Abstract
DNA double-strand breaks pose a significant threat to cell survival and must be repaired. In higher eukaryotes, such damage is repaired efficiently by non-homologous end joining (NHEJ). Within this pathway, XRCC4 and XLF fulfill key roles required for end joining. Using DNA-binding and -bridging assays, combined with direct visualization, we present evidence for how XRCC4–XLF complexes robustly bridge DNA molecules. This unanticipated, DNA Ligase IV-independent bridging activity by XRCC4–XLF suggests an early role for this complex during end joining, in addition to its more well-established later functions. Mutational analysis of the XRCC4–XLF C-terminal tail regions further identifies specialized functions in complex formation and interaction with DNA and DNA Ligase IV. Based on these data and the crystal structure of an extended protein filament of XRCC4–XLF at 3.94 Å, a model for XRCC4–XLF complex function in NHEJ is presented.
INTRODUCTION
DNA double-strand breaks (DSBs) pose a serious threat to genomic stability. DSBs result from exposure to exogenous sources such as ionizing radiation, but can also result from normal cellular processes including intermediates of both V(D)J recombination and class switch recombination in developing lymphocytes (1,2). Multiple mechanisms exist to repair DSBs but in mammalian cells, Non-Homologous End Joining (NHEJ) is the predominant pathway utilized to repair two-ended DSBs. The core set of proteins required for NHEJ includes the Ku heterodimer, the catalytic subunit of the DNA-dependent protein kinase (DNA-PKcs), XRCC4, XLF/Cernunnos and DNA Ligase IV. Ku initially binds DSBs and recruits DNA-PKcs, generating the DNA-PK complex (3). Synapsis of two DNA-PK complexes regulates, via autophosphorylation, processing of damaged DNA ends by Artemis and other DNA modifying enzymes (4–7). XRCC4–DNA Ligase IV can then ligate the two DNA strands together. XLF directly interacts with XRCC4 and stimulates XRCC4–DNA Ligase IV joining of DNA ends although the mechanistic basis for XLF's stimulatory effect is not understood (8–10). Both XLF−/− and XRCC4−/− mammalian cells exhibit increased sensitivity to ionizing radiation coupled with serious defects in DSB repair (11,12). Furthermore, XRCC4 deficiency in mice results in embryonic lethality (13). Although humans with XLF deficiency display growth retardation, and immunodeficiency (14), XLF deficiency in mice results in only a modest immunodeficient phenotype with no effect on growth (15). Alt and colleagues (16) have recently shown, however, that mice deficient in both XLF and ATM have a much more severe NHEJ phenotype consistent with functional redundancy of XLF and ATM and suggest that like ATM, XLF may also facilitate stabilization or bridging of DNA ends at a DSB.
Still, how an XRCC4–XLF complex functions during repair is not well-understood. Although they interact with one another, only XRCC4 interacts directly with DNA Ligase IV even though XLF stimulates ligation of non-cohesive DNA ends (8,10,17). Together these findings have led to the idea that XRCC4 and XLF act late during NHEJ, in particular promoting the final ligation step. More recently, however, live-cell imaging demonstrated that recruitment of XLF and XRCC4 to damaged nuclear sites is rapid and dependent on the presence of Ku but not DNA-PKcs (18,19). Furthermore, Roy et al. (52) report that perturbations of the XRCC4–XLF interaction result in DSB repair deficits including reduced frequency of coding-end joining during V(D)J recombination but not signal-end joining. Taken together, these data suggest that XRCC4 and XLF may have multiple functions during NHEJ, including their well-characterized late step in ligation and also an earlier step that may be independent of DNA Ligase IV.
Individual crystal structures of XRCC4 and XLF reveal their structural similarity, forming homodimers with nearly identical N-terminal head domains and long α-helical tails (15,20,21). Mutational analysis within the head domains identified residues R64, L65 and L115 of XLF, and K65 and K99 of XRCC4 as critical to XRCC4–XLF complex formation (20,22). Given the location of these residues and the dimeric nature of the proteins, it was proposed that XRCC4–XLF could form extended filament-like structures (20,22). This model has gained further support through recent structural analyses demonstrating that filaments of alternating XRCC4–XLF dimers can be formed in vitro (23–26). The functional significance of these structures is not yet understood. Here, we determine a role for XRCC4–XLF independent of DNA Ligase IV in bridging DSBs. XRCC4–XLF complexes are also shown to exist in two distinct states, one of which mediates DNA bridging. We further identify regions of XLF and XRCC4 important for DNA binding and demonstrate that these regions make independent contributions to XRCC4–XLF function in DNA bridging. In particular, the C-terminal tails of XLF are essential for XRCC4–XLF complex–DNA interactions, while the corresponding region of XRCC4 appears to regulate formation of the nucleoprotein complex required for DNA bridging. Sequestration of XRCC4 C-terminal tails through binding of DNA LigaseIV tandem BRCT domains disrupts this species, ablating DNA bridging. Based on these data and the crystal structure of an extended protein filament of XRCC4–XLF at 3.94 Å, a model for the mechanism of XRCC4–XLF association within NHEJ is presented.
MATERIALS AND METHODS
XLF and XRCC4 expression vector construction
All XLF and XRCC4 expression plasmids, with the exception of XRCC41–157, were from previously described work (20,21,27). See Supplementary Data for further details.
Protein expression and purification
XLF, XRCC4 and BRCT domains, wild-type and all mutations, were expressed and purified as previously described (20,21). XRCC41–157 was expressed in M9 SeMET growth media kit (Medicilon Inc.).
Electrophoretic mobility shift assays for DNA binding
DNA preparation is described in Supplementary Data. Binding reactions (20 µl) contained 100 ng of DNA, 20 mM HEPES pH 8, 0.5 mM EDTA, 1 mM DTT, 5% glycerol and 120 mM KCl after addition of proteins at the indicated concentrations. Reactions were incubated at room temperature for 30 min and resolved by electrophoresis in a 0.8% agarose gel in Tris–borate buffer (89 mM Tris–base, 89 mM boric acid, 2 mM EDTA pH 8.3) at 80 V/15 cm for 60 or 90 min. Gels were stained in Tris–borate buffer supplemented with 0.5 µg/ml ethidium bromide (30 min) and destained in deionized water (2 h).
DNA bridging
DNA bridging was carried out as illustrated in Figure 2A. Full details are provided in Supplementary Data.
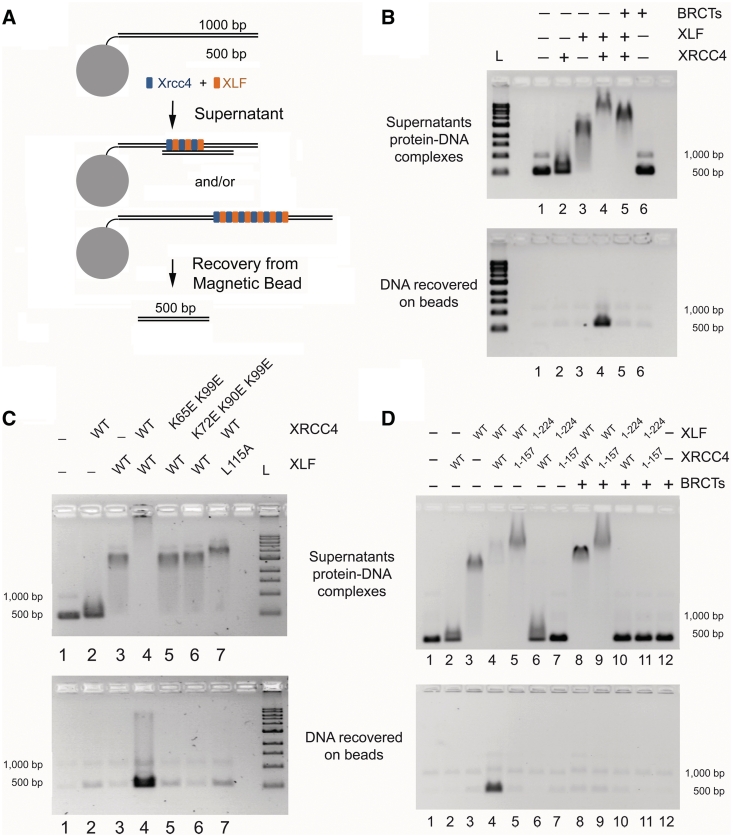
Figure 2.
Bridging of DNA molecules by XRCC4–XLF. (A) Schematic of DNA-bridging assay. Proteins were incubated with magnetic beads linked to 1000-bp DNA and free 500-bp DNA. Beads were separated from supernatant and analyzed separately for presence of the 500-bp DNA. (B) an amount of 200 ng each of 1000- and 500-bp DNA fragments were incubated with XRCC4 (2 µM), XLF (2 µM) or DNA Ligase IV tandem BRCT domains (BRCTs, 2 µM). Top panel shows the analysis of the protein–DNA complexes in the supernatants. Bottom panel shows the recovery of DNA species on the beads. L = 1 kb DNA ladder (NEB). (C) Bridging assays performed as in (B) with mutants preventing XRCC4–XLF filament formation. (D) Bridging assays performed as in (B). XRCC41–157 and XLF1–224 are truncated proteins lacking C-terminal tails and DNA-binding activity.
Scanning force microscopy
DNA substrates and XRCC4–XLF complexes were prepared as described in Supplementary Data. Images were obtained on a NanoScope IV SFM (Digital Instruments; Santa Barbara, CA) operating in tapping mode in air with a type E scanner. Silicon Nanotips were from Digital Instruments (Nanoprobes). The length and height measurements were done with NanoScope software v 5.12 (Digital Instruments; Santa Barbara, CA, USA). Although absolute dimensions in SFM are distorted by tip convolution, relative size and separation between objects can be used very accurately.
Crystallization and data collection of XRCC41−157–XLF1−224
Crystals were grown using the hanging drop vapor diffusion method. A 1 μl protein solution (50 μM XLF1−224: 100 μM XRCC41−157 in 20 mM Tris, pH 8, 200 mM KCl, 1 mM EDTA, 10 mM DTT and 10% glycerol) was combined with microseeded crystallization solution (0.8 μl of 1.8 M tri-ammonium citrate, pH 8) and additives (0.2 μl each 0.1 M barium chloride dihydrate and 2.0 M sodium thiocyanate). Crystals were dehydrated over 4 M ammonium sulfate, pH 7, prior to a 3-h soak in phasing solution before freezing (1 μl 0.5 mM tantalum bromide and 0.5 μl of 60% PEG 8000). Full crystallization details are described elsewhere (28). Diffraction data were collected at NSLS, X25 (Brookhaven, NY, USA). Coordinates for the structure have been deposited to the PDB, accession code 3RWR.
Structure determination and model refinement
Data was processed with HKL2000 (29). Phases were determined using MR-SAD, followed by density modification with Phenix. An XRCC4–XLF docking model was used as a search model (22). Eight tantalum bromide sites were found. Iterative rounds of model building and refinement were carried out using Phenix, Wincoot and CNS v1.3 with DEN refinement (30–33). DEN refinement was chosen based on its ability to refine lower resolution structures. Interacting regions were analyzed using ZMM (34). All structural figures were generated using PyMol (The PyMOL Molecular Graphics System, Version 1.2r2, Schrödinger, LLC). Buried surface area calculations were carried out through the PISA server, v.1.18 (35).
RESULTS
Identification of regions required for XRCC4–XLF complex-dependent DNA binding
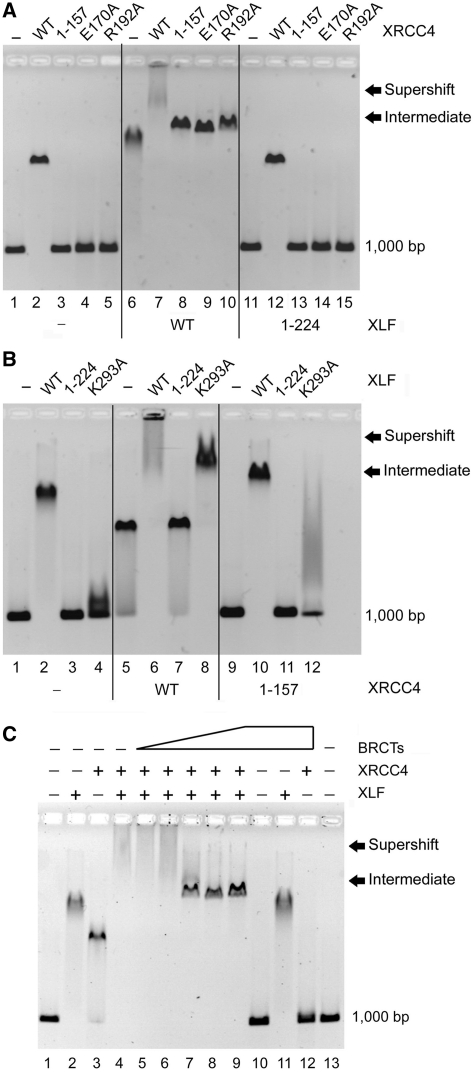
Here we have localized the DNA-binding interface of XRCC4 and XLF to residues 157–200 and 224–299, respectively (Figure 1A, lane 3 and Figure 1B, lane 3). Further mutational analysis within XRCC4 identified amino acids E170 and R192 as necessary for DNA binding (Figure 1A, lanes 4–5). These residues fall in a highly conserved segment of XRCC4 also responsible for homo-tetramerization and interaction with DNA Ligase IV-tandem BRCT domains (BRCTs) (Supplementary Figure S1). Several additional residues located between 157 and 200 were also implicated in DNA binding (Supplementary Table S1). Like XRCC4, DNA binding of XLF resides in its tail region (20). This region of XLF has also been shown to mediate XRCC4–XLF parallel filament arrangements (25). Alanine substitution at K293 decreased the overall DNA-binding capacity of XLF (Figure 1B, lane 4), localizing XLF–DNA-binding activity to a highly conserved lysine cluster at the C-terminus, similar to reports for XLF's yeast homolog, Nej1 (36) (Supplementary Figure S2). Additional DNA-binding sites within the head region of XLF have been identified by HDX (Hydrogen Deuterium Exchange) experiments (25); however, based on our studies with XLF1–224 the head region alone does not support stable DNA binding (Figure 1).
Figure 1.
XRCC4–XLF DNA binding. (A) XRCC4 wild-type (WT) and mutants (8 µM) were incubated with 100 ng of DNA with or without 2 µM XLF (WT and mutants), analyzed by EMSA. (B) WT XLF and its mutants (2 µM) were incubated with 100 ng of DNA with or without 8 µM XRCC4 (WT and mutants). (C) Effect of DNA Ligase IV tandem BRCT domains on XRCC4–XLF–DNA complex formation. XRCC4 (8 µM) and XLF (2 µM) were incubated with 100 ng DNA fragments in the presence of increasing amounts of DNA Ligase IV tandem BRCT domains (BRCTs, 0, 0.5, 1, 2, 4–8 µM).
We next tested DNA-binding activity under conditions where XRCC4–XLF complexes are formed (20) and observed a super-shifted protein–DNA complex that migrated as a diffuse smear up into the well of the gel suggesting formation of a heterogeneous collection of large nucleoprotein complexes (Figure 1A, lane 7; Figure 1B, lane 6). Interestingly, when XRCC4–DNA-binding mutants were present, only an intermediate gel-shift was observed (Figure 1A, lanes 8–10). This intermediate species migrated faster than the heterogeneous super-shifted species, but slower than XLF– or XRCC4–DNA complexes on their own. Similar to XLF– and XRCC4–DNA complexes, the intermediate species migrated as a discrete, homogeneous complex. Like the super-shifted complex, the intermediate complex is also dependent on the presence of both XRCC4 and XLF.
Unlike XRCC4, XLF truncations and point mutants unable to bind DNA, in complex with wild-type XRCC4, produced either no shift (XLF1–224), or an intermediate shift (XLF–K293A) (Figure 1B) indicating that the DNA-binding activity of XLF but not XRCC4 is necessary for XRCC4–XLF–DNA association. Complex observed in lane 8 is likely due to a partial DNA-binding defect of XLF–K293A (Figure 1B, lane 3 versus 4). DNA binding in the C-terminal region of XLF is clearly more important for XRCC4–XLF complex–DNA interactions than that of XRCC4 (Figure 1B, lane 7 versus Figure 1A, lane 8). Disruption of DNA binding by C-terminal deletion of XLF fully ablates XRCC4–XLF complex–DNA association, while XRCC4–DNA binding within the complex is dispensable, as the intermediate species is still observed. Furthermore, previously identified mutations (20) that prevent XRCC4–XLF interaction fail to bind DNA beyond individual wild-type proteins, indicating XRCC4–XLF complexes are required to form both intermediate and super-shifted species (Supplementary Figure S3).
DNA Ligase IV and DNA-binding regions of XRCC4 modulate XRCC4–XLF complex–DNA interactions
Residues 155–195 of XRCC4 interact with DNA Ligase IV BRCTs, including direct contact with R192 (37). Here we show that R192 and other residues within this region also mediate interaction with DNA (Supplementary Table S1) (38). Accordingly, simultaneous incubation with BRCTs precluded XRCC4's interaction with DNA (Figure 1C, lane 12) suggesting these binding events are indeed mutually exclusive. Consistent with this observation, the super-shifted species was converted to the intermediate species in the presence of equimolar concentrations of the tandem BRCT domains of DNA Ligase IV (Figure 1C, lanes 7–9). The intermediate species is not disrupted, however, even at the highest concentrations of BRCTs tested, and is reminiscent of the complex observed in Figure 1A and B with wild-type XLF and XRCC4 truncations and point mutants unable to bind DNA. The observation of multiple nucleoprotein complexes (super-shifted and intermediate) suggests that XRCC4–XLF complexes can associate with DNA in at least two distinct ways reflecting changes in shape and/or number of XRCC4–XLF complexes and DNA.
XRCC4–XLF complexes bridge DNA molecules
During DSB repair, DNA ends are maintained in close proximity by higher order nucleoprotein complexes (39). The finding that XRCC4–XLF complexes interact with DNA, suggests these structures may be important for bridging DNA during repair. To test this hypothesis, we followed the experimental design outlined in Figure 2A. If XRCC4–XLF bridges the 1000- and 500-bp DNA, then the 500-bp DNA will be recovered from streptavidin-coated beads. XLF and XRCC4 on their own were unable to bridge DNA (Figure 2B, lanes 2–3). However, in the presence of both XRCC4 and XLF, 500-bp DNA was efficiently recovered from the beads, reflecting the ability of XRCC4–XLF complexes to bridge DNA molecules (Figure 2B, lane 4). XLF and XRCC4 mutants that abolish XRCC4–XLF interaction did not support DNA bridging, suggesting that XRCC4–XLF complex mediates bridging activity (Figure 2C).
Similar to the effect seen in DNA binding, DNA bridging by XRCC4–XLF was also disrupted by the presence of BRCTs (Figure 2B, lane 5). Since the intermediate species was still present in the supernatant when bridging was abolished, it appears that formation of the heterogeneous, super-shifted species is required for DNA bridging. One would further expect disruption of DNA bridging by other factors that alter the C-terminal tail of XRCC4 such as XRCC4 truncations and point mutations unable to bind DNA, or phosphorylation by DNA-PK, as illustrated by Roy et al. (52).
As expected, complex-dependent DNA bridging was abolished when an XRCC4 C-terminal truncation unable to bind DNA was assayed for bridging capabilities (Figure 2D). Any truncation of either XLF or XRCC4 that fully disrupts DNA binding also prevented DNA bridging (Figure 2D). In particular, reactions carried out with XRCC41–157 do not bridge DNA, but are able to support complex–DNA binding, even though XRCC41–157 is unable to bind DNA alone (Figure 2D, lanes 5 and 9). Together these results demonstrate that XRCC4 tails are not required for complex-dependent DNA binding, yet are still necessary for DNA bridging, which requires formation of a heterogeneous super-shifted nucleoprotein complex.
Direct visualization of higher-order XRCC4–XLF nucleoprotein complexes
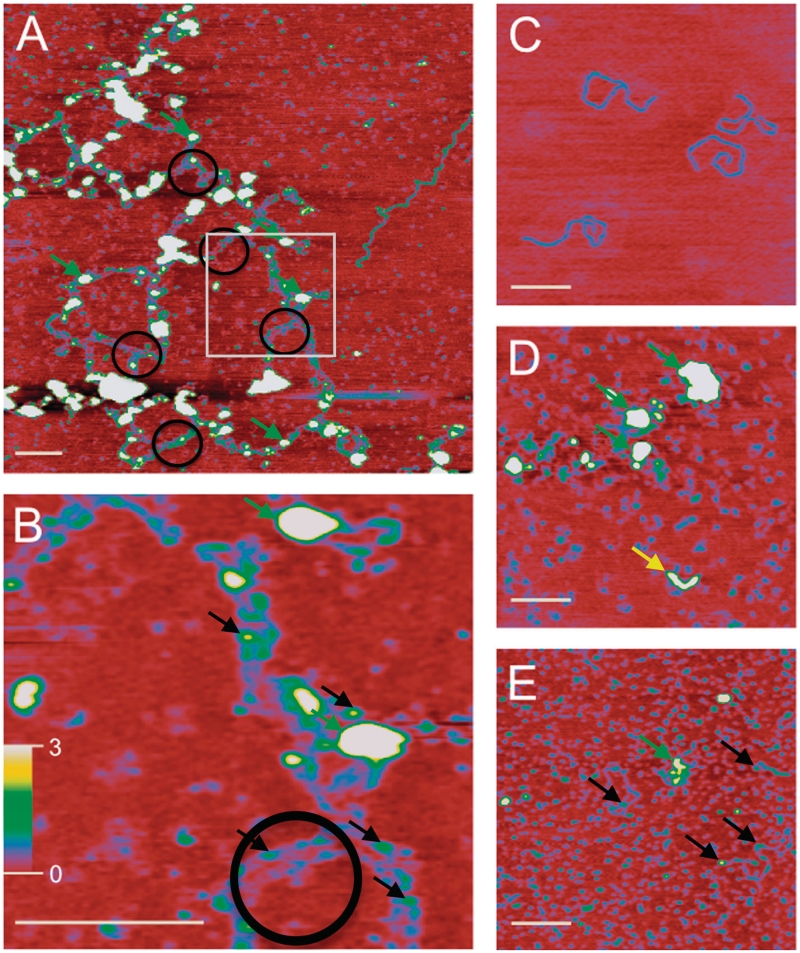
Observation of multiple nucleoprotein complexes during DNA binding and bridging studies suggests that at least two distinct higher order protein–DNA complexes are formed by XRCC4–XLF. To further investigate this possibility, we visualized such complexes using Scanning Force Microscopy (SFM), under conditions identical to DNA-binding and -bridging experiments.
XRCC4 and XLF were imaged independently and appeared as monodisperse objects (data not shown). DNA alone displayed dimensions characteristic for this type of SFM imaging (1821-bp DNA measured 6000 × 180 × 2 Å) (Figure 3C). When XRCC4 and XLF were incubated together in the absence of DNA, monodisperse objects were still observed (Figure 3D, small blue objects), however additional, larger, irregular species (Figure 3D, green arrows, 30–60 Å high and 400–800 Å wide) and elongated objects (Figure 3D, yellow arrow, ∼1300 Å long, 20–25 Å high and 300 Å wide) were also observed. Addition of DNA to XRCC4–XLF generated large protein–DNA networks (Figure 3A and B). DNA molecules were identified based on similar width and height to the DNA only control (Figure 3C). XRCC4–XLF complexes arranged DNA in two distinct ways. First, multiple DNAs were aligned end-to-end as indicated by complexes that were several fold longer than the length of the individual 1.8-kb DNA fragments (compare DNA in Figure 3C and A). In addition, DNA molecules were found aligned in parallel with an even spacing of ∼200 Å (Figure 3A and B, black circles). Given the regularity and extensive occurrence of these parallel DNA arrangements it is likely they reflect specialized repair complexes. Each of these DNA arrangements appear connected by XRCC4–XLF complexes that appear as objects higher and wider than DNA alone (Figure 3A and B, green and black arrows). The largest of these XRCC4–XLF complexes were ∼30–60 Å high and 400–800 Å wide (Figure 3A and B, green arrows) while the smaller XRCC4–XLF complexes were 4–7 Å high and 110–180 Å wide (Figure 3B, black arrows). Either or both of these DNA–protein (DNA end-to-end or parallel) arrangements may be essential for the DNA-bridging activity observed in Figure 2.
Figure 3.
SFM analysis of protein–DNA networks. (A) Large protein–DNA networks observed in XRCC4–XLF DNA-binding reactions. Nucleoprotein complexes appear as higher wider objects (∼30–60 Å high and 400–800 Å wide, green arrows). Such complexes were typical and observed on all 16 of the 2 × 2 micron images collected for this sample. Protein-induced parallel bridging of DNA molecules is evident (black circles) and occurs in many places along the long complex. Extended DNA networks indicate DNA molecules are positioned end-to-end. White box indicates area enlarged in (B). (B) Enlargement of (A) highlighting parallel DNA molecules. Spacing between parallel DNA molecules is ∼200 Å. Smaller nucleoprotein complexes, measuring 4–7 Å high and 110–180 Å wide, are also evident (Figure 3B, black arrows). (C) The 1.8-kb linear DNA, contour length 6000 Å, width 180 Å and height 2 Å. (D) Image of XRCC4–XLF complexes in absence of DNA. Small protein complexes, likely dimers, tetramers and small multimers, appear as blue uniform objects distributed over the surface. Larger protein complexes (30–60 Å high and 400–800 Å wide) and elongated forms were also present and are indicated by green and yellow arrows, respectively. Elongated structures (yellow arrow) measuring ∼1300 Å long, 20–25 Å high and 300 Å wide, were observed with a frequency of approximately one for every 2 × 2 micron image inspected. (E) Addition of DNA Ligase IV BRCT domains disrupts nucleoprotein networks. Smaller protein complexes similar to (D) are uniformly distributed and appear as small blue objects. DNA molecules associated with protein complexes are also observed (25–60 Å high and 400–800 Å wide, green arrow) with a frequency of approximately one for every 2 × 2 micron image inspected. Smaller nucleoprotein complexes (4–7 Å high and 110–180 Å wide, black arrows) were observed at higher frequency. (A) is 2 × 2 microns. (B) is 500 × 500 nm. (C–E) are 1 × 1 microns. In all images the white bar is 2000 Å long and height is indicated by color (0–3 nm red to yellow/white, scale bar in panel B). Note that biomolecule dimensions are distorted in SFM images. X–Y dimensions increase due to tip convolution and Z (height) decreases relative to tip surface and tip molecule interactions. For instance DNA, that is 20 Å wide and 20 Å high based on crystal structure of B-form, typically measures 200 Å wide and 2–5 Å high in our SFM images. Relative size and separation between objects can be used very accurately.
Consistent with results from bridging assays (Figure 2B), addition of BRCTs fully disrupted the bridged DNA networks observed via SFM in Figure 3A, without ablating protein–DNA associations (Figure 3E, green and black arrows). The majority of protein–DNA complexes observed in the presence of BRCT domains measured ∼4–7 Å high and 110–180 Å wide (black arrows), making them roughly equivalent in size to the smaller nucleoprotein complexes observed in the absence of BRCT domains (Figure 3B, black arrows). Larger complexes observed in the presence of BRCT domains (Figure 3E, green arrow) remained similar in size to those indicated by green arrows in Figure 3A, B and D.
Structure of the XRCC41−157–XLF1−224 complex
To further investigate the structural relationship(s) of XRCC4–XLF, we crystallized domains of these proteins responsible for complex formation. To facilitate crystallization, flexible C-terminal regions were removed from full-length XRCC4 (1–336) and XLF (1–299). XRCC41–157–XLF1–224 crystals diffracted to ∼3 Å resolution; however, anisotropy limited the data to 3.94 Å resolution. Phase information was obtained using a combination of molecular replacement and single-wavelength anomalous diffraction (MR-SAD). Representative electron density is provided in Supplementary Figure S4. XRCC41–157–XLF1–224 crystals grew in spacegroup C2 with one extremely long axis (∼750 Å) and contained six dimers of each protein in the asymmetric unit. The structure was refined to R and Rfree values of 26.2 and 32.9, respectively (PDB 3RWR). Table 1 lists data collection and refinement statistics.
Table 1.
Crystallographic data and refinement statistics
| Data collection | |
| Wavelength (Å) | 1.2536 |
| Space group | C2 |
| Cell parameters (Å) | a = 745.4, b = 149.6, c = 80.5 α = γ = 90, β = 94.7 |
| Molecules in A.U. | 24 |
| Resolution range (Å)a | 50.0–3.9 (4.2–3.9) |
| Unique reflections | 78 630 |
| Data redundancya | 6.7 (6.6) |
| Completeness (%)a | 97.6 (98.4) |
| I/σ(I)a | 11.7 (2.1) |
| Rmerge (%)a | 15.2 (96.3) |
| Mosaicity | 0.35 |
| Wilson scaling B factor (Å2) | 168.55 |
| Model and refinement | |
| Resolution range (Å) | 50.0–3.94 |
| Rwork (%) | 27.0 |
| Rfree (%) | 32.6 |
| Reflections observed | 73 366 |
| Reflection test set | 3886 |
| Number of protein atoms | 3464 |
| RMSD bond lengths (Å) | 0.0028 |
| RMSD bond angles (Å) | 0.77 |
| Average B factor (Å2) | 176.5 |
aStatistics for the highest data resolution shell are shown in parentheses.
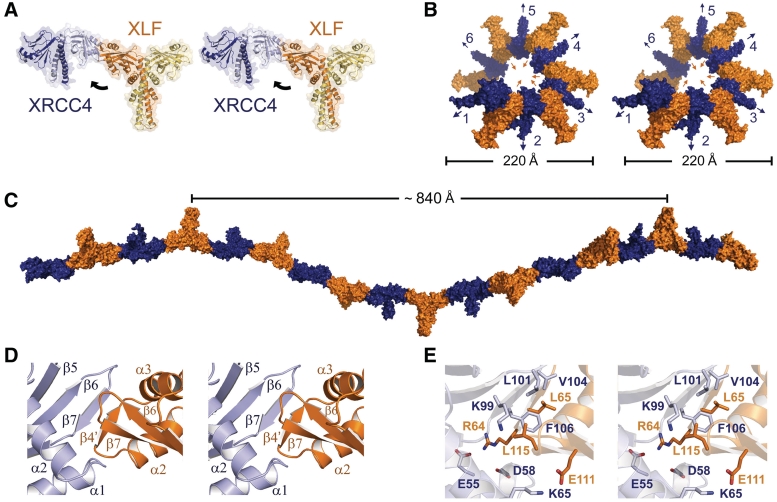
All dimers of XRCC41–157 and XLF1–224 within the asymmetric unit were part of continuous filaments extending the length of the crystal. Each left-handed helical filament was composed of alternating XRCC41–157 and XLF1–224 dimers with an ∼30° offset (Figure 4A). The filament diameter is ∼220 Å, with an internal pore of ∼70 Å, toward which both proteins’ N-terminal heads are directed. For XRCC41–157, the C-terminal tail region extends outward while the analogous region of XLF1–224 is directed toward the inner pore (Figure 4B and C), however, since both of these tail regions are highly flexible it is possible that they may not continue in a linear trajectory.
Figure 4.
Crystal structure of the XRCC41–157–XLF1–224 complex. (A) Interaction of XLF1–224 homodimer (orange) with an adjacent XRCC41–157 homodimer (blue) as seen in the crystal structure. Black arrow illustrates ∼30° offset between homodimers. (B) XRCC41–157–XLF1–224 filament. XRCC41–157 homodimers are numbered. Colored arrows indicate directionality of XRCC41–157 (blue) and XLF1–224 (orange) C-terminal tails. Filament diameter is indicated in Å. (C) XRCC41–157–XLF1–224 filament, rotated 90° clockwise from (B). Length of one filament revolution is indicated in Å. (D) XRCC41–157–XLF1–224 head-to-head interface. (E) Key amino acids directly involved in XRCC41–157–XLF1–224 head-to-head interaction.
Closer inspection of the XRCC41–157–XLF1–224 structure reveals formation of a β-sheet between strands β5-7 of XRCC41–157 and β4′ of XLF1–224 (Figure 4D). In the absence of XRCC41–157, β4′ forms a loop between α2 and α3 of the helix–turn–helix in XLF1–224, indicating that small conformational changes result from complex formation. The XRCC41–157–XLF1–224 interface is further strengthened through direct amino acid interactions (Figure 4E). All amino acids involved in this interface are listed in Supplementary Table S2. L65 and L115 of XLF make key contributions by sandwiching a highly conserved XRCC4 F106 (Supplementary Figure S1 and S2). In addition, K99 of XRCC4 makes hydrophobic interactions with L115 of XLF, through its aliphatic chain and also forms a hydrogen bond with the carbonyl oxygen of S113 in XLF (Figure 4E). These interactions are consistent with recently reported mutational and structural analyses (21,23–27).
DISCUSSION
The filamentous structure of XRCC4–XLF observed here is consistent with recently reported structures of these proteins (24–26). Despite these structures having been determined under different conditions, with differently sized protein truncations and in different crystal packing environments, little difference is observed in the interaction interface between XRCC4 and XLF. Combined with supportive biochemical analysis, mounting structural data strongly suggests that the interface observed here reflects a biologically relevant XRCC4–XLF complex. Despite these similarities, interesting differences were observed in the relative helical pitch associated with different filament structures (720, 760, 840 and 860 Å) (24–26). The ability of XRCC4–XLF filaments to accommodate changes in pitch indicates these structures possess elastic potential that one would expect for scaffolding molecules associated with bridging DNA. In addition, altered pitch results in variable internal pore sizes that could accommodate other molecules ranging in size from dsDNA to an entire nucleosome (24–26). Precedence for functional protein filaments in DNA repair is seen in NHEJ's counterpart, Homologous Recombination (HR). HR utilizes a nucleoprotein filament conserved in humans (RAD51), bacteria (RecA) and archaea (RadA)(40–42). RAD51–RadA forms protein filaments through a β-strand polymerization motif, where the β-strand of one RAD51–RadA protein binds to the β-sheet of an adjacent RAD51–RadA monomer (43,44). In a similar manner, RecA forms a continuous β-sheet across monomers within its growing filament. As an isolated protein, however, the interacting β-strand exists as a flexible loop (45). This is identical to what occurs in the loop region of XLF, when in contact with XRCC4 (Figure 4D, β4′). Furthermore, RadA, like XRCC4–XLF, also forms a left-handed protein helix to bind DNA (43). The crystal structure of DNA-bound RecA further shows that RecA uses an extended filament structure to wrap around DNA (46).
DNA binding by XRCC4 and XLF is reported to occur in a highly cooperative manner (27), suggesting a protein filament may form along DNA similar to what is observed with RecA or RAD51. In our structure, the DNA-binding region of XLF is situated toward the inner helical pore (Figure 4B), which has sufficient room (70 Å) to accommodate both DNA and the XLF DNA binding domain (residues 224–299) suggesting that XRCC4–XLF may wrap around DNA. However, it is equally possible for DNA to be wrapped around a filament as suggested by Hammel et al. (24). This mode of interaction is remarkably similar to how histone-like nucleoid structuring protein, H-NS, has been observed to bind DNA (47).
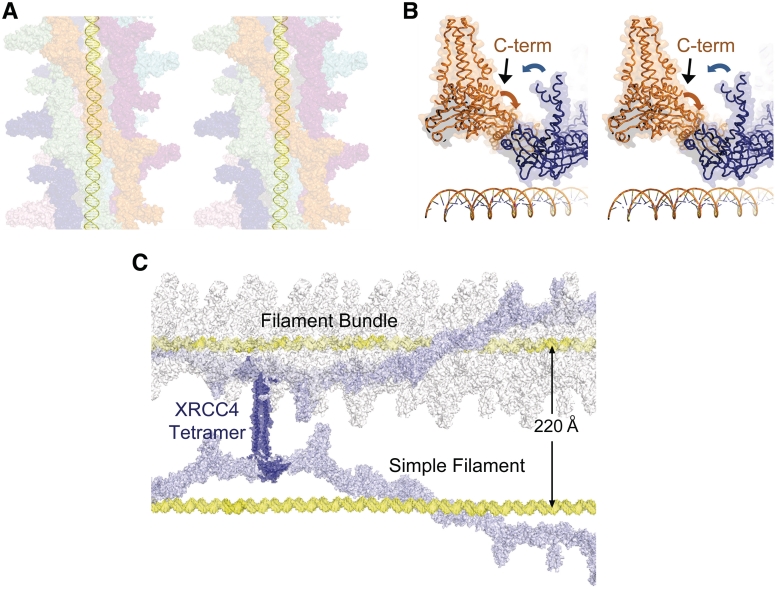
Results from DNA-binding studies presented here indicate two modes of protein–DNA interaction (an intermediate and heterogeneous super-shifted species), suggesting nucleoprotein filaments exist in more than one structural state. Of these, only the super-shifting species correlated with DNA bridging. XRCC4–XLF nucleoprotein complexes were further characterized via SFM and shown to interact with DNA in two general arrangements. The larger protein complexes observed in Figure 3 (green arrow) greatly exceed the size of a single XRCC4–XLF filament, consistent with multiple filament bundles observed recently both crystallographically and by SAXs analysis (24–26). Further support for this arrangement comes from direct visualization by transmission electron microscopy (24). In this way, DNA could be fully coated by multiple adjacent filaments (Figure 5A). Evidence for this arrangement is also observed in the crystal structure presented here and elsewhere (26). In particular, when modeled within the context of a filament bundle, the C-terminal tails of some XRCC4 dimers are directed toward the head domains of XLF in an adjacent filament (Figure 5B). This arrangement may explain why XRCC4 tails are required for formation of the larger nucleoprotein complexes capable of bridging DNA (Figures 2 and 3). Alteration of these tails through mutation, binding of BRCTs as observed in a previous structure with XRCC4 (37) or DNA-PK phosphorylation [Roy et al. (52)] could disrupt interactions between adjacent filaments resulting in smaller, less complex nucleoprotein assemblies, perhaps corresponding to the intermediate species observed in Figures 1 and 2, which are unable to bridge DNA. Given the structural information currently available (24–26) it is not obvious how BRCT binding to XRCC4 alone would disrupt single or multiple XRCC4–XLF filament(s). Current structures of XRCC4–XLF complexes lack the BRCT-binding region of XRCC4 making it difficult to assess a precise mechanism for BRCT-mediated disruption of the XRCC4–XLF bridging complex. However, results from SFM suggest alignment of DNA molecules both end-to-end and parallel to one another, which may account for disruption of XRCC4–XLF by BRCT domains. Interestingly, the same tail region of XRCC4 that is required for DNA bridging also corresponds to its homo-tetramerization region (38). It is therefore possible, that through homo-tetramerization, adjacent filament bundles bound to DNA could be bridged (Figure 5C). The spacing observed between parallel DNA molecules in SFM (∼200 Å) correlates well with those predicted in this model (Figure 5C). Since tetramerization and BRCT binding are mutually exclusive, addition of equimolar amounts of BRCT domains would be expected to disrupt all interactions dependent on XRCC4 tails, as was observed via SFM (Figure 3). It is important to note that the data presented here and resulting model are both compatible with DNA being present on the interior or exterior of XRCC4–XLF filaments, and perhaps even on the interior and the exterior at the same time. The model for XRCC4–XLF complexes presented in Figure 5 suggests that the filament observed within the asymmetric unit of the crystal structure presented here reflects a biologically relevant structure. At this time, however, no direct evidence exists to support filament formation in vivo.
Figure 5.
Model of XRCC4–XLF filaments bound to DNA. (A) Multiple adjacent filaments bound to DNA (yellow). Each color is a separate filament. DNA runs through the pore of the XRCC4–XLF filament. (B) Tails of an XRCC41–157 homodimer (blue) point toward the N-terminus of XLF1–224 (orange), in an adjacent filament. (C) XRCC4 dimers associate into a tetramer through C-terminal tails (PDB 1FU1). DNA-bridging models illustrating single and complex filaments. Two DNA molecules coated in a simple or multi-filament bundle are bridged through XRCC4 C-terminal tails.
DNA-bridging activity of XRCC4–XLF is consistent with a role in stabilizing DNA ends during NHEJ. Interestingly, binding of DNA Ligase IV apparently causes these bridging-competent complexes to be remodelled such that they no longer stably bridge DNA. If XRCC4 is available in excess over DNA Ligase IV, both types of complexes (XRCC4–XLF and XRCC4–Ligase IV) should be able to exist simultaneously. Furthermore, at substoichiometric concentrations of DNA Ligase IV (BRCT domains) XRCC4–XLF super-shifted complexes remain intact and are able to bridge DNA (Figure 1 and data not shown). Together this suggests that XRCC4–XLF multifilament complexes may persist when bound by a limited number of DNA Ligase IV molecules.
Stable bridging of DNA ends by XRCC4–XLF complexes is further supported by recent in vivo analysis suggesting XRCC4–XLF complex assembly is necessary for repair [Roy et al. (52)]. In NHEJ, Ku and DNA-PKcs have been implicated in synapsis of DNA ends (48). However recent studies show rapid, DNA-PKcs-independent recruitment of XLF to damaged sites by Ku that is further stabilized at DNA breaks by the recruitment of XRCC4 suggesting XRCC4–XLF plays an early role in repair (18,19). Collectively these results imply that Ku may initially recruit XLF to the damaged site and nucleate XRCC4–XLF-bridging complex formation along DNA (18,19,24–26). Regulation of this DNA-bridging complex (Figures 2 and 3) occurs via modifications to the C-terminus of XRCC4 (summarized in Figure 6). Interestingly, Roy et al. further demonstrate that DNA-PK hyper-phosphorylation within the same region of XRCC4 also modulates DNA-bridging activity (52). There are now several proteins reported to bridge DNA ends (Ku, ATM and MRN) (49–51). It will be interesting to determine how each of these proteins contribute to repair via their bridging activity of various types of DNA damage in vivo.
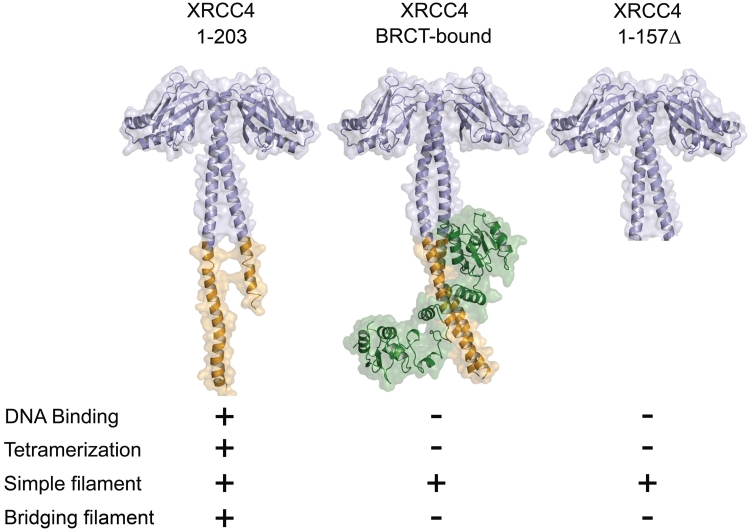
Figure 6.
Summary of the structural states of XRCC4. Structural states of XRCC4 are indicated with their associated function (PDB 1FU1 and PDB 3II6; 21,23,37,38).
Together this work presents new insight into the mechanism for how XRCC4–XLF complexes function in NHEJ. The ability of XRCC4–XLF to stably bridge DNA ends is consistent with previous observations and provides a mechanistic basis for how XLF stimulates XRCC4–DNA Ligase IV non-cohesive end joining (8–10). How these structures assemble with one another and other NHEJ factors for repair in vivo will require further investigation.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online: Supplementary Tables 1 and 2, Supplementary Figures 1–4 and Supplementary methods.
FUNDING
USA National Cancer Institute [5PO1CA09584 to C.W.]; the Netherlands Organization of Scientific Research, Chemical Sciences Division [VICI 700.56.441 to C.W.]; the Association for International Cancer Research [09-0633 to M.M.]; the Association pour la Recherche sur le Cancer [A09/2/5075 to M.M.]; the Canadian Institutes of Health Research [MOP-89903 to M.S.J.]; and the National Science and Engineering Research Council Canadian Graduate Scholarship to S.N.A. Funding for open access charge: Canadian Institutes of Health Research.
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
The authors would like to thank Dr M. Morar, McMaster University for structural discussion, and X-25 beamline staff, NSLS, Brookhaven National Laboratory.
REFERENCES
- 1.Kuzminov A. Single-strand interruptions in replicating chromosomes cause double-strand breaks. Proc. Natl Acad. Sci. USA. 2001;98:8241–8246. doi: 10.1073/pnas.131009198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Roth DB, Nakagima PB, Menetski JP, Bosma MJ, Gellert M. V(D)J recombination in mouse thymocytes: double-strand breaks near T cell receptor delta rearrangement signals. Cell. 1992;69:41–53. doi: 10.1016/0092-8674(92)90117-u. [DOI] [PubMed] [Google Scholar]
- 3.Gottlieb TM, Jackson SP. The DNA-dependent protein kinase: requirement for DNA ends and association with Ku antigen. Cell. 1993;72:131–142. doi: 10.1016/0092-8674(93)90057-w. [DOI] [PubMed] [Google Scholar]
- 4.Ding Q, Reddy YV, Wang W, Woods T, Douglas P, Ramsden DA, Lees-Miller SP, Meek K. Autophosphorylation of the catalytic subunit of the DNA-dependent protein kinase is required for efficient end processing during DNA double-strand break repair. Mol. Cell Biol. 2003;23:5836–5848. doi: 10.1128/MCB.23.16.5836-5848.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.McElhinny SA, Havener JM, Garcia-Diaz M, Juárez R, Bebenek K, Kee BL, Blanco L, Kunkel TA, Ramsden DA. A gradient of template dependence defines distinct biological roles for family × polymerases in nonhomologous end joining. Mol. Cell. 2005;19:357–366. doi: 10.1016/j.molcel.2005.06.012. [DOI] [PubMed] [Google Scholar]
- 6.Cui X, Yu Y, Gupta S, Cho YM, Lees-Miller SP, Meek K. Autophosphorylation of DNA-dependent protein kinase regulates DNA end processing and may also alter double-strand break repair pathway choice. Mol. Cell Biol. 2005;25:10842–10852. doi: 10.1128/MCB.25.24.10842-10852.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Goodarzi AA, Yu Y, Riballo E, Douglas P, Walker SA, Ye R, Härer C, Marchetti C, Morrice N, Jeggo PA, et al. DNA-PK autophosphorylation facilitates Artemis endonuclease activity. EMBO J. 2006;25:3880–3889. doi: 10.1038/sj.emboj.7601255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ahnesorg P, Smith P, Jackson SP. XLF interacts with the XRCC4-DNA ligase IV complex to promote DNA nonhomologous end-joining. Cell. 2006;2:301–313. doi: 10.1016/j.cell.2005.12.031. [DOI] [PubMed] [Google Scholar]
- 9.Grawunder U, Wilm M, Wu X, Kulesza P, Wilson TE, Mann M, Lieber MR. Activity of DNA ligase IV stimulated by complex formation with XRCC4 protein in mammalian cells. Nature. 1997;388:492–495. doi: 10.1038/41358. [DOI] [PubMed] [Google Scholar]
- 10.Tsai CJ, Kim SA, Chu G. XLF promotes the ligation of mismatched and noncohesive DNA ends. Proc. Natl Acad. Sci. USA. 2007;19:7851–7856. doi: 10.1073/pnas.0702620104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Giaccia AJ, Denko N, MacLaren R, Mirman D, Waldren C, Hart I, Stamato TD. Human chromosome 5 complements the DNA double-strand break-repair deficiency and gamma-ray sensitivity of the XR-1 hamster variant. Am. J. Hum. Genet. 1990;47:459–469. [PMC free article] [PubMed] [Google Scholar]
- 12.Zha S, Alt FW, Cheng HL, Brush JW, Li G. Defective DNA repair and increased genomic instability in Cernunnos-XLF-deficient murine ES cells. Proc. Natl Acad. Sci. USA. 2007;104:4518–4523. doi: 10.1073/pnas.0611734104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gao Y, Sun Y, Frank KM, Dikkes P, Fujiwara Y, Seidl KJ, Sekiguchi JM, Rathbun GA, Swat W, Wang J, et al. A critical role for DNA end-joining proteins in both lymphogenesis and neurogenesis. Cell. 1998;95:891–902. doi: 10.1016/s0092-8674(00)81714-6. [DOI] [PubMed] [Google Scholar]
- 14.Buck D, Malivert L, de Chasseval R, Barraud A, Fondanèche MC, Sanal O, Plebani A, Stéphan JL, Hufnagel M, le Deist F, et al. Cernunnos, a novel nonhomologous end-joining factor, is mutated in human immunodeficiency with microcephaly. Cell. 2006;124:287–299. doi: 10.1016/j.cell.2005.12.030. [DOI] [PubMed] [Google Scholar]
- 15.Li Y, Chirgadze DY, Bolanos-Garcia VM, Sibanda VM, Sibanda BL, Davies OR, Ahnesorg P, Jackson SP, Blundell TL. Crystal structure of human XLF/Cernunnos reveals unexpected differences from XRCC4 with implications for NHEJ. EMBO J. 2008;27:290–300. doi: 10.1038/sj.emboj.7601942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zha S, Guo C, Boboila C, Oksenych V, Cheng HL, Zhang Y, Wesemann DR, Yuen G, Patel H, Goff PH, et al. ATM damage response and XLF repair factor are functionally redundant in joining DNA breaks. Nature. 2011;469:250–254. doi: 10.1038/nature09604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Critchlow SE, Bowater RP, Jackson SP. Mammalian DNA double-strand break repair protein XRCC4 interacts with DNA ligase IV. Curr. Biol. 1997;7:588–598. doi: 10.1016/s0960-9822(06)00258-2. [DOI] [PubMed] [Google Scholar]
- 18.Yano K, Morotomi-Yano K, Lee KJ, Chen DJ. Functional significance of the interaction with Ku in DNA double-strand break recognition of XLF. FEBS Lett. 2011;585:841–846. doi: 10.1016/j.febslet.2011.02.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yano K, Morotomi-Yano K, Wang SY, Uematsu N, Lee KJ, Asaithamby A, Weterings E, Chen DJ. Ku recruits XLF to DNA double-strand breaks. EMBO Rep. 2008;9:91–96. doi: 10.1038/sj.embor.7401137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Andres SN, Modesti M, Tsai CJ, Chu G, Junop MS. Crystal structure of human XLF: a twist in nonhomologous DNA end-joining. Mol. Cell. 2007;28:1093–1101. doi: 10.1016/j.molcel.2007.10.024. [DOI] [PubMed] [Google Scholar]
- 21.Junop MS, Modesti M, Guarné A, Ghirlando R, Gellert M, Yang W. Crystal structure of the Xrcc4 DNA repair protein and implications for end joining. EMBO J. 2000;22:5962–5970. doi: 10.1093/emboj/19.22.5962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Malivert L, Ropars V, Nunez M, Drevet P, Miron S, Faure G, Guerois R, Mornon JP, Revy P, Charbonnier JB, et al. Delineation of the Xrcc4-interacting region in the globular head domain of cernunnos/XLF. J. Biol. Chem. 2010;285:26475–26483. doi: 10.1074/jbc.M110.138156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hammel M, Yu Y, Fang S, Lees-Miller SP, Tainer JA. XLF regulates filament architecture of the XRCC4-ligaseIV complex. Structure. 2010;11:1431–1442. doi: 10.1016/j.str.2010.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ropars V, Drevet P, Legrand P, Baconnais S, Amram J, Faure G, Márquez JA, Piétrement O, Guerois R, Callebaut I, et al. Structural characterization of filaments formed by human XRCC4-Cernunnos/XLF complex involved in nonhomologous DNA end-joining. Proc. Natl Acad. Sci. USA. 2011;108:12663–12668. doi: 10.1073/pnas.1100758108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hammel M, Rey M, Yu Y, Mani RS, Classen S, Liu M, Pique ME, Fang S, Mahaney BL, Weinfeld M, et al. XRCC4 interactions with XRCC4-like factor (XLF) create an extended grooved scaffold for DNA ligation and double-strand break repair. J. Biol. Chem. 2011;286:32638–32650. doi: 10.1074/jbc.M111.272641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wu Q, Ochi T, Matak-Vinkovic D, Robinson CV, Chirgadze DY, Blundell TL. Non-homologous end-joining partners in a helical dance: structural studies of XLF-XRCC4 interactions. Biochem. Soc. Trans. 2011;39:1387–1392. doi: 10.1042/BST0391387. [DOI] [PubMed] [Google Scholar]
- 27.Modesti M, Hesse JE, Gellert M. DNA binding of Xrcc4 protein is associated with V(D)J recombination but not with stimulation of DNA ligase IV activity. EMBO J. 1999;18:2008–2018. doi: 10.1093/emboj/18.7.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Andres SN, Junop MS. Crystallization and preliminary X-ray diffraction analysis of the human XRCC4-XLF complex. Acta Cryst. 2011;F67:1399–1402. doi: 10.1107/S1744309111033549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Otwinowski Z, Minor W. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 30.Adams PD, Afonine PV, Bunkóczi G, Chen VB, Davis IW, Echols N, Headd JJ, Hung LW, Kapral GJ, Grosse-Kunstleve RW, et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Cryst. 2010;D66:213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Brunger AT. Version 1.2 of the crystallography and NMR system. Nat. Protoc. 2007;2:2728–2733. doi: 10.1038/nprot.2007.406. [DOI] [PubMed] [Google Scholar]
- 32.Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Cryst. 2004;D60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 33.Schroeder GF, Levitt M, Brunger AT. Super-resolution biomolecular crystallography with low-resolution data. Nature. 2010;464:1218–1222. doi: 10.1038/nature08892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhorov BS. Vector method for calculating derivatives of energy of atom-atom interactions of complex molecules according to generalized coordinates. J. Struct. Chem. 1981;22:4–8. [Google Scholar]
- 35.Krissinel E, Henrick K. Inference of macromolecular assemblies from crystalline state. J. Mol. Biol. 2007;372:774–797. doi: 10.1016/j.jmb.2007.05.022. [DOI] [PubMed] [Google Scholar]
- 36.Sulek M, Yarrington R, McGibbon G, Boeke JD, Junop M. A critical role for the C-terminus of Nej1 protein in Lif1p association, DNA binding and non-homologous end-joining. DNA Repair. 2007;6:1805–1818. doi: 10.1016/j.dnarep.2007.07.009. [DOI] [PubMed] [Google Scholar]
- 37.Wu PY, Frit P, Meesala S, Dauvillier S, Modesti M, Andres SN, Huang Y, Sekiguchi J, Calsou P, Salles B, et al. Structural and functional interaction between the human DNA repair proteins DNA Ligase IV and XRCC4. Mol. Cell. Biol. 2009;29:3163–3172. doi: 10.1128/MCB.01895-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Modesti M, Junop MS, Ghirlando R, van de Rakt M, Gellert M, Yang W, Kanaar R. Tetramerization and DNA ligase IV interaction of the DNA double-strand break repair protein XRCC4 are mutually exclusive. J. Mol. Biol. 2003;334:215–228. doi: 10.1016/j.jmb.2003.09.031. [DOI] [PubMed] [Google Scholar]
- 39.Dobbs TA, Tainer JA, Lees-Miller SP. A structural model for regulation of NHEJ by DNA-PKcs autophosphorylation. DNA Repair. 2010;9:1307–1314. doi: 10.1016/j.dnarep.2010.09.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Di Capua E, Engel A, Stasiak A, Koller T. Characterization of complexes between recA protein and duplex DNA by electron microscopy. J. Mol. Biol. 1982;157:87–103. doi: 10.1016/0022-2836(82)90514-9. [DOI] [PubMed] [Google Scholar]
- 41.Galkin VE, Wu Y, Zhang XP, Qian X, He Y, Yu X, Heyer WD, Luo Y, Egelman EH. The Rad51/RadA N-terminal domain activates nucleoprotein filament ATPase activity. Structure. 2006;14:983–992. doi: 10.1016/j.str.2006.04.001. [DOI] [PubMed] [Google Scholar]
- 42.Sandler SJ, Satin LH, Samra HS, Clark AJ. RecA-like genes from three archaean species with putative protein products similar to Rad51 and Dmc1 proteins of the yeast Saccharomyces cerevisiae. Nucleic Acids Res. 1996;24:2125–2132. doi: 10.1093/nar/24.11.2125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Chen LT, Ko TP, Chang YC, Lin KA, Chang CS, Wang AH, Wang TF. Crystal structure of the left-handed archael RadA helical filament: identification of a functional motif for controlling quaternary structures and enzymatic functions of RecA family proteins. Nucleic Acids Res. 2007;35:1781–1801. doi: 10.1093/nar/gkl1131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Conway AB, Lynch TW, Zhang Y, Fortin GS, Fung CW, Symington LS, Rice PA. Crystal structure of a Rad51 filament. Nat. Struct. Mol. Biol. 2004;11:791–796. doi: 10.1038/nsmb795. [DOI] [PubMed] [Google Scholar]
- 45.Datta S, Prabu MM, Vaze MB, Ganesh N, Chandra NR, Muniyappa K, Vijayan M. Crystal structures of Mycobacterium tuberculosis RecA and its complex with ADP-AlF (4): implications for decreased ATPase activity and molecular aggregation. Nucleic Acids Res. 2000;28:4964–4973. doi: 10.1093/nar/28.24.4964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Chen Z, Yang H, Pavletich NP. Mechanism of homologous recombination from RecA-ssDNA/dsDNA structures. Nature. 2008;453:489–494. doi: 10.1038/nature06971. [DOI] [PubMed] [Google Scholar]
- 47.Arold ST, Leonard PG, Parkinson GN, Ladbury JE. H-NS forms a superhelical protein scaffold for DNA condensation. Proc. Natl Acad. Sci. USA. 2010;107:15728–15732. doi: 10.1073/pnas.1006966107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.deFazio LG, Stansel RM, Griffith JD, Chu G. Synapsis of DNA ends by DNA-dependent protein kinase. EMBO J. 2002;21:3192–3200. doi: 10.1093/emboj/cdf299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Chen L, Trujillo K, Ramos W, Sung P, Tomkinson AE. Promotion of Dnl4-catalyzed DNA end-joining by the Rad50/Mre11/Xrs2 and Hdf1/Hdf2 complexes. Mol. Cell. 2011;8:1105–1115. doi: 10.1016/s1097-2765(01)00388-4. [DOI] [PubMed] [Google Scholar]
- 50.Bredemeyer AL, Sharma GG, Huang CY, Helmink BA, Walker LM, Khor KC, Nuskey B, Sullivan KE, Pandita TK, Bassing CH, et al. ATM stabilizes DNA double-strand-break complexes during V(D)J recombination. Nature. 2006;442:466–470. doi: 10.1038/nature04866. [DOI] [PubMed] [Google Scholar]
- 51.Ramsden DA, Gellert M. Ku protein stimulates DNA end joining by mammalian DNA ligases: a direct role for Ku in repair of DNA double-strand breaks. EMBO J. 1998;17:609–614. doi: 10.1093/emboj/17.2.609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Roy S, Andres SN, Vergnes A, Neal JA, Xu Y, Yu Y, Lees-Miller SP, Junop M, Modesti M, Meek K. XRCC4's interaction with XLF is required for coding (but not signal) end joining. Nucl. Acids Res. 2012;40:1684–1694. doi: 10.1093/nar/gkr1315. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.