Abstract
It is largely recognized that microRNAs (miRNAs) function to silence gene expression by targeting 3′UTR regions. However, miRNAs have also been implicated to positively-regulate gene expression by targeting promoter elements, a phenomenon known as RNA activation (RNAa). In the present study, we show that expression of mouse Cyclin B1 (Ccnb1) is dependent on key factors involved in miRNA biogenesis and function (i.e. Dicer, Drosha, Ago1 and Ago2). In silico analysis identifies highly-complementary sites for 21 miRNAs in the Ccnb1 promoter. Experimental validation identified three miRNAs (miR-744, miR-1186 and miR-466d-3p) that induce Ccnb1 expression in mouse cell lines. Conversely, knockdown of endogenous miR-744 led to decreased Ccnb1 levels. Chromatin immunoprecipitation (ChIP) analysis revealed that Ago1 was selectively associated with the Ccnb1 promoter and miR-744 increased enrichment of RNA polymerase II (RNAP II) and trimethylation of histone 3 at lysine 4 (H3K4me3) at the Ccnb1 transcription start site. Functionally, short-term overexpression of miR-744 and miR-1186 resulted in enhanced cell proliferation, while prolonged expression caused chromosomal instability and in vivo tumor suppression. Such phenotypes were recapitulated by overexpression of Ccnb1. Our findings reveal an endogenous system by which miRNA functions to activate Ccnb1 expression in mouse cells and manipulate in vivo tumor development/growth.
INTRODUCTION
Small RNA including short interfering RNA (siRNA), microRNA (miRNA) and Piwi-interacting RNA (piRNA) have emerged as master regulators of gene expression and play important roles in diverse biological processes and diseases (1). miRNAs are endogenous 20- to 24-nt small RNAs transcribed from the genome as primary miRNAs, which are processed into mature miRNAs by ribonuclease (RNase) III family members Drosha and Dicer (2). By interacting with members of the Argonaute (Ago) subfamily of proteins (1), miRNAs mainly target homologous sites in 3′ untranslated regions (UTRs) to suppress translation (3) and/or degrade mRNA in a mechanism known as RNA interference (RNAi). Cases in which miRNAs target 5′-UTRs (4,5), coding regions (6), promoters (7,8), or sequences downstream of gene termini (9) to silence gene expression have also been reported, raising the possibility of multiple modes of action for miRNAs. Complexity of miRNA-mediated gene regulation is further expanded by observations that miRNAs can positively affect gene expression. For example, miR-122 can enhance hepatitis C viral (HCV) gene replication by targeting 5′-non-coding elements in the HCV genome (10,11). miR-369-3 has been shown to activate mRNA translation by targeting AU-rich elements in 3′-UTRs under serum starvation (12,13). miR-10a has also been shown to enhance translation by interacting with the 5′-UTR of ribosomal protein mRNAs (14).
The potential impact of small RNA species, especially miRNAs, on gene transcription, epigenetic memory and genome integrity is largely an under-explored territory. Previously, we and others have shown that exogenous dsRNAs targeting promoter sequences can either suppress (15) or activate gene expression (16–20) by mechanisms respectively referred to as transcriptional gene silencing (TGS) and RNA activation (RNAa). Sequencing and bioinformatics analyses have found that a majority of miRNAs are imported back into the nucleus after maturation (21) with an abundance of putative miRNA target sites in gene promoters (7,22) to suggest that miRNAs may also function in TGS and/or RNAa. Indeed, miR-320—transcribed from the promoter of the POLR3D gene—has been shown to suppress PLOR3D transcription in cis (7), whereas miR-373 has been shown to function in trans by activating the expression of E-cadherin and cold shock domain-containing protein C2 (CSDC2) through promoter sites (23). Recently, Younger et al. (8) reported another trans-acting miRNA (miR-423-5p) that functioned to suppress progesterone receptor (PR) gene transcription by targeting an antisense non-coding RNA overlapping the PR promoter. So far, studies have not followed up on the endogenous functions of these promoter-targeting miRNAs specifically in the context of cancer.
Lines of evidence show that some miRNAs act as oncogenes (24) or tumor suppressor genes (25) by modulating molecular pathways involved in human cancer. It is reasonable to speculate that endogenous miRNAs may be involved in tumorigenesis via a nuclear function that impacts gene transcription and epigenetic states. In the present study, we show that the expression of mouse Cyclin B1 (Ccnb1) is partially dependent on the miRNA machinery (i.e. Drosha, Dicer, Ago1 and Ago2) in immortal mouse cell lines. In silico analysis identifies multiple miRNAs with putative sites complementary to sequence in the Ccnb1 promoter. Subsequent experimentation reveals that several miRNAs (e.g. miR-744 and miR-1186) can induce Ccnb1 expression and enhance cell proliferation; however, prolonged overexpression causes chromosomal alterations and in vivo tumor suppression. Our results reveal an endogenous function for miRNA in gene activation and cancer cell growth.
MATERIALS AND METHODS
siRNA and synthetic miRNA
siRNAs against mouse Drosha, Dicer, Ago1, Ago2 and Ccnb1 were designed using BLOCK-iT™ RNAi Designer Program (Invitrogen). Synthetic miRNA mimics were purchased from Qiagen (miScript miRNA mimics). miRNA seed mutants and biotinylated miR-744 were synthesized by Sigma. All siRNA and miRNA sequences are listed in Supplementary Table S3.
Plasmids
Genomic sequences encompassing pre-miR-1186 and pre-miR-744 were amplified from DNA isolated from NIH/3T3 cells and cloned into the lentiviral expression vector pPS-EF1-copGFP-LCS via the Clone-itTM Ligase free system (System Biosciences) to generate the miRNA overexpression constructs. Mouse Ccnb1 was amplified from cDNA samples and cloned into the lentiviral cDNA expression vector pCDH-EF1-MCS-T2A-copGFP (System Biosciences) via the EcoRI and BamHI sites to generate a Ccnb1 overexpression construct. Vectors pIRESneo-FLAG/HA-Ago1 and pIRESneo-FLAG/HA-Ago2 (Addgene plasmids 10 820 and 10 822) were used to establish Ago1 (NIH/3T3-Ago1) and Ago2 (NIH/3T3-Ago2) stable cell lines, respectively.
miRNA target prediction
miRNA targets on Ccnb1 promoter were predicted using the miRanda program (26) run on a Linux operating system. One kilobase of the mouse Ccnb1 promoter sequence was retrieved from the Ensembl genome database (www.ensembl.org), while mouse miRNA sequences were retrieved from miRBase (www.mirbase.org). The Ccnb1 promoter sequence was scanned against all known mouse miRNAs to identify putative target sites. An alignment score of 140 was set as the positive prediction threshold. The program's output results were further processed by a custom Perl script to sort target prediction by alignment score and derive statistical information.
Cell culture and transfection
NIH/3T3 cells (ATCC) were maintained in high-glucose (4.5 g/l) Dulbecco's modified Eagle medium (UCSF Cell Culture Core) supplemented with penicillin G (100 U/ml), streptomycin (100 µg/ml), 10% bovine calf serum (Atlanta) in a humidified atmosphere of 5% CO2 maintained at 37°C. TRAMP C1 (mouse prostate adenocarcinoma) cells were originally derived from the TRAMP (transgenic adenocarcinoma mouse prostate) mouse model and cultured as previously described (27). NIH/3T3 or TRAMP C1 cells (~1 × 104) were transfected with synthetic siRNA or miRNA mimics by reverse transfection using Lipofectamine RNAiMax (Invitrogen) according to manufacturer's protocol. NIH/3T3-eGFP, NIH/3T3-Ago1 and NIH/3T3-Ago2 stable cell lines were established by standard transfection, selection, and propagation protocols. Briefly, single colonies were subcultured following selection by G418. Ago1 and Ago2 expression was confirmed by immunoblot analysis.
Lentivirus-based overexpression
Lentivirus particles were generated by transfecting 293FT cells (Invitrogen) with lentivirus vector and packaging plasmids according to the instructions provided with the ViraPowerTM Lentiviral Expression System (Invitrogen). NIH/3T3 and TRAMP C1 cells were infected twice with the viral supernatant harvested at 48 and 72 h following transfection and positively transduced (GFP positive) cells were sorted by FACS (UCSF Laboratory of Cell Analysis).
RNA extraction and real-time PCR analysis
Total RNA was isolated by using the RNeasy Mini Kit (Qiagen) and reverse transcribed with MMLV reverse transcriptase (Promega) in conjunction with oligo(dT) primers. The resulting cDNA samples were subjected to real-time PCR analysis using gene-specific primers (sequences are available in Supplementary Table S3). Small RNA was extracted by using the miReasy Mini Kit (Qiagen). Reverse transcription reactions were performed using the Taqman miRNA reverse transcription system (Applied Biosystems). Expression levels of each miRNA were detected by Taqman miRNA assays (Applied Biosystems).
Immunoblotting
Cells were washed with PBS and lysed in RIPA buffer (50 mM Tris, pH 7.4, 150 mM NaCl, 1% Triton X-100, 0.5% deoxycholate, 0.1% SDS, protease inhibitor cocktail and phosphatase inhibitor) for 15 min at 4°C. Lysates were clarified by centrifugation for 15 min at 14 000 rpm and supernatants were collected. Protein concentration in the soluble fraction was determined by BCA protein assay (Thermo Scientific). Equal amounts of protein were resolved on SDS–PAGE gel and transferred to nitrocellulose membrane. Blots were blocked overnight with 5% nonfat dry milk in TBST. Specific proteins were detected by using SuperSignal West Pico Chemiluminescent reagents (Thermo Scientific). The following antibodies were used at the indicated dilutions: anti-Ccnb1 V152 (Cell Signaling) at 1:1000, anti-HA 6E2 (Cell Signaling) at 1:1000, anti-Histone H3 (Cell signaling) at 1:1000, anti-Phospho-Histone H3 (Ser10) (Cell Signaling) at 1:500, anti-Tubulin (Sigma) at 1:1000 and anti-Topoisomerase I (Santa Cruz) at 1:500. Anti-β-actin (Sigma) at 1:3000 was used as a loading control.
Subcellular fractionation
Cytoplasmic and nuclear fractions were prepared by using the NE-PER Nuclear and Cytoplasmic Extraction Reagents (Thermo Scientific). Immunodetection of β-tubulin and Topoisomerase I were used as cytoplasmic and nuclear markers, respectively.
Chromatin immunoprecipitation
NIH/3T3-Ago1, NIH/3T3-Ago2, or NIH/3T3-eGFP cells were cultured to ~70-80% confluency in 10-cm plates. Cells were fixed with 1% formaldehyde in serum-free media for 10 min at room temperature. Formaldehyde crosslinking was quenched with 125 µM glycine (final concentration). Cells were washed in PBS and nuclei were prepared as previously described (28). Cell suspension was sonicated 12 times in 10-s pulses to yield DNA fragments with an average length of ~500 bp. Lysates were pre-cleared with Protein A/G sepharose beads (GE Healthcare) and subsequently incubated with or without 5 µl of anti-HA antibody (Cell Signaling) at 4°C overnight. Samples omitting antibody were used as negative controls. Fresh beads were then added for 2 h to immunoprecipitate chromatin and sequentially washed in low salt, high salt, LiCl and TE buffer. DNA/protein complexes were eluted and crosslinking was reversed by heating overnight at 65°C. Samples were subsequently treated with Proteinase K followed by phenol/chloroform extraction and RNase A treatment. DNA fragments were then purified in nuclease free water using Qiaquick PCR purification columns (Qiagen). For RNAP II and H3K4me3 ChIP experiments, NIH/3T3 cells were transfected at 100 nM dsControl or miR-744 for 3 days, while TRAMP-C1-miR-EV and TRAMP-C1-miR-744 cells were cultured to ~70–80% confluency in 10 cm plates prior to harvest. Antibodies to RNAP II and H3K4me3 were purchased from Millipore. For ChIP–immunoblot analysis, NIH/3T3-Ago1 cells were transfected with 100 nM biotin-labeled miR-744 or dsControl for 3 days. Immunoprecipitation was performed with Dynabeads® M-280 Streptavidin (invitrogen) followed by immunoblotting with an anti-HA antibody.
ChIP and quantitative PCR analysis (ChIP–qPCR)
Primers covering the 2-kb promoter region of mouse Ccnb1 were designed using Primer Express v2.0 (Applied Biosystems). Each PCR reaction was performed in triplicate in 10 µl reaction volumes containing 2 µl of ChIP–enriched or input DNA, Power SYBR Green master mix (Applied Biosystems), and region specific primer sets. Target amplification and detection was performed by the ABI Prism 7500 Real-Time System. Fold enrichment = 2−(delta CTexpt – delta CTcontrol) where delta CT = CTIP – CTno antibody control, expt = Ccnb1 promoter, control = negative control region. Specificity of the PCR reactions was confirmed by electrophoresis and dissociation curve analysis. Primer sequences are listed in Supplementary Table S3.
Propidium iodide staining and cell cycle analysis
Cells were trypsinized and centrifuged at 1000 rpm for 5 min at 4°C. Cell pellets were resuspended in 500 µl cold saline GM solution (6.1 mM glucose, 1.5 mM NaCl, 5.4 mM KCl, 1.5 mM Na2HPO4, 0.9 mM KH2PO4, 0.5 mM EDTA). The resuspended cells were fixed in 1.5 ml cold 100% ethanol for at least 2 h at 4°C. Cells were washed twice with cold PBS and resuspended in 1 ml propidium iodide staining solution (50 µg/ml propidium iodide, 10 mg/ml RNase A, 0.1% Triton X-100, 0.1% sodium citrate in PBS) and incubated at room temperature for 30 min. The stained cells were immediately analyzed on a FACSCaliber flow cytometer (UCSF Laboratory of Cell Analysis) for relative DNA content. Collected data was evaluated using ModFit software (Verity Software House, Topsham ME).
Cell proliferation and doubling time analysis
TRAMP C1 cells (1 × 104) expressing empty vector, miR-744, miR-1186, or Ccnb1 cDNA were seeded in six-well plates. The cells were trypsinized and viable cells were counted on Days 1–5. Doubling time was calculated from four independent experiments using an online doubling time calculator (http://www.doubling-time.com/compute.php). For the growth rescue experiment, TRAMP-C1-miR-EV and TRAMP-C1-miR-744 cells were transfected with 50 nM dsControl or siCcnb1 in a 96-well format. Cell proliferation was measured by using the CellTiter96 Aqueous One Solution cell proliferation assay kit (Promega) according to manufacturer's protocol.
Karyotyping
Parental or TRAMP C1 sublines were treated with 10 ng/ml Colcemid (Invitrogen) in culture medium overnight at 37°C. Cells were harvested and G-banded according to the standard mouse karyotyping protocol (28). The G-banded chromosomes were analyzed using CytoVision system (Applied Imaging) and karyotyped according to the standard mouse chromosome nomenclature (29,30).
Xenograft model
TRAMP C1 stable cell lines were grown to ~80% confluency in 15-cm dishes, trypsinized, and counted. Cells (2 × 106) in 200 µl PBS containing 50% matrigel were injected into the right lower flank of 4-week-old male athymic (nu/nu) mice (Simonsen Laboratories). The mice were monitored for tumor volume, body weight, and overall health for 35 days. Digital calipers were used to monitor tumor size and volume utilizing the formula (width × width × length)/2. Each experimental and control group contained six mice. At the end of the animal study, all mice were sacrificed according to protocol approved by the Institutional Animal Care and Use Committee (IACUC). Body and tumor weight were recorded following tumor resection.
Statistical analysis
Data was analyzed using the StatView program (SAS Institute Inc., Cary, NC). The χ2 test was used to assess the significance of the differences between proportions. The difference in continuous variables between treatments was assessed by Student's t-test or ANOVA. In all cases, P-values < 0.05 were considered statistically significant.
RESULTS
Ccnb1 expression depends on components of the miRNA pathway
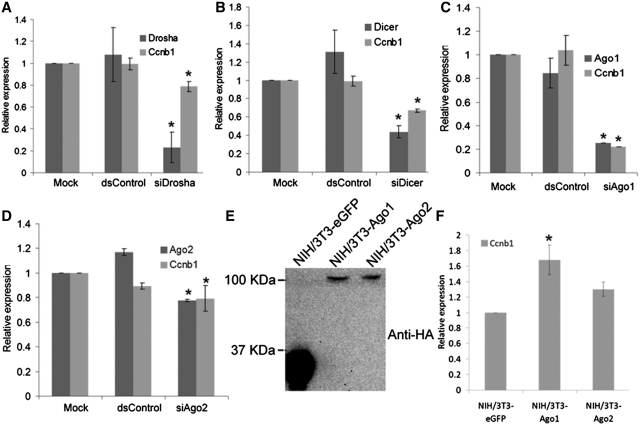
Canonical biogenesis of functionally mature miRNA is dependent, in part, on ribonuclease III family members Drosha and Dicer (2). It is anticipated that the expression of genes positively-regulated by endogenous miRNAs may be sensitive to perturbation in miRNA maturation. In support, RNAa facilitated by pre-miRNA has been shown to be dependent on Dicer for gene activation (23). To identify putative examples of endogenous miRNA-mediated gene activation, we depleted Drosha or Dicer by RNAi in NIH/3T3 (mouse embryonic fibroblast) cells and screened for transcripts cooperatively depleted with Drosha or Dicer by real-time PCR (data not shown). Surprisingly, depletion of the miRNA biogenesis components downregulated a large number of cell cycle genes, including Cyclin B1 (Ccnb1), implying that miRNAs may positively regulate cell-cycle genes. As shown in Figure 1A and B, knockdown of Drosha and Dicer depleted Ccnb1 mRNA expression by ~20% and ~35%, respectively.
Figure 1.
Ccnb1 expression depends on components of the miRNA pathway. (A–D) NIH/3T3 cells were transfected at 50 nM with the indicated siRNAs for 72 h. dsControl served as non-specific control duplex. Mock samples were transfected in the absence of siRNA. Relative expression levels were quantified by real-time PCR using gene-specific primer sets. Values were normalized to β-actin. (E) Establishment of NIH/3T3-Ago1 and NIH/3T3-Ago2 cell lines. Expression of HA-tagged Ago1 and Ago2 was confirmed by immunoblot analysis using an antibody specific to the HA epitope (anti-HA). NIH/3T3-eGFP cells served as a control cell line. (F) Expression levels of Ccnb1 were determined by real-time PCR in NIH/3T3-Ago1 and NIH/3T3-Ago2 cell lines relative to NIH/3T3-eGFP cells. Values were normalized to β-actin. All data represents mean ± SE of three independent experiments; *P < 0.05.
The function of miRNAs largely depends on the Ago subfamily of proteins (Ago1 through Ago4), which function as components of the miRNA effector ribonucleoprotein (miRNP). Ago1 and Ago2 have been shown to mediate dsRNA-guided transcriptional gene silencing (31,32) and activation (33,34) suggesting a universal role for Ago proteins in most, if not all, mechanisms of small RNA-guided gene regulation. To examine whether Ago1 and Ago2 are also required in miRNA-mediated Ccnb1 gene activation, we transiently knocked down Ago1 or Ago2 in NIH/3T3 cells by siRNA (siAgo1 or siAgo2) and examined Ccnb1 expression. siAgo1 resulted in >70% knockdown of Ago1 and caused a corresponding ~70% reduction in Ccnb1 mRNA level as detected at 72 h following siAgo1 transfection (Figure 1C). Despite the low efficiency of Ago2 knockdown, siAgo2 also caused a subtle reduction (~20%) in Ccnb1 mRNA levels (Figure 1D). Conversely, we established stable lines from NIH/3T3 cells that expressed HA-tagged human Ago1 or Ago2 (NIH/3T3-Ago1 or NIH/3T3-Ago2) (Figure 1E) and examined Ccnb1 expression. As shown in Figure 1F, Ccnb1 mRNA was increased by ~1.7-fold in NIH/3T3-Ago1 cells as compared to the control NIH/3T3 cell line expressing only enhanced GFP (NIH/3T3-eGFP). Ccnb1 mRNA was also moderately induced (~1.3-fold) by Ago2 overexpression (Figure 1F). Taken together, these results indicate that key components of miRNA biogenesis and the miRNP effector complex positively regulate Ccnb1 expression.
miRNA-mediated activation of Ccnb1 expression
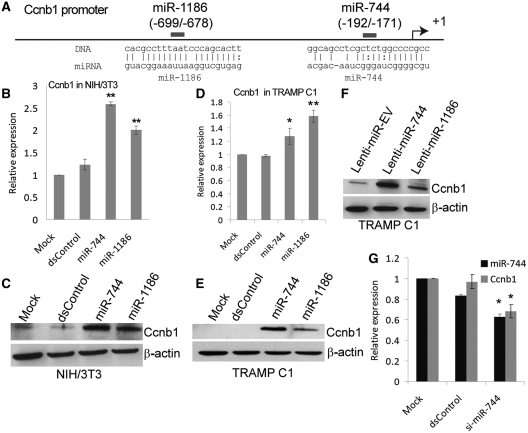
To identify candidate miRNA(s) that may be involved in positively regulating Ccnb1 expression, we used the miRanda algorithm (35) to scan 1-kb of Ccnb1 promoter sequence for potential target sites complementary to known mouse miRNAs included in the miRBASE database (www.mirbase.org). By setting the prediction stringency level to medium-high, in silico analysis identified a total of 22 putative targets for 21 miRNAs on the Ccnb1 promoter (Supplementary Table S1). The top four miRNAs with the highest alignment scores including miR-1186, miR-744, miR-1196 and miR-1962 were selected for further validation. These miRNAs have significant sequence homology with the Ccnb1 promoter both in their ‘seed’ (nucleotide 2–8) and extended regions (Supplementary Table S1), but do not possess putative target sites in either the 3′- or 5′-UTR of the Ccnb1 transcript. We transfected synthetic miRNA mimics of the four candidate miRNAs or a control miRNA (dsControl) into NIH/3T3 cells and assessed Ccnb1 expression levels five days later. Compared to control treatments, miR-744 and miR-1186 caused a 2.6- and 2-fold induction in Ccnb1 mRNA levels, respectively (Figure 2A and B). However, transfection of miR-1196 or miR-1962 had little-to-no effect on Ccnb1 transcript levels (Supplementary Figure S1A and B). At Day 5, cellular levels of miR-744 and miR-1186 were increased >300-fold following transfection of their respective mimics compared to controls (Supplementary Figure S2A and B). The induction of Ccnb1 by miR-744 and miR-1186 was further confirmed by immunoblot analysis in NIH/3T3 cells (Figure 2C). Sequence requirement was investigated by mutating the “seed” region of both miR-744 and miR-1186. The mutation in both miRNAs (miR-744-MM and miR-1186-MM) abolished their ability to activate Ccnb1 expression (Supplementary Table S3, Supplementary Figure S3A and B). In contrast, a miR-744 variant (miR-744-Perfect) with improved complementary to the predicted miR-744 target site did not suffer from reduced activity; rather, it retained function and readily activated Ccnb1 expression in a manner similar to miR-744 (Supplementary Figure S3A and C).
Figure 2.
miRNAs targeting the Ccnb1 promoter induce Ccnb1 expression. (A) Schematic representation of the Ccnb1 promoter and miRNA target locations relative to the TSS. (B and D) NIH/3T3 or TRAMP C1 cells were transfected with 100 nM miRNA control (dsControl), miR-744, or miR-1186 for 5 days. Mock samples were transfected in the absence of miRNA. Ccnb1 mRNA levels were analyzed by real-time RT–PCR and normalized to β-actin levels. (C and E) NIH/3T3 or TRAMP C1 cells were transfected as in (B and D). Proteins levels of Ccnb1 and β-actin were evaluated by immunoblot analysis. Detection of β-actin served as a loading control. (F) Ccnb1 and β-actin protein levels were determined by immunoblot analysis in stable TRAMP C1 cells infected with either lenti-miR-744 or lenti-miR-1186 virus particles. Cells stably expressing the empty vector (Lenti-miR-EV) were established as a control. (G) NIH/3T3 cells were transfected with 100 nM dsControl or si-miR-744 for 72 h. miR-744 levels were evaluated by real-time RT–PCR and standardized to U6 levels, while Ccnb1 expression was normalized to β-actin. All data represents mean ± SE of three independent experiments; *P < 0.05, **P < 0.005.
miR-744 and miR-1186 were selected for subsequent studies given that both miRNAs induced Ccnb1 expression. miR-744 potentially targets position −192 in the Ccnb1 promoter relative to the transcription start site (TSS) with an overall sequence identity of 93% (Figure 2A and Supplementary Table S1). This site, in particular the sequence complementary to the ‘seed’ region, is also conserved among seven mammalian species (Supplementary Figure S4). miR-1186 targets a repetitive region on the Ccnb1 promoter at position −699 with a 94% sequence identity (Figure 2A and Supplementary Table S1). We further transfected miR-744 and miR-1186 mimics into the TRAMP C1 cells and observed a ~1.3- and ~1.6-fold induction in Ccnb1 mRNA levels, respectively (Figure 2D). Immunoblot analysis confirmed that Ccnb1 protein expression was also induced by both miRNAs in TRAMP C1 cells (Figure 2E).
To determine if constitutive expression of miRNA could also induce Ccnb1 expression, we stably expressed precursor miR-744 (Lenti-miR-744) and miR-1186 (Lenti-miR-1186) in TRAMP C1 cells via lentiviral-mediated transduction. Lenti-miR-744 and Lenti-miR-1186 caused a respective increase in the levels of mature miR-744 and miR-1186 by ~10- and 1.5-fold as compared to the empty vector control (Lenti-miR-EV) (Supplementary Figure S5A and B). As shown in Figure 2F, Ccnb1 protein levels were induced in both miR-744 and miR-1186 stable expressing cell lines suggesting naturally-processed miRNAs also induce Ccnb1.
To verify if downregulation of Ccnb1 also correlated with depletion of miRNA levels following perturbation of Dicer or Drosha, we evaluated endogenous miR-744 levels and found a 30–60% reduction in mature miR-744 following siDicer and siDrosha treatments, respectively (Supplementary Figure S6A). Ago proteins have been shown to stabilize mature miRNAs leading to an increase in their abundance (36). Indeed, overexpression of either Ago1 or Ago2 caused a subtle increase in the levels of mature miR-744 (Supplementary Figure S6B) suggesting Ccnb1 induction upon Ago1 or Ago2 overexpression may result, in part, from a stabilization of miR-744. Knockdown of Ago1 or Ago2 did not cause any significant reduction in the steady-state levels of mature miR-744 (Supplementary Figure S6A). As such, declines in Ccnb1 following siAgo1 and siAgo2 treatments occurred regardless of miR-744 levels suggesting Ago proteins may function as mediators of miRNA-guided Ccnb1 transcription.
To further demonstrate a correlation between miR-744 depletion and Ccnb1 downregulation, we used a pool of two siRNAs (si-miR-744) to knockdown endogenous miR-744 expression. The siRNAs were designed to target the pre-miR-744 loop sequence, which decreased miR-744 levels by ~40% compared to control siRNA (dsControl) treatments in NIH/3T3 cells (Figure 2G). Concurrently, si-miR-744 also decreased Ccnb1 mRNA expression by ~32% (Figure 2G).
Although knockdown of miR-744 correlated with Ccnb1 downregulation, other miRNAs (i.e. miR-1186) may also function to positively-regulate Ccnb1 expression. To determine if other miRNAs from the target predication list (Supplementary Table S1) also influenced Ccnb1 expression, we transfected NIH/3T3 cells with miR-466d-3p. Although miR-466 d-3p is ranked 19th on the list by alignment score, it possesses a higher sequence complementarity within its ‘seed’ and extended regions to its putative target site in the Ccnb1 promoter compared to other higher ranking miRNAs (Supplementary Table S1). Transfection of miR-466d-3p caused ~1.5-fold increase in Ccnb1 mRNA and protein expression (Supplementary Figure S1A, C and D). Taken together, these findings indicate that Ccnb1 transcription may be positively-regulated by multiple miRNAs, which may have physiological context in disease and miRNA dysregulation.
Ago1 is associated with Ccnb1 promoter
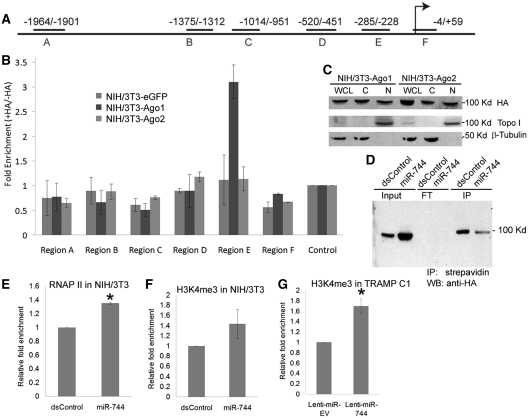
Since Ccnb1 expression depends on Ago1 and Ago2, we reasoned that they may be recruited to the Ccnb1 promoter. To test this view, we performed scanning ChIP to examine the association of Ago1 and/or Ago2 at the mouse Ccnb1 promoter in stable NIH/3T3 cell lines overexpressing HA-tagged Ago1 (NIH/3T3-Ago1) or Ago2 (NIH/3T3-Ago2) by using an HA-specific antibody. Stable NIH/3T3 cells expressing HA-tagged eGFP (NIH/3T3-eGFP) were used as a negative control. Six primer pairs encompassing select regions (Region A–F) across 2-kb of the mouse Ccnb1 promoter were designed to amplify DNA pulled down by the HA-tagged Ago proteins (Figure 3A). We quantitatively determined enrichment of HA-Ago1 and HA-Ago2 at the selected regions by real-time PCR. All data was normalized to an intergenic control (Control) region anticipated to have no Ago protein association. A ~3-fold enrichment of HA-Ago1 was detected in Region E (−285/−228) of the Ccnb1 promoter, which was just proximal to the putative miR-744-binding site (−192/−171) (Figure 3B). No significant HA-Ago2 enrichment was detected in the scanned promoter regions in NIH/3T3-Ago2 cells (Figure 3B). Subcellular fractionation experiments also revealed that both HA-tagged Ago1 and Ago2 localize to the nuclear and cytosolic fractions in the NIH/3T3 sublines (Figure 3C). Furthermore, ChIP–immunoblot analyses in NIH/3T3-Ago1 cells transfected with biotin-labeled miR-744 revealed HA-Ago1 was present in the streptavidin pulldown indicating that miR-744 is bound to Ago1 (Figure 3D). This data indicates HA-Ago1 is enriched at the Ccnb1 promoter, found in the nucleus, and can associate with exogenous miR-744.
Figure 3.
Ago1 associates with the proximal promoter of mouse Ccnb1. (A) Schematic illustration of the primer amplicons (Regions A–F) used for scanning ChIP analysis of 2 kb of the proximal Ccnb1 promoter. Locations are shown relative to the TSS. (B) ChIP experiments were performed with an anti-HA antibody in NIH/3T3-Ago1, NIH/3T3-Ago2 and NIH/3T3-eGFP cells to examine Ago1/2 enrichment within the Ccnb1 promoter. Real-time PCR was utilized to quantify fold enrichment of Ago1 and/or Ago2 at designated primer regions relative to no antibody controls. Data was normalized to an intergenic region (Control) devoid of Ago binding. (C) Subcellular localization of Ago1 and Ago2. Whole-cell lysate (WCL), nuclear (N) and cytoplasmic (C) extracts were analyzed by immunoblot analysis in NIH/3T3-Ago1 and NIH/3T3-Ago2 cell lines. HA-tagged Ago1 and Ago2 were detected using an anti-HA antibody. Topoisomerase I (Topo I) and β-tubulin served as nuclear and cytoplasmic markers, respectively. (D) Biotinylated miR-744 associates with Ago1. NIH/3T3-Ago1 cells were transfected with biotinylated dsControl or miR-744 at 100 nM for 3 days. Immunoprecipitation (IP) was performed with streptavidin Dynabeads followed by immunoblotting (IB) with an anti-HA antibody to detect HA-tagged Ago1. FT: flowthrough, Input: 10% input chromatin. (E and F) Enrichment of RNAP II and H3K4me3 at the Ccnb1 promoter. NIH/3T3 cells were transfected with 100 nM dsControl or miR-744 for 72 h. ChIP–qPCR was utilized to quantify enrichment of RNAP II and H3K4me3 at the Ccnb1 TSS (Region F). Fold changes are relative to dsControl treatments and normalized to amplification values in IgG control samples. (G) ChIP–qPCR analysis of H3K4me3 at the Ccnb1 TSS in TRAMP C1 sublines expressing either lenti-miR-EV or lenti-miR-744. Relative enrichment was calculated as in (E and F). Error bars represent mean ± SE of two independent experiments; *P < 0.05.
Upregulation of Ccnb1 by miR-744 correlates with increased RNAP II occupancy and H3K4me3 at Ccnb1 promoter
Transcription activation is associated with RNA polymerase II (RNAP II) enrichment and epigenetic modulation such as histone H3 lysine 4 tri-methylation (H3K4me3) at gene promoters (19,23,37). To determine if miR-744 facilitates such changes at the Ccnb1 promoter, we performed ChIP assays using antibodies specific to RNAP II and H3K4me3. Following miR-744 transfection, ~1.4-fold increase in RNAP II occupancy was observed at the Ccnb1 TSS compared to dsControl (Figure 3E). H3K4me3 levels were also increased at the Ccnb1 TSS in miR-744 transfected NIH/3T3 cells and Lenti-miR-744 stable TRAMP C1 cells (Figure 3F and G). Taken together, these results suggest that miR-744-induced Ccnb1 expression involves RNAP II enrichment and epigenetic changes associated with active transcription. In support, nuclear run-on experiments show direct transcriptional upregulation of Ccnb1 in Lenti-miR-744 stable TRAMP C1 cells as compared to controls (Supplementary Figure S7).
We also generated two artificial reporter systems in which ~3.2 kb of the Ccnb1 promoter was cloned upstream of either GFP or luciferase to further investigate the specificity of Ccnb1 activation. The Ccnb1 promoter-GFP chimera was stably integrated into NIH/3T3 cells, while the luciferase construct was transiently transfected with miRNA. As shown in Supplementary Figure S8, neither miR-744 nor miR-1186 significantly increased the activity of either exogenous reporter system suggesting additional elements other than simple sequence complementarity are required for gene induction.
miR-744 and miR-1186 overexpression enhance cell proliferation
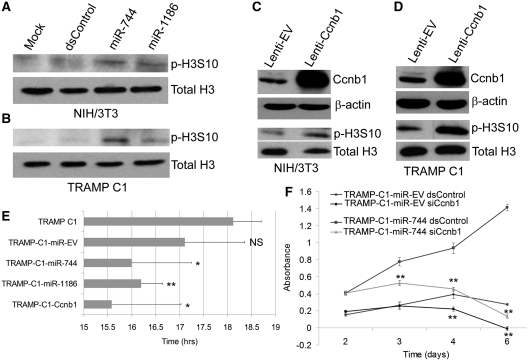
Phenotypically, transfection of miR-744 and miR-1186 mimics into NIH/3T3 and TRAMP C1 cells induced histone H3 phosphorylation at serine 10 (p-H3S10), a marker for cell proliferation that correlates with chromosome condensation during mitosis (Figure 4A and B). Cyclin B1 in complex with Cdk2 controls the G2-M transition of the cell cycle and is frequently overexpressed in a variety of cancer cells to promote proliferation (38,39). To determine if Ccnb1 overexpression recapitulates miR-744 and/or miR-1186 results, we established stable sublines from NIH/3T3 and TRAMP C1 cells by utilizing a lentiviral system to overexpress mouse Cyclin B1 (Lenti-Ccnb1). These sublines also exhibited elevated p-H3S10 levels as compared to control cells expressing an empty vector (Lenti-EV) (Figure 4C and D). Phenotypically, TRAMP C1 cells infected with Lenti-Ccnb1 (TRAMP-C1-Ccnb1), Lenti-miR744 (TRAMP-C1-miR-744), or Lenti-miR1186 (TRAMP-C1-miR-1186) exhibited a shorter doubling time compared to the TRAMP C1 parental line or cells expressing an empty vector control (TRAMP-C1-miR-EV) (Figure 4E).
Figure 4.
miR-744 and miR-1186 promote phosphorylation of histone H3 and enhance in vitro cell proliferation. (A and B) NIH/3T3 and TRAMP C1 cells were transfected with 100 nM dsControl, miR-744 or miR-1186 for 5 days. Mock samples were transfected in the absence of miRNA. Total H3 and phosphorylated H3S10 (p-H3S10) levels were examined by immunoblot analysis using specific antibodies. (C and D) Total H3 and p-H3S10 levels were immunodetected in stable NIH/3T3 and TRAMP C1 sublines infected with either lenti-Ccnb1 or lenti-EV viral particles. (E) The doubling time of TRAMP C1 cells expressing empty vector (TRAMP-C1-miR-EV), miR-744 (TRAMP-C1-miR-744), miR-1186 (TRAMP-C1-miR-1186), or Ccnb1 (TRAMP-C1-Ccnb1) was evaluated over the course of 5 days. The average doubling time (hrs) ± SE of independent experiments is plotted in the corresponding bar graph; *P < 0.05, **P < 0.01, NS: not significant compared to the TRAMP C1 parental cell line. (F) TRAMP-C1-miR-EV or TRAMP-C1-miR-744 cells were transfected with 50 nM dsControl or siCcnb1. Cell proliferation was quantified at each day utilizing the CellTiter96 Aqueous One Solution. Error bars represents SE from four independent transfections. Statistical significance was determined between dsControl and siCcnb1 treatments within each subline; *P < 0.05, **P < 0.01.
To further confirm that Ccnb1 activation is responsible for enhanced cell proliferation by miR-744, we performed a rescue experiment by utilizing RNAi to knockdown Ccnb1 in TRAMP-C1-miR-744 cells. Compared to dsControl treatments, Ccnb1 siRNA (siCcnb1) effectively depleted Cyclin B1 levels in both TRAMP-C1-miR-EV and TRAMP-C1-miR-744 cells (Supplementary Figure S9). However, knockdown of Ccnb1 in TRAMP-C1-miR-744 cells blocked the enhanced proliferative effects associated with miR-774 overexpression (Figure 4F). These findings suggest that miR-744 and miR-1186 have a proliferative function that is, in part, dependent on activation of Ccnb1.
miR-744 and miR-1186 induce cytogenetic changes in TRAMP C1 cells
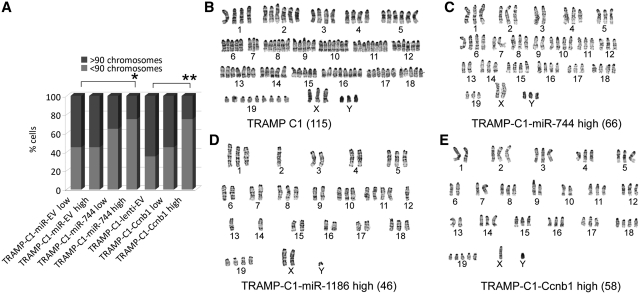
It has also been reported that overexpression of Ccnb1 can result in chromosome instability (CIN) (40). To determine if prolonged expression of Ccnb1, miR-744 and/or miR-1186 affects the cytogenetic profile in mouse prostate cancer cells in a manner similar to Ccnb1, we performed karyotype analysis in TRAMP-C1-miR-744, TRAMP-C1-miR-1186 and TRAMP-C1-Ccnb1 cells. Twenty metaphase spreads from each cell line, as well as the parental TRAMP C1 cell line, were subjected to cytogenetic characterization by G-banding.
As previously reported (41), TRAMP C1 cells were highly aneuploid (Figure 5A and B) composed of two major karyotypically discrete cell populations with multiple marker chromosomes of unknown origin (Supplementary Table S2). No cells possessed normal karyotypes; rather, a majority of the cells (75%) contained a chromosome number in the range of 91–130, while 25% contained 51–90 chromosomes (Supplementary Table S2 and Figure 5B). Consistent with this finding, analysis of DNA content by flow cytometry in cells stained with PI revealed that TRAMP C1 cells contained two major cycling populations with differential DNA content referred to as the low and high distributions (Supplementary Figure S10, top panel). Forward versus side scatter plots showed that TRAMP C1 cells possessed one phenotypically homogenous population excluding the possibility of a mixture of two different cell types (Supplementary Figure S10, bottom panel).
Figure 5.
miR-744, miR-1186 and Ccnb1 induce conserved cytogenetic changes in TRAMP C1 cells. (A) Twenty randomly selected cells from the parental TRAMP C1 or indicated derived sublines were subjected to G-banded karyotyping. Percent cell population (% cells) with chromosome numbers >90 or <90 are indicated in the bar graph; *P < 0.05, **P < 0.001. (B–E) Representative chromosome spreads from parental TRAMP C1 (B), TRAMP-C1-miR-744 high (C), TRAMP-C1-miR-1186 high (D), or TRAMP-C1-Ccnb1 high (E) cells. Denoted in parentheses is the total number of chromosomes found in corresponding spread.
To closely compare the cytogenetic differences between the cell lines, we calculated the percentage of analyzed cells with chromosome numbers >90 or <90 (Figure 5A). Compared to empty vector control cells (TRAMP-C1-miR-EV), miR-744 overexpressing cells (TRAMP-C1-miR-744) exhibited a progressive loss of chromosomes with increased passages. As shown in Figure 5A–C and Supplementary Table S2, 45% of TRAMP-C1-miR-EV cells (either low or high passage) contained chromosome numbers between 51 and 90, while 65% of the early passage (TRAMP-C1-miR-744 low, passage <10) and 75% of the late passage miR-744 overexpressing cells (TRAMP-C1-miR-744 high, passage > 30) (P < 0.05) contained chromosome numbers <90. Furthermore, overexpression of miR-744 significantly increased the number of marker chromosomes (rearranged chromosomes that lost identities of origins) (P < 0.03) (Supplementary Table S2). Late passage miR-1186 overexpressing cells (TRAMP-C1-miR-1186 high, passage >30) also showed a reduction in total chromosome number where only 10% of the analyzed cells contained more than 110 chromosomes and 10% contained chromosome numbers in the lowest range (chromosome number = 31–50) (Supplementary Table S2 and Figure 5D). Similarly, analysis of DNA content by flow cytometry revealed that overexpression of either miR-744 or miR-1186 promoted a shift from a population with high DNA content to low DNA content (Supplementary Figure S10, top panel).
Analysis of TRAMP-C1-Ccnb1 cells also revealed a similar pattern of cytogenetic changes in a passage-dependent manner (Figure 5A and E). Compared to empty vector control (TRAMP-C1-lenti-EV) where 35% of the cells contained chromosome numbers <90, 45% of the early passage (TRAMP-C1-Ccnb1 low, passage <5) and 75% of the late passage Ccnb1 overexpressing cells (TRAMP-C1-Ccnb1 high, passage >10) contained 51–90 chromosomes (Figure 5A and Supplementary Table S2) (P < 0.001). It is noteworthy that TRAMP-C1-Ccnb1 cells required fewer passages to reach a similar degree of CIN found in high passage miR-744 overexpressing cells suggesting that high levels of Ccnb1 accelerate genomic instability (Supplementary Table S2). Taken together, these results suggest that prolonged expression of miR-744 and miR-1186 induced cytogenetic changes in TRAMP C1 cells in a manner similar to Ccnb1 overexpression.
Prolonged overexpression of miR-744 and miR-1186 inhibits tumorigenesis in vivo
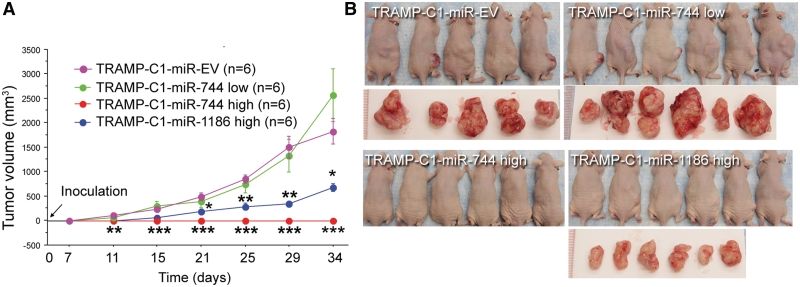
Given that overexpression of Ccnb1 plays a role in tumorigenesis (42,43), we decided to evaluate the tumorigenicity of miR-744 and miR-1186 in a mouse subcutaneous xenograft model. Previous studies showed that TRAMP C1 cells form tumors readily in vivo (27). Indeed, six out of six mice inoculated with TRAMP-C1-miR-EV control cells developed tumors within 2 weeks. Comparatively, mice injected with early passage miR-744 overexpressing cells (TRAMP-C1-miR744 low, passage number <10) formed tumors with an average size larger than the control group by the end of the study (P = 0.06) (Figure 6A) and appeared to be more aggressive (tumors from two mice invaded the spinal column) (Figure 6B). Strikingly, mice injected with late passage miR-744 overexpressing cells (TRAMP-C1-miR744 high, passage number >30) did not give rise to any tumors at the injection site (Figure 6A and B). Late passage miR-1186 overexpressing cells (TRAMP-C1-miR1186 high, passage number >30) also formed significantly smaller tumors than the control group (P < 0.005) (Figure 6A and B). Collectively, our data suggests that short-term expression of miR-744 and/or miR-1186 may promote in vivo tumor growth through Ccnb1 activation; however, prolonged activation of Ccnb1 may cause chromosomal instability in TRAMP C1 cells and inhibit tumor development/growth. In support, high levels of genetic instability have been reported to have a tumor suppressive effect in certain context (44,45).
Figure 6.
Prolonged overexpression of miR-744 or miR-1186 alters in vivo tumor growth. (A) Cells (2 × 106) from the indicated stable TRAMP C1 sublines were injected into the right lower flank of male athymic mice and monitored for 35 days. Subcutaneous tumor dimensions were recorded using calipers at the indicated time points (days) following initial inoculation. Tumor volume was calculated according to the formula (width × width × length)/2. Data is plotted as the mean volume ± SE; *P < 0.05; **P < 0.01; ***P < 0.001. (B) Photographs of inoculated mice and corresponding resected tumors from each group at time of sacrifice on Day 35.
DISCUSSION
In the present work, we demonstrate that the expression of Ccnb1 depends on components of miRNA biogenesis (Drosha and Dicer) and function (Ago1 and Ago2) in mouse cells. We identify several miRNAs (e.g. miR-744, miR-1186, etc.) with predicted target sites in the Ccnb1 promoter that induce Ccnb1 expression. Mechanistically, we indicate that Ago1 and Ago2 are abundant in the nucleus consistent with previous observations (46). Ago1 is localized to the proximal promoter region of Ccnb1 and miR-744 promotes RNAP II and H3K4me3 enrichment at the Ccnb1 TSS. Phenotypically, Ccnb1 overexpression has been reported to promote tumorigenesis and function as a putative oncogene in a variety of cancers (42,43,47–49). As such, short-term overexpression of miR-744 or miR-1186 led to enhanced tumor cell proliferation in a manner similar to vector-mediated overexpression of Ccnb1. However, long-term expression in TRAMP C1 mouse cells altered chromosome composition suggesting prolonged Ccnb1 activation may cause chromosomal instability. Xenografts derived from early passage cells overexpressing miR-744 also possessed enhanced tumorigenicity, while cells from late passage populations displayed poor tumor growth in vivo. Our data suggests that short-term expression of miR-744 and/or miR-1186 may promote in vivo tumor growth through Ccnb1 activation; however, prolonged activation of Ccnb1 may cause chromosomal instability and inhibit tumor growth. This study reveals a physiologically relevant role for miRNA-mediated gene activation, which may be implicated in cancer initiation and/or development.
miRNAs are known to participate in a wide variety of cellular functions by suppressing gene expression via binding complementary sites in the 3′-UTR of mRNA sequences. Target predication in gene transcripts is largely dependent on sequence complementarity within the ‘seed’ region and target conservation across multiple species (50,51). However, without experimental validation, such computational predictions are not always bona fide targets (50). Predicting miRNA target sites in gene promoters is even more challenging due to a lack of relevant experimental data and general poor conservation of promoters across different species. However, by repurposing target prediction algorithms (e.g. miRanda) to scan promoter sequence for sites complementary to miRNAs, numerous putative targets can be identified with reasonably high score (22,52). As such, it has been postulated that miRNAs may also play a regulatory role in fine-tuning gene transcription in the nucleus (52,53).
In silco analysis utilizing the miRanda algorithm (35) revealed several miRNAs with sites highly-complementary to sequence in the Ccnb1 promoter. Of the five miRNAs tested, three resulted in Ccnb1 induction. Additionally, we previously reported two artificial dsRNAs with perfect complementary to target sites located at −313 and −597 relative to the TSS that also activated Ccnb1 expression (18). The ability to simply activate gene expression cannot exclude the possibility that miRNAs/dsRNAs are functioning through a canonical mechanism of gene silencing and Ccnb1 induction is a consequence of downregulation of other regulatory genes. If taken into consideration that each functioning miRNA and dsRNA possesses a different ‘seed’ sequence and targets non-overlapping regions, the likelihood that each of the five duplexes induces Ccnb1 expression via suppression of common or different upstream regulatory genes is potentially low. Their only apparent similarity is that they all putatively target the same gene promoter. ChIP analysis did reveal an enrichment of Ago1 at the Ccnb1 promoter in vicinity close to the putative miR-744 target site providing a direct linking between the miRNA machinery and Ccnb1 promoter. However, miRNAs failed to increase Ccnb1 promoter activity in two reporter systems including a stably integrated Ccnb1 promoter-GFP chimera and transiently transfected luciferase reporter. We reason that promoter environment plays a critical role in determining susceptibility to gene activation mediated by miRNA. The cloned promoter may not fully recapitulate context of the endogenous environment. For instance, chromatin state, nucleosome positioning and/or the presence of potential regulatory non-coding transcript(s) may not be preserved in the exogenous promoters. Models have implicated overlapping antisense and/or promoter-derived cryptic non-coding RNAs as target molecules for facilitating RNAa and TGS (8,19,34,52,54,55). Such transcripts may derive from sites outside the cloned promoter (e.g. intragenic regions), which would be missing in the reporter constructs.
We and others have previously shown that RNAa mediated by synthetic dsRNA with perfect complementary to promoter sequence requires Ago2 for optimal activity (33,34,56). However, gene knockdown and overexpression experiments performed within this study revealed Ago1 had greater impact on Ccnb1 expression than Ago2 in context to miRNA-mediated Ccnb1 induction. ChIP analysis further indicated a selective enrichment of only Ago1 protein at the Ccnb1 promoter. Mescalchin et al. (57) has shown that Ago2 is primarily involved in siRNA-mediated silencing pathways, while Ago1 and other family members are primarily involved in miRNA-mediated gene regulation. This segregation in Ago protein function may also remain for mechanisms of transcriptional regulation mediated by miRNA or perfect dsRNA duplexes. In support, Ago1 has been reported to be involved in miRNA-mediated TGS (31).
Prostate cancer progression is accompanied by accumulating genetic alterations and CIN (58). As such, features associated with karyotypic instability have been identified in human prostate cancer cell lines (59,60). Similarly, the mouse prostate TRAMP C1 cell line also showed complex chromosomal abnormalities including polyploidy, aneuploidy and structural rearrangements indicative of chromosome/genome instability. It is noteworthy to point out that Ccnb1 overexpression appeared to enhance CIN in TRAMP C1 cells by promoting further numerical and structural alterations including loss of chromosomes (aneuploidy) and gain of additional marker chromosomes. Interestingly, miR-744 and miR-1186 also lead to similar chromosomal irregularities suggesting miRNAs (i.e. miR-744) might impair chromosome segregation/maintenance machinery via activation of Ccnb1 and render cancer cells more susceptible to cytogenetic alterations over time
It has been proposed that aneuploidy promotes tumorigenesis (45). However, studies have also shown it may function as a tumor suppressor in context to some cells (61). We made an unexpected finding in that high passage cells overexpressing miR-744 or miR-1186 possessed reduced tumorigenicity in the TRAMP C1 xenograft model. We speculate that the cytogenetic alteration resulting from Ccnb1 activation may have reduced viability and hampered TRAMP C1 cell growth. Cancer cells possess a dynamically balanced genome in order to maintain a growth advantage. The cytogenetic alteration caused by miRNA or Ccnb1 overexpression may disturb this balance resulting in a loss of some tumorigenic features.
In summary, the present study provides evidence that miRNA-mediated upregulation of gene expression functions in physiological context of cancer within a mouse model. We show activation of Ccnb1 by miRNA alters the growth potential of TRAMP C1 cells both in vitro and in vivo. Short-term overexpression of miR-744 or miR-1186 leads to enhanced cell proliferation, while long-term expression alters chromosome composition and negatively impacts tumor growth. We speculate that endogenous miRNA may facilitate RNAa in cancer to fine-tune gene expression and manipulate cancer cell phenotype. Further study is needed to identify additional examples and elucidate mechanism(s) by which miRNAs directly activate gene expression and oncogenic pathways.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online: Supplementary Tables S1–S3, Supplementary Figures S1–S10, Supplementary References [62–64].
FUNDING
National Institutes of Health (1R01GM090293-0109 to L-C.L.); Department of Defense (W81XWH-08-1-0260 to L-C.L., W81XWH-10-1-0505 to V.H.); National Institutes of Health/National Cancer Institute, University of California, San Francisco SPORE Special Program of Research Excellence (P50CA89520 to M.S.). Funding for open access charge: Department of Defense (W81XWH-08-1-0260).
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
The authors would like to thank Dr Marcella Fasso for generously donating the TRAMP C1 cells with permission from Dr Norman Greenberg. In addition, Dr Margaret Hough kindly provided additional mouse Ccnb1 promoter reporter constructs for comparative sequencing analysis and confirmation of reporter results.
REFERENCES
- 1.Czech B, Hannon GJ. Small RNA sorting: matchmaking for Argonautes. Nat. Rev. Genet. 2011;12:19–31. doi: 10.1038/nrg2916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 3.Ambros V. microRNAs: tiny regulators with great potential. Cell. 2001;107:823–826. doi: 10.1016/s0092-8674(01)00616-x. [DOI] [PubMed] [Google Scholar]
- 4.Lytle JR, Yario TA, Steitz JA. Target mRNAs are repressed as efficiently by microRNA-binding sites in the 5′ UTR as in the 3′ UTR. Proc. Natl Acad. Sci. USA. 2007;104:9667–9672. doi: 10.1073/pnas.0703820104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lee I, Ajay SS, Yook JI, Kim HS, Hong SH, Kim NH, Dhanasekaran SM, Chinnaiyan AM, Athey BD. New class of microRNA targets containing simultaneous 5′-UTR and 3′-UTR interaction sites. Genome Res. 2009;19:1175–1183. doi: 10.1101/gr.089367.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tay Y, Zhang J, Thomson AM, Lim B, Rigoutsos I. MicroRNAs to Nanog, Oct4 and Sox2 coding regions modulate embryonic stem cell differentiation. Nature. 2008;455:1124–1128. doi: 10.1038/nature07299. [DOI] [PubMed] [Google Scholar]
- 7.Kim DH, Saetrom P, Snove O, Jr, Rossi JJ. MicroRNA-directed transcriptional gene silencing in mammalian cells. Proc. Natl Acad. Sci. USA. 2008;105:16230–16235. doi: 10.1073/pnas.0808830105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Younger ST, Corey DR. Transcriptional gene silencing in mammalian cells by miRNA mimics that target gene promoters. Nucleic Acids Res. 2011;39:5682–5691. doi: 10.1093/nar/gkr155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Younger ST, Corey DR. Transcriptional regulation by miRNA mimics that target sequences downstream of gene termini. Mol. Biosyst. 2011;7:2383–2388. doi: 10.1039/c1mb05090g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jopling CL, Yi M, Lancaster AM, Lemon SM, Sarnow P. Modulation of hepatitis C virus RNA abundance by a liver-specific MicroRNA. Science. 2005;309:1577–1581. doi: 10.1126/science.1113329. [DOI] [PubMed] [Google Scholar]
- 11.Roberts AP, Lewis AP, Jopling CL. miR-122 activates hepatitis C virus translation by a specialized mechanism requiring particular RNA components. Nucleic Acids Res. 2011;39:7716–7729. doi: 10.1093/nar/gkr426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Vasudevan S, Steitz JA. AU-Rich-Element-Mediated Upregulation of Translation by FXR1 and Argonaute 2. Cell. 2007;128:1105–1118. doi: 10.1016/j.cell.2007.01.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Vasudevan S, Tong Y, Steitz JA. Switching from repression to activation: microRNAs can up-regulate translation. Science. 2007;318:1931–1934. doi: 10.1126/science.1149460. [DOI] [PubMed] [Google Scholar]
- 14.Orom UA, Nielsen FC, Lund AH. MicroRNA-10a binds the 5′UTR of ribosomal protein mRNAs and enhances their translation. Mol. Cell. 2008;30:460–471. doi: 10.1016/j.molcel.2008.05.001. [DOI] [PubMed] [Google Scholar]
- 15.Morris KV, Chan SW, Jacobsen SE, Looney DJ. Small interfering RNA-induced transcriptional gene silencing in human cells. Science. 2004;305:1289–1292. doi: 10.1126/science.1101372. [DOI] [PubMed] [Google Scholar]
- 16.Li LC, Okino ST, Zhao H, Pookot D, Place RF, Urakami S, Enokida H, Dahiya R. Small dsRNAs induce transcriptional activation in human cells. Proc. Natl Acad. Sci. USA. 2006;103:17337–17342. doi: 10.1073/pnas.0607015103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Janowski BA, Younger ST, Hardy DB, Ram R, Huffman KE, Corey DR. Activating gene expression in mammalian cells with promoter-targeted duplex RNAs. Nat. Chem. Biol. 2007;3:166–173. doi: 10.1038/nchembio860. [DOI] [PubMed] [Google Scholar]
- 18.Huang V, Qin Y, Wang J, Wang X, Place RF, Lin G, Lue TF, Li LC. RNAa is conserved in mammalian cells. PLoS One. 2010;5:e8848. doi: 10.1371/journal.pone.0008848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Matsui M, Sakurai F, Elbashir S, Foster DJ, Manoharan M, Corey DR. Activation of LDL receptor expression by small RNAs complementary to a noncoding transcript that overlaps the LDLR promoter. Chem. Biol. 2010;17:1344–1355. doi: 10.1016/j.chembiol.2010.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wang J, Place RF, Huang V, Wang X, Noonan EJ, Magyar CE, Huang J, Li LC. Prognostic Value and Function of KLF4 in Prostate Cancer: RNAa and Vector-Mediated Overexpression Identify KLF4 as an Inhibitor of Tumor Cell Growth and Migration. Cancer Res. 2010;70:10182–10191. doi: 10.1158/0008-5472.CAN-10-2414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Liao JY, Ma LM, Guo YH, Zhang YC, Zhou H, Shao P, Chen YQ, Qu LH. Deep sequencing of human nuclear and cytoplasmic small RNAs reveals an unexpectedly complex subcellular distribution of miRNAs and tRNA 3′ trailers. PLoS ONE. 2010;5:e10563. doi: 10.1371/journal.pone.0010563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Younger ST, Pertsemlidis A, Corey DR. Predicting potential miRNA target sites within gene promoters. Bioorg. Med. Chem. Lett. 2009;19:3791–3794. doi: 10.1016/j.bmcl.2009.04.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Place RF, Li LC, Pookot D, Noonan EJ, Dahiya R. MicroRNA-373 induces expression of genes with complementary promoter sequences. Proc. Natl Acad. Sci. USA. 2008;105:1608–1613. doi: 10.1073/pnas.0707594105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.He L, Thomson JM, Hemann MT, Hernando-Monge E, Mu D, Goodson S, Powers S, Cordon-Cardo C, Lowe SW, Hannon GJ, et al. A microRNA polycistron as a potential human oncogene. Nature. 2005;435:828–833. doi: 10.1038/nature03552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Johnson SM, Grosshans H, Shingara J, Byrom M, Jarvis R, Cheng A, Labourier E, Reinert KL, Brown D, Slack FJ. RAS is regulated by the let-7 microRNA family. Cell. 2005;120:635–647. doi: 10.1016/j.cell.2005.01.014. [DOI] [PubMed] [Google Scholar]
- 26.Enright AJ, John B, Gaul U, Tuschl T, Sander C, Marks DS. MicroRNA targets in Drosophila. Genome Biol. 2003;5:R1. doi: 10.1186/gb-2003-5-1-r1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Foster BA, Gingrich JR, Kwon ED, Madias C, Greenberg NM. Characterization of prostatic epithelial cell lines derived from transgenic adenocarcinoma of the mouse prostate (TRAMP) model. Cancer Res. 1997;57:3325–3330. [PubMed] [Google Scholar]
- 28.Shang Y, Hu X, DiRenzo J, Lazar MA, Brown M. Cofactor dynamics and sufficiency in estrogen receptor-regulated transcription. Cell. 2000;103:843–852. doi: 10.1016/s0092-8674(00)00188-4. [DOI] [PubMed] [Google Scholar]
- 29.Nagy A, Gertsenstein M, Vintersten K, Behringer R. Karyotyping mouse cells. Cold Spring Harb. Protoc. 2008;2008 doi: 10.1101/pdb.prot4706. doi:10.1101/pdb.prot4706. [DOI] [PubMed] [Google Scholar]
- 30.Nesbitt MN, Francke U. A system of nomenclature for band patterns of mouse chromosomes. Chromosoma. 1973;41:145–158. doi: 10.1007/BF00319691. [DOI] [PubMed] [Google Scholar]
- 31.Kim DH, Villeneuve LM, Morris KV, Rossi JJ. Argonaute-1 directs siRNA-mediated transcriptional gene silencing in human cells. Nat. Struct. Mol. Biol. 2006;13:793–797. doi: 10.1038/nsmb1142. [DOI] [PubMed] [Google Scholar]
- 32.Janowski BA, Huffman KE, Schwartz JC, Ram R, Nordsell R, Shames DS, Minna JD, Corey DR. Involvement of AGO1 and AGO2 in mammalian transcriptional silencing. Nat. Struct. Mol. Biol. 2006;13:787–792. doi: 10.1038/nsmb1140. [DOI] [PubMed] [Google Scholar]
- 33.Li LC, Okino ST, Zhao H, Pookot D, Place RF, Urakami S, Enokida H, Dahiya R. Small dsRNAs induce transcriptional activation in human cells. Proc. Natl Acad. Sci. USA. 2006;103:17337–17342. doi: 10.1073/pnas.0607015103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chu Y, Yue X, Younger ST, Janowski BA, Corey DR. Involvement of argonaute proteins in gene silencing and activation by RNAs complementary to a non-coding transcript at the progesterone receptor promoter. Nucleic Acids Res. 2010;38:7736–7748. doi: 10.1093/nar/gkq648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.John B, Enright AJ, Aravin A, Tuschl T, Sander C, Marks DS. Human MicroRNA targets. PLoS Biol. 2004;2:e363. doi: 10.1371/journal.pbio.0020363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Diederichs S, Haber DA. Dual role for argonautes in microRNA processing and posttranscriptional regulation of microRNA expression. Cell. 2007;131:1097–1108. doi: 10.1016/j.cell.2007.10.032. [DOI] [PubMed] [Google Scholar]
- 37.Turunen MP, Lehtola T, Heinonen SE, Assefa GS, Korpisalo P, Girnary R, Glass CK, Vaisanen S, Yla-Herttuala S. Efficient regulation of VEGF expression by promoter-targeted lentiviral shRNAs based on epigenetic mechanism: a novel example of epigenetherapy. Circ. Res. 2009;105:604–609. doi: 10.1161/CIRCRESAHA.109.200774. [DOI] [PubMed] [Google Scholar]
- 38.Soria JC, Jang SJ, Khuri FR, Hassan K, Liu D, Hong WK, Mao L. Overexpression of cyclin B1 in early-stage non-small cell lung cancer and its clinical implication. Cancer Res. 2000;60:4000–4004. [PubMed] [Google Scholar]
- 39.Agarwal R, Gonzalez-Angulo AM, Myhre S, Carey M, Lee JS, Overgaard J, Alsner J, Stemke-Hale K, Lluch A, Neve RM, et al. Integrative analysis of cyclin protein levels identifies cyclin b1 as a classifier and predictor of outcomes in breast cancer. Clin. Cancer Res. 2009;15:3654–3662. doi: 10.1158/1078-0432.CCR-08-3293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sarafan-Vasseur N, Lamy A, Bourguignon J, Le Pessot F, Hieter P, Sesboue R, Bastard C, Frebourg T, Flaman JM. Overexpression of B-type cyclins alters chromosomal segregation. Oncogene. 2002;21:2051–2057. doi: 10.1038/sj.onc.1205257. [DOI] [PubMed] [Google Scholar]
- 41.Somers KD, Brown RR, Holterman DA, Yousefieh N, Glass WF, Wright GL, Jr, Schellhammer PF, Qian J, Ciavarra RP. Orthotopic treatment model of prostate cancer and metastasis in the immunocompetent mouse: efficacy of flt3 ligand immunotherapy. Int. J. Cancer. 2003;107:773–780. doi: 10.1002/ijc.11464. [DOI] [PubMed] [Google Scholar]
- 42.Song Y, Zhao C, Dong L, Fu M, Xue L, Huang Z, Tong T, Zhou Z, Chen A, Yang Z, et al. Overexpression of cyclin B1 in human esophageal squamous cell carcinoma cells induces tumor cell invasive growth and metastasis. Carcinogenesis. 2008;29:307–315. doi: 10.1093/carcin/bgm269. [DOI] [PubMed] [Google Scholar]
- 43.Yuan J, Yan R, Kramer A, Eckerdt F, Roller M, Kaufmann M, Strebhardt K. Cyclin B1 depletion inhibits proliferation and induces apoptosis in human tumor cells. Oncogene. 2004;23:5843–5852. doi: 10.1038/sj.onc.1207757. [DOI] [PubMed] [Google Scholar]
- 44.Weaver BA, Silk AD, Montagna C, Verdier-Pinard P, Cleveland DW. Aneuploidy acts both oncogenically and as a tumor suppressor. Cancer Cell. 2007;11:25–36. doi: 10.1016/j.ccr.2006.12.003. [DOI] [PubMed] [Google Scholar]
- 45.Holland AJ, Cleveland DW. Boveri revisited: chromosomal instability, aneuploidy and tumorigenesis. Nat. Rev. Mol. Cell Biol. 2009;10:478–487. doi: 10.1038/nrm2718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Robb GB, Brown KM, Khurana J, Rana TM. Specific and potent RNAi in the nucleus of human cells. Nat. Struct. Mol. Biol. 2005;12:133–137. doi: 10.1038/nsmb886. [DOI] [PubMed] [Google Scholar]
- 47.Aaltonen K, Amini RM, Heikkila P, Aittomaki K, Tamminen A, Nevanlinna H, Blomqvist C. High cyclin B1 expression is associated with poor survival in breast cancer. Br. J. Cancer. 2009;100:1055–1060. doi: 10.1038/sj.bjc.6604874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Arinaga M, Noguchi T, Takeno S, Chujo M, Miura T, Kimura Y, Uchida Y. Clinical implication of cyclin B1 in non-small cell lung cancer. Oncol Rep. 2003;10:1381–1386. [PubMed] [Google Scholar]
- 49.Li JQ, Kubo A, Wu F, Usuki H, Fujita J, Bandoh S, Masaki T, Saoo K, Takeuchi H, Kobayashi S, et al. Cyclin B1, unlike cyclin G1, increases significantly during colorectal carcinogenesis and during later metastasis to lymph nodes. Int. J. Oncol. 2003;22:1101–1110. [PubMed] [Google Scholar]
- 50.Saito T, Saetrom P. MicroRNAs—targeting and target prediction. Nat. Biotechnol. 2010;27:243–249. doi: 10.1016/j.nbt.2010.02.016. [DOI] [PubMed] [Google Scholar]
- 51.Zhang Y, Verbeek FJ. Comparison and integration of target prediction algorithms for microRNA studies. J. Integr. Bioinform. 2010;7:1–13. doi: 10.2390/biecoll-jib-2010-127. [DOI] [PubMed] [Google Scholar]
- 52.Portnoy V, Huang V, Place R, Li LC. Small RNA and transcriptional upregulation. Wiley Interdiscip Rev RNA. 2011;2:748–760. doi: 10.1002/wrna.90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Li LC. The multifaceted small RNAs. RNA Biol. 2008;5:61–64. doi: 10.4161/rna.5.2.5989. [DOI] [PubMed] [Google Scholar]
- 54.Schwartz JC, Younger ST, Nguyen NB, Hardy DB, Monia BP, Corey DR, Janowski BA. Antisense transcripts are targets for activating small RNAs. Nat. Struct. Mol. Biol. 2008;15:842–848. doi: 10.1038/nsmb.1444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Napoli S, Pastori C, Magistri M, Carbone GM, Catapano CV. Promoter-specific transcriptional interference and c-myc gene silencing by siRNAs in human cells. EMBO J. 2009;28:1708–1719. doi: 10.1038/emboj.2009.139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Place RF, Noonan EJ, Foldes-Papp Z, Li LC. Defining features and exploring chemical modifications to manipulate RNAa activity. Curr. Pharm. Biotechnol. 2010;11:518–526. doi: 10.2174/138920110791591463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Mescalchin A, Detzer A, Weirauch U, Hahnel MJ, Engel C, Sczakiel G. Antisense tools for functional studies of human Argonaute proteins. RNA. 2010;16:2529–2536. doi: 10.1261/rna.2204610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Brothman AR. Cytogenetics and molecular genetics of cancer of the prostate. Am. J. Med. Genet. 2002;115:150–156. doi: 10.1002/ajmg.10694. [DOI] [PubMed] [Google Scholar]
- 59.Pan Y, Kytola S, Farnebo F, Wang N, Lui WO, Nupponen N, Isola J, Visakorpi T, Bergerheim US, Larsson C. Characterization of chromosomal abnormalities in prostate cancer cell lines by spectral karyotyping. Cytogenet. Cell Genet. 1999;87:225–232. doi: 10.1159/000015432. [DOI] [PubMed] [Google Scholar]
- 60.Beheshti B, Karaskova J, Park PC, Squire JA, Beatty BG. Identification of a high frequency of chromosomal rearrangements in the centromeric regions of prostate cancer cell lines by sequential giemsa banding and spectral karyotyping. Mol. Diagn. 2000;5:23–32. doi: 10.1007/BF03262019. [DOI] [PubMed] [Google Scholar]
- 61.Weaver BA, Cleveland DW. The aneuploidy paradox in cell growth and tumorigenesis. Cancer Cell. 2008;14:431–433. doi: 10.1016/j.ccr.2008.11.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Greenberg ME, Bender TP. Identification of newly transcribed RNA. Curr. Protoc. Mol. Biol. 2007 doi: 10.1002/0471142727.mb0410s78. Chapter 4: p. Unit 4 10. [DOI] [PubMed] [Google Scholar]
- 63.Hirayoshi K, Lis TS. Nuclear run-on assays: assessing transcription by measuring density of engaged RNA polymerases. Methods Enzymol. 1999;304:351–362. doi: 10.1016/s0076-6879(99)04021-5. [DOI] [PubMed] [Google Scholar]
- 64.Love JD, Minton KW. Screening of lambda library for differentially expressed genes using in vitro transcripts. Anal. Biochem. 1985;150:429–441. doi: 10.1016/0003-2697(85)90532-9. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.