. 2004;431:338–342. doi: 10.1038/nature02872. [DOI] [PubMed] [Google Scholar]
- 12.Zhou D, He QS, Wang C, Zhang J, Wong-Staal F. RNA interference and potential applications. Curr. Top. Med. Chem. 2006;6:901–911. doi: 10.2174/156802606777303630. [DOI] [PubMed] [Google Scholar]
- 13.Qin XF, An DS, Chen IS, Baltimore D. Inhibiting HIV-1 infection in human T cells by lentiviral-mediated delivery of small interfering RNA against CCR5. Proc. Natl Acad. Sci. USA. 2003;100:183–188. doi: 10.1073/pnas.232688199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tiscornia G, Singer O, Ikawa M, Verma IM. A general method for gene knockdown in mice by using lentiviral vectors expressing small interfering RNA. Proc. Natl Acad. Sci USA. 2003;100:1844–1848. doi: 10.1073/pnas.0437912100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ter Brake O, Berkhout B. Lentiviral vectors that carry anti-HIV shRNAs: problems and solutions. J. Gene Med. 2007;9:743–750. doi: 10.1002/jgm.1078. [DOI] [PubMed] [Google Scholar]
- 16.Sen G, Wehrman TS, Myers JW, Blau HM. Restriction enzyme-generated siRNA (REGS) vectors and libraries. Nat. Genet. 2002;36:183–189. doi: 10.1038/ng1288. [DOI] [PubMed] [Google Scholar]
- 17.Shirane D, Sugao K, Namiki S, Tanabe M, Iino M, Hirose K. Enzymatic production of RNAi libraries from cDNAs. Nat. Genet. 2004;36:190–196. doi: 10.1038/ng1290. [DOI] [PubMed] [Google Scholar]
- 18.Luo B, Heard AD, Lodish HF. Small interfering RNA production by enzymatic engineering of DNA (SPEED) Proc. Natl Acad. Sci. USA. 2004;101:5494–5499. doi: 10.1073/pnas.0400551101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhou D, Wang C, Zhang J, Bliesath J, He QS, Ke N, Yu D, Li Q, Zhang L, Wong-Staal F. Generation of shRNA pool library: a revision of the biological technique from the viewpoint of chemistry. ChemBioChem. 2008;9:1365–1367. doi: 10.1002/cbic.200800049. [DOI] [PubMed] [Google Scholar]
- 20.Schopman NC, Liu YP, Konstantinova P, ter Brake O, Berkhout B. Optimization of shRNA inhibitors by variation of the terminal loop sequence. Antiviral. Res. 2010;86:204–211. doi: 10.1016/j.antiviral.2010.02.320. [DOI] [PubMed] [Google Scholar]
- 21.He QC, Tang HL, Zhang J, Zhang LH, Wong-Staal F, Zhou DM. Comparison of RNAi approaches for validation of human RNA helicase A as an essential factor in hepatitis C virus replication. J. Viro. Methods. 2008;154:216–219. doi: 10.1016/j.jviromet.2008.08.005. [DOI] [PubMed] [Google Scholar]
- 22.Zhang J, Wang C, Ke N, Bliesath J, Chionis J, He QS, Li QX, Chatterton JE, Wong-Staal F, Zhou D. A more efficient RNAi inducible system for tight regulation of gene expression inmammalian cells and xenograft animals. RNA. 2007;13:1375–1383. doi: 10.1261/rna.520707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kawasaki H, Taira K. Short hairpin type of dsRNAs that are controlled by tRNA(Val) promoter significantly induce RNAi-mediated gene silencing in the cytoplasm of human cells. Nucleic Acids Res. 2003;31:700–707. doi: 10.1093/nar/gkg158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hu X, Hipolito S, Lynn R, Abraham V, Ramos S, Wong-Staal F. Relative gene-silencing efficiencies of small interfering RNAs targeting sense and antisense transcripts from the same genetic locus. Nucleic Acids Res. 2004;32:4609–4617. doi: 10.1093/nar/gkh790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Shi R, Chiang VL. Facile means for quantifying microRNA expression by real-time PCR. Biotechnology. 2005;39:519–524. doi: 10.2144/000112010. [DOI] [PubMed] [Google Scholar]
- 26.Kim SW, Li Z, Moore PS, Monaghan AP, Chang Y, Nichols M, John B. A sensitive non-radioactive northern blot method to detect small RNAs. Nucleic Acids Res. 2010;38:e98. doi: 10.1093/nar/gkp1235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Benes V, Castoldi M. Expression profiling of microRNA using real-time quantitative PCR, how to use it and what is available. Methods. 2010;50:244–249. doi: 10.1016/j.ymeth.2010.01.026. [DOI] [PubMed] [Google Scholar]
- 28.Yi R, Doehle BP, Qin Y, Macara IG, Cullen BR. Overexpression of exportin 5 enhances RNA interference mediated by short hairpin RNAs and microRNAs. RNA. 2005;11:220–226. doi: 10.1261/rna.7233305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cook AG, Fukuhara N, Jinek M, Conti E. Structures of the tRNA export factor in the nuclear and cytosolic states. Nature. 2009;461:60–65. doi: 10.1038/nature08394. [DOI] [PubMed] [Google Scholar]
- 30.Okada C, Yamashita E, Lee SJ, Shibata S, Katahira J, Nakagawa A, Yoneda Y, Tsukihara T. A high-resolution structure of the pre-microRNA nuclear export machinery. Science. 2009;326:1275–1279. doi: 10.1126/science.1178705. [DOI] [PubMed] [Google Scholar]
- 31.Paddison PJ, Caudy AA, Bernstein E, Hannon GJ, Conklin DS. Short hairpin RNAs (shRNAs) induce sequence-specific silencing in mammalian cells. Genes Dev. 2002;6:948–958. doi: 10.1101/gad.981002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Holen T, Amarzguioui M, Babaie E, Prydz H. Similar behaviors of single-strand and double-strand siRNAs suggests they act through a common RNAi pathway. Nucleic Acids Res. 2003;31:2401–2407. doi: 10.1093/nar/gkg338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Martinez J, Patkaniowska A, Urlaub H, Luhrmann R, Tuschl T. Single-stranded antisense siRNAs guide target RNA cleavage in RNAi. Cell. 2002;110:563–574. doi: 10.1016/s0092-8674(02)00908-x. [DOI] [PubMed] [Google Scholar]
- 34.Schwarz DS, Hutvagner G, Du T, Xu Z, Aronin N, Zamore PD. Asymmetry in the assembly of the RNAi enzyme complex. Cell. 2003;115:199–208. doi: 10.1016/s0092-8674(03)00759-1. [DOI] [PubMed] [Google Scholar]
- 35.Khvorova A, Reynolds A, Jayasena SD. Functional siRNAs and miRNAs exhibit strand bias. Cell. 2003;115:209–216. doi: 10.1016/s0092-8674(03)00801-8. [DOI] [PubMed] [Google Scholar]
- 36.Ahlquist P. RNA-dependent RNA polymerase, viruses, and RNA silencing. Science. 2002;296:1270–1273. doi: 10.1126/science.1069132. [DOI] [PubMed] [Google Scholar]
- 37.Pacha RF, Ahlquist P. Use of bromovirus RNA3 hybrids to study template specificity in viral RNA amplification. J. Virol. 1991;65:3693–3703. doi: 10.1128/jvi.65.7.3693-3703.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ishikawa M, Janda M, Krol MA, Ahlquist P. In vivo DNA expression of functional brome mosaic virus RNA replicons in Saccharomyces cerevisiae. J. Virol. 1997;71:7781–7790. doi: 10.1128/jvi.71.10.7781-7790.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Xia XG, Zhou H, Ding H, Affar EB, Shi Y, Xu Z. An enhanced U6 promoter for synthesis of short hairpin RNA. Nucleic Acids Res. 2003;31:e100. doi: 10.1093/nar/gng098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kretschmer-Kazemi FR, Sczakiel G. The activity of siRNA in mammalian cells is related to structural target accessibility: a comparison with antisense oligonucleotides. Nucleic Acids Res. 2003;31:4417–4424. doi: 10.1093/nar/gkg649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Westerhout EM, Berkhout B. A systematic analysis of the effect of target RNA structure on RNA interference. Nucleic Acids Res. 2007;35:4322–4330. doi: 10.1093/nar/gkm437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wunsche W, Sczakiel G. The activity of siRNA in mammalian cells is related to the kinetics of siRNA-target recognition in vitro: mechanistic implications. J. Mol. Biol. 2005;345:203–209. doi: 10.1016/j.jmb.2004.10.041. [DOI] [PubMed] [Google Scholar]
- 43.Poluri A, Sutton RE. Titers of HIV-based vectors encoding shRNAs are reduced by a dicer-dependent mechanism. Mol. Ther. 2008;16:378–386. doi: 10.1038/sj.mt.6300370. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.