Abstract
MicroRNA-21 (miR-21) is thought to be an oncomir because it promotes cancer cell proliferation, migration, and survival. miR-21 is also expressed in normal cells, but its physiological role is poorly understood. Recently, it has been found that miR-21 expression is rapidly induced in rodent hepatocytes during liver regeneration after two-thirds partial hepatectomy (2/3 PH). Here, we investigated the function of miR-21 in regenerating mouse hepatocytes by inhibiting it with an antisense oligonucleotide. To maintain normal hepatocyte viability and function, we antagonized the miR-21 surge induced by 2/3 PH while preserving baseline expression. We found that knockdown of miR-21 impaired progression of hepatocytes into S phase of the cell cycle, mainly through a decrease in levels of cyclin D1 protein, but not Ccnd1 mRNA. Mechanistically, we discovered that increased miR-21 expression facilitated cyclin D1 translation in the early phase of liver regeneration by relieving Akt1/mTOR complex 1 signaling (and thus eIF-4F–mediated translation initiation) from suppression by Rhob. Our findings reveal that miR-21 enables rapid hepatocyte proliferation during liver regeneration by accelerating cyclin D1 translation.
Introduction
Evidence is rapidly accumulating in support of a prominent role for microRNA-21 (miR-21) in cancer. miR-21 is overexpressed in almost all types of cancer and has been shown to promote cancer cell proliferation, migration, and survival (1–3). Yet little is currently known about the physiological functions of miR-21, even though it is expressed in many normal tissues.
In the normal adult liver, most miR-21 expression stems from hepatocytes (4). While typically quiescent, adult hepatocytes have the ability to proliferate after liver tissue injury or loss (5, 6). Recently, we and others found that hepatocyte proliferation after two-thirds partial hepatectomy (2/3 PH) is accompanied by increased expression of miR-21 in mice (4, 7) and rats (8). miR-21 is the microRNA (miRNA) most significantly induced by 2/3 PH in mouse liver, and its expression rises and peaks as hepatocytes exit the G0 phase of the cell cycle and progress through the G1 phase (4, 7). Furthermore, we previously reported that entry into S phase, which is normally precisely timed, is delayed in hepatocytes lacking all miRNAs (4). These findings suggest that the surge in miR-21 expression occurring during liver regeneration functions to promote critical cell cycle events leading up to S phase.
To test this hypothesis, we investigated the effects of miR-21 deficiency on liver regeneration after 2/3 PH. Because miR-21 expression levels are high in the quiescent hepatocytes of the normal liver (4), we reasoned that complete miR-21 depletion by genetic deletion may disturb normal hepatocyte physiology, which may confound analyses of miR-21’s role in cell cycle regulation. Therefore, we took an alternative approach and antagonized specifically the miR-21 surge induced by 2/3 PH in hepatocytes with a miR-21 antisense oligonucleotide (miR-21–ASO).
We found that timing the in vivo application of miR-21–ASO so that it antagonized the initial phase of induced miR-21 expression after 2/3 PH prevented cyclin D1 translation in hepatocytes, leading to impaired progression through G1 and into S phase. Our results show that miR-21 functions in normal liver regeneration to promote cyclin D1 translation by activating mechanistic target of rapamycin (mTOR) complex 1 (mTORC1), which relieves assembly of the eukaryotic translation initiation factor–4F (eIF-4F) complex from inhibition by eIF-4E–binding protein 1 (4E-BP1). This function of miR-21 entailed direct inhibition of the Ras homolog gene family member B (Rhob), which led to activation of thymoma viral proto-oncogene 1 (Akt1) and mTORC1 as its downstream mediator. Our findings suggest that induction of miR-21 expression in the early phase of liver regeneration functions to accelerate hepatocyte proliferation by facilitating cyclin D1 translation, a mechanism that may also be effective in other regenerative cell types and cancer cells.
Results
miR-21–ASO is effective in timed and dosed antagonism of miR-21 in the regenerating liver.
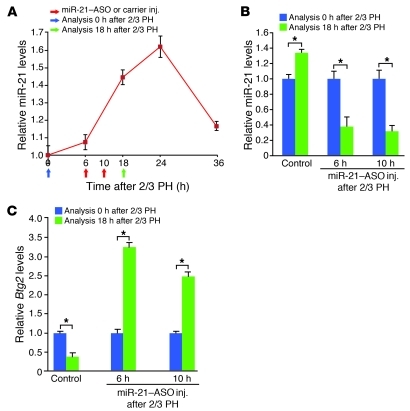
Cell cycle entry and progression of hepatocytes after 2/3 PH occur not only rapidly, but also in a synchronized and timed fashion (5, 6). Most hepatocytes have entered S phase by 36 hours after 2/3 PH in adult male C57BL/6 mice (9). Confirming our previous findings (4), 2/3 PH in such mice caused increased miR-21 expression that was detectable at 6 hours, peaked between 18 and 24 hours, and returned to almost normal levels by 36 hours after the surgery (Figure 1A). The timing of the miR-21 surge suggests that it plays a role in the regulation of cell cycle events preceding S phase, a hypothesis that is indirectly supported by our previous finding of delayed S phase entry after 2/3 PH in hepatocytes lacking all miRNAs (4).
Figure 1. miR-21–ASO injected into the tail vein facilitates inhibition of increased miR-21 expression and derepression of its target genes after 2/3 PH.
(A) Time course of miR-21 expression after 2/3 PH. miR-21 levels were determined by qRT-PCR. Arrows indicate time points of miR-21–ASO or carrier injection and liver analysis relative to 2/3 PH. Liver samples obtained by 2/3 PH (Analysis 0 hours after 2/3 PH) were used to define baseline levels in the quiescent liver. (B) qRT-PCR showed that miR-21–ASO injections at 6 or 10 hours after 2/3 PH were similarly effective in antagonizing the peak of the surge in miR-21 expression after 2/3 PH. (C) Repression of Btg2 mRNA levels in livers of control mice after 2/3 PH was reversed in mice injected with miR-21–ASO at 6 or 10 hours after 2/3 PH. At least 3 mice were analyzed for each time point and treatment. Control mice were injected with carrier. Data represent mean ± SEM. *P < 0.05.
To determine whether and how miR-21 contributes to regulation of the early phase of liver regeneration, we performed 2/3 PH in mice incapable of increased miR-21 expression in hepatocytes. We reasoned that fully depleting miR-21 could prohibit unbiased analysis of miR-21’s effect on hepatocyte proliferation, because miR-21 is expressed at high levels in quiescent hepatocytes and little is known about its role in cellular homeostasis (4). Thus, we aimed to antagonize the miR-21 surge occurring in the liver after 2/3 PH while maintaining physiological miR-21 expression levels.
For this purpose, we generated a miR-21–ASO stabilized with locked nucleic acids (LNAs), similar to a miR-122–ASO that was recently reported to efficiently inhibit this highly abundant miRNA in hepatocytes in vivo (10, 11). To establish timed and dosed miR-21 inhibition in vivo, we determined the onset, extent, and duration of changes in liver miR-21 expression after tail vein injection of the miR-21–ASO. A single dose of 25 μg/g body weight miR-21–ASO decreased liver miR-21 levels 4-fold by 6 hours after injection, suppression that increased with time and lasted for at least 36 hours (Supplemental Figure 1A; supplemental material available online with this article; doi: 10.1172/JCI46039DS1).
Given that miR-21–ASO inhibited miR-21 in the liver rapidly and progressively, we decided to inject it after 2/3 PH in order to antagonize the surge in miR-21 expression, but avoid complete knockdown of miR-21 during the early phase of liver regeneration. We aimed at suppressing miR-21 to uninduced levels at 18 hours after 2/3 PH, that is, at the peak of the rapid surge in miR-21 expression (Figure 1A). We reasoned that, due to increased portal vein flow, uptake of intravenously injected miR-21–ASO into hepatocytes after 2/3 PH could be even more efficient than uptake into hepatocytes in the normal liver. Therefore, we used a single dose of 25 μg/g body weight miR-21–ASO as in normal mice, but tested 2 injection time points, 6 and 10 hours after 2/3 PH (Figure 1A). However, we found that the levels of liver miR-21 suppression at 18 hours after 2/3 PH were indistinguishable between the 2 time points (Figure 1B). Residual miR-21 levels in livers of these mice were within 50% of that of control mice (injected with the carrier NaCl 0.9%), which resulted in normal liver function tests (data not shown). Considering that miR-21 is induced approximately 2-fold after 2/3 PH, this finding suggests an almost linear relationship between miR-21–ASO dose and miR-21 suppression in both regenerating and normal liver (Figure 1B and Supplemental Figure 1A). Importantly, we found that mRNA levels of B cell translocation gene 2 (Btg2), a known miR-21 target gene that is normally repressed at 18 hours after 2/3 PH (4), were derepressed in mice injected at either of the 2 time points (Figure 1C). These results establish that miR-21–ASO can be used to specifically antagonize the increased expression of miR-21 and its downstream effects in the regenerating liver.
Induced miR-21 expression is needed for cyclin D1 translation in the early phase of liver regeneration.
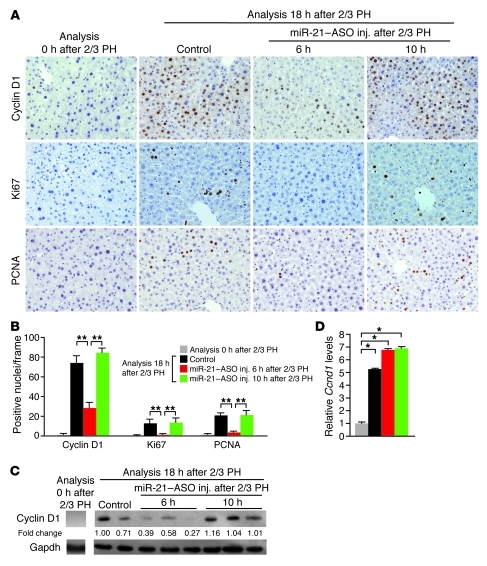
Next, we analyzed the livers of the mice injected with miR-21–ASO for expression of cell cycle phase–specific markers by immunostaining (Figure 2, A and B). One of the earliest cell cycle events after 2/3 PH is induction of cyclin D1 expression by extracellular mitogenic signals (5, 6). Cyclin D1 controls transition of hepatocytes through checkpoints in G1 phase. Subsequent sequential activation of cyclins E1, A2, and B1 allows hepatocytes to progress into late G1, S, G2, and M phase. As expected, many hepatocytes in control mice expressed cyclin D1, indicative of progress through G1 phase, at 18 hours after 2/3 PH. A small subset of the cells was already in late G1 or S phase, as evidenced by positive staining for Ki67 or proliferating cell nuclear antigen (PCNA), respectively. In contrast, cyclin D1, Ki67, and PCNA expression was rarely or not detectable in hepatocytes of mice injected with miR-21–ASO at 6 hours after 2/3 PH. This finding suggests that cyclin D1 expression and G1 phase transition of hepatocytes after 2/3 PH depend on induced miR-21 expression.
Figure 2. Inhibition of miR-21 decreases levels of cyclin D1 protein but not mRNA in hepatocytes after 2/3 PH.
(A) Immunostainings showed that hepatocytes were normally quiescent but proliferated at 18 hours after 2/3 PH in control mice. Hepatocytes of mice injected with miR-21–ASO at 6 hours, but not at 10 hours, after 2/3 PH failed to express markers of progression through G1 (cyclin D1 and Ki67, brown) and into S (Ki67 and PCNA, brown) phase of the cell cycle. Original magnification, ×200. (B) Quantification of hepatocytes expressing markers of cell cycle progression. For each immunostaining, approximately 1,500 hepatocytes (250 per frame) were analyzed per time point and treatment. (C) Immunoblotting showed failure to increase cyclin D1 protein levels in livers of mice injected with miR-21–ASO at 6 hours, but not at 10 hours, after 2/3 PH. Numbers indicate protein levels relative to controls. Gapdh was analyzed as a loading control. (D) qRT-PCR showed that miR-21–ASO injection did not interfere with induction of liver Ccnd1 transcription after 2/3 PH. At least 3 mice were analyzed for each time point and treatment. No differences were observed between control mice injected with carrier at 6 versus 10 hours after 2/3 PH. Data represent mean ± SEM. *P < 0.05, **P < 0.01.
However, when we analyzed miR-21–ASO–injected mice at 10 hours after 2/3 PH, we found that their hepatocytes stained normally for all 3 cell cycle markers (Figure 2, A and B). This finding was surprising because miR-21 levels were indistinguishable between livers of mice injected with miR-21–ASO at 10 versus 6 hours after 2/3 PH (Figure 1B). Considering that miR-21–ASO derepressed miR-21 target genes in hepatocytes by 8 hours after tail vein injection (Figure 1C), this finding reveals that the miR-21 surge promotes the expression of cyclin D1 before and around 14 hours after 2/3 PH, but is no longer needed by about 18 hours.
To confirm the discrepancy in cyclin D1 expression between mice injected with miR-21–ASO at 6 versus 10 hours after 2/3 PH, we analyzed their livers using immunoblotting and quantitative RT-PCR (qRT-PCR). In control mice, the induction of Ccnd1 mRNA after 2/3 PH mirrored that of miR-21, and Ccnd1 mRNA was rapidly translated into protein (Supplemental Figure 1B, Figure 1A, and Figure 2C). After treatment with miR-21–ASO, cyclin D1 protein levels were normal in mice injected at 10 hours after 2/3 PH, but low in mice injected at 6 hours after 2/3 PH (Figure 2C). Ccnd1 mRNA levels, however, were identical in livers of mice injected at the 2 time points (Figure 2D). The uncoupling of Ccnd1 mRNA and protein levels in mice injected with miR-21–ASO at 6 hours after 2/3 PH indicated that miR-21 either promotes the translation or prevents the degradation of cyclin D1.
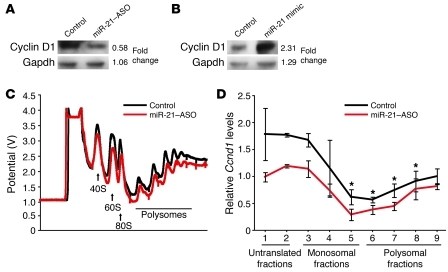
The activity of the enzyme mainly responsible for degradation of cyclin D1, glycogen synthase kinase 3 β (Gsk3b), is inhibited by phosphorylation (12). Because Gsk3b was expressed normally and did not show inhibitory phosphorylation in miR-21–ASO–injected mice at 18 hours after 2/3 PH (Supplemental Figure 2A), we reasoned that the effect of miR-21 on cyclin D1 expression is due to promotion of translation. To test this hypothesis, we determined whether cyclin D1 translation is miR-21 dependent. First, we investigated whether altering the levels of miR-21 in Hepa1,6 mouse hepatoma cells affects cyclin D1 protein levels. For this purpose, we transfected the cells with miR-21–ASO or miR-21 mimic (Figure 3, A and B, and Supplemental Figure 2, B and C). We found that inhibiting miR-21 decreased cyclin D1 protein levels, whereas adding miR-21 increased them. Next, we performed polysome analysis to determine whether miR-21 acts on cyclin D1 translation (13). We fractionated cytoplasmic lysates from Hepa1,6 cells transfected with miR-21–ASO and control cells by sucrose density gradient centrifugation and distinguished fractions containing no ribosomes (untranslated fractions), single ribosomes (monosomal fractions), or multiple associated ribosomes (polysomal fractions) (Figure 3C and Supplemental Figure 2D). Using qRT-PCR, we found that overall Ccnd1 mRNA levels in unfractionated RNA of Hepa1,6 cells were not altered by miR-21–ASO transfection (Supplemental Figure 2E), which was in accordance with our findings in vivo (Figure 2D). However, Ccnd1 mRNA levels were decreased in RNA isolated from polysomal fractions of miR-21–depleted Hepa1,6 cells, which are most actively translated (Figure 3D). Viewed together, our results show that the miR-21 surge induced by 2/3 PH functions to facilitate translation of cyclin D1 in the early phase of liver regeneration.
Figure 3. miR-21 regulates cyclin D1 translation.
(A) Transfection of 40 nM miR-21–ASO decreased cyclin D1 levels in Hepa1,6 cells approximately 2-fold. (B) Transfection of 40 nM miR-21 mimic increased cyclin D1 levels in Hepa1,6 cells approximately 2-fold. Results are representative of 3 separate experiments. (C) Elution profile of fractionated cytoplasmic lysates from Hepa1,6 cells transfected with miR-21–ASO or nontargeting ASO (Control). Untranslated fractions, containing 40S or 60S ribosomal subunits, were eluted first, followed by monosomal fractions containing 80S single intact ribosomes and polysomal fractions containing multiple associated ribosomes. Absorbance readings at 254 nm were automatically converted into potential values, which were plotted against the eluted fractions. (D) qRT-PCR showed that Ccnd1 mRNA levels in RNA isolated from the actively translated polysomal fractions were lower in miR-21–ASO–transfected cells than in control cells. Relative levels of Ccnd1 mRNA in fractions were determined by normalization to Gapdh. These values were further normalized to Ccnd1 mRNA levels in unfractionated samples to account for potential differences in starting cell numbers. Data represent mean ± SEM. *P < 0.05.
Hepatocyte entry into S phase after 2/3 PH is delayed when promotion of cyclin D1 translation by miR-21 is absent.
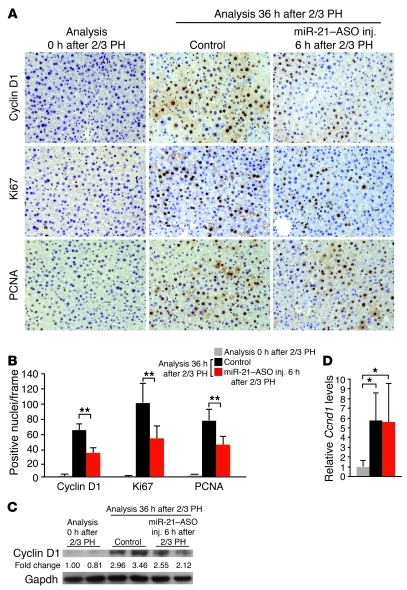
Considering that cyclin D1 initiates the cyclin activation cascade after 2/3 PH (5, 6), we next asked whether impaired cyclin D1 translation due to miR-21 inhibition limits the ability of hepatocytes to progress beyond the restriction point in late G1 and enter S phase. As expected, many hepatocytes in control mice continued to express cyclin D1 at 36 hours after 2/3 PH (Figure 4, A and B). Staining for Ki67 and PCNA both showed a large number of positive hepatocytes, indicating that many hepatocytes had not only progressed to late G1 but had already entered S phase in these mice. In contrast, Ki67 and PCNA staining showed significantly fewer hepatocytes in S phase in mice injected with miR-21–ASO at 6 hours after 2/3 PH than in control mice (Figure 4, A and B), which we confirmed by BrdU labeling (Supplemental Figure 3, A and B).
Figure 4. Inhibition of miR-21 delays S phase entry of hepatocytes after 2/3 PH.
(A and B) Quantification of Ki67 and PCNA immunostainings (both brown) showed that many hepatocytes had entered S phase at 36 hours after 2/3 PH in control mice. Significantly fewer hepatocytes stained positive for these markers or cyclin D1 (all brown) in mice injected with miR-21–ASO. For each immunostaining, approximately 1,500 hepatocytes (250 per frame) were analyzed per time point and treatment. Original magnification, ×200. (C) Immunoblotting showed lower cyclin D1 protein levels at 36 hours after 2/3 PH in livers of mice injected with miR-21–ASO compared with controls. Numbers indicate protein levels relative to time point 0 hours after 2/3 PH. Gapdh was analyzed as a loading control. (D) qRT-PCR showed that miR-21–ASO injection did not interfere with induction of liver Ccnd1 transcription at 36 hours after 2/3 PH. At least 3 mice were analyzed for each time point and treatment. miR-21–ASO was injected at 6 hours after 2/3 PH. Control mice were injected with carrier at 6 hours after 2/3 PH. Data represent mean ± SEM. *P < 0.05, **P < 0.01.
In accordance with our findings at 18 hours after 2/3 PH, cyclin D1 protein levels were lower in miR-21–ASO–injected mice than in control mice (Figure 4C and Figure 2C). Ccnd1 mRNA levels remained equal (Figure 4D and Figure 2D), which showed that cyclin D1 translation was still impaired. To ascertain that decreased cyclin D1 levels were responsible for impaired S phase entry of hepatocytes in miR-21–ASO–injected mice after 2/3 PH, we directly inhibited cyclin D1 with a Ccnd1-ASO. Tail vein injection of Ccnd1-ASO at 6 hours after 2/3 PH markedly blunted the increase in cyclin D1 mRNA and protein levels normally observed at 36 hours after 2/3 PH (Supplemental Figure 4, A and B). Quantification of BrdU-labeled hepatocytes in Ccnd1-ASO–injected mice revealed that cyclin D1 deficiency impaired S phase entry to a similar degree as miR-21 deficiency (Supplemental Figure 4, C and D). The result suggests that miR-21 acts mainly through cyclin D1 to promote hepatocyte proliferation in the early phase of liver regeneration.
To further ascertain that the hepatocyte cell cycle defect observed in miR-21–ASO–injected mice is due to miR-21 deficiency, and not toxicity caused by the ASO, we generated miR-21–MM–ASO, a control ASO that differs from miR-21–ASO in 4 mismatched base pairs. As was done with miR-21–ASO, we injected miR-21–MM–ASO into the tail vein of mice at 6 hours after 2/3 PH and analyzed their livers at 18 or 36 hours after the surgery. As expected, we found that the modification prohibited miR-21–MM–ASO from binding and inhibiting miR-21, as was evident from unaltered levels of miR-21 and its target genes (Supplemental Figure 5A). Furthermore, injection of miR-21–MM–ASO did not alter cyclin D1 protein levels or hepatocyte cell cycle entry and progression during liver regeneration (Supplemental Figure 5, B–F). These results rule out miR-21–unrelated effects as the cause of impaired cyclin D1 translation and cell cycle progression in hepatocytes of mice injected with miR-21–ASO.
Interestingly, we noted that the difference in liver cyclin D1 protein levels between control and miR-21–ASO–injected mice grew smaller with time after 2/3 PH (Figure 4C and Figure 2C), indicating that cyclin D1 translation was improving. Because miR-21 was still repressed and its target genes derepressed (Supplemental Figure 3, C and D), the finding suggested that increased miR-21 levels were no longer essential for translation of cyclin D1 at 36 hours after 2/3 PH. To investigate the possibility that impaired S phase entry was eventually compensated for in miR-21–ASO–injected mice, we analyzed mice at later time points after 2/3 PH. We found that the number of BrdU-labeled hepatocytes was similar between miR-21–ASO–injected and control mice at 72 hours after 2/3 PH (Supplemental Figure 6, A and B). Moreover, both groups showed the same number of hepatocytes staining positive for phosphorylated histone H3, a marker of mitosis (Supplemental Figure 6, A and C). In miR-21–ASO–injected mice, miR-21 continued to be inhibited, although its levels had increased 2-fold compared with 36 hours after 2/3 PH (Supplemental Figure 6D). These findings indicated that miR-21–depleted hepatocytes had overcome the G1 phase arrest and transitioned through S and into M phase. Moreover, normal levels of mitosis at 72 hours after 2/3 PH suggested that hepatocytes in miR-21–ASO–injected mice had compensated for the delay in S phase entry. This interpretation was supported by the finding that the extent of liver mass restoration was similar between miR-21–ASO–injected and control mice at this time point (Supplemental Figure 6E). Indeed, at 192 hours after 2/3 PH, when normal liver regeneration is complete, miR-21–ASO–injected mice had the same ratio of liver to body weight as control mice (Supplemental Figure 7A). In addition, hepatocytes in miR-21–ASO–injected mice had returned to normal quiescence and expressed almost normal levels of miR-21 by that time (Supplemental Figure 7, B–D).
In findings similar to ours, a previous study reported delayed, but not permanently blocked, S phase entry in Ccnd1–/– mice after hepatomitogen application (14). The study suggested that overexpression of cyclin E can compensate for the lack of cyclin D1 and facilitate cell cycle progression. Thus, we decided to determine the expression levels of cyclins downstream of cyclin D1 in livers of miR-21–ASO–injected mice after 2/3 PH. We found that Ccne1 was 8-fold higher in miR-21–ASO–injected mice than in control mice at 18 hours after 2/3 PH and continued to be overexpressed until 72 hours (Supplemental Figure 8A). Ccna2 and Ccnb1 were also overexpressed in miR-21–ASO–injected mice, most likely as a consequence of Ccne1 overexpression (Supplemental Figure 8, B and C). These results suggest that cyclin E1 overexpression, potentially aided by emerging translation of cyclin D1, can overcome the G1 phase arrest in miR-21–depleted hepatocytes.
miR-21 suppresses Rhob in the regenerating liver.
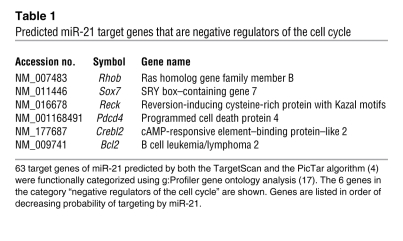
Next, we sought to identify the mechanism of how miR-21 facilitates cyclin D1 translation in the early phase of liver regeneration. Few instances have been reported in which a miRNA directly enhances the translation of a target gene (15, 16). Because we failed to detect the necessary miR-21 binding sites in the message of Ccnd1 (data not shown), we reasoned that miR-21 promotes cyclin D1 translation indirectly by targeting a cell cycle inhibitor. Thus, we used gene ontology analysis (17) to identify candidate genes that negatively regulate the cell cycle among 63 genes previously predicted to be highly probable targets of miR-21 (Table 1 and ref. 4).
Table 1 .
Predicted miR-21 target genes that are negative regulators of the cell cycle
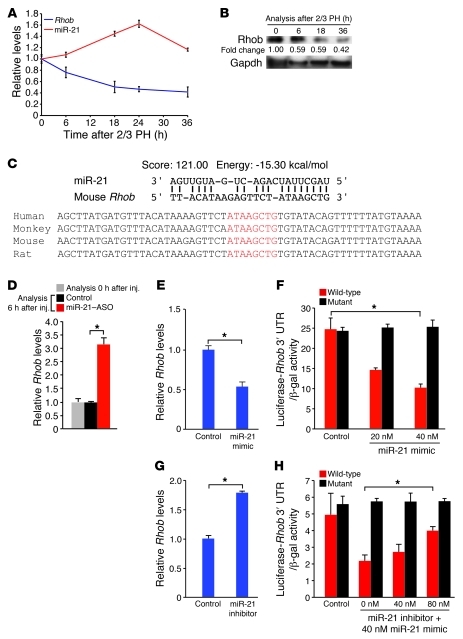
One of the candidate genes, programmed cell death protein 4 (Pdcd4), is an established target of miR-21 in cancer cells (18). We found that miR-21 also targeted Pdcd4 in normal liver, as was evident from increased Pdcd4 mRNA levels in livers of mice at 6 hours after tail vein injection of miR-21–ASO (Supplemental Figure 9A). Moreover, we found that the surge in miR-21 expression induced by 2/3 PH in normal mice was associated with a decrease in Pdcd4 mRNA levels (Supplemental Figure 9B). However, Pdcd4 protein levels were increased at 6 hours after 2/3 PH and only moderately decreased at 18 hours after 2/3 PH (Supplemental Figure 9C), which suggests that Pdcd4 is not effectively suppressed by miR-21 in the regenerating normal liver. Because of this result and, more importantly, previous findings that Pdcd4 overexpression does not affect cyclin D1 mRNA or protein levels (19), we reasoned that Pdcd4 derepression is not the cause of impaired cyclin D1 translation in hepatocytes of miR-21–ASO–injected mice. Therefore, we focused our investigations on Rhob, the candidate gene with the highest probability of targeting by miR-21 (Table 1). Rhob is a GTPase that functions as a tumor suppressor by inhibiting cancer cell proliferation, migration, and survival (20). We found that both Rhob mRNA and protein levels were inversely correlated with the expression of miR-21 after 2/3 PH (Figure 5, A and B). Bioinformatic analysis revealed that the 3′ untranslated region (UTR) of Rhob contains a highly probable miR-21 binding site that is conserved in mammalian species (Figure 5C). In support of this prediction, we found that tail vein injection of miR-21–ASO into mice caused increased Rhob mRNA levels in the liver 6 hours later (Figure 5D). To prove that the inverse correlation between miR-21 and Rhob expression in vivo was due to direct interaction, we investigated whether miR-21 targets Rhob in Hepa1,6 cells in vitro (Figure 5, E–H). Indeed, we found that Rhob mRNA levels and the activity of a luciferase reporter gene linked to the 3′ UTR of mouse Rhob decreased or increased in response to transfection of miR-21 mimic or inhibitor, respectively. Moreover, we found that the Rhob 3′ UTR sequence complimentary to the miR-21 seed sequence was essential for targeting by miR-21. The conservation of Rhob targeting by miR-21 is further supported by a recent study showing that miR-21 promotes the defining features of the metastatic phenotype, migration, elongation, and invasion, by inhibiting the human gene in a breast cancer cell line (21). Viewed together, our findings suggest that Rhob is directly suppressed by the surge in miR-21 expression occurring in the early phase of liver regeneration.
Figure 5. miR-21 induction in liver regeneration decreases Rhob expression by direct targeting.
(A and B) Inverse correlation of miR-21 and Rhob expression levels after 2/3 PH. miR-21 induction after 2/3 PH was associated with decreased Rhob mRNA (A) and Rhob protein (B) levels. Numbers indicate protein level relative to time point 0 hours after 2/3 PH. Gapdh was analyzed as a loading control. (C) High prediction score and favorable binding energy suggested that miR-21 targets the 3′ UTR of Rhob. The complementary sequence in the Rhob 3′ UTR and the seed region of miR-21 (red letters) is conserved among mammalian species. (D) Liver Rhob mRNA levels increased 6 hours after miR-21–ASO injection. Control mice were injected with carrier. At least 3 mice were analyzed per time point and treatment. (E–H) Direct inhibition of Rhob by miR-21. (E) Transfection of miR-21 mimic into Hepa1,6 cells decreased Rhob mRNA. (F) The activity of a luciferase reporter gene linked to the Rhob 3′ UTR was inhibited by miR-21 mimic in a dose-dependent fashion. Mutation of the Rhob 3′ UTR sequence complimentary to the miR-21 seed sequence prevented inhibition by miR-21 mimic. (G) Transfection of miR-21 inhibitor into Hepa1,6 cells increased Rhob mRNA. (H) miR-21 inhibitor effectively restored the activity of a luciferase reporter gene linked to the Rhob 3′ UTR in Hepa1,6 cells transfected with miR-21 mimic. Mutation of the Rhob 3′ UTR sequence complimentary to the miR-21 seed sequence prevented this effect. Data represent mean ± SEM. *P < 0.05.
Rhob inhibits cyclin D1 translation by preventing Akt1-mediated activation of mTORC1.
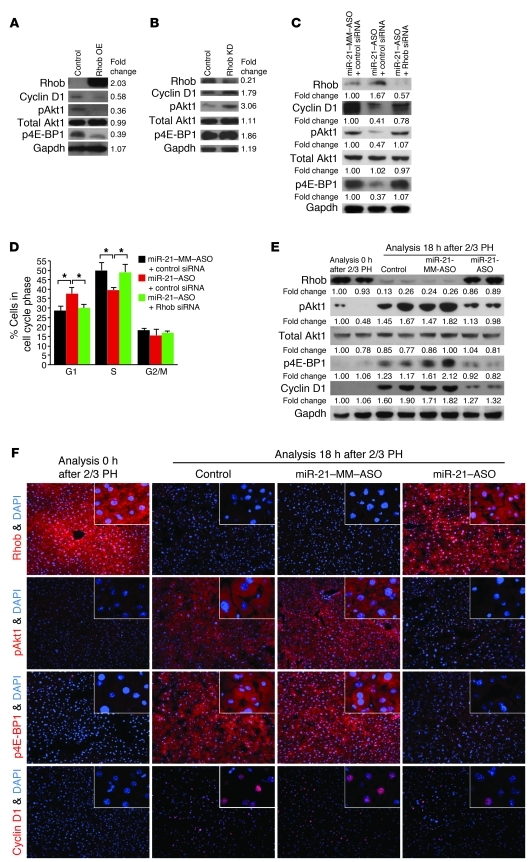
Next, we investigated whether Rhob inhibition is responsible for miR-21’s effect on cyclin D1 translation. Although Rhob had not been implicated in the regulation of cyclin D1 expression, several lines of evidence suggested this possibility. First, Rhob inhibits activating phosphorylation of Akt1 (22, 23). Then, activated Akt1 regulates cyclin D1 expression through mTORC1 (24, 25). Finally, mTORC1 promotes assembly of the eIF-4F complex, which mediates translation initiation of genes like Ccnd1 (24, 26). In fact, mTORC1 has been shown to be needed for eIF-4F assembly and cyclin D1 translation in hepatocytes after 2/3 PH (27, 28). Further considering that Akt1 activation is critical for the early phase of liver regeneration (29), we hypothesized that miR-21 facilitates rapid cyclin D1 translation in liver regeneration by relieving Akt1, and thus mTORC1, from inhibition by Rhob. To test this hypothesis, we overexpressed Rhob by transient plasmid transfection in Hepa1,6 cells that normally express Rhob at low levels (Figure 6A and Supplemental Figure 10A). We used immunoblotting to determine the effects on cyclin D1 and Akt1. Moreover, as a readout for mTORC1-mediated activation of eIF-4F–dependent translation initiation, we measured phosphorylation of 4E-BP1. mTORC1 initiates 4E-BP1 phosphorylation, which disables its ability to bind and inhibit eIF-4E and thus facilitates eIF-4F complex assembly (24, 26).
Figure 6. miR-21 promotes cyclin D1 translation in liver regeneration by relieving Akt1-mediated activation of mTORC1 from suppression by Rhob.
(A) Immunoblotting showed that Rhob overexpression (OE) plasmid transfection into Hepa1,6 cells decreased levels of cyclin D1, Akt1 activated by phosphorylation at Ser473 (pAkt1), and 4E-BP1 inhibited by phosphorylation at Thr37 and/or Thr46 (p4E-BP1). Cells transfected with empty overexpression plasmid were used as control. (B) Additional Rhob knockdown (KD) with a shRNA plasmid increased cyclin D1, pAkt1, and p4E-BP1 levels. (C) Immunoblotting showed that miR-21–ASO transfection into Hepa1,6 cells increased Rhob and decreased cyclin D1, pAkt1, and p4E-BP1 levels. Additional Rhob knockdown with siRNA restored cyclin D1, pAkt1, and p4E-BP1 levels. (D) Cell cycle phase distribution analysis by flow cytometry showed Rhob-dependent accumulation of miR-21–ASO–transfected cells in G1 phase. Cells transfected with miR-21–MM-ASO and negative control siRNA were used as control. Numbers indicate protein levels relative to control. Results are representative of 3 separate experiments (Supplemental Figure 12, A–C). (E) Immunoblotting showed failure to decrease Rhob and increase pAkt1, p4E-BP1, and cyclin D1 levels in the liver after 2/3 PH in mice injected with miR-21–ASO compared with mice injected with carrier (Control) or miR-21–MM–ASO. Liver samples were obtained by 2/3 PH and 18 hours later. Numbers indicate protein levels relative to time point 0 hours after 2/3 PH. (F) Confirmation of the immunoblotting results by immunostaining (red). Original magnification, ×200; ×400 (insets). At least 3 mice were analyzed for each time point and treatment (Supplemental Figure 12D). Carrier, miR-21–MM–ASO, or miR-21–ASO was injected at 6 hours after 2/3 PH. Gapdh was analyzed as a loading control. Data represent mean ± SEM. *P < 0.05.
We found that overexpression of Rhob decreased cyclin D1 protein levels and levels of Akt1 activated by phosphorylation (Figure 6A). Moreover, as evidence for impaired mTORC1 activity, we found decreased inhibitory phosphorylation of 4E-BP1. In accordance with the hypothesized mechanism, Rhob overexpression decreased cyclin D1 protein but not Ccnd1 mRNA levels (Supplemental Figure 10B). To confirm these results, we transfected Hepa1,6 cells with both the plasmid overexpressing Rhob and a plasmid expressing shRNA inhibiting Rhob. This resulted in decreased Rhob mRNA and protein levels, which caused increased levels of cyclin D1 protein and activated Akt1 (Figure 6B and Supplemental Figure 10C). As expected, phosphorylation of 4E-BP1 was increased in response to Akt1 activation. Moreover, Ccnd1 mRNA levels were not altered and thus dissociated from cyclin D1 protein levels (Supplemental Figure 10D). Rhob overexpression or knockdown similarly inhibited or activated another target of mTORC1, S6 kinase 1 (Supplemental Figure 10, E and F). These results reveal that Rhob inhibits eIF-4F–mediated initiation of cyclin D1 translation by preventing activation of Akt1 and its downstream mediator, mTORC1.
miR-21 promotes cyclin D1 translation and cell cycle progression in early liver regeneration by suppressing Rhob.
To further confirm that miR-21’s ability to promote cyclin D1 translation was mediated by Rhob, we inhibited both miR-21 and Rhob in Hepa1,6 cells and examined cyclin D1 protein levels. We found that the decreased cyclin D1 protein levels present in cells transfected with miR-21–ASO alone normalized after additional Rhob knockdown (Figure 6C and Supplemental Figure 10G), which establishes Rhob as the critical target of miR-21 in promoting cyclin D1 translation. As expected, miR-21 knockdown also impaired Akt1/mTORC1 activity, whereas additional Rhob knockdown restored normal levels. We also analyzed the cell cycle phase distribution of these cells to determine whether promotion of cyclin D1 translation by Rhob inhibition is indeed the mechanism by which miR-21 faciliates rapid progression through the G1 phase of the cell cycle (Figure 6D). We found that miR-21 knockdown impaired G1 phase exit and S phase entry, and that additional Rhob knockdown restored normal cell cycle progression, which confirms Rhob as the critical target of miR-21 in promotion of G1 phase progression.
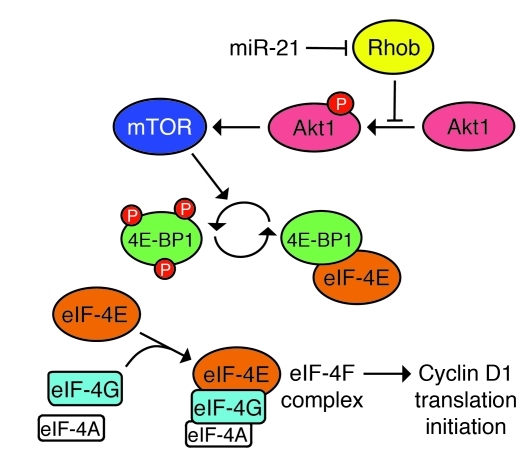
Finally, we determined whether miR-21 prevents Rhob from inhibiting Akt1/mTORC1 signaling and thus cyclin D1 translation in early liver regeneration. Our initial analyses showed that miR-21–ASO injection at 6 hours after 2/3 PH impaired expression of cyclin D1 in the liver at 18 and 36 hours after 2/3 PH (Figure 2, A–C, and Figure 4, A–C). When we repeated this experiment, we found that 2/3 PH suppressed Rhob, activated Akt1, inhibited 4E-BP1, and induced cyclin D1 in control mice injected with carrier or miR-21–MM–ASO 6 hours after 2/3 PH, but not in mice injected with miR-21–ASO (Figure 6, E and F, and Supplemental Figure 10H). To ascertain that miR-21 regulates Akt1/mTORC1 signaling in liver regeneration through Rhob, we analyzed the expression of phosphatase and tensin homolog (PTEN) because inhibition of PTEN by miR-21 was previously shown to cause Akt1 activation in cancer cells (30, 31). We found that miR-21 targeted PTEN in normal liver, as was evident from increased Pten mRNA levels at 6 hours after tail vein injection of miR-21–ASO into mice (Supplemental Figure 11A). However, we also found that PTEN mRNA and protein levels markedly increased in the liver after 2/3 PH in normal mice (Supplemental Figure 11, B and C), which shows that the miR-21 surge induced by 2/3 PH does not effectively antagonize the expression of PTEN in early liver regeneration. In addition, the finding revealed that accumulation of PTEN does not prevent Akt1 activation in liver regeneration. Viewed together, our results exclude suppression of PTEN by miR-21 as the reason for Akt1 and mTORC1 activation in liver regeneration and confirm our hypothesis that miR-21 facilitates rapid translation of cyclin D1 by relieving Akt1 and its mediator, mTORC1, from inhibition by Rhob (Figure 7).
Figure 7. Proposed mechanism by which miR-21 promotes cyclin D1 translation in liver regeneration.
By directly inhibiting Rhob expression, miR-21 facilitates Akt1-mediated activation of mTORC1, which promotes cyclin D1 translation initiation. This effect of mTORC1 involves phosphorylation of 4E-BP1, which disables its inhibitory binding of eIF-4E and allows assembly of the eIF-4F complex that additionally contains eIF-4G and eIF-4A (22, 24, 25).
Discussion
Previously, we and others observed a surge in miR-21 expression in G1 phase in hepatocytes after 2/3 PH (4, 7, 8). Here, we investigated the function of induced miR-21 expression in liver regeneration by specifically antagonizing the miR-21 surge, but not baseline miR-21 expression, with miR-21–ASO, a short LNA-stabilized antisense oligonucleotide inhibitor of miR-21. This partial knockdown strategy revealed that increased miR-21 expression facilitates cyclin D1 translation in early liver regeneration. Cyclin D1 translation is mediated by the eIF-4F translation initiation complex, which is activated by Akt1/mTORC1 signaling (24, 26). Our results showed that miR-21 activates Akt1/mTORC1-mediated cyclin D1 translation by inhibiting Rhob. While Rhob has been shown to inhibit Akt1 (22, 23), to our knowledge, it was not previously known that this effect of Rhob results in inhibition of cyclin D1 translation.
Our results further showed that promotion of cyclin D1 translation by miR-21 is important for rapid G1 phase progression and S phase entry of hepatocytes after 2/3 PH. In mice in which the miR-21 surge was antagonized by miR-21–ASO, the number of hepatocytes in S phase at 36 hours after 2/3 PH was markedly reduced compared with controls. However, hepatocyte cell cycle progression was not permanently blocked in miR-21–ASO–injected mice, and the cells eventually entered S phase and restored the lost liver mass. Impaired hepatocyte proliferation, particularly if it is caused by single gene deficiencies, is typically compensated for in liver regeneration. Compensation is the result of redundant signaling pathways providing the missing function, which leads to delayed rather than failed liver mass restoration (32). A previous study suggested that overexpression of cyclin E can compensate for lack of cyclin D1 in Ccnd1–/– mice, thereby facilitating normal hepatocyte proliferation (14). In accordance with this result, we found that cyclin E1 and downstream cyclins involved in liver regeneration were overexpressed as early as 18 hours after 2/3 PH, when cyclin D1 protein deficiency was most prominent in miR-21–depleted hepatocytes. In addition, we observed that cyclin D1 translation was slowly improving between 18 and 36 hours after 2/3 PH. Because miR-21 was still depleted at 36 hours after 2/3 PH, the finding suggests miR-21–independent cyclin D1 translation as another mechanism that helps rescue liver regeneration in miR-21–ASO–injected mice.
Our finding that miR-21 promoted cyclin D1 translation in liver regeneration by activating Akt1/mTORC1 prompted us to investigate whether PTEN, a miR-21 target known to inhibit Akt1 (30, 31), is also involved in this process. After performing 2/3 PH on normal mice, we found that PTEN protein accumulated during G1 phase despite the surge in miR-21 expression induced by 2/3 PH. The finding revealed that accumulation of PTEN does not prevent Akt1 activation in normal liver regeneration. Moreover, the finding showed that suppression of PTEN by miR-21 is not involved in Akt1 activation and thus cyclin D1 translation in early liver regeneration. Nevertheless, miR-21’s previously reported ability to activate Akt1 by inhibiting PTEN in cancer cells (30, 31), taken together with our present results, suggests the intriguing possibility that miR-21 acts as a central regulator of Akt1/mTORC1 signaling in other contexts.
Because miRNAs can target many genes, the function of miR-21 in liver regeneration is likely not limited to promotion of cyclin D1 translation. In fact, we and others have previously identified Btg2 and Pellino 1, which are regulators of FoxM1b and NF-κB activity, respectively, as targets of miR-21 during liver regeneration after 2/3 PH (4, 7). In addition, our present results suggest that Pdcd4 is moderately suppressed at 18 hours after 2/3 PH, and it is possible that in the future, other miR-21 targets will be identified as playing a role in liver regeneration.
However, our finding that the hepatocyte cell cycle defect in miR-21–ASO–injected mice phenocopied that of mice in which cyclin D1 was suppressed with Ccnd1-ASO suggests that miR-21’s effect on hepatocyte cell cycle progression in early liver regeneration is mainly mediated by cyclin D1. Importantly, derepression of Btg2, Pellino 1, or Pdcd4 cannot explain the lack of cyclin D1 expression in hepatocytes of miR-21–ASO–injected mice. Hepatocyte-specific deficiency of the Btg2 target FoxM1b leads to impaired activity of cyclin-dependent kinase complexes involving cyclin E1, A2, and B1, but not D1, after 2/3 PH in mice (33). Moreover, Btg2 overexpression in human hepatoma cells decreases mRNA and protein levels of cyclin B1, but not cyclin D1 (34). Similarly, Pdcd4 overexpression fails to suppress cyclin D1 mRNA or protein levels (19). Because Pellino 1 activates NF-κB signaling (35), its derepression in hepatocytes of miR-21–ASO–injected mice would be expected to accelerate, not delay, cell cycle progression (32). Thus, the evidence points to Rhob as the critical target of miR-21 in promotion of Akt1/mTORC1-mediated cyclin D1 translation and cell cycle progression of hepatocytes in early liver regeneration.
Our finding that miR-21–ASO can be used to partially inhibit miR-21 and derepress its target genes within 8 hours of intravenous injection has important implications. First, it highlights a method of miRNA inhibition in hepatocytes that avoids unspecific changes due to compensation or adaptation, which can mask or confound the phenotype caused by deficiency of the miRNA. Compensation of impaired hepatocyte proliferation by redundant signaling pathways occurs not instantaneously, but typically within a day after 2/3 PH (32). The rapid onset of miR-21 inhibition by miR-21–ASO limits the time available for activation of redundant pathways and may therefore be more effective than genetic deletion for studies of liver regeneration. In addition, limiting the extent of miRNA inhibition in hepatocytes may help to maintain homeostasis and avoid secondary changes and adaptation. Second, our study showed that the rapid onset of miR-21–ASO’s effect allows the dynamic processes that occur during liver regeneration to be dissected: miR-21 and Ccnd1 expression are concomitantly induced after 2/3 PH, and our results revealed that promotion of cyclin D1 translation is critical for the early phase of liver regeneration, when Ccnd1 mRNA levels are still low, but not for later phases. Finally, our results raise the possibility of using oligonucleotide mimics of miR-21 for therapy of liver failure. Provided that miR-21 mimics enter hepatocytes and exert their function as quickly as miR-21–ASO, they may be effective in accelerating progression of hepatocytes through G1 and into S phase, which is critical for survival from liver injury (36).
In conclusion, our results revealed that the increased expression of miR-21 during liver regeneration functions to kick-start cyclin D1 translation in hepatocytes. This effect of miR-21 was mediated by a mechanism that integrated inhibition of Rhob by miR-21 with a function of Rhob that we believe to be previously unrecognized, the suppression of eIF-4F–mediated translation initiation through suppression of Akt1/mTORC1 signaling. Our results, together with miR-21’s ability to activate Akt1 by inhibiting PTEN (30, 31), suggest miR-21 as a key regulator of mTORC1. Moreover, our finding that miR-21 derepressed eIF-4F, which mediates the rate-limiting ribosomal binding step of translation initiation, revealed its profound impact on cell proliferation. In liver regeneration, acceleration of cyclin D1 translation by miR-21 facilitated rapid progression of hepatocytes through G1 and into S phase. This effect of miR-21 may also be involved in the rapid proliferation of other regenerative cell types and cancer cells.
Methods
miR-21–ASO and miR-21–MM–ASO generation.
Both oligonucleotides were produced by Exiqon. To prevent toxicity and facilitate efficient cellular uptake, a short (<16 mer) design was chosen. Both oligonucleotides had a fully phosphorothioate-modified backbone. The sequence of miR-21–ASO is TCAGTCTGATAAGCT. High target affinity was ensured by LNA modifications. miR-21–MM–ASO is identical to miR-21–ASO except for 4 base pair changes that prevented binding to miR-21; its sequence is TCAGTATTAGCAGCT. Both oligonucleotides were purified by reverse-phase high-performance liquid chromatography and lyophilized.
miR-21–ASO and miR-21–MM–ASO intravenous injection.
Lyophilized oligonucleotide was resuspended in NaCl 0.9% to a final concentration of 30 μg/μl. A dose of 25 μg/g body weight oligonucleotide in a total volume of 100 μl NaCl 0.9% was injected via the tail vein into 8- to 12-week-old male C57BL/6 mice (Jackson Laboratory). Control mice were injected with 100 μl NaCl 0.9% (carrier).
2/3 PH.
Two-thirds of the liver was surgically removed under isoflurane anesthesia as previously described (37).
qRT-PCR.
Total RNA was isolated with the miRNAeasy kit (Qiagen) and treated with DNase I (Ambion) to eliminate genomic DNA. Superscript III reverse transcription reagent (Invitrogen) was used to generate cDNA using 1 μg total RNA. PCR amplification was performed as previously described (4). Primers for qRT-PCR were designed using Beacon Designer (Premier Biosoft). miRNA isolation, amplification, and analysis were performed as previously described, including normalization to sno202RNA (4). Relative changes in mRNA and miRNA expression were determined using the 2–ΔΔCt method (38).
Immunostaining.
Paraffin-embedded liver samples were sectioned and stained with the antibodies rabbit anti-cyclin D1 (NeoMarkers), mouse anti-PCNA (Biosource), and rabbit anti-Ki67 (Lab Vision) at 1:100 dilutions. Immunostainings of sections of frozen liver samples embedded in optimum cutting temperature compound (Tissue-Tek; Sakura Finetek) were performed with rabbit anti-Rhob (Santa Cruz Biotechnology), rabbit anti–Akt1 phosphorylated at Ser473 (Cell Signaling), rabbit anti–4E-BP1 phosphorylated at Thr37 and/or Thr46 (Cell Signaling), and rabbit anti-cyclin D1 (Neomarkers) antibodies at 1:100 dilutions. For fluorescence microscopy, the secondary antibody goat anti-rabbit conjugated with Alexa Fluor 594 (Invitrogen) was used at 1:500 dilution. Nuclear DNA was stained with 300 nM DAPI (Millipore).
Immunoblotting.
Liver samples were immediately frozen in liquid nitrogen and homogenized in 20 mM Tris buffer, pH 7.4, containing 150 mM NaCl, 1% Triton X-100, 10 mM EDTA, and Complete Protease Inhibitor Cocktail (Roche Applied Science). After homogenization, tissue extracts were centrifuged at 14,000 g at 4°C, and supernatants were collected. Protein concentration was measured using the Bradford assay (Biorad). 50 μg protein per well was loaded on 13% sodium dodecyl sulfate–polyacrylamide gels. After electrophoresis, gels were electrotransferred onto polyvinylidene fluoride membranes (Biorad). Membranes were blocked with 5% BSA in Tris-buffered saline with 0.1% Tween 20 (Sigma-Aldrich; TBST) and incubated with primary antibodies in 5% BSA in TBST. In addition to the antibodies listed above, rabbit anti-Rhob (Cell Signaling), rabbit anti-Akt1 (Cell Signaling), and rabbit anti-Gapdh (Santa Cruz Biotechnology) antibodies were used. After 3 washes with TBST, membranes were incubated with the secondary antibody goat anti-rabbit-HRP (Jackson ImmunoResearch) at 1:10,000 dilutions in 5% BSA in TBST and developed with ECL or ECLplus Western Chemiluminescent HRP substrate (both Pierce). Signal intensities were quantified and normalized to Gapdh using ImageJ.
miRNA mimic and inhibitor transfection.
miR-21 mimic or hairpin inhibitor (both Dharmacon), miR-21–ASO, or miR-21–MM–ASO was introduced into Hepa1,6 cells at a final concentration of 20, 40, or 80 nM. Hepa1,6 cells were plated in 24-well plates (Corning; 5 × 104 cells/well) and transfected 24 hours later using Lipofectamine 2000 (Invitrogen). Equal concentrations of double-stranded or single-stranded oligonucleotides sense or antisense to cel-miR-67 were used as nontargeting controls for miR-21 mimic or inhibitor, respectively.
Polysome analysis.
Isolation and analysis of polysomal fractions was performed as previously described (13), with some modifications. Hepa1,6 cells (ATCC) were seeded into 15-cm tissue culture plates and transfected at 50% confluency with 40 nM miR-21–ASO using Lipofectamine 2000 and harvested 48 hours later. Control cells were transfected with 40 nM nontargeting ASO. Cells were lysed in ice-cold buffer A (10 mM Tris-HCl [pH 8], 140 mM NaCl, 1.5 mM MgCl2, 0.25% NP-40, and 0.5% Triton X-100 supplemented with 40 U/ml RNase inhibitor [Promega], 150 μg/ml cycloheximide, and 20 mM dithiothreitol) for 40 minutes on ice. Lysates were centrifuged at 10,000 g for 10 minutes at 4°C, and supernatants of equal volume were loaded onto 10%–60% sucrose gradients in buffer B (25 mM Tris-HCl, 25 mM NaCl, and 1.5 mM MgCl2). Sucrose density gradient centrifugation was carried out at 100,000 g for 3 hours at 4°C using an SW41Ti rotor (Beckman). Fractions were collected using an ISCO gradient fraction collector. RNA from each fraction and unfractionated samples was isolated using TRIzol (Invitrogen), purified using Purelink RNA mini kit (Invitrogen), and treated with Turbo DNase (Ambion) before reverse transcription. Ccnd1 mRNA levels in fractionated and unfractionated RNA were determined by qRT-PCR and were normalized to Gapdh expression. To account for potential variations in overall Ccnd1 mRNA levels between samples caused by differences in cell numbers, Ccnd1 mRNA levels of fractionated RNA were further normalized to Ccnd1 mRNA levels of the respective unfractionated RNA. Unless otherwise specified, reagents were from Sigma-Aldrich.
Rhob overexpression.
Rhob cDNA was amplified from mouse liver total RNA and cloned into pIRES-GFP (Clontech) to generate the Rhob OE plasmid. Hepa1,6 cells were plated in 6-well plates (Corning; 5 × 105 cells/well) and transfected 24 hours later with 3 μg Rhob OE plasmid using Lipofectamine 2000. Rhob expression was determined using qRT-PCR and immunoblotting 48 hours later.
Rhob inhibition.
shRNA sequences for mouse Rhob were designed using the Hush-27 algorithm (Origene) and cloned into pGFP-V-RS (Origene). Hepa1,6 cells were plated in 6-well plates (5 × 105 cells/well) and transfected 24 hours later with 2 μg pGFP-V-RS containing Rhob-shRNA and 2 μg Rhob OE plasmid using Fugene HD (Roche). Cells were collected 72 hours later, and Rhob expression was analyzed using qRT-PCR and immunoblotting. To determine the consequences of knockdown of both miR-21 and Rhob, Hepa1,6 cells were plated in 6-well plates (5 × 105 cells/well) and transfected 24 hours later with miR-21–ASO and Rhob siRNA or negative control siRNA (both Qiagen) at final concentrations of 40 or 10 nM, respectively. Cells transfected with miR-21–MM–ASO and negative control siRNA at 40 and 10 nM final concentrations were used as controls. Cells were collected 24 hours later for analysis by qRT-PCR and immunoblotting.
Luciferase assay.
Rhob 3′ UTR was amplified from mouse genomic DNA and cloned into the pMIR-REPORT vector (Ambion). To generate the mutant Rhob 3′ UTR, mutations were introduced into the sequence complimentary to the miR-21 seed sequence using the QuickChange Site-directed Mutagenesis Kit (Stratagene). Constructs were validated by sequencing. Hepa1,6 cells were plated in 24-well plates (5 × 104 cells/well) and transfected 24 hours later with miR-21 mimic or inhibitor and 30 ng of the pMIR-REPORT vector containing the wild-type or mutant Rhob 3′ UTR and 30 ng of the pSV-β-Galactosidase Control Vector (Promega) to monitor transfection efficiencies. 24 hours after transfection, luciferase and β-galactosidase activities were measured using the Luciferase Assay System and Beta-Glo Assay System (both Promega) in a Synergy 2 Microplate Reader (BioTek Instruments). Luciferase activities were normalized to β-galactosidase activities for each well.
Cell cycle phase distribution analysis.
Hepa1,6 cells were plated in 6-well plates (1 × 106 cells/well) and cultured for 24 hours before the indicated oligonucleotides were transfected at the same final concentrations as for immunoblotting. 24 hours after transfection, cells were fixed by incubation in ice-cold 70% ethanol for at least 16 hours. After fixation, 2 × 105 cells were stained in 10 μg/ml propidium iodide (Sigma-Aldrich) for 30 minutes at room temperature in the dark. Flow cytometry was performed on a LSR II, and data were analyzed by CellQuest software (both BD Biosciences).
Statistics.
Statistical significance was determined with 2-tailed Student’s t test. A P value less than 0.05 was considered significant.
Study approval.
All procedures involving mice were approved by the Institutional Animal Care and Use Committee at UCSF.
Supplementary Material
Acknowledgments
This work was supported by grants to H. Willenbring from the California Institute for Regenerative Medicine, American Liver Foundation, and American Society of Transplantation. R. Ng was supported by a scholarship from the Agency of Science Technology and Research (A*STAR) Singapore. G.R. Roll was supported by a fellowship from the NIH NRSA Hepatology Training Grant at UCSF. The authors thank Sandra Huling for immunostainings; Donghui Wang for tail vein injections; Morgan Truitt, Andrew Hsieh, and Davide Ruggero for help with polysome analysis; Erich Koller for Ccnd1-ASO; and Montgomery Bissell and Pamela Derish for critical reading of the manuscript.
Footnotes
Conflict of interest: The authors have declared that no conflict of interest exists.
Citation for this article: J Clin Invest. 2012;122(3):1097–1108. doi:10.1172/JCI46039.
References
- 1.Krichevsky AM, Gabriely G. miR-21: a small multi-faceted RNA. J Cell Mol Med. 2009;13(1):39–53. doi: 10.1111/j.1582-4934.2008.00556.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Selcuklu SD, Donoghue MT, Spillane C. miR-21 as a key regulator of oncogenic processes. Biochem Soc Trans. 2009;37(Pt 4):918–925. doi: 10.1042/BST0370918. [DOI] [PubMed] [Google Scholar]
- 3.Garzon R, Marcucci G, Croce CM. Targeting microRNAs in cancer: rationale, strategies and challenges. Nat Rev Drug Discov. 2010;9(10):775–789. doi: 10.1038/nrd3179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Song G, et al. MicroRNAs control hepatocyte proliferation during liver regeneration. Hepatology. 2010;51(5):1735–1743. doi: 10.1002/hep.23547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fausto N, Campbell JS, Riehle KJ. Liver regeneration. Hepatology. 2006;43(2 suppl 1):S45–S53. doi: 10.1002/hep.20969. [DOI] [PubMed] [Google Scholar]
- 6.Michalopoulos GK. Liver regeneration. J Cell Physiol. 2007;213(2):286–300. doi: 10.1002/jcp.21172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Marquez RT, Wendlandt E, Galle CS, Keck K, McCaffrey AP. MicroRNA-21 is upregulated during the proliferative phase of liver regeneration, targets Pellino-1, and inhibits NF-kappaB signaling. Am J Physiol Gastrointest Liver Physiol. 2010;298(4):G535–G541. doi: 10.1152/ajpgi.00338.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Castro RE, et al. Identification of microRNAs during rat liver regeneration after partial hepatectomy and modulation by ursodeoxycholic acid. Am J Physiol Gastrointest Liver Physiol. 2010;299(4):G887–G897. doi: 10.1152/ajpgi.00216.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mitchell C, et al. Heparin-binding epidermal growth factor-like growth factor links hepatocyte priming with cell cycle progression during liver regeneration. J Biol Chem. 2005;280(4):2562–2568. doi: 10.1074/jbc.M412372200. [DOI] [PubMed] [Google Scholar]
- 10.Elmen J, et al. LNA-mediated microRNA silencing in non-human primates. Nature. 2008;452(7189):896–899. doi: 10.1038/nature06783. [DOI] [PubMed] [Google Scholar]
- 11.Elmen J, et al. Antagonism of microRNA-122 in mice by systemically administered LNA-antimiR leads to up-regulation of a large set of predicted target mRNAs in the liver. Nucleic Acids Res. 2008;36(4):1153–1162. doi: 10.1093/nar/gkm1113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Alao JP. The regulation of cyclin D1 degradation: roles in cancer development and the potential for therapeutic invention. Mol Cancer. 2007;6:24. doi: 10.1186/1476-4598-6-24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bellodi C, Kopmar N, Ruggero D. Deregulation of oncogene-induced senescence and p53 translational control in X-linked dyskeratosis congenita. EMBO J. 2010;29(11):1865–1876. doi: 10.1038/emboj.2010.83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ledda-Columbano GM, Pibiri M, Concas D, Cossu C, Tripodi M, Columbano A. Loss of cyclin D1 does not inhibit the proliferative response of mouse liver to mitogenic stimuli. Hepatology. 2002;36(5):1098–1105. doi: 10.1053/jhep.2002.36159. [DOI] [PubMed] [Google Scholar]
- 15.Vasudevan S, Tong Y, Steitz JA. Switching from repression to activation: microRNAs can up-regulate translation. Science. 2007;318(5858):1931–1934. doi: 10.1126/science.1149460. [DOI] [PubMed] [Google Scholar]
- 16.Orom UA, Nielsen FC, Lund AH. MicroRNA-10a binds the 5′UTR of ribosomal protein mRNAs and enhances their translation. Mol Cell. 2008;30(4):460–471. doi: 10.1016/j.molcel.2008.05.001. [DOI] [PubMed] [Google Scholar]
- 17.Reimand J, Kull M, Peterson H, Hansen J, Vilo J. g:Profiler–a web-based toolset for functional profiling of gene lists from large-scale experiments. Nucleic Acids Res. 2007;35(Web Server issue):W193–W200. doi: 10.1093/nar/gkm226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Frankel LB, Christoffersen NR, Jacobsen A, Lindow M, Krogh A, Lund AH. Programmed cell death 4 (PDCD4) is an important functional target of the microRNA miR-21 in breast cancer cells. J Biol Chem. 2008;283(2):1026–1033. doi: 10.1074/jbc.M707224200. [DOI] [PubMed] [Google Scholar]
- 19.Goke R, Barth P, Schmidt A, Samans B, Lankat-Buttgereit B. Programmed cell death protein 4 suppresses CDK1/cdc2 via induction of p21(Waf1/Cip1). Am J Physiol Cell Physiol. 2004;287(6):C1541–C1546. doi: 10.1152/ajpcell.00025.2004. [DOI] [PubMed] [Google Scholar]
- 20.Karlsson R, Pedersen ED, Wang Z, Brakebusch C. Rho GTPase function in tumorigenesis. Biochim Biophys Acta. 2009;1796(2):91–98. doi: 10.1016/j.bbcan.2009.03.003. [DOI] [PubMed] [Google Scholar]
- 21.Connolly EC, Van Doorslaer K, Rogler LE, Rogler CE. Overexpression of miR-21 promotes an in vitro metastatic phenotype by targeting the tumor suppressor RHOB. Mol Cancer Res. 2010;8(5):691–700. doi: 10.1158/1541-7786.MCR-09-0465. [DOI] [PubMed] [Google Scholar]
- 22.Flynn P, Mellor H, Casamassima A, Parker PJ. Rho GTPase control of protein kinase C-related protein kinase activation by 3-phosphoinositide-dependent protein kinase. J Biol Chem. 2000;275(15):11064–11070. doi: 10.1074/jbc.275.15.11064. [DOI] [PubMed] [Google Scholar]
- 23.Bousquet E, et al. Loss of RhoB expression promotes migration and invasion of human bronchial cells via activation of AKT1. Cancer Res. 2009;69(15):6092–6099. doi: 10.1158/0008-5472.CAN-08-4147. [DOI] [PubMed] [Google Scholar]
- 24.Gingras AC, Kennedy SG, O’Leary MA, Sonenberg N, Hay N. 4E-BP1, a repressor of mRNA translation, is phosphorylated and inactivated by the Akt(PKB) signaling pathway. Genes Dev. 1998;12(4):502–513. doi: 10.1101/gad.12.4.502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gera JF, et al. AKT activity determines sensitivity to mammalian target of rapamycin (mTOR) inhibitors by regulating cyclin D1 and c-myc expression. . J Biol Chem. 2004;279(4):2737–2746. doi: 10.1074/jbc.M309999200. [DOI] [PubMed] [Google Scholar]
- 26.Fingar DC, Richardson CJ, Tee AR, Cheatham L, Tsou C, Blenis J. mTOR controls cell cycle progression through its cell growth effectors S6K1 and 4E-BP1/eukaryotic translation initiation factor 4E. Mol Cell Biol. 2004;24(1):200–216. doi: 10.1128/MCB.24.1.200-216.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nelsen CJ, Rickheim DG, Tucker MM, Hansen LK, Albrecht JH. Evidence that cyclin D1 mediates both growth and proliferation downstream of TOR in hepatocytes. J Biol Chem. 2003;278(6):3656–3663. doi: 10.1074/jbc.M209374200. [DOI] [PubMed] [Google Scholar]
- 28.Goggin MM, Nelsen CJ, Kimball SR, Jefferson LS, Morley SJ, Albrecht JH. Rapamycin-sensitive induction of eukaryotic initiation factor 4F in regenerating mouse liver. Hepatology. 2004;40(3):537–544. doi: 10.1002/hep.20338. [DOI] [PubMed] [Google Scholar]
- 29.Jackson LN, et al. PI3K/Akt activation is critical for early hepatic regeneration after partial hepatectomy. Am J Physiol Gastrointest Liver Physiol. 2008;294(6):G1401–G1410. doi: 10.1152/ajpgi.00062.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Meng F, Henson R, Wehbe-Janek H, Ghoshal K, Jacob ST, Patel T. MicroRNA-21 regulates expression of the PTEN tumor suppressor gene in human hepatocellular cancer. Gastroenterology. 2007;133(2):647–658. doi: 10.1053/j.gastro.2007.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Iliopoulos D, Jaeger SA, Hirsch HA, Bulyk ML, Struhl K. STAT3 activation of miR-21 and miR-181b-1 via PTEN and CYLD are part of the epigenetic switch linking inflammation to cancer. Mol Cell. 2010;39(4):493–506. doi: 10.1016/j.molcel.2010.07.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Michalopoulos GK. Liver regeneration after partial hepatectomy: critical analysis of mechanistic dilemmas. Am J Pathol. 2010;176(1):2–13. doi: 10.2353/ajpath.2010.090675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wang X, Kiyokawa H, Dennewitz MB, Costa RH. The Forkhead Box m1b transcription factor is essential for hepatocyte DNA replication and mitosis during mouse liver regeneration. Proc Natl Acad Sci U S A. 2002;99(26):16881–16886. doi: 10.1073/pnas.252570299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Park TJ, et al. TIS21 negatively regulates hepatocarcinogenesis by disruption of cyclin B1-Forkhead box M1 regulation loop. Hepatology. 2008;47(5):1533–1543. doi: 10.1002/hep.22212. [DOI] [PubMed] [Google Scholar]
- 35.Jiang Z, Johnson HJ, Nie H, Qin J, Bird TA, Li X. Pellino 1 is required for interleukin-1 (IL-1)-mediated signaling through its interaction with the IL-1 receptor-associated kinase 4 (IRAK4)-IRAK-tumor necrosis factor receptor-associated factor 6 (TRAF6) complex. J Biol Chem. 2003;278(13):10952–10956. doi: 10.1074/jbc.M212112200. [DOI] [PubMed] [Google Scholar]
- 36.Clavien PA, Petrowsky H, DeOliveira ML, Graf R. Strategies for safer liver surgery and partial liver transplantation. N Engl J Med. 2007;356(15):1545–1559. doi: 10.1056/NEJMra065156. [DOI] [PubMed] [Google Scholar]
- 37.Mitchell C, Willenbring H. A reproducible and well-tolerated method for 2/3 partial hepatectomy in mice. Nat Protoc. 2008;3(7):1167–1170. doi: 10.1038/nprot.2008.80. [DOI] [PubMed] [Google Scholar]
- 38.Schmittgen TD, Livak KJ. Analyzing real-time PCR data by the comparative C(T) method. Nat Protoc. 2008;3(6):1101–1108. doi: 10.1038/nprot.2008.73. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.