Abstract
Initial steps in the synthesis of functional tRNAs require 5′- and 3′-processing of precursor tRNAs (pre-tRNAs), which in yeast mitochondria are achieved by two endonucleases, RNase P and RNase Z. In this study, using a combination of detergent-free Blue Native Gel Electrophoresis, proteomics and in vitro testing of pre-tRNA maturation, we reveal the physical association of these plus other mitochondrial activities in a large, stable complex of 136 proteins. It contains a total of seven proteins involved in RNA processing including RNase P and RNase Z, five out of six subunits of the mitochondrial RNA degradosome, components of the fatty acid synthesis pathway, translation, metabolism and protein folding. At the RNA level, there are the small and large rRNA subunits and RNase P RNA. Surprisingly, this complex is absent in an oar1Δ deletion mutant of the type II fatty acid synthesis pathway, supporting a recently published functional link between pre-tRNA processing and the FAS II pathway—apparently by integration into a large complex as we demonstrate here. Finally, the question of mt-RNase P localization within mitochondria was investigated, by GFP-tracing of a known protein subunit (Rpm2p). We find that about equal fractions of RNase P are soluble versus membrane-attached.
INTRODUCTION
To synthesize functional tRNAs, precursor tRNAs (pre-tRNA) need to be processed at their 5′- and 3′-termini. An almost ubiquitous RNase P is responsible for endonucleolytic 5′-processing (1). Until the discovery that human and Arabidopsis mitochondrial RNase P are protein-only enzymes (2,3), these activities were thought to be always ribonucleo-proteins, composed of a catalytic RNA plus one or several proteins, throughout Archaea, Bacteria and eukaryotes (4–9). Processing of tRNA 3′-termini is achieved by exonucleolytic and/or endonucleolytic activities, depending on the organism and the cellular compartment (10–14). In Escherichia coli, processing of 3′-termini requires a multi-step process initiated by an endonucleotic cleavage, followed by exonucleolytic trimming (11). In other bacteria and most eukaryotes (including nuclear, mitochondria, and plastid) and in all Archaea, 3′-tRNA processing is an endonucleolytic cleavage catalyzed by tRNase Z that exists in two forms, a long form of 750–930 amino acids only present in Eukarya (tRNase ZL), and a short form of 280–360 amino acids (tRNaseZ S) (15).
Most information on mitochondrial (mt) pre-tRNA processing comes from studies of Saccharomyces cerevisiae. A highly purified form of mitochondrial RNase P (mt-RNase P) has been obtained by lysis of mitochondria in the presence of detergent and high salt, and a series of chromatographic steps. It contains the nucleus-encoded Rpm2p protein (Rpm2p; 119 kDa), and only an incomplete set of RNA fragments covering the mtDNA-encoded RPM1 RNA subunit (mt-P RNA) of 427 nt (16–18). Yet, given that yeast mt-P RNA has a highly reduced RNA structure compared to its cytosolic counterpart (19), we strongly expect that native mt-RNase P contains more than just one protein, to compensate for the lack of RNA structure [for comparison, cytosolic RNase P contains nine proteins; (20)]. Previous studies revealed that Rpm2p is involved not only in mt-RNase P activity, but also effects mitochondrial import (21), fermentative growth (22), and transcriptional activation of several nucleus-encoded mitochondrial components (23). In addition (and most curiously), tRNA processing intersects with the type II fatty acid synthesis pathway (24). Disruption of any enzyme in the FAS II pathway leads to a defective mt-RNase P (24). One possibility to explain this observation is that there is a structural association between mt-RNase P and FAS II that is impaired with deletion of the FAS II subunits. In fact, this led us to the hypothesis that a super-structure may combine some or all of the above-mentioned mitochondrial functions into a large physical unit. Indications that RNase P may be associated with other functions also come from mt-RNase P purification experiments. In early isolation steps, its activity co-fractionates with the tRNA 3′-processing activity (18), and purification to homogeneity is achieved at an extremely low yield, despite the use of high salt and detergent. In yeast mitochondria, 3′-tRNA processing is an endonucleotic cleavage (25), which is accomplished by a multifunctional RNase Z (11,26,27). In yeast, both the nuclear and mitochondrial forms of RNase Z are encoded by a nuclear gene (TRZ1) (15). Again, this enzyme is also implicated in ribosomal RNA maturation (26).
Controlling RNA levels (balance between RNA synthesis and degradation) is vital for both the regulation and functioning of the mitochondrial system (28–30). RNA degradation is mediated principally by (arguably) small multiprotein complexes, like the exosome in the cytoplasm of eukaryotes (31) or the degradosome in bacteria (32). The activity responsible for RNA turnover in mitochondria is a 3′- to 5′-processive exoribonuclease (33), which is organized in an RNA degradosome complex (mtEXO), as first described in S. cerevisiae (34). The highly purified mtEXO complex is composed of only two protein subunits, the exoribonuclease (35), and an NTP-dependent RNA helicase (related to the DExH superfamily) (36). Yet, the degradosome apparently associates with the mitochondrial ribosome, as mtEXO co-purifies with ribosomal proteins that are difficult to remove (34). These observations reinforce the hypothesis that the RNA degradosome is part of a large super-structure that associates with a variety of mitochondrial functions.
In this study, we demonstrate that mt-RNase P and tRNA Z activities are part of a large ribo-nucleoprotein complex, which also includes the RNA degradosome, five additional RNA processing proteins, plus other mitochondrial functions. We further show that the biogenesis of this complex is impaired in a mutant that is deficient in type II fatty acid synthesis, rationalizing its known pleiotropic tRNA processing phenotype discussed above.
MATERIALS AND METHODS
Cell culture and mitochondrial isolation
Saccharomyces cerevisiae (BY 4743) was kindly provided by Dr S. Michnick (Université de Montréal), and yeast GFP (Green Fluorescent Protein) constructs used in this study (MATa his3Δ1 leu2Δ0 met15Δ0 ura3Δ0 RPM2-GFP::HIS3) by Dr J. Vogel (McGill). Cells were grown to an optical density of 1.5–2.5 in a medium containing 1% yeast extract, 2% peptone, 3% glycerol and 1% galactose, pH 5.0 (YEPGal). Mitochondrial purification followed previously published procedures with slight modifications (37). Yeast cell walls were removed by digestion with glucanase, spheroplasts were disrupted by osmotic shock, and a crude mitochondrial fraction was isolated by differential centrifugation. Mitochondria were resuspended in ‘washing buffer’ (600 mM sucrose, 0.2 mM PMSF and 10 mM MOPS/KOH, pH 7.2) and further purified by centrifugation (60 min at 134 000g) through a discontinuous sucrose gradient (concentrations from top to bottom, 25%, 36% and 60%; 5 ml each). For a second purification step, the mitochondrial fraction was collected and mixed with about four times its volume of 80% sucrose, layered at the bottom of a discontinuous sucrose gradients, and purified by centrifuged at 134 000g for 120 min. Intact mitochondria move upwards (‘flotation’), and accumulate at the interface between the 36% and 60% sucrose layers. For enhanced purity, this two-step purification of the mitochondria was repeated twice. Purified mitochondria were resuspended in buffer (600 mM sucrose, 0.2 mM PMSF and 10 mM Tricine/KOH, pH 7.2) and pelleted at 14 000g for 15 min. Usually, mitochondria were directly processed in further steps, but they may also be shock-frozen and kept at −80°C until use.
Extraction of mitochondria soluble matrix proteins and membrane fractions
For the extraction of matrix proteins, we adapted a previously published procedure (37,38). Mitochondria were re-suspended at 10 mg/ml in breaking buffer (600 mM sucrose, 20 mM HEPES–KOH, pH 7.4, 10 mM EDTA) and incubated for 30 min on ice in nine volumes of 20 mM HEPES–KOH, pH 7.4, 0.5 mM EDTA and 1 mM PMSF. We used three alternative ways for rupturing mitochondria, which only differ in yield but not in banding pattern or protein composition of supercomplexes. The gentlest method is to burst mitochondria through osmotic shock, by abruptly adding 10 volumes buffer. Alternatively, the mitochondrial suspension is adjusted to a final sucrose concentration of 0.45 M, incubated for 30 min on ice, and then homogenized in a Potter homogenizer for 90 s. The third method, for maximum yield of matrix proteins, involves mechanical disruption of mitochondria by shaking with an equal ratio of 125–212 µ and 425–600 µ glass beads (Sigma), in three rounds of 20 s each at 4°C. After a clarifying centrifugation at 12 000g for 15 min, the supernatant was subjected to centrifugation at 100 000g, for 2 h at 4°C. The pellet containing mitochondrial membranes was discarded, and the supernatant was subjected to a second centrifugation at 120 000g, for 30 min at 4°C. The matrix protein fraction (supernatant) was mostly used immediately, but may be stored at −80°C after shock-freezing. Protein concentrations were determined with the Bradford protein assay. A mitochondrial membrane fraction was prepared as described previously (39,40).
Tracking of GFP-tagged marker proteins
Soluble matrix protein complexes (1.5 mg) isolated from an Rpm2-GFP construct were separated by ultracentrifugation (16 h at 20 000g, Beckman SW41 rotor) on discontinuous glycerol gradients (10–70% glycerol, in a buffer containing 20 mM HEPES–KOH pH 7.9, 50 mM KCl, 1.5 mM MgCl2 and 1 mM DTT). 0.7 ml fractions were collected and analyzed for protein concentration (Bradford protein assay) and relative GFP fluorescence intensity (excitation at 488 nm, light emission between 500 and 540 nm).
Purification of mt-RNase P activity
To purify the native RNase P complex, we developed a preparative Blue Native Column Electrophoresis (NEC), which permits large-scale purification of native protein complexes. It is based on the regular blue native gel electrophoresis (BN–PAGE) protocol (41–44), using instead of a slab gel a cylindrical running chamber filled with polyacrylamide gel (e.g. 7%). The different concentrations of native polyacrylamide gel were prepared as previously published (41–44). The mitochondrial extracts (~250 µg) were separated at constant voltage (140 V and 9 mA, overnight, in a cold room 4°C), using the same electrophoresis buffers as in regular BN–PAGE. Samples were collected every 30 min from a 200 µl dialysis-cup placed at the outlet of the column. Collected fractions were assayed for mt-RNase P activity and analyzed for complexes composition with BN–PAGE.
In vitro preparation and radio-labeling of pre-tRNAproline
The mitochondrial Reclinomonas americana pre-tRNAproline substrate for the RNase P assays was prepared by in vitro transcription and purified as described previously (45). The tRNAproline DNA ligated into pFBS/EcoRV (2.9 kb) vector with T4 DNA ligase (Roche) and amplified by PCR using 5′-GAAATTAATACGACTCACTATAGGGTAACGTACTTAATGTAAAAGGTT-3′, 5′-TGGTCGGGATGACGTGATTTGAACA-3′ and 5′-TCACTAAAGGGAACAAAAGCTGGGT-3′ primers produce respectively 5′-leader pre-tRNAproline and, 5′-leader and 3′-termini pre-tRNAproline. The two amplified tRNAproline DNAs were used separately for two in vitro radio-labeled transcriptions. Two micrograms of each amplified tRNAproline DNA were used for tRNAproline transcription with Invitrogen T7 RNA polymerase (2 u/µl) and α P32 ATP (10 mCi/ml) (Perkin Elmer) as previously described (45). After an overnight incubation at 37°C, loading buffer was added and the samples were heated at 75°C for 2 min before loading on a 9% polyacrylamide/8 M urea gel (4 h, 200 V at room temperature). The 117 nt (5′-leader pre-tRNAproline) and 148 nt (5′-leader and 3′-termini pre-tRNAproline) RNA bands were cut out of the gel and incubated over night at 37°C in a 300 µl buffer extraction (30 µl of 1% SDS and 270 µl of H202). After phenol–chloroform extraction and ethanol precipitation, the precursor RNA was 5′-labeled with α-ATP32.
Activity assay of mt-RNase P
To test the mt-RNase P activity we used the same procedure as described previously (46). Radio-labeled pre-tRNAproline (2000 cpm) was dissolved in 1× PA buffer (50 mM Tris–HCl, pH 7.5, 100 mM NH4Cl, 10 mM MgCl2) and incubated 30 min at 37°C, in a 15 -µl mixture reaction. After incubation, 10 μl of loading buffer was added, and the sample was heated at 75°C for 2 min before loading on a 9% polyacrylamide/8 M urea gel (4 h, 200 V at room temperature). The gels were then either exposed to a Kodak film (~12 h) or a Biorad molecular imaging screen K (2 h).
The M1 RNA ribozyme was used as positive control for 5′-tRNA processing. The expression plasmid carrying the E. coli M1 RNA gene (provided by Sidney Altman) was used for in vitro transcription. For all experimental information for M1 RNA transcription and purification see (45). The primers for amplification of the rnpB gene were 5′-GAAATTAATACGACTCACTATAGGGAAGCTGACCAGACAGTCGC-3′ and 5′AGGTGAAACTGACCGATAAGCC 3′. The M1 RNA was activated before use, by heating (65°C for 5 min), and slow cooling to room temperature in 1× PA buffer (50 mM Tris–HCl, pH 7.5, 100 mM NH4Cl, 10 mM MgCl2). Radio-labeled pre-tRNAproline (2.000 cpm) was incubated at 37°C in 10 mM Tris–HCl, pH 7.5, 100 mM MgCl2, 100 mM NH4Cl, 4% PEG, in the presence of ~10 nM M1 RNA. The total volume of the reaction is 15 μl. After 30 min, 10 μl of loading buffer was added, and the sample was heated at 75°C for 2 min before loading on a 9% polyacrylamide/8 M urea gel.
Identification of mt-RNase P RNA (RPM1) by RT–PCR
Endogenous RNA from the identified MRT complex was purified using the ‘RNeasy plus’ kit by QIAGEN, and RT–PCR assays were performed on 10 ng RNA with AMV reverse transcriptase (cDNA synthesis). The cDNA was amplified with the Expand High Fidelity PCR system provided by Roche. The PCR primers for RPM1 RNA amplification are 5′-TAATAGGAAAGTCATAAATAT-3′, 5′-GTAATATAT ATATATATATTGGAATAG-3′ and 5′-TTATATTATTATACAGAAATA-3′. For RT–PCR the same primers of the PCR were used in addition to 5′-AGAAATAATATTATAAATAAAATATAT-3′, 5′-GGATATTATATTATAAGCA-3′ and 5′-AAGCATATTTCTGTATAATAA-3′. The expected PCR products have lengths of 51, 71, 52, 387 and 400 nt.
Blue native gel electrophoresis
Preparation of samples and of 4–14% BN–PAGE gel followed previously published procedures (41–44), except that detergents were omitted (their effect was only tested in supercomplex stability tests). Approximately 150 µg protein was loaded per well, and electrophoretic separation was performed in a Höfer (18 × 16 cm) electrophoresis chamber, at 140 V and 9 mA, overnight at 4°C.
Analysis of supercomplex composition by mass spectrometry
To identify protein complexes that form discrete bands in BN–PAGE, complex bands were cut from the gel and submitted to liquid chromatography tandem mass spectrometry (LC–MS/MS) analysis (47,48). In-gel tryptic digestion and LC–MS/MS analysis was performed by a service at the Université de Montréal (IRIC), including functional annotation by Mascot (49).
RESULTS AND DISCUSSION
Purification of mt-RNase P without the use of detergents
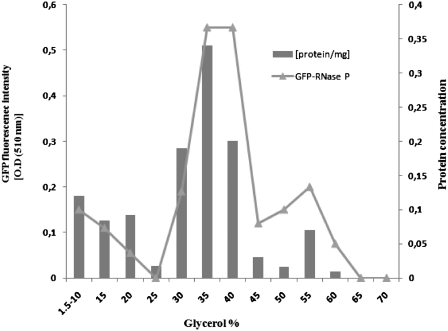
To preserve the integrity of a postulated large complex that combines mt-RNase P with other mitochondrial functions, we have developed purification procedures that do not make use of detergents or other highly disruptive conditions as previously applied [such as detergent, high salt, EDTA, heat shock etc; (16–18)]. For this we start with highly purified yeast mitochondria, gently disrupt the organelles and extract a soluble matrix fraction by centrifugation, which represents ~57% of total mitochondrial proteins. When separating the soluble extract on a glycerol gradient (10–70%; for details see ‘Materials and Methods’ section), most of the material moves into the glycerol phase, in support of the idea that most soluble proteins are organized in large complexes. In fact, tracing of the protein subunit of mt-RNase P (Rpm2p) by in vivo GFP fluorescence labeling reveals that about half (54%) of the GFP-Rpm2p is in the soluble mitochondrial matrix fraction, and that most of it (81%) is associated with a high-molecular-weight complex as it migrates far into the glycerol gradient (35–40%; Figure 1). This gradient fraction also contains the bulk of RNase P activity (Supplementary Figure S1). GFP-labeled cells grow normally on non-fermentable substrates, indicating that the GFP fusion does not lead to functional disturbances.
Figure 1.
Density-based fractionation of mitochondrial GFP-Rpm2p. After fractionation on a discontinuous 10–70% glycerol gradient, relative GFP fluorescence absorbance/fraction identify most Rpm2-GFP with high-molecular-weight complexes.
As gradient procedures have only limited resolution, we set out to find an alternative, non-disruptive and preparative purification procedure for large protein complexes. This led us to develop a variant of BN–PAGE (50), in which all steps are performed under physiological conditions (e.g. pH 7), and in the absence of detergents (named non-denaturing polyacrylamide gel electrophoresis column, or NEC; for details see ‘Materials and Methods’ section). Instead of using a slab gel, preparative NEC separates in a large column with a continuous polyacrylamide gel, and fractions are continuously recovered in a dialysis cup as they leave the column. Tracking of mt-RNase P by in vitro cleavage of a mitochondrial pre-tRNAproline precursor identifies ~70% of the total mt-RNase P activity in fractions 7–9 (with a total of 20 fractions; Figure 2A). To investigate the purity of fractions 7–9, they were separated by a second, regular slab-gel NEC that has more resolution than preparative NEC, and at a gel concentration that is optimal for separation close to the complex's size range. Several complexes were identified in all three fractions, but only one of them occurs regularly (Figure 3A), which is the putative mt-RNase P-containing complex. Therefore, for further large-scale purification, fractions 7, 8 and 9 were pooled and processed on a second preparative NEC column at a higher polyacrylamide concentration (7.5%). Under this condition, mt-RNase P activity elutes exclusively in fraction 38 (for a total of 50 samples; Figure 2B and Supplementary Figure S2). Slab-gel NEC analysis confirms that it contains a single complex (Figure 3A), which remains stable following treatments with DNase, RNase and to some degree (~50%) with a mild detergent (digitonin; Figure 3B). Taken together, we argue that this complex closely represents the in vivo organization of a set of soluble mitochondrial matrix proteins, combining a variety of RNA maturation and other functions as shown in the following experiments.
Figure 2.
Purification of mt-RNase P from the mitochondrial matrix extract. Purification and assays conditions are given in ‘Materials and Methods’ section. Mt-RNase P activities in (A) correspond to a separation on a 5.5% NEC column and (B) on 7.5% NEC slab gel. Aliquots from each fraction were assayed for RNase P activity. The negative control (C–) is without and the positive control (+M1) with E. coli M1 RNA. The RNA corresponding to 5′-leader pre-tRNAproline (117 nt), tRNAproline (78 nt) and the removed 5′-leader sequence (39 nt) are indicated. Mt-RNase P activity was found in fractions 7, 8 and 9 in preparative NEC (A), and in fraction 38 only in a subsequent regular NEC separation (B). Note that the activity test of fraction 38 (B) was performed with more material, leading to almost complete processing of the tRNA precursor, but also to partial, unspecific cleavage of the mature tRNA into smaller fragments. With less material, the processing pattern is similar to that in (A) (Supplementary Figure S2).
Figure 3.
A homogenous, stable RMT complex. A total of 50 µg protein was loaded per lane, and after overnight electrophoresis (140 V and 9 mA; 4°C) the gel was either stained with silver nitrate (BioRad silver staining kit) in (A), and Serva Blue G250 in (B). (A) Aliquots of fractions 7, 8 and 9 (with mt-RNase P activity) from a 5.5% preparative NEC were analyzed by slab-gel NEC (4–14%) for complex composition. (B) The RMT complex (fraction 38 from 7.5% NEC) is to some degree (~50%) not dissociated by treatment (30 min) with detergent [digitonin/protein ratio (2 g/g)].
Association of 5′- and 3′-tRNA processing activities
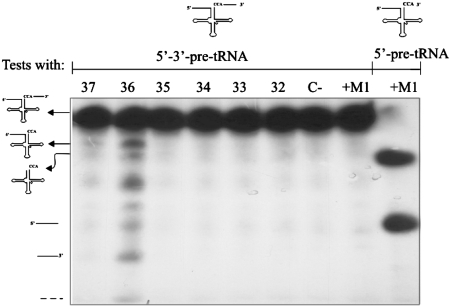
The processing activity of the purified complex was subsequently tested in an in vitro assay, with a different pre-tRNAproline substrate that has both 5′-leader and 3′-trailer extensions. We find essentially two forms of processed RNAs, the mature tRNAproline and an intermediate pre-tRNAproline with a 5′-leader (Figure 4), revealing the presence of a 3′-tRNA processing that is somewhat more effective than RNase P. This result is consistent with the previously observed co-fractionation of RNase P and RNase Z in early steps of yeast mt-RNase P purification (18).
Figure 4.
Association of mt-RNase P with 3′-tRNA processing activity. The RMT complex is assayed for in vitro processing of 5′-leader and 3′-termini pre-tRNAproline. Assay conditions are given in ‘Materials and Methods’ section. The negative control (C–) is without addition of M1 RNA and the positive control is with M1 RNA (+M1). The RNA corresponding to 5′-leader and 3′-terminus pre-tRNAproline (148 nt), 5′-leader pre-tRNAproline (117 nt), tRNAproline (78 nt), 5′-leader (39 nt), and 3′-trailer (31 nt) sequences are indicated. Activities of the combined mt-RNase P and 3′-tRNA processing activity were found in fractions 36 and 37.
Presence of RPM1 RNA and Rpm2p protein in the complex
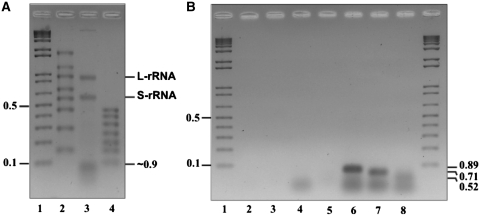
To further characterize the homogeneity and composition of this tRNA processing complex, we tracked the presence of the known yeast mt-RNase P core RNA and protein components (i.e. the RPM1 RNA subunit and Rpm2p). Proteomic analysis by mass spectrometry (LC–MS/MS, see ‘Materials and Methods’ section) shows that the Rpm2p is present only in this complex, together with a variety of other proteins (see below). Likewise, an extraction and analysis of the endogenous RNA of the purified complex demonstrate the presence of the two (un-degraded) small and large subunit rRNAs, as well as several RNAs of ~70–90 nt (Figure 5A). As expected, an RNA with the predicted size of an intact RPM1 RNA (427 nt) is absent, which is in agreement with previous investigations on purified yeast mt-RNaseP [e.g. (16)], in which only fragments of RPM1 RNA are detected. In fact, consistent with these studies, we find by RT–PCR and sequencing the same two RPM1 RNA fragments of 52, 89 and 71 nt (Figure 5B and Supplementary Figure S3). Our results thus confirm that an intact RPM1 RNA subunit is not required for RNase P activity in vitro (16), and that the mt-RNase P core subunits are integrated within a large complex when isolated under non-denaturing conditions.
Figure 5.
Two rRNAs subunits and fragments of RPM1 RNA are components of the RMT complex. (A) Agarose gel electrophoresis of the RNA extracted from the RMT complex. The prominent small and large S-rRNA) (L-rRNA) rRNA subunits are indicated (S-rRNA and L-rRNA). Lane 1, DNA size marker kb+ Lane 2, RNA size marker RiboRuler High Range; Lane 3, RNA extracted from RMT complex; Lane 4, RNA size marker RiboRuler Low Range. (B) RPM1 RNA of the RMT complex was reverse transcribed and PCR amplified using different primer combinations. Lane 1, DNA size marker kb+ lane 2, RNA control without RT–PCR amplification; lane 3, negative control with primers; lanes 4 to 8 RT–PCR products of 89, 71 and 51 nt were identified. The diffuse RNAs migrating close to 35 bp in lanes 4, 6, 7 and 8 are primers and/or primer dimers.
Protein composition of the mt-RNase P (RMT) complex
Proteomic analysis with liquid chromatography and tandem mass spectrometry (LC–MS/MS) reveals the presence of an unexpected high number of proteins (130) in the purified ‘native’ RNase P/RNase Z complex (confirmed by three independent analyses; Supplementary Table S1). These proteins are implicated in numerous functions including (as expected) RNA processing, but also metabolism (e.g. TCA cycle), translation (two aminoacyl–tRNA synthetases; two translation elongation factors; an incomplete set of ribosomal proteins but un-degraded forms of the large and small rRNA subunits (Figure 5A), and others (e.g. mitochondrial genome maintenance and chaperons) (Supplementary Table S1). In the following we will refer to this complex as the ‘RMT complex’ (for RNA processing, metabolism, translation complex). A total of 564 physical protein–protein interactions are listed among proteins of the RMT complex in the BioGRID and SGD databases, in support of their organization in a complex.
Besides Rpm2p and tRNA Z that are involved in 5′- and 3′-tRNA maturation, we find five out of six subunits (Mtr4p; Dead-box family of ATP dependent helicase; ribosomal proteins Mrp1p, Mrpl3p, Mrpl35p and Mrpl40) of the RNA degradosome complex, as previously characterized by others (34). The remaining second catalytic subunit of the RNA degradosome (3′–5′ exoribonuclease, Dss1p) was identified only once in the three LC-MS/MS experiments, which is consistent with its low level of cellular expression [only about 1000 molecules/cell (51)]. The RNA degradosome is responsible for RNA turnover (34), but loss of function of any of its subunits also results in accumulation of RNA precursors with abnormal 5′- and 3′-termini, and stalled mitochondrial translation (33,34). Finally, the presence of five further proteins involved in rRNA and mRNA processing and RNA modification demonstrates a structural integration of most mitochondrial RNA processing activities (Nop7p; involved in rRNA processing, Mrm1p; ribose methyltransferase of the mitochondrial 21S rRNA, Hsh155p; mRNA-binding protein, Ngl1p; putative endonuclease and Prp22p; DEAH-box RNA-dependent ATPase/ATP-dependent RNA helicase).
That the RMT complex contains 22 ribosomal proteins of the large subunit and 13 of the small subunit is in agreement with the existing data that show an association of a highly purified RNA degradosome complex with four ribosomal proteins (Mrpl3p, Mrpl35p, Mrpl40 and Mrp1p) (34). Depending on the adopted experimental procedure, the mitochondrial ribosome contains about 70 proteins (52,53). The presence of only 35 mito-ribosomal proteins in the RMT complex falls short of about one half of the protein components, yet contains the un-degraded large and small subunit rRNAs (Figure 5A). This may be interpreted in two ways: (i) the RMT complex results from dissociation of a large membrane-attached complex that contains the intact ribosome. The dissociation may be caused by experimental manipulation (e.g. mechanical extraction procedure). It has been reported that the mitochondrial ribosome is attached to the inner membrane (40), which would be consistent with our result that mt-RNase P and the RMT complex is partially membrane-attached. Alternatively, (ii) the RMT complex represents a biogenesis intermediate, with an incomplete set of ribosomal proteins that are being added to the rRNA subunits.
The finding of metabolic proteins (e.g. the complete TCA cycle) and chaperons in this complex is consistent with the observed co-purification of metabolic (TCA cycle) and ribosomal proteins in enriched human mt-RNase P fractions (2). Interestingly, the RMT complex also contains several chaperons that are involved in protein maturation, modification, targeting and assembly steps.
Lack of RMT complex in a FAS II deletion mutant
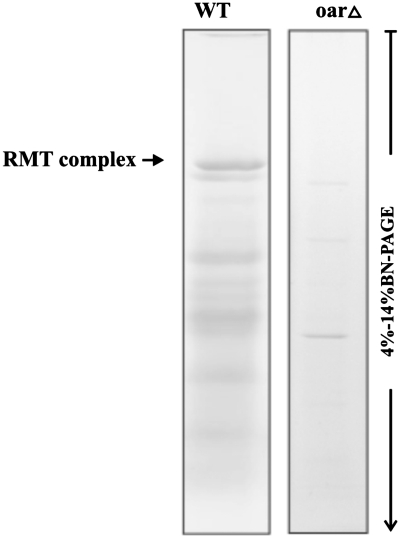
A recent investigation of yeast mutants demonstrates that deficiency in any enzyme of the mitochondrial fatty acid type II biosynthetic pathway (FASII) leads to impaired 5′-tRNA processing in yeast mitochondria and an overall pleiotropic phenotype (24). Our data reveal the presence of one regular component of the pathway (Oar1p; 3-Oxoacyl-[acyl-carrier-protein] reductase; FASII biosynthesis) in the RMT complex. Two other FAS II enzymes (Htd2p and Mct1p) were also identified, but only once in the three LC-MS/MS experiments. This is probably due to their relatively low level of cellular expression (51). Consistent with the above observations, the RMT complex is no longer discernable in an Oar1 deletion mutant, and the amount of other complexes is strongly reduced (Figure 6). Our data support the interpretation that lack of FAS II enzymes indirectly affects RNAse P assembly and its activity (24), as well as enzymes in the TCA cycle (24,54). As a consequence of lacking mature tRNAs and strongly reduced mitochondrial translation, FAS 2 mutants have a pleiotropic phenotype.
Figure 6.
Biogenesis and stability of the RMT complex are affected in the yeast oar1Δ mutant. Soluble mitochondrial multiprotein complexes (50 µg of total protein) of the yeast wild strain and oar1Δ mutant were separated on linear 4–14% BN–PAGE, overnight (140 V and 9 mA; 4°C). Gels were stained with Serva Blue G250.
A membrane-attached form of mt-RNase P?
Previous publications suggest that yeast mt-RNase P is difficult to purify without the aid of detergents (16–18), and at least partially membrane-bound. Unfortunately, measuring mt-RNase P activity of mitochondrial membrane fractions is difficult because of rapid unspecific degradation of the pre-tRNA. We have therefore tracked mt-RNase P quantities by localization of its known GFP-labeled protein subunit (Rpm2p), and find that ~54% of GFP-Rpm2p is in the soluble mitochondria matrix fraction. With a mild detergent (1% digitonin) the amount of soluble GFP-Rpm2p increases to only ~68%, suggesting that the RMT complex has indeed a strong tendency for membrane attachment, but we clearly confirm that more than half is soluble, integrated with the RMT complex.
CONCLUSION
Here we show that native mt-RNase P and RNase Z activities are organized in a stable RMT supercomplex in yeast mitochondria, associated with the RNA degradosome, other RNA processing activities and several other mitochondrial functions including fatty acid synthesis, the complete TCA cycle and components of the translation machinery including a ribosome with both rRNAs but a reduced set of ribosomal proteins. Our results rationalize and further extend the surprising finding by others, that a deficiency in any component of the fatty acid type II biosynthetic pathway leads to impaired tRNA processing (24), and that deletion of genes coding for components of the RNA degradosome lead to pleiotropic defects in RNA processing and protein translation (33,34). In addition, our demonstration of stable (detergent-resistant), structural organization of metabolic activities in the RMT complex signals an end to the common belief that most metabolic enzymes are small separate enzymes that interact only casually (by diffusion) from within a pool.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online: Supplementary Table S1, Supplementary Figures S1–S3.
FUNDING
National Science and Engineering Research Council (NSERC) of Canada; Funding for open access charge: NSERC Canada.
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
The authors thank Nihade El Kraimi for discussions and valuable comments on the manuscript.
REFERENCES
- 1.Lai LB, Vioque A, Kirsebom LA, Gopalan V. Unexpected diversity of RNase P, an ancient tRNA processing enzyme: challenges and prospects. FEBS Lett. 2010;584:287–296. doi: 10.1016/j.febslet.2009.11.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Holzmann J, Frank P, Loffler E, Bennett KL, Gerner C, Rossmanith W. RNase P without RNA: identification and functional reconstitution of the human mitochondrial tRNA processing enzyme. Cell. 2008;135:462–474. doi: 10.1016/j.cell.2008.09.013. [DOI] [PubMed] [Google Scholar]
- 3.Gobert A, Gutmann B, Taschner A, Gossringer M, Holzmann J, Hartmann RK, Rossmanith W, Giege P. A single Arabidopsis organellar protein has RNase P activity. Nat. Struct. Mol. Biol. 2010;17:740–744. doi: 10.1038/nsmb.1812. [DOI] [PubMed] [Google Scholar]
- 4.Walker SC, Engelke DR. A protein-only RNase P in human mitochondria. Cell. 2008;135:412–414. doi: 10.1016/j.cell.2008.10.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Guerrier-Takada C, Gardiner K, Marsh T, Pace N, Altman S. The RNA moiety of ribonuclease P is the catalytic subunit of the enzyme. Cell. 1983;35:849–857. doi: 10.1016/0092-8674(83)90117-4. [DOI] [PubMed] [Google Scholar]
- 6.Pannucci JA, Haas ES, Hall TA, Harris JK, Brown JW. RNase P RNAs from some Archaea are catalytically active. Proc. Natl Acad. Sci. USA. 1999;96:7803–7808. doi: 10.1073/pnas.96.14.7803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kikovska E, Svard SG, Kirsebom LA. Eukaryotic RNase P RNA mediates cleavage in the absence of protein. Proc. Natl Acad. Sci. USA. 2007;104:2062–2067. doi: 10.1073/pnas.0607326104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cherayil B, Krupp G, Schuchert P, Char S, Soll D. The RNA components of Schizosaccharomyces pombe RNase P are essential for cell viability. Gene. 1987;60:157–161. doi: 10.1016/0378-1119(87)90223-x. [DOI] [PubMed] [Google Scholar]
- 9.Waugh DS, Pace NR. Complementation of an RNase P RNA (rnpB) gene deletion in Escherichia coli by homologous genes from distantly related eubacteria. J. Bacteriol. 1990;172:6316–6322. doi: 10.1128/jb.172.11.6316-6322.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Apirion D, Miczak A. RNA processing in prokaryotic cells. Bioessays. 1993;15:113–120. doi: 10.1002/bies.950150207. [DOI] [PubMed] [Google Scholar]
- 11.Morl M, Marchfelder A. The final cut. The importance of tRNA 3′-processing. EMBO Rep. 2001;2:17–20. doi: 10.1093/embo-reports/kve006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Garber RL, Altman S. In vitro processing of B. mori transfer RNA precursor molecules. Cell. 1979;17:389–397. doi: 10.1016/0092-8674(79)90165-x. [DOI] [PubMed] [Google Scholar]
- 13.Engelke DR, Gegenheimer P, Abelson J. Nucleolytic processing of a tRNAArg-tRNAAsp dimeric precursor by a homologous component from Saccharomyces cerevisiae. J. Biol. Chem. 1985;260:1271–1279. [PubMed] [Google Scholar]
- 14.Mayer M, Schiffer S, Marchfelder A. tRNA 3′ processing in plants: nuclear and mitochondrial activities differ. Biochemistry. 2000;39:2096–2105. doi: 10.1021/bi992253e. [DOI] [PubMed] [Google Scholar]
- 15.Zhao Z, Su W, Yuan S, Huang Y. Functional conservation of tRNase ZL among Saccharomyces cerevisiae, Schizosaccharomyces pombe and humans. Biochem. J. 2009;422:483–492. doi: 10.1042/BJ20090743. [DOI] [PubMed] [Google Scholar]
- 16.Morales MJ, Wise CA, Hollingsworth MJ, Martin NC. Characterization of yeast mitochondrial RNase P: an intact RNA subunit is not essential for activity in vitro. Nucleic Acids Res. 1989;17:6865–6881. doi: 10.1093/nar/17.17.6865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Morales MJ, Dang YL, Lou YC, Sulo P, Martin NC. A 105-kDa protein is required for yeast mitochondrial RNase P activity. Proc. Natl Acad. Sci. USA. 1992;89:9875–9879. doi: 10.1073/pnas.89.20.9875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.AME J-C, Martin RP. France: Université de Strasbourg; 1993. Thèse de doctorat: Etude des mécanismes de maturation des précurseurs de tRNA dans la mitochondrie de levure: RNase P et 3′ pre-tRNase mitochondriales. [Google Scholar]
- 19.Seif ER, Forget L, Martin NC, Lang BF. Mitochondrial RNase P RNAs in ascomycete fungi: lineage-specific variations in RNA secondary structure. RNA. 2003;9:1073–1083. doi: 10.1261/rna.5880403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chamberlain JR, Lee Y, Lane WS, Engelke DR. Purification and characterization of the nuclear RNase P holoenzyme complex reveals extensive subunit overlap with RNase MRP. Genes Dev. 1998;12:1678–1690. doi: 10.1101/gad.12.11.1678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kassenbrock CK, Gao GJ, Groom KR, Sulo P, Douglas MG, Martin NC. RPM2, independently of its mitochondrial RNase P function, suppresses an ISP42 mutant defective in mitochondrial import and is essential for normal growth. Mol. Cell Biol. 1995;15:4763–4770. doi: 10.1128/mcb.15.9.4763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Stribinskis V, Gao GJ, Ellis SR, Martin NC. Rpm2, the protein subunit of mitochondrial RNase P in Saccharomyces cerevisiae, also has a role in the translation of mitochondrially encoded subunits of cytochrome c oxidase. Genetics. 2001;158:573–585. doi: 10.1093/genetics/158.2.573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Stribinskis V, Heyman HC, Ellis SR, Steffen MC, Martin NC. Rpm2p, a component of yeast mitochondrial RNase P, acts as a transcriptional activator in the nucleus. Mol. Cell Biol. 2005;25:6546–6558. doi: 10.1128/MCB.25.15.6546-6558.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Schonauer MS, Kastaniotis AJ, Hiltunen JK, Dieckmann CL. Intersection of RNA processing and the type II fatty acid synthesis pathway in yeast mitochondria. Mol. Cell Biol. 2008;28:6646–6657. doi: 10.1128/MCB.01162-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chen JY, Martin NC. Biosynthesis of tRNA in yeast mitochondria. An endonuclease is responsible for the 3′-processing of tRNA precursors. J. Biol. Chem. 1988;263:13677–13682. [PubMed] [Google Scholar]
- 26.Papadimitriou A, Gross HJ. Pre-tRNA 3′-processing in Saccharomyces cerevisiae. Purification and characterization of exo- and endoribonucleases. Eur. J. Biochem. 1996;242:747–759. doi: 10.1111/j.1432-1033.1996.0747r.x. [DOI] [PubMed] [Google Scholar]
- 27.Vogel A, Schilling O, Spath B, Marchfelder A. The tRNase Z family of proteins: physiological functions, substrate specificity and structural properties. Biol. Chem. 2005;386:1253–1264. doi: 10.1515/BC.2005.142. [DOI] [PubMed] [Google Scholar]
- 28.Rogowska AT, Puchta O, Czarnecka AM, Kaniak A, Stepien PP, Golik P. Balance between transcription and RNA degradation is vital for Saccharomyces cerevisiae mitochondria: reduced transcription rescues the phenotype of deficient RNA degradation. Mol. Biol. Cell. 2006;17:1184–1193. doi: 10.1091/mbc.E05-08-0796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Borowski LS, Szczesny RJ, Brzezniak LK, Stepien PP. RNA turnover in human mitochondria: more questions than answers? Biochim. Biophys. Acta. 2010;1797:1066–1070. doi: 10.1016/j.bbabio.2010.01.028. [DOI] [PubMed] [Google Scholar]
- 30.Chernyakov I, Whipple JM, Kotelawala L, Grayhack EJ, Phizicky EM. Degradation of several hypomodified mature tRNA species in Saccharomyces cerevisiae is mediated by Met22 and the 5′-3′ exonucleases Rat1 and Xrn1. Genes Dev. 2008;22:1369–1380. doi: 10.1101/gad.1654308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Butler JS. The yin and yang of the exosome. Trends Cell. Biol. 2002;12:90–96. doi: 10.1016/s0962-8924(01)02225-5. [DOI] [PubMed] [Google Scholar]
- 32.Carpousis AJ, Vanzo NF, Raynal LC. mRNA degradation. A tale of poly(A) and multiprotein machines. Trends Genet. 1999;15:24–28. doi: 10.1016/s0168-9525(98)01627-8. [DOI] [PubMed] [Google Scholar]
- 33.Gagliardi D, Stepien PP, Temperley RJ, Lightowlers RN, Chrzanowska-Lightowlers ZM. Messenger RNA stability in mitochondria: different means to an end. Trends Genet. 2004;20:260–267. doi: 10.1016/j.tig.2004.04.006. [DOI] [PubMed] [Google Scholar]
- 34.Dziembowski A, Piwowarski J, Hoser R, Minczuk M, Dmochowska A, Siep M, van der Spek H, Grivell L, Stepien PP. The yeast mitochondrial degradosome. Its composition, interplay between RNA helicase and RNase activities and the role in mitochondrial RNA metabolism. J. Biol. Chem. 2003;278:1603–1611. doi: 10.1074/jbc.M208287200. [DOI] [PubMed] [Google Scholar]
- 35.Dmochowska A, Golik P, Stepien PP. The novel nuclear gene DSS-1 of Saccharomyces cerevisiae is necessary for mitochondrial biogenesis. Curr. Genet. 1995;28:108–112. doi: 10.1007/BF00315775. [DOI] [PubMed] [Google Scholar]
- 36.Stepien PP, Margossian SP, Landsman D, Butow RA. The yeast nuclear gene suv3 affecting mitochondrial post-transcriptional processes encodes a putative ATP-dependent RNA helicase. Proc. Natl Acad. Sci. USA. 1992;89:6813–6817. doi: 10.1073/pnas.89.15.6813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zahedi RP, Sickmann A, Boehm AM, Winkler C, Zufall N, Schonfisch B, Guiard B, Pfanner N, Meisinger C. Proteomic analysis of the yeast mitochondrial outer membrane reveals accumulation of a subclass of preproteins. Mol. Biol. Cell. 2006;17:1436–1450. doi: 10.1091/mbc.E05-08-0740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Meisinger C, Sommer T, Pfanner N. Purification of Saccharomcyes cerevisiae mitochondria devoid of microsomal and cytosolic contaminations. Anal. Biochem. 2000;287:339–342. doi: 10.1006/abio.2000.4868. [DOI] [PubMed] [Google Scholar]
- 39.Daum G, Bohni PC, Schatz G. Import of proteins into mitochondria. Cytochrome b2 and cytochrome c peroxidase are located in the intermembrane space of yeast mitochondria. J. Biol. Chem. 1982;257:13028–13033. [PubMed] [Google Scholar]
- 40.Liu M, Spremulli L. Interaction of mammalian mitochondrial ribosomes with the inner membrane. J. Biol. Chem. 2000;275:29400–29406. doi: 10.1074/jbc.M002173200. [DOI] [PubMed] [Google Scholar]
- 41.Schagger H. Native electrophoresis for isolation of mitochondrial oxidative phosphorylation protein complexes. Methods Enzymol. 1995;260:190–202. doi: 10.1016/0076-6879(95)60137-6. [DOI] [PubMed] [Google Scholar]
- 42.Schagger H, von Jagow G. Blue native electrophoresis for isolation of membrane protein complexes in enzymatically active form. Anal. Biochem. 1991;199:223–231. doi: 10.1016/0003-2697(91)90094-a. [DOI] [PubMed] [Google Scholar]
- 43.Wittig I, Braun HP, Schagger H. Blue native PAGE. Nat Protoc. 2006;1:418–428. doi: 10.1038/nprot.2006.62. [DOI] [PubMed] [Google Scholar]
- 44.Wittig I, Schagger H. Electrophoretic methods to isolate protein complexes from mitochondria. Methods Cell Biol. 2007;80:723–741. doi: 10.1016/S0091-679X(06)80033-6. [DOI] [PubMed] [Google Scholar]
- 45.Jacob Y, Seif E, Paquet PO, Lang BF. Loss of the mRNA-like region in mitochondrial tmRNAs of jakobids. RNA. 2004;10:605–614. doi: 10.1261/rna.5227904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Guerrier-Takada C, Altman S. Inactivation of gene expression using ribonuclease P and external guide sequences. Methods Enzymol. 2000;313:442–456. doi: 10.1016/s0076-6879(00)13028-9. [DOI] [PubMed] [Google Scholar]
- 47.Wessels HJ, Vogel RO, van den Heuvel L, Smeitink JA, Rodenburg RJ, Nijtmans LG, Farhoud MH. LC-MS/MS as an alternative for SDS-PAGE in blue native analysis of protein complexes. Proteomics. 2009;9:4221–4228. doi: 10.1002/pmic.200900157. [DOI] [PubMed] [Google Scholar]
- 48.Fandino AS, Rais I, Vollmer M, Elgass H, Schagger H, Karas M. LC-nanospray-MS/MS analysis of hydrophobic proteins from membrane protein complexes isolated by blue-native electrophoresis. J. Mass Spectrom. 2005;40:1223–1231. doi: 10.1002/jms.903. [DOI] [PubMed] [Google Scholar]
- 49.Perkins DN, Pappin DJ, Creasy DM, Cottrell JS. Probability-based protein identification by searching sequence databases using mass spectrometry data. Electrophoresis. 1999;20:3551–3567. doi: 10.1002/(SICI)1522-2683(19991201)20:18<3551::AID-ELPS3551>3.0.CO;2-2. [DOI] [PubMed] [Google Scholar]
- 50.Wittig I, Schagger H. Features and applications of blue-native and clear-native electrophoresis. Proteomics. 2008;8:3974–3990. doi: 10.1002/pmic.200800017. [DOI] [PubMed] [Google Scholar]
- 51.Ghaemmaghami S, Huh WK, Bower K, Howson RW, Belle A, Dephoure N, O'Shea EK, Weissman JS. Global analysis of protein expression in yeast. Nature. 2003;425:737–741. doi: 10.1038/nature02046. [DOI] [PubMed] [Google Scholar]
- 52.Gan X, Kitakawa M, Yoshino K, Oshiro N, Yonezawa K, Isono K. Tag-mediated isolation of yeast mitochondrial ribosome and mass spectrometric identification of its new components. Eur. J. Biochem. 2002;269:5203–5214. doi: 10.1046/j.1432-1033.2002.03226.x. [DOI] [PubMed] [Google Scholar]
- 53.Smits P, Smeitink JA, van den Heuvel LP, Huynen MA, Ettema TJ. Reconstructing the evolution of the mitochondrial ribosomal proteome. Nucleic Acids Res. 2007;35:4686–4703. doi: 10.1093/nar/gkm441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Schonauer MS, Kastaniotis AJ, Kursu VA, Hiltunen JK, Dieckmann CL. Lipoic acid synthesis and attachment in yeast mitochondria. J. Biol. Chem. 2009;284:23234–23242. doi: 10.1074/jbc.M109.015594. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.