Abstract
Glioblastoma (GBM), a uniformly lethal brain cancer, is characterized by diffuse invasion and abnormal activation of multiple receptor tyrosine kinase (RTK) signaling pathways, presenting a major challenge to effective therapy. The activation of many RTK pathways is regulated by extracellular heparan sulfate proteoglycans (HSPG), suggesting these molecules may be effective targets in the tumor microenvironment. In this study, we demonstrated that the extracellular sulfatase, SULF2, an enzyme that regulates multiple HSPG-dependent RTK signaling pathways, was expressed in primary human GBM tumors and cell lines. Knockdown of SULF2 in human GBM cell lines and generation of gliomas from Sulf2–/– tumorigenic neurospheres resulted in decreased growth in vivo in mice. We found a striking SULF2 dependence in activity of PDGFRα, a major signaling pathway in GBM. Ablation of SULF2 resulted in decreased PDGFRα phosphorylation and decreased downstream MAPK signaling activity. Interestingly, in a survey of SULF2 levels in different subtypes of GBM, the proneural subtype, characterized by aberrations in PDGFRα, demonstrated the strongest SULF2 expression. Therefore, in addition to its potential as an upstream target for therapy of GBM, SULF2 may help identify a subset of GBMs that are more dependent on exogenous growth factor–mediated signaling. Our results suggest the bioavailability of growth factors from the microenvironment is a significant contributor to tumor growth in a major subset of human GBM.
Introduction
Glioblastoma (GBM) is the most common malignant brain tumor of adults, with a median survival of less than 1 year (1). The disease is characterized by invasion of the tumor into the adjacent brain parenchyma and by the abnormal activation of receptor tyrosine kinase (RTK) signaling pathways.
However, despite the testing of a number of chemotherapeutic modalities targeting known GBM signaling pathways, only limited clinical success has been achieved. One explanation for the limited efficacy of targeted therapeutics may be that GBM is driven by the summation of multiple signaling inputs (2). Thus, effective therapeutic strategies may require a more comprehensive understanding of tumor signaling, including modulation by its microenvironment (3), a known regulator of lethal characteristics of other cancers (4). The identification of distinct GBM subtypes, based on expression and genomic and proteomic data (3, 5–8), supports the notion that GBM is a heterogeneous disease with different patterns of abnormal signaling.
RTK signaling pathways regulate many aspects of tumorigenesis, including cell growth and proliferation. In GBM, abnormal activation of these pathways can be driven by altered ligand availability and altered receptor levels. Indeed, the second most commonly amplified gene in GBM is PDGFRα (8–11). Overexpression of its ligand, PDGF-B, mediates the oncogenic influence of TGF-β in human GBM (12) and can drive tumorigenesis in murine models for glioma (13–15). Once released from the cell, growth factors can be sequestered by the extracellular microenvironment. Mechanisms regulating their postsynthetic availability are just beginning to be elucidated, but include enzymatic release from the extracellular matrix (ECM), thus allowing the growth factors to be available for signaling or alternatively to diffuse away. Because GBMs diffusely invade into the surrounding brain parenchyma, ligand availability in the tumor microenvironment, an underappreciated factor, may be a critical determinant of RTK signaling pathway activity and tumor growth.
Heparan sulfate proteoglycans (HSPGs) are ubiquitously produced by most animal cells and are a major component of the extracellular environment in normal brain and GBM (16, 17). Present on the cell surface and in the ECM, HSPGs play a key role in a number of biological processes based on their ability to bind and regulate the activity of diverse molecules including chemokines and growth factors, such as PDGF, VEGF, and FGF, in many tissues, including the brain (18–22). Indeed, HSPG binding can sequester ligands and decrease signaling, such as with the Wnts, or it can act as a coreceptor and actually promote receptor signaling, such as with FGF2 and VEGF (23–25). HSPGs consist of a protein core and heparan sulfate (HS) chains containing repeating disaccharide units of glucuronic/iduronic acid and glucosamine. The fine structure of HSPG is highly modified through a combination of posttranslational modifications, including sulfation on the N, 3-O, and 6-O positions of glucosamine and the 2-O position of the uronic acid units (26). The sulfation pattern of the HS chains is a major determinant of the specificity and the affinity of ligand interactions (27, 28). Changes in this pattern can alter growth factor bioavailability and thus influence cell signaling during development and disease. Pertinent to the present study, 6-O–sulfation is pivotal for the binding of many growth factors to HSPGs and is critical for normal development (29–31).
Although a number of enzymes control HSPG biosynthesis, the extracellular sulfatases, or SULFs, are unique because they modify the sulfation status of HSPG postsynthetically in the cellular microenvironment. By removing internal 6-O–sulfates of glucosamine on HS chains in regions of high overall sulfation, the SULFs can regulate the activity of HSPG-interacting ligands dynamically in the extracellular environment (23). In humans, 2 extracellular sulfatases, SULF1 and SULF2, have been characterized (32). First identified for their role in Wnt-dependent signaling during muscle development in quail (23), the SULFs regulate the in vivo activity of a number of HSPG-interacting ligands. In addition to Wnts, these include glial cell line–derived neurotrophic factor (GDNF), sonic hedgehog (SHH), VEGF, FGF2, and Noggin (23, 24, 33–39). Both Sulf1 and Sulf2 are highly expressed in the central nervous system and help regulate SHH signaling and neurite outgrowth (36, 40, 41).
In tumorigenesis, SULFs may serve either tumor-promoting or tumor-inhibiting functions, depending on the dominant signaling pathway(s) active in a given tumor (42). In human hepatocellular, breast, pancreatic, and non–small cell lung carcinoma, SULF2 is upregulated and promotes tumorigenesis (43–45). In the latter 2 cases, SULF2 exerts its growth-promoting effects via increased Wnt signaling. In contrast, in SULF-negative human ovarian adenocarcinoma cell lines, overexpression of SULF1 results in decreased FGF2 and heparin-binding EGF-like growth factor (HB-EGF) signaling (46).
GBM is driven by the abnormal activation of RTK signaling pathways. We hypothesized that GBM uses the extracellular SULFs to manipulate the tumor microenvironment and affect tumorigenesis. We tested this hypothesis in human GBM cell lines and in an orthotopic murine model for high-grade glioma (47, 48) by altering SULF2 expression and examining the effects on tumor growth and activation of critical growth factor signaling pathways. In this study, we also explored SULF expression levels in human GBM. Our findings indicate SULF2 expression may contribute to the pathogenesis of an important subset of human GBM.
Results
SULF2 protein is expressed in 50% of human GBM.
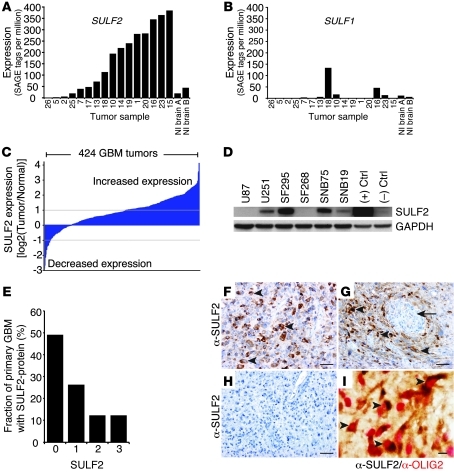
By in silico analysis of human expression data (49), we found elevated expression of SULF2 in GBM (Figure 1A). Using a stringent cutoff of a 10-fold increase in SULF2 SAGE tags over levels in normal brain to define high SULF2 levels, 7 of 16 GBMs (including both primary and xenograft tumors) had increased SULF2 expression. In contrast, SULF1 expression was not altered in most tumors (Figure 1B). Strikingly, in an independent set of 424 primary human GBM tumors, SULF2 expression was increased in 46% (197/424) of tumors relative to normal brain (Figure 1C). Consistent with the expression data, we found robust expression of SULF2 protein in 4 of 6 human high-grade astrocytoma cell lines (Figure 1D). Furthermore, immunohistochemistry on an independent set of 57 primary human GBM tumors demonstrated SULF2 protein in tumor cells in 50% of tumors (29/57 tumors) (Figure 1E). In a majority of the tumors, SULF2-positive tumor cells were widely distributed throughout the tumors and many coexpressed OLIG2 (Figure 1, F–I).
Figure 1. SULF2 expression in human GBM.
(A and B) In silico analysis of SULF2 and SULF1 expression in 16 human GBM tumor samples (49). Each bar represents normalized expression (y axis), as number of SAGE tags per million tags, for each patient tumor listed on the x axis. Expression in normal (Nl) brain is shown in each graph. (C) Increased SULF2 expression in 197/424 (46%) primary GBM tumors, log2(tumor/normal) greater than 1.0 (fold change of tumor versus normal greater than or equal to 2.0). See also Supplemental Figure 1 and Supplemental Table 1. (D) Western blot analysis of 6 human high-grade astrocytoma cell lines for SULF2 (~135 kDa). 293T cells with or without expression of mSULF2 were used as positive (+) and negative (–) controls. (E) Distribution of SULF2 protein expression in 57 primary human GBM tumors by immunohistochemistry. The percentage of SULF2-positive tumor cells was scored from no positive cells (score 0) to more than 75% of tumor cells positive (score 3) (see Methods). (F–I) Representative images from 2 SULF2-positive tumors (F, G, and I) and a SULF2-negative tumor (H). SULF2-positive (brown) tumor cells (F) were widely distributed except in occasional tumors that displayed a more prominent perivascular distribution (G). Many SULF2-positive (brown) tumor cells were also OLIG2-positive (red). See also Supplemental Figure 2. (I) Examples of SULF2-positive tumor cells (arrowheads) and microvascular proliferation characteristic of GBM (arrow). Scale bars: 50 μm (F–H); 10 μm (I).
The SULF2 gene resides within a region of chromosomal amplification in GBM on chromosome 20q13 (50). In 18 of 372 primary human GBM tumors analyzed, the region containing SULF2 was amplified, and in 12 of these tumors (67%), there was increased SULF2 expression (Supplemental Figure 1A and Supplemental Table 1; supplemental material available online with this article; doi: 10.1172/JCI58215DS1). Furthermore, in the human glioma cell lines, the line with the most abundant SULF2 protein also had amplification of SULF2 (Supplemental Table 2). We found no uniform trend between expression levels of SULF2 and TP53 or WT1, 2 transcription factors implicated in regulating SULF2 (51, 52) in glioma cell lines and primary human GBMs (Supplemental Figure 1, B–E).
SULF2 confers a growth advantage to human GBM cells.
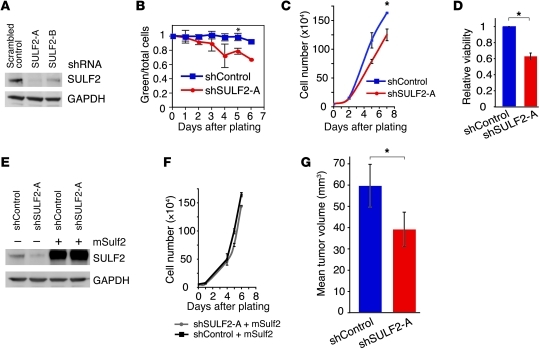
We transduced U251 cells, which contain moderate levels of SULF2 protein (Figure 1D), with either 1 of 2 validated SULF2 shRNAs (43) or a scrambled control shRNA (Figure 2A). Transduced cells also expressed EGFP, allowing for enrichment by flow cytometry. Over time in culture, however, the SULF2-positive cells outgrew the SULF2-knockdown cells, as reflected by a decrease in the ratio of EGFP-positive to total cells (Figure 2B). Selective growth of EGFP-negative cells was not observed in scrambled control shRNA cultures. Thus, cells were sorted for EGFP and used immediately for in vitro and in vivo assays. SULF2 knockdown resulted in a significant decrease in cell viability (Figure 2, C and D). This decrease was largely rescued by overexpression of murine Sulf2 (mSulf2) (Figure 2, E and F), as demonstrated by increased SULF2 protein (Figure 2E). A similar decrease in cell viability with SULF2 knockdown was observed in SF295 and SNB75 cells (Supplemental Figure 7, A and B).
Figure 2. SULF2 confers a growth advantage to human GBM cells in vitro and in vivo.
(A) Knockdown of SULF2 in U251 cells by 2 different shRNA constructs; SULF2 is decreased by 83% with shRNA SULF2-A and 55% with SULF2-B compared with the scrambled shRNA control. (B) In vitro growth of EGFP-positive SULF2-A shRNA (shSULF2-A) and scrambled shRNA control (shControl) cells demonstrated a selective growth advantage of SULF2-expressing cells over cells with SULF2 knockdown, as demonstrated by a decreased ratio of GFP-positive to total cells over time. *P < 0.01; n = 3. (C and D) Decreased growth and cell viability of SULF2-A shRNA cells versus scrambled shRNA control cells, as determined by counting live cells over time (*P < 0.00005; n = 3) (C) and by the colorimetric MTT viability assay, viable cell number normalized to control, day 5 after plating (*P < 0.00005; n = 5 independent experiments) (D). (E) Overexpression of mSulf2 in cells with SULF2 knockdown and in scrambled shRNA control cells. (F) Restoration of control growth with overexpression of mSulf2 in SULF2-A shRNA–containing cells (n = 3). (G) Mean tumor volume (mm3); subcutaneous flank transplant (11 days, *P < 0.05; n = 10 mice per group). (B, C, D, F, and G) Results are depicted as mean ± SEM.
Likewise, we observed a growth advantage of SULF2-positive U251 cells in vivo following subcutaneous transplant of SULF2-knockdown cells into nude mice (Figure 2G). SULF2-knockdown resulted in smaller tumor volumes in subcutaneous tumors. Decreased tumor cell growth in vitro and in vivo with SULF2 knockdown suggested a role for SULF2 in human GBM growth.
Sulf2 is expressed in a murine model for high-grade glioma.
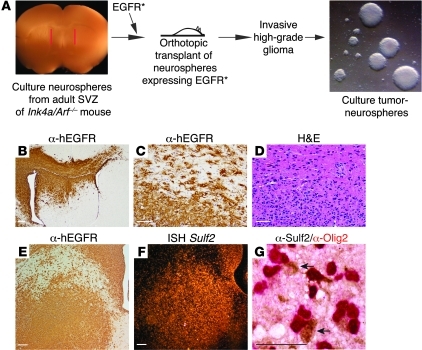
To model invasive aspects of the adult disease further, we adapted a murine model for high-grade glioma, based on the genetic manipulation of embryonic neural progenitor/stem cells (47, 48). We cultured adult neural progenitor cells from the subventricular zone (SVZ) of 11–week-old Ink4a/Arf–/– mice, transduced them with EGFRvIII (EGFR*), a constitutively active variant of EGFR derived from a human GBM (53), cultured them as tumorigenic neurospheres, and transplanted these cells orthotopically into host mice (Figure 3A). Both of these genetic alterations are common in adult human GBM. Within 7 weeks, 100% of mice developed highly invasive high-grade glial tumors (Figure 3, B–D).
Figure 3. Sulf2 expression in a murine model for high-grade glioma.
(A) Schema for generating high-grade, invasive glioma from adult neural progenitor cells. (B–D) Invasive, high-grade tumors generated from tumorigenic neurospheres immunostained for human EGFR and H&E. Scale bars: 300 μm (B); 50 μm (C and D). Similar to human GBM, in B, the tumor invades across the corpus callosum. (E and F) Tumor cells are hEGFR positive and exhibit robust Sulf2 expression by in situ hybridization (ISH) for the Sulf2 transcript. Scale bars: 200 μm. (G) Expression of Sulf2 protein (brown) in Olig2-positive (red) tumor cells in a primary murine tumor (arrow). Scale bar: 50 μm. See also Supplemental Figures 2 and 3.
We collected the tumors at the time of sacrifice for biochemical analysis or we cultured the tumor cells as tumor neurospheres (tumor-NS), as this enriches for tumor-initiating cells and retains the molecular properties of the parental tumor (54). Indeed, we observed that the tumorigenicity of tumor-NS increased as compared with the parental tumorigenic neurospheres, as reflected by a 38% decrease in median survival following orthotopic transplant of tumor-NS (median survival 23 days vs. 37 days; P < 0.0001).
Similar to human GBM, Sulf2 expression was readily detected in the murine tumor cells in vivo by in situ hybridization and immunohistochemistry (Figure 3, E–G, and Supplemental Figure 2). Tumor cells were identified by morphology and expression of hEGFR. Sulf2 protein was present in a subpopulation of the tumor cells including Olig2-positive cells (84.5% ± 3.2%; mean ± SEM, n = 4), occasional GFAP-positive cells, and Nestin-positive cells (Supplemental Figures 2 and 3). These data indicate that highly invasive, high-grade gliomas generated from adult neural progenitor cells have abundant expression of Sulf2 and are a useful model to study Sulf2 function.
Sulf2 confers increased tumorigenicity and proliferation.
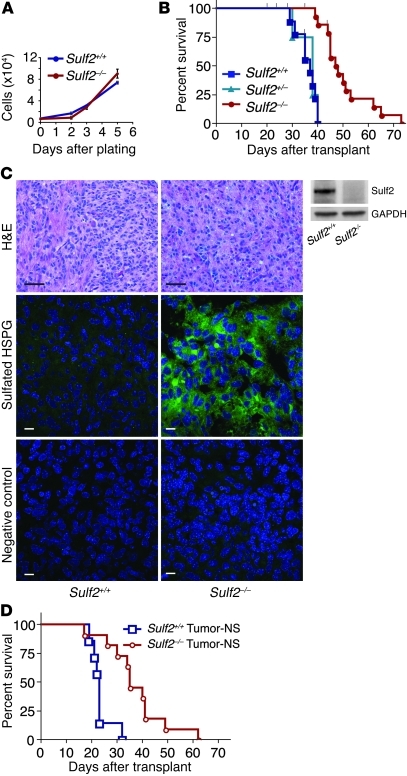
To establish Sulf2 function in glioma, we generated tumorigenic neurospheres from double-transgenic mice that were Ink4a/Arf–/– and either wild type, heterozygous, or null for Sulf2 (55). As expected, the extent of HSPG sulfation was greater in Sulf2–/– versus Sulf2+/+ neurospheres, as measured by a phage-display antibody whose binding to HSPGs depends on 6-O–sulfation (ref. 56, Supplemental Figure 4, A and B; relative MFI 2.3 versus 1.0; P < 0.05, n = 2). When tumor-NS were grown in minimal medium with only EGF and FGF2, however, Sulf2–/– and Sulf2+/+ cells had similar in vitro growth (Figure 4A). Strikingly, following orthotopic transplant, we observed a significant delay in tumor development from Sulf2–/– tumorigenic neurospheres versus those that were Sulf2+/+ or Sulf2+/–. The median survival of mice transplanted with 3 independent Sulf2–/– lines was 48 days (n = 14) compared with the median survival of mice transplanted with Sulf2+/+ cells (37 days; n = 9) or Sulf2+/– cells (38 days; n = 4; P < 0.001) (Figure 4B). In addition to prolonged mouse survival, Sulf2–/– tumors were 23% smaller than Sulf2+/+ tumors (mean ratio of tumor weight to body weight was 0.014 ± 0.00034 versus 0.018 ± 0.0014; ± SEM, P < 0.05; Supplemental Figure 4C). Although Sulf2+/+ and Sulf2–/– tumors had similar histologic appearance, only tumor-NS from Sulf2+/+ tumors expressed Sulf2 protein, and this was associated with decreased HSPG sulfation relative to Sulf2–/– tumors (Figure 4C and Supplemental Figure 4). These data support a functional requirement for Sulf2 in optimal gliomagenesis in the context of the brain microenvironment.
Figure 4. Prolonged survival conferred by ablation of Sulf2 in tumorigenic neurospheres.
(A) Similar in vitro growth of Sulf2+/+;Ink4a/Arf–/– or Sulf2–/–;Ink4a/Arf–/– tumorigenic neurospheres when cultured under nonadherent conditions (n = 3; mean ± SEM). (B) Kaplan-Meier survival analysis. Mice transplanted with Sulf2–/– cells have prolonged survival (median survival of 48 days) relative to mice transplanted with Sulf2+/+ or Sulf2+/– cells (median survival 37 days and 38 days, respectively, P < 0.001 for Sulf2–/– [n = 14] versus Sulf2+/+ [n = 9] or Sulf2+/– [n = 4]). 3 independent Sulf2–/– tumor progenitor lines were analyzed. Censored animals (black ticks) indicate individual mice sacrificed for tumor analysis prior to signs of tumor. (C) Similar tumor histology in Sulf2+/+ and Sulf2–/– tumors (H&E) despite absence of Sulf2 protein in Sulf2–/– tumor-NS (right panels, Western blot). Sulf2–/– tumors exhibit increased sulfated HSPGs (RB4CD12) compared with Sulf2+/+ tumors. Negative control antibody (MPB49) does not bind HSPG. Scale bars: 40 μm (H&E); 10 μm (HSPG, control). (D) Kaplan-Meier survival analysis demonstrating mice transplanted with tumor-NS isolated from Sulf2–/– tumors (median survival, 35 days) retain prolonged survival relative to those with tumor-NS from Sulf2+/+ tumors (median survival, 23 days). P < 0.001 for Sulf2–/– tumor-NS (n = 11) versus Sulf2+/+ tumor-NS (n = 7). See also Supplemental Figure 4.
Because Sulf2–/– tumor progenitor cells exhibited a marked delay in tumor growth relative to controls, we wondered whether these late-forming Sulf2–/– tumors had escaped Sulf2 dependence. However, when we isolated tumor-NS from mouse tumors and retransplanted them, the Sulf2–/– cells maintained their dependence on Sulf2 and exhibited significantly (P < 0.001) delayed growth (median survival, 35 days; n = 11) relative to Sulf2+/+ cells (median survival, 23 days; n = 7) (Figure 4D), thus indicating that the growth disadvantage conferred by genetic ablation of Sulf2 is durable in vivo.
Sulf2–/– tumors had a greater than 2-fold decrease in the number of proliferating tumor cells as compared with Sulf2+/+ tumors (n = 4 for Sulf2–/– and n = 6 for Sulf2+/+; P < 0.05) (Figure 5, A–C). There was no difference in the number of tumor cells undergoing apoptosis, as determined by cleaved caspase-3 immunostaining (data not shown). These data demonstrate that ablation of Sulf2 function in vivo results in decreased tumor cell proliferation, decreased tumor growth, and prolonged survival.
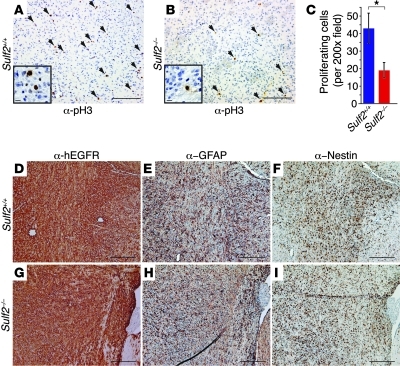
Figure 5. Decreased tumor cell proliferation in the absence of Sulf2 in vivo.
(A and B) Tumor cell proliferation, as determined by immunostaining for phospho-histone H3 (a-pH3), was greater in Sulf2+/+ tumors than in Sulf2–/– tumors. Representative pH3-positive cells indicated by arrows. Scale bars: 100 μm. Insets highlight representative positive cells. (C) Quantified proliferation data; number of mean pH3-positive cells per ×200 field per mouse in Sulf2+/+ tumors was 2-fold greater than in Sulf2–/– tumors (42.9 ± 8.7 versus 19.0 ± 4.4 [± SEM] for Sulf2+/+ [n = 4] and Sulf2–/– [n = 6], respectively). *P < 0.05. (D–I) In contrast, Sulf2+/+ and Sulf2–/– tumors exhibit similar expression of differentiation markers. Tumors, highlighted by hEGFR (D and G), express both GFAP (E and H) and Nestin (F and I). Scale bars: 60 μm. See also Supplemental Figure 5.
We found no significant differences in tumor cell morphology or differentiation between Sulf2–/– and Sulf2+/+ tumors. Both tumor types contained similar proportions of GFAP-positive and Nestin-positive tumor cells (Figure 5, D–I, and Supplemental Figure 5, A–D), and they were negative for NG2, a marker of oligodendrocyte differentiation (Supplemental Figure 5, E and F). In addition, although VEGF binds HSPG and is sensitive to SULF action (57, 58), we did not observe a phenotype resembling altered VEGF signaling (59), including differences in vascular morphology or tumor cell invasion, when we ablated Sulf2 (Supplemental Figure 5, G and H).
SULF2 alters the activity of multiple RTKs in human GBM.
Since we observed a SULF2-mediated growth phenotype in brain tumor cells, we hypothesized that SULF2 may act to alter the activity of important signaling pathways in human GBM. In carcinoma, SULFs have been implicated in abnormal Wnt signaling (43–45). Wnt signaling has previously been implicated in U251 growth (60). However, we observed no SULF2-dependent increase in canonical β-catenin–dependent transcriptional activity, as detected by a TCF/LEF reporter assay (Supplemental Figure 6E).
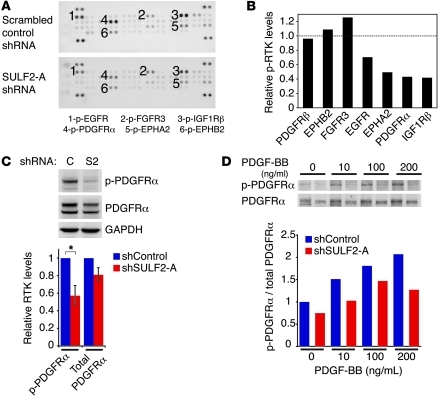
Instead, we observed that several RTKs were influenced by knockdown of SULF2 in U251 cells (Figure 6A), with a greater than 50% reduction in the activity of 3 RTKs of significance in GBM, PDGFRα, IGF1Rβ, and EPHA2 (Figure 6B). Moreover, EGFR activity was decreased by 30%. This did not reflect a global decrease in RTK activation, but a selective decrease in the activity of specific RTKs. Indeed, the closely related PDGFRβ and EPHB2 exhibited similar degrees of phosphorylation in cells with or without SULF2 knockdown. Furthermore, FGFR3 showed increased phosphorylation with SULF2 knockdown, a finding consistent with the ability of the SULFs to decrease FGF2-mediated signaling (61).
Figure 6. SULF2 alters activity of several RTKs in human GBM cells.
(A) RTK phosphorylation in U251 cells expressing SULF2-A shRNA or scrambled control shRNA. Individual RTKs are spotted in duplicate, and the identities of specific receptors are indicated. Positive control spots are located at the corners. See also Supplemental Figure 6. (B) Relative levels of phosphorylated RTKs in cells with knockdown of SULF2 normalized to cells with scrambled shRNA control. Duplicate spots were averaged. Data are representative of 2 independent experiments. (C) Phosphorylated and total PDGFRα levels in cells with SULF2 knockdown (S2) and scrambled shRNA control (C). Western blots were probed for GAPDH as a loading control. Results are means normalized to levels in scrambled shRNA control cells ± SEM (n = 4 independent experiments). *P < 0.01. (D) Knockdown of SULF2 decreases PDGFRα activation in response to PDGF-BB (10, 100, 200 ng/ml) stimulation. Relative levels of phosphorylated to total PDGFRα normalized to levels in unstimulated scrambled shRNA control cells. Data are representative of 2 independent experiments.
To determine whether these effects were generally true, we knocked down SULF2 in another high-grade astrocytoma cell line, SNB75, and again performed human phospho-RTK antibody arrays. As with U251 cells, SULF2 knockdown resulted in alterations in RTK activity, including decreased phosphorylation of PDGFRα and decreased cell viability (Supplemental Figure 6, A–C, and Supplemental Figure 7B). Together, these data suggest that SULF2 modulates the activity of several RTKs in GBM.
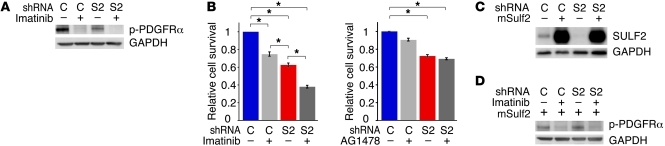
Because PDGFR is a major signaling pathway in human GBM (3, 8), we sought to validate the regulation of this RTK pathway by SULF2. Consistent with our array results, cells with knockdown of SULF2 exhibited a 43% decrease in phosphorylated PDGFRα (Figure 6C) and a modest decrease of 19% in total PDGFRα levels. Furthermore, SULF2 knockdown resulted in dramatically reduced activation of PDGFRα upon the addition of PDGF ligands (Figure 6D and Supplemental Figure 6D). Using imatinib mesylate, a RTK inhibitor with activity against PDGFRα, we demonstrated an additive decrease in both PDGFRα phosphorylation and cell survival following SULF2 knockdown (Figure 7, A and B). Similar effects were observed in SNB75 cells (Supplemental Figure 7). Overexpression of mSulf2 protein rescued the decreased activity of PDGFRα in cells with knockdown of SULF2 in an imatinib mesylate–dependent manner (Figure 7, C and D). Imatinib mesylate did not decrease the activation of EGFR (Supplemental Figure 6F).
Figure 7. Decreased tumor cell viability conferred by knockdown of SULF2 and inhibition of PDGFR signaling.
(A) PDGFRα phosphorylation is decreased by imatinib mesylate (9 μM) in both scrambled shRNA control and SULF2-A shRNA–containing cells by Western blot. (B) Knockdown of SULF2 in combination with inhibition of PDGFRα by imatinib mesylate (9 μM) results in decreased cell viability. This effect was not observed with inhibition of EGFR signaling by AG1478 (10 μM). *P < 0.005. (C) Overexpression of mouse SULF2 (mSULF2) in control and SULF2-A shRNA–containing cells by Western blot. (D) Overexpression of mSULF2 restores PDGFRα activity in cells with knockdown of human SULF2 in an imatinib mesylate–dependent manner. All data are representative of 2 independent experiments done in quadruplicate, and data are presented as mean ± SEM. C, scrambled shRNA control; S2, SULF2-A shRNA.
Ablation of Sulf2 confers decreased activation of PDGFRα and downstream signaling pathways in murine high-grade glioma.
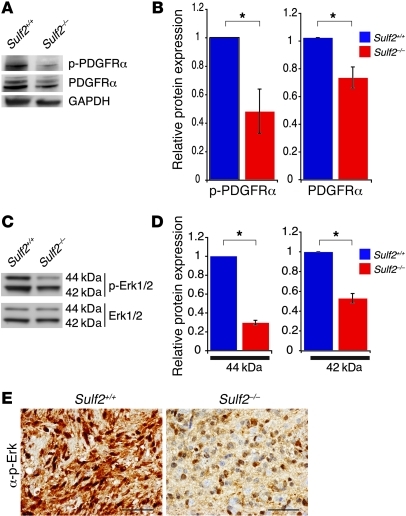
Similar to our results in 2 human astrocytoma cell lines, Sulf2 regulated PDGFRα activity in our murine tumors. In Sulf2–/– tumor-NS, we observed a marked reduction in activation of PDGFRα, with a slight reduction in total PDGFRα levels (Figure 8, A and B). Sulf2–/– tumor-NS stimulated with PDGF-BB also had reduced activation of PDGFRα relative to Sulf2+/+ tumor-NS (Supplemental Figure 8, A and B). In contrast, there was no decrease in activation of EGFR in Sulf2–/– tumor-NS stimulated with EGF relative to Sulf2+/+ tumor-NS (Supplemental Figure 8C). Although the tumor-prone cells expressed EGFRvIII, they still responded to growth factor stimulation in vitro (Supplemental Figure 8D).
Figure 8. Sulf2 alters the activity of PDGFRα in murine tumor-NS.
(A) Phosphorylated and total PDGFRα levels in Sulf2+/+ and Sulf2–/– tumor-NS. Western blots were probed for GAPDH as a loading control. (B) Quantification of p-PDGFRα and total PDGFRα levels in tumor-NS from Sulf2+/+ and Sulf2–/– cells normalized to mean ± SEM (n = 3 independent experiments). *P < 0.05. (C) SULF2 also affected the activity of downstream signaling pathways. Phosphorylated and total Erk1/2 (p44/p42) levels in Sulf2+/+ and Sulf2–/– tumor-NS. (D) The relative mean ratio of phosphorylated Erk to total Erk levels in Sulf2+/+ and Sulf2–/– tumor-NS normalized to Sulf2+/+ levels ± SEM (n = 3 independent experiments). *P < 0.005. (E) Sulf2+/+ tumors had more prominent phosphorylated Erk immunostaining relative to Sulf2–/– tumors. Scale bars: 50 μm.
Phosphorylation of PDGFRα results in the activation of downstream signaling pathways, including the MAPK pathway, also known to be important in human GBM. Accordingly, we observed that Sulf2–/– tumor-NS had decreased activation of the MAPK family members Erk1/2 (p44/p42) (Figure 8, C and D). Furthermore, in Sulf2+/+ tumors, we observed greater p-Erk immunostaining (2.5 ± 0.2, n = 10) than in Sulf2–/– tumors (1.6 ± 0.3, n = 11; P < 0.05) (Figure 8E). Together these data demonstrate a role for Sulf2 in modulating the activity of RTKs and downstream signaling pathways in high-grade glioma.
SULF2 expression is associated with the proneural subtype of GBM characterized by abnormalities in the PDGFRα-signaling pathway.
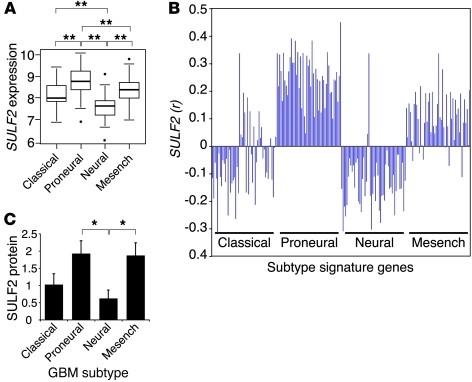
Since we found that SULF2 alters ligand-mediated RTK activity in GBM, we hypothesized that SULF2 may be enriched in a specific molecular GBM subtype. In human tumors of different subtypes, we observed a striking difference in SULF2 expression levels using data from The Cancer Genome Atlas (TCGA) ( 9A). Interestingly, SULF2 was most highly expressed in the proneural subtype of GBM (n = 173, P < 0.005), which is characterized by alterations in PDGFRα signaling. Indeed, SULF2 expression was associated with the expression of signature genes for the proneural GBM subtype (Figure 9B; Supplemental Table 3). There was also a less robust but positive association between expression of SULF2 and genes that are characteristic of the mesenchymal GBM subtype. Consistent with SULF2 expression, immunohistochemistry for SULF2 in 28 subtyped GBMs demonstrated that proneural and mesenchymal subtypes had the most abundant SULF2 protein (Figure 9C). Interestingly, the mesenchymal subtype, including the designated signature genes, is characterized by the upregulation of genes associated with the ECM and angiogenesis (7, 8). These data suggest SULF2 may help identify functional subsets of GBM.
Figure 9. SULF2 expression is associated with the proneural GBM subtype.
(A) SULF2 expression by human GBM subtype. Box plots show the median (range) normalized SULF2 expression levels were 8.1 (6.9–9.5), 8.9 (7.0–10.2), 7.8 (6.6–9.2), and 8.5 (7.1–9.9) for the classical, proneural, neural, and mesenchymal (mesench) subtypes, respectively (n = 173 tumors); **P < 0.005. The values within the box represent the lower quartile (Q1), median, and the upper quartile (Q3) of the distribution. The horizontal bars at the 2 ends are the smallest and largest nonoutlier observations. The circles beyond the horizontal bars represent outlying cases, defined as 1.5 times the interquartile range (Q3–Q1), below Q1 or above Q3. (B) Similarity (Pearson correlation, r) between SULF2 expression and the expression of 50 genes characterized as signature genes for each of the previously defined GBM subtypes (8). A positive coefficient denotes a positive relationship between SULF2 expression and expression of the gene of interest on the x axis (n = 202 tumors). In silico analysis for A and B was performed on expression data from the TCGA Data Portal. (C) SULF2 protein expression in primary human GBM samples of different subtypes. Tissue microarrays of previously subtyped human tumors were immunostained for SULF2 and scored. Data are represented as mean ± SEM for the classical, proneural, neural, and mesenchymal GBM subtypes, respectively (n = 28 tumors total). *P < 0.05.
Discussion
GBM pathogenesis depends on abnormal activation of RTK signaling pathways, in common with many cancers. Our data demonstrate that GBM utilizes SULF2 for optimal ligand-mediated RTK activation. We observed, first, that SULF2 protein is expressed in 50% of human GBM and in a murine model for high-grade glioma. Second, tumors generated from Sulf2–/– progenitor cells were smaller with decreased proliferation, resulting in prolonged survival. Third, we establish that SULF2 alters the activity of several RTKs, including PDGFRα, which may explain the SULF2 dependency for optimal tumor growth. Finally, high SULF2 expression was strongly associated with the proneural GBM subtype. Together, these data support what we believe is a novel role for SULF2 and identify a potential mechanism of regulating GBM growth.
In view of its regulation of multiple RTK signaling pathways in GBM, SULF2 is likely upstream of many of the currently targeted cell signaling pathways. Therefore, in patients with ligand-dependent tumors, inhibition of SULF2 may be a useful therapeutic strategy in combination with other treatments in GBM, such as inhibition of PDGFRα. Interestingly, the HS mimetic, PI-88, inhibits SULF function (56) and has been tested in a number of cancer clinical trials (62). Originally designed for their ability to inhibit heparanase, PI-88 and other HS mimetics have antiproliferative and antiangiogenic properties (63–65). These activities and our findings in human GBM make these potentially promising new therapeutic agents.
SULF2 regulates RTK signaling in high-grade glioma.
Our RTK activity profiling suggests SULF2 expression might have multiple and diverse effects on GBM. The 3 RTKs most affected by SULF2 knockdown, PDGFRα, IGF1Rβ, and EPHA2, each may play important roles in GBM. Increased PDGFRα signaling can drive tumor initiation, is important for tumor maintenance, and increases tumor cell proliferation (3, 8, 9, 13, 14, 66). Abnormal IGF1R signaling in GBM is associated with increased proliferation and tumor cell invasion (67, 68). In addition, activation of IGF1R may confer resistance to EGFR inhibitors in glioma (69). Finally, EPHA2 is an RTK commonly expressed in GBM and is thought to play important roles in tumor cell invasiveness (70–72). Although both WNT and VEGF signaling can be influenced by SULF2, these factors did not appear to play a major role in our SULF2-dependent phenotype.
Multiple lines of evidence suggest a key role for SULF2 in PDGFRα signaling. First, we found that SULF2 mediated differences in PDGFRα activity in multiple human glioma cell lines and in a murine model for high-grade glioma. Second, we observed that SULF2 is highly expressed in human tumors of the proneural GBM subtype, characterized by alterations in PDGFRα signaling, supporting a functional role for SULF2 in PDGFRα signaling in vivo. Our murine brain tumor model is based on the genetic manipulation of neural progenitor cells, and the PDGFRα pathway is implicated in progenitor cell gliomagenesis. PDGF stimulation of progenitor cells in the neonatal and adult brain can generate tumors or tumor-like growth (13, 14, 73–75). In addition, Sulf2 is one of a number of candidate genes identified in a screen for genes that, when activated by insertional mutagenesis, promote PDGFB-driven gliomas (76). Furthermore, Sulf2 expression is increased in many PDGFB-driven tumors regardless of the insertion site (77). Finally, PDGFs interact with HSPGs, and this interaction is influenced by 6-O–sulfation (18, 78, 79).
In addition to potential effects on PDGF bioavailability, SULF2 may alter PDGFRα activation via other mechanisms. Indeed, VEGF-A, a factor known to bind heparin/HSPGs in a SULF-dependent manner (18, 78, 79), can directly activate PDGFRα (80). Furthermore, we observed a slight but consistent decrease in the total levels of PDGFRα. This decrease could be due to alterations in the activity of other RTK signaling pathways or in the composition of the ECM, given that PDGF receptor levels are affected by alterations in hedgehog pathway activity (81) and alterations in the composition or amount of HSPGs (82). Thus, there may be multiple mechanisms by which SULF2 alters the activity of critical RTK signaling pathways in GBM.
Tumor interactions with the microenvironment in GBM.
There is accumulating evidence that the complex interplay between brain tumor cells and their microenvironment can have profound implications on tumorigenesis and invasion (58, 83). SULF modifies HSPGs in the tumor microenvironment and influences exogenous ligand availability (23, 38, 57). As GBM is a heterogeneous disease, SULF2 expression may help identify tumor subtypes with a greater dependence on ligand-mediated signaling, such as with PDGF. Consistent with this idea, we found that SULF2 affected ligand-mediated activation of PDGFRα and that SULF2 expression in human GBM most strongly correlates with the proneural GBM subtype, which is primarily ligand driven (3). As SULF2 expression was not exclusive to the proneural subtype, it will be interesting to determine whether SULF2 function differs between subtypes.
Identification of the specific pathways and mechanisms of pathway activation critical in individual tumors is anticipated to lead to improved therapy for GBM. We have identified SULF2 as a mechanism by which GBM increases the activity of several RTKs, including PDGFRα, to promote tumorigenesis. In addition to its potential as a therapeutic target in GBM, SULF2 may help to identify tumors that are more dependent on exogenous ligand–mediated signaling.
Methods
Antibodies and reagents.
We purchased antibodies against the following: p44/42 MAPK, phospho-p44/42 MAPK (Thr202, Tyr204), GAPDH, and β-actin (Cell Signaling); PDGFRα (Millipore); EGFR (Santa Cruz Biotechnology Inc.), phospho-EGFR (R&D Systems); GFAP and hEGFR (Dako); Nestin, Olig2, and NG2 (Chemicon/Millipore International); and phospho-PDGFRα (Tyr742), phospho-EGFR (Tyr1068), and human RTK Proteome Profiler (R&D Systems). The monoclonal antibody against Sulf2 (2B4) has been previously described (44). RB4CD12 was a gift of Toin van Kuppevelt (Nijmegen Medical Center, Nijmegen, Netherlands). Imatinib mesylate was purchased from United States Biological. DMEM, Neurobasal Medium, B-27 supplement, N2 supplement, recombinant human EGF, and recombinant human PDGF-BB were purchased from Invitrogen. Recombinant human PDGF-AA (R&D Systems), puromycin (Sigma-Aldrich), AG1478 (Calbiochem; EMD Bioscience), and FGF2 (Chemicon/Millipore Corp.) were also used.
U251 and U87 cells were obtained from the UCSF Brain Tumor Research Center Tissue Bank. Retroviral vectors were used to transduce cells with EGFRvIII (EGFR*) (47) and the Sulf2 shRNAs and scrambled control shRNA (43). Mouse Sulf2 cDNA (mSulf2), as previously described (32), was subcloned into a lentiviral expression vector expressing a fluorescent marker (84). SNB-75, SF295, and SF268 cells, obtained from the DCTD Tumor/Cell Line Repository, were maintained in DMEM plus 10% FCS, and protein lysates were harvested after 4 days of growth in 0.05% FCS.
Cell culture and virus production.
Murine neurospheres were cultured as previously described (48). Briefly, neural progenitor cells were isolated from the SVZ of 11-week-old male Sulf2–/–;Ink4a/Arf–/– mice and Sulf2+/– and Sulf2+/+;Ink4a/Arf–/– male littermates. Cells were cultured on nonadherent plates (Ultralow Attachment Plates; Corning Inc.) in Neurobasal Medium plus B27 plus N2 with 20 ng/ml EGF and 20 ng/ml FGF2. Cells were passaged once per week and plated at equivalent densities (2 × 104 cells per well of a 6-well plate). Following transduction, neurospheres were maintained in puromycin (2 μg/ml). Transduced cells were passaged at least 2 to 3 times, but fewer than 7 times prior to transplant.
Neurosphere growth rates were determined by averaging the total number of viable cells per well at various times after plating (n = 3 wells per time point). For analysis of PDGFRα activation, tumors were cultured as neurospheres, and 12–15 hours prior to collection, cells were either stimulated with 2% FCS or PDGF-BB (100 ng/ml).
Human glioma cell lines were cultured in DMEM plus 10% FCS. Growth rates of human glioma cell lines were determined by CellTiter 96 NonRadioactive Cell Proliferation Assay (Promega) or by averaging the total number of viable cells per well and the total number of viable GFP-positive cells per well at various times after plating (n = 3–5 wells per cell line per time point). For experiments involving addition of imatinib mesylate (9 μM) or AG1478 (5 μM), 4 × 104 cells were plated per well of a 48-well plate, drug was added on day 2, and cells were collected on day 5. Counting was done using a hemocytometer and trypan blue to exclude dead cells. Transduced cells expressing mSULF2 and a fluorescent marker were sorted by flow cytometry prior to use. For protein analysis, cells were grown for a minimum of 4 days prior to stimulating or harvesting total cell lysates.
Quantitative RT-PCR.
Real-time PCR with SYBR green detection was performed using a Mastercycler ep realplex real-time PCR system (Eppendorf) and Faststart Universal SYBR Green Master Mix (Rox; Roche Applied Science) as follows: 10 minutes at 95°C for initial denaturation, followed by 40 cycles at 95°C for 15 seconds and 60°C for 1 minute. After each cycle, fluorescence was measured. Each run included a melting curve for each primer set to determine the correct response of the primers, and only values with the correct melting curves were used.
Mouse procedures.
Sulf2 mutant mice containing a gene trap insertion in the sixth intron of the Sulf2 locus were generated as previously described (55). Ink4a/Arf–/– mice were obtained from the National Cancer Institute Mouse Repository and bred to the FVB/n background. Immunocompromised 6-week-old male Icr-SCID homozygous mice obtained from Taconic Inc. were used for neurosphere transplants. Neurospheres were dissociated and resuspended in HBSS at a concentration of 200,000 viable cells/μl, and 2 μl were injected into the striatum (48). Mice were monitored daily and were sacrificed at the onset of neurological signs or, if no overt signs developed, upon loss of 20% or more of their peak body weight. The brain to body weight ratio was determined at 35 days after transplant (n = 3 Sulf2+/+ and n = 4 Sulf2–/– mice). For subcutaneous transplant of U251, cells were sorted for EGFP, briefly cultured, and resuspended in HBSS at a concentration of 250,000 viable cell/μl; 2 μl were injected. Tumors were measured (length/width/height) every other day using a caliper (0.1 cm), and mice were weighed 3 times a week. Tumor volume was calculated based on the equation (π/6) (length × width × height).
To account for potential loss of EGFP-expressing cells during the brief in vitro passage, a cohort of mice were transplanted with equal percentages of EGFP-positive cells (n = 3 mice per group) with similar results.
Histology, immunohistochemistry, and immunoblotting.
To collect tissue for immunohistochemical analysis, mice were perfused with 4% PFA, and the tissue was fixed overnight in 4% PFA, rinsed in PBS, and stored in 70% ethanol until further processing. Histological analysis of tumor tissue was performed on H&E-stained sections. Formalin-fixed paraffin-embedded sections of human GBM were obtained from the UCSF Brain Tumor Research Center Tissue Bank. Immunohistochemistry was performed according to standard methods, and immunostaining for SULF2 was performed both by hand (44) and on the Ventana Medical Systems Benchmark XT using the iView detection system. To score for human SULF2 and p-ERK1/2 immunostaining, the percentage of tumor cells that stained positive was noted: 0 denoted no positive tumor cells; 1+ indicated 1%–25%; 2+ indicated 26%–75%; and 3+ indicated more than 75%. To evaluate murine phospho-histone H3 immunostaining, 3 to 9 200× fields were examined per mouse, and the mean number of positive cells per field was calculated per mouse. All histologic and immunohistologic analyses were performed without knowledge of the Sulf2 status of the tumor.
Human phospho-RTK arrays (Proteome Profiler; R&D Systems) were incubated with 200 μg of protein lysate from human glioma cells with and without SULF2 knockdown. Western blotting and probing for GAPDH confirmed equal protein loading. Arrays and Western blots were analyzed using ImageJ software. For arrays, the integrated densities of duplicate spots were averaged and normalized to the values for the control spots. For Western blots, relative integrated densities of the desired band were normalized to the loading control (GAPDH or β-actin), averaged across experiments, and then normalized to control.
Sulf2 in situ hybridization.
Radioactive in situ hybridization for Sulf2 was performed on paraffin-embedded sections using probes containing nucleotides 1043 to 1726 from the mouse Sulf2 cDNA (55). Briefly, plasmids were linearized with the appropriate restriction enzymes to transcribe either sense or anti-sense 35S-labeled riboprobes (Promega). Paraffin sections placed on acid-etched, 3-aminopropyl-triethoxysilase–treated slides were washed at a final stringency of 65°C in 2xSSC, dipped in emulsion, and exposed for 1–2 weeks. DNA was counterstained with Hoechst 33342.
In silico analyses.
For correlation of SULF2 gene expression and GBM subtype (Figure 9), we used data available through the TCGA Data Portal (85) ( http://cancergenome.nih.gov, accessed on April 15, 2010). For Figure 1C, Figure 9, A–C, Supplemental Figure 1, A, D, and E, and Supplemental Table 1, we accessed the TCGA Data Portal on January 17, 2011. Fold change for SULF2 transcript was calculated as the ratio of gene expression values of tumor to normal, with the normal sample value used as baseline. A ratio of log2(tumor/normal) greater than or equal to 1.0 (a fold change of tumor vs. normal greater than or equal to 2) was considered increased, and a ratio of log2(tumor/normal) less than or equal to –1.0 was considered decreased. SULF2 was considered amplified, if the value of log2(tumor/normal) was greater than or equal to 0.5 (a fold change of tumor copy number vs. normal copy number greater than or equal to 1.41). SAGE expression data included xenograft tumors (tumor numbers 26, 5, 2, 25, 7, 17, 13, 18, 10, 14, 19, 1, 20, 16, 23, 15) and primary tumors (tumor numbers 10, 20, and 23).
Statistics.
A 2-tailed t test was used to compare mean values except where noted. P < 0.05 was considered statistically significant. For Kaplan-Meier survival analysis, groups were compared using the log-rank test. For Figure 9, Wilcoxon’s rank-sum test was used to test for pairwise differences between GBM subtypes in SULF2 expression, with the Holm adjusted P values for multiple comparisons. Multiple comparisons in Supplemental Figures 4 and 8 were performed using 1-way ANOVA, and post hoc analyses were based on Tukey’s test.
Study approval.
All procedures were performed according to protocols approved by the University of California Committee on Research (San Francisco, California, USA). All experimental protocols involving mice were approved by the UCSF Institutional Animal Care and Use Committee. Deidentified formalin-fixed paraffin embedded sections of human GBM were obtained from the UCSF Brain Tumor Research Center Tissue Bank.
Supplementary Material
Acknowledgments
We thank Mark S. Singer for helpful advice, Cynthia Cowdrey for assistance with obtaining human tissue, and G.J.P. Dijkgraaf for helpful discussions. This work was supported by funds from the NIH (K08 NS063456 to J.J. Phillips; UCSF Brain Tumor SPORE CA097257 to Z. Werb; CA076002, CA057621, and NS039278 to Z. Werb; and AI053194 to S.D. Rosen) and a Tobacco-Related Disease Research Program of the University of California grant (17RT-0216 to S.D. Rosen). This work was also funded in part by a generous gift from the Preuss Foundation. RB4CD12 was a gift of Toin van Kuppevelt of Nijmegen Medical Center, the Netherlands. The work was also funded in part by a Susan Resnick Fisher/American Brain Tumor Association fellowship and a generous gift from the Preuss Foundation.
Footnotes
Conflict of interest: The authors have declared that no conflict of interest exists.
Citation for this article: J Clin Invest. 2012;122(3):911–922. doi:10.1172/JCI58215.
Emmanuelle Huillard’s present address is: Centre de Recherche de l’Institut du Cerveau et de la Moelle ωpinière, Paris, France.
Anna Ward’s present address is: University of Arizona College of Medicine, Phoenix, Phoenix, Arizona, USA.
David H. Lum’s present address is: Department of Oncological Sciences, Huntsman Cancer Institute, University of Utah, Salt Lake City, Utah, USA.
Mei-Yin Polley’s present address is: National Cancer Institute, Biometric Research Branch, Rockville, Maryland, USA.
References
- 1.Louis DN, et al. The 2007 WHO classification of tumours of the central nervous system. Acta Neuropathol. 2007;114(2):97–109. doi: 10.1007/s00401-007-0243-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Stommel JM, et al. Coactivation of receptor tyrosine kinases affects the response of tumor cells to targeted therapies. Science. 2007;318(5848):287–290. doi: 10.1126/science.1142946. [DOI] [PubMed] [Google Scholar]
- 3.Brennan C, et al. Glioblastoma subclasses can be defined by activity among signal transduction pathways and associated genomic alterations. PLoS One. 2009;4(11):e7752. doi: 10.1371/journal.pone.0007752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kessenbrock K, Plaks V, Werb Z. Matrix metalloproteinases: regulators of the tumor microenvironment. Cell. 2010;141(1):52–67. doi: 10.1016/j.cell.2010.03.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Colman H, et al. A multigene predictor of outcome in glioblastoma. Neuro Oncol. 2010;12(1):49–57. doi: 10.1093/neuonc/nop007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mischel PS, et al. Identification of molecular subtypes of glioblastoma by gene expression profiling. Oncogene. 2003;22(15):2361–2373. doi: 10.1038/sj.onc.1206344. [DOI] [PubMed] [Google Scholar]
- 7.Phillips HS, et al. Molecular subclasses of high-grade glioma predict prognosis, delineate a pattern of disease progression, and resemble stages in neurogenesis. Cancer Cell. 2006;9(3):157–173. doi: 10.1016/j.ccr.2006.02.019. [DOI] [PubMed] [Google Scholar]
- 8.Verhaak RG, et al. Integrated genomic analysis identifies clinically relevant subtypes of glioblastoma characterized by abnormalities in PDGFRA, IDH1, EGFR, and NF1. Cancer Cell. 2010;17(1):98–110. doi: 10.1016/j.ccr.2009.12.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hermanson M, et al. Platelet-derived growth factor and its receptors in human glioma tissue: expression of messenger RNA and protein suggests the presence of autocrine and paracrine loops. Cancer Res. 1992;52(11):3213–3219. [PubMed] [Google Scholar]
- 10.Ozawa T, et al. PDGFRA gene rearrangements are frequent genetic events in PDGFRA-amplified glioblastomas. Genes Dev. 2010;24(19):2205–2218. doi: 10.1101/gad.1972310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Guha A, Dashner K, Black PM, Wagner JA, Stiles CD. Expression of PDGF and PDGF receptors in human astrocytoma operation specimens supports the existence of an autocrine loop. Int J Cancer. 1995;60(2):168–173. doi: 10.1002/ijc.2910600206. [DOI] [PubMed] [Google Scholar]
- 12.Bruna A, et al. High TGFbeta-Smad activity confers poor prognosis in glioma patients and promotes cell proliferation depending on the methylation of the PDGF-B gene. Cancer Cell. 2007;11(2):147–160. doi: 10.1016/j.ccr.2006.11.023. [DOI] [PubMed] [Google Scholar]
- 13.Dai C, Celestino JC, Okada Y, Louis DN, Fuller GN, Holland EC. PDGF autocrine stimulation dedifferentiates cultured astrocytes and induces oligodendrogliomas and oligoastrocytomas from neural progenitors and astrocytes in vivo. Genes Dev. 2001;15(15):1913–1925. doi: 10.1101/gad.903001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Uhrbom L, Hesselager G, Nister M, Westermark B. Induction of brain tumors in mice using a recombinant platelet-derived growth factor B-chain retrovirus. Cancer Res. 1998;58(23):5275–5279. [PubMed] [Google Scholar]
- 15.Liu KW, et al. SHP-2/PTPN11 mediates gliomagenesis driven by PDGFRA and INK4A/ARF aberrations in mice and humans. J Clin Invest. 2011;121(3):905–917. doi: 10.1172/JCI43690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Steck PA, et al. Altered expression and distribution of heparan sulfate proteoglycans in human gliomas. Cancer Res. 1989;49(8):2096–2103. [PubMed] [Google Scholar]
- 17.Watanabe A, et al. Expression of syndecans, a heparan sulfate proteoglycan, in malignant gliomas: participation of nuclear factor-kappaB in upregulation of syndecan-1 expression. J Neurooncol. 2006;77(1):25–32. doi: 10.1007/s11060-005-9010-3. [DOI] [PubMed] [Google Scholar]
- 18.Smith EM, Mitsi M, Nugent MA, Symes K. PDGF-A interactions with fibronectin reveal a critical role for heparan sulfate in directed cell migration during Xenopus gastrulation. Proc Natl Acad Sci U S A. 2009;106(51):21683–21688. doi: 10.1073/pnas.0902510106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Feyzi E, Lustig F, Fager G, Spillmann D, Lindahl U, Salmivirta M. Characterization of heparin and heparan sulfate domains binding to the long splice variant of platelet-derived growth factor A chain. . J Biol Chem. 1997;272(9):5518–5524. doi: 10.1074/jbc.272.9.5518. [DOI] [PubMed] [Google Scholar]
- 20.Ono K, Hattori H, Takeshita S, Kurita A, Ishihara M. Structural features in heparin that interact with VEGF165 and modulate its biological activity. Glycobiology. 1999;9(7):705–711. doi: 10.1093/glycob/9.7.705. [DOI] [PubMed] [Google Scholar]
- 21.Kreuger J, Salmivirta M, Sturiale L, Gimenez-Gallego G, Lindahl U. Sequence analysis of heparan sulfate epitopes with graded affinities for fibroblast growth factors 1 and 2. J Biol Chem. 2001;276(33):30744–30752. doi: 10.1074/jbc.M102628200. [DOI] [PubMed] [Google Scholar]
- 22.Ashikari-Hada S, Habuchi H, Kariya Y, Itoh N, Reddi AH, Kimata K. Characterization of growth factor-binding structures in heparin/heparan sulfate using an octasaccharide library. J Biol Chem. 2004;279(13):12346–12354. doi: 10.1074/jbc.M313523200. [DOI] [PubMed] [Google Scholar]
- 23.Dhoot GK, Gustafsson MK, Ai X, Sun W, Standiford DM, Emerson CP., Jr Regulation of Wnt signaling and embryo patterning by an extracellular sulfatase. Science. 2001;293(5535):1663–1666. doi: 10.1126/science.293.5535.1663. [DOI] [PubMed] [Google Scholar]
- 24.Ai X, Do AT, Lozynska O, Kusche-Gullberg M, Lindahl U, Emerson CP., Jr QSulf1 remodels the 6-O sulfation states of cell surface heparan sulfate proteoglycans to promote Wnt signaling. J Cell Biol. 2003;162(2):341–351. doi: 10.1083/jcb.200212083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gallagher JT. Heparan sulfate: growth control with a restricted sequence menu. J Clin Invest. 2001;108(3):357–361. doi: 10.1172/JCI13713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bernfield M, et al. Functions of cell surface heparan sulfate proteoglycans. Annu Rev Biochem. 1999;68:729–777. doi: 10.1146/annurev.biochem.68.1.729. [DOI] [PubMed] [Google Scholar]
- 27.Esko JD, Lindahl U. Molecular diversity of heparan sulfate. J Clin Invest. 2001;108(2):169–173. doi: 10.1172/JCI13530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Habuchi H, Habuchi O, Kimata K. Sulfation pattern in glycosaminoglycan: does it have a code? Glycoconj J. 2004;21(1-2):47–52. doi: 10.1023/B:GLYC.0000043747.87325.5e. [DOI] [PubMed] [Google Scholar]
- 29.Bink RJ, et al. Heparan sulfate 6-o-sulfotransferase is essential for muscle development in zebrafish. . J Biol Chem. 2003;278(33):31118–31127. doi: 10.1074/jbc.M213124200. [DOI] [PubMed] [Google Scholar]
- 30.Bulow HE, Hobert O. Differential sulfations and epimerization define heparan sulfate specificity in nervous system development. Neuron. 2004;41(5):723–736. doi: 10.1016/S0896-6273(04)00084-4. [DOI] [PubMed] [Google Scholar]
- 31.Kamimura K, et al. Drosophila heparan sulfate 6-O-sulfotransferase (dHS6ST) gene. Structure, expression, and function in the formation of the tracheal system. J Biol Chem. 2001;276(20):17014–17021. doi: 10.1074/jbc.M011354200. [DOI] [PubMed] [Google Scholar]
- 32.Morimoto-Tomita M, Uchimura K, Werb Z, Hemmerich S, Rosen SD. Cloning and characterization of two extracellular heparin-degrading endosulfatases in mice and humans. J Biol Chem. 2002;277(51):49175–49185. doi: 10.1074/jbc.M205131200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ai X, Kitazawa T, Do AT, Kusche-Gullberg M, Labosky PA, Emerson CP., Jr SULF1 and SULF2 regulate heparan sulfate-mediated GDNF signaling for esophageal innervation. Development. 2007;134(18):3327–3338. doi: 10.1242/dev.007674. [DOI] [PubMed] [Google Scholar]
- 34.Dai Y, et al. HSulf-1 and HSulf-2 are potent inhibitors of myeloma tumor growth in vivo. J Biol Chem. 2005;280(48):40066–40073. doi: 10.1074/jbc.M508136200. [DOI] [PubMed] [Google Scholar]
- 35.Viviano BL, Paine-Saunders S, Gasiunas N, Gallagher J, Saunders S. Domain-specific modification of heparan sulfate by Qsulf1 modulates the binding of the bone morphogenetic protein antagonist Noggin. J Biol Chem. 2004;279(7):5604–5611. doi: 10.1074/jbc.M310691200. [DOI] [PubMed] [Google Scholar]
- 36.Danesin C, et al. Ventral neural progenitors switch toward an oligodendroglial fate in response to increased Sonic hedgehog (Shh) activity: involvement of Sulfatase 1 in modulating Shh signaling in the ventral spinal cord. J Neurosci. 2006;26(19):5037–5048. doi: 10.1523/JNEUROSCI.0715-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Freeman SD, Moore WM, Guiral EC, Holme AD, Turnbull JE, Pownall ME. Extracellular regulation of developmental cell signaling by XtSulf1. Dev Biol. 2008;320(2):436–445. doi: 10.1016/j.ydbio.2008.05.554. [DOI] [PubMed] [Google Scholar]
- 38.Lamanna WC, Frese MA, Balleininger M, Dierks T. Sulf loss influences N-, 2-O-, and 6-O-sulfation of multiple heparan sulfate proteoglycans and modulates fibroblast growth factor signaling. J Biol Chem. 2008;283(41):27724–27735. doi: 10.1074/jbc.M802130200. [DOI] [PubMed] [Google Scholar]
- 39.Fujita K, et al. HpSulf, a heparan sulfate 6-O-endosulfatase, is involved in the regulation of VEGF signaling during sea urchin development. Mech Dev. 2010;127(3–4):235–245. doi: 10.1016/j.mod.2009.12.001. [DOI] [PubMed] [Google Scholar]
- 40.Kalus I, et al. Differential involvement of the extracellular 6-O-endosulfatases Sulf1 and Sulf2 in brain development and neuronal and behavioural plasticity. J Cell Mol Med. 2009;13(11-12):4505–4521. doi: 10.1111/j.1582-4934.2008.00558.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Nagamine S, Koike S, Keino-Masu K, Masu M. Expression of a heparan sulfate remodeling enzyme, heparan sulfate 6-O-endosulfatase sulfatase FP2, in the rat nervous system. Brain Res Dev Brain Res. 2005;159(2):135–143. doi: 10.1016/j.devbrainres.2005.07.006. [DOI] [PubMed] [Google Scholar]
- 42.Rosen SD, Lemjabbar-Alaoui H. Sulf-2: an extracellular modulator of cell signaling and a cancer target candidate. Expert Opin Ther Targets. 2010;14(9):935–949. doi: 10.1517/14728222.2010.504718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Nawroth R, van Zante A, Cervantes S, McManus M, Hebrok M, Rosen SD. Extracellular sulfatases, elements of the Wnt signaling pathway, positively regulate growth and tumorigenicity of human pancreatic cancer cells. PLoS One. 2007;2(4):e392. doi: 10.1371/journal.pone.0000392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lemjabbar-Alaoui H, et al. Sulf-2, a heparan sulfate endosulfatase, promotes human lung carcinogenesis. Oncogene. 2009;29(5):635–646. doi: 10.1038/onc.2009.365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lai JP, et al. Sulfatase 2 up-regulates glypican 3, promotes fibroblast growth factor signaling, and decreases survival in hepatocellular carcinoma. Hepatology. 2008;47(4):1211–1222. doi: 10.1002/hep.22202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lai J, et al. Loss of HSulf-1 up-regulates heparin-binding growth factor signaling in cancer. J Biol Chem. 2003;278(25):23107–23117. doi: 10.1074/jbc.M302203200. [DOI] [PubMed] [Google Scholar]
- 47.Bachoo RM, et al. Epidermal growth factor receptor and Ink4a/Arf: convergent mechanisms governing terminal differentiation and transformation along the neural stem cell to astrocyte axis. Cancer Cell. 2002;1(3):269–277. doi: 10.1016/S1535-6108(02)00046-6. [DOI] [PubMed] [Google Scholar]
- 48.Ligon KL, et al. Olig2-regulated lineage-restricted pathway controls replication competence in neural stem cells and malignant glioma. Neuron. 2007;53(4):503–517. doi: 10.1016/j.neuron.2007.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Parsons DW, et al. An integrated genomic analysis of human glioblastoma multiforme. Science. 2008;321(5897):1807–1812. doi: 10.1126/science.1164382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wiltshire RN, et al. Comparative genomic hybridization analysis of astrocytomas: prognostic and diagnostic implications. J Mol Diagn. 2004;6(3):166–179. doi: 10.1016/S1525-1578(10)60507-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Langsdorf A, et al. Expression regulation and function of heparan sulfate 6-O-endosulfatases in the spermatogonial stem cell niche. Glycobiology. 2010;21(2):152–161. doi: 10.1093/glycob/cwq133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Chau BN, et al. Identification of SULF2 as a novel transcriptional target of p53 by use of integrated genomic analyses. Cancer Res. 2009;69(4):1368–1374. doi: 10.1158/0008-5472.CAN-08-2742. [DOI] [PubMed] [Google Scholar]
- 53.Nishikawa R, et al. A mutant epidermal growth factor receptor common in human glioma confers enhanced tumorigenicity. Proc Natl Acad Sci U S A. 1994;91(16):7727–7731. doi: 10.1073/pnas.91.16.7727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Lee J, et al. Tumor stem cells derived from glioblastomas cultured in bFGF and EGF more closely mirror the phenotype and genotype of primary tumors than do serum-cultured cell lines. Cancer Cell. 2006;9(5):391–403. doi: 10.1016/j.ccr.2006.03.030. [DOI] [PubMed] [Google Scholar]
- 55.Lum DH, Tan J, Rosen SD, Werb Z. Gene trap disruption of the mouse heparan sulfate 6-O-endosulfatase gene, Sulf2. Mol Cell Biol. 2007;27(2):678–688. doi: 10.1128/MCB.01279-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Hossain MM, et al. Direct detection of HSulf-1 and HSulf-2 activities on extracellular heparan sulfate and their inhibition by PI-88. Glycobiology. 2010;20(2):175–186. doi: 10.1093/glycob/cwp159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Xu D, Fuster MM, Lawrence R, Esko JD. Heparan sulfate regulates VEGF165- and VEGF121-mediated vascular hyperpermeability. J Biol Chem. 2011;286(1):737–745. doi: 10.1074/jbc.M110.177006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Uchimura K, et al. HSulf–2, an extracellular endoglucosamine–6–sulfatase, selectively mobilizes heparin–bound growth factors and chemokines: effects on VEGF, FGF–1, and SDF–1. BMC Biochem. 2006;7:2. doi: 10.1186/1471-2091-7-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Du R, et al. HIF1alpha induces the recruitment of bone marrow-derived vascular modulatory cells to regulate tumor angiogenesis and invasion. Cancer Cell. 2008;13(3):206–220. doi: 10.1016/j.ccr.2008.01.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Pu P, et al. Downregulation of Wnt2 and beta-catenin by siRNA suppresses malignant glioma cell growth. Cancer Gene Ther. 2009;16(4):351–361. doi: 10.1038/cgt.2008.78. [DOI] [PubMed] [Google Scholar]
- 61.Wang S, et al. QSulf1, a heparan sulfate 6-O-endosulfatase, inhibits fibroblast growth factor signaling in mesoderm induction and angiogenesis. Proc Natl Acad Sci U S A. 2004;101(14):4833–4838. doi: 10.1073/pnas.0401028101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Khasraw M, et al. Multicentre phase I/II study of PI-88, a heparanase inhibitor in combination with docetaxel in patients with metastatic castrate-resistant prostate cancer. Ann Oncol. 2010;21(6):1302–1307. doi: 10.1093/annonc/mdp524. [DOI] [PubMed] [Google Scholar]
- 63.Johnstone KD, et al. Synthesis and biological evaluation of polysulfated oligosaccharide glycosides as inhibitors of angiogenesis and tumor growth. J Med Chem. 2010;53(4):1686–1699. doi: 10.1021/jm901449m. [DOI] [PubMed] [Google Scholar]
- 64.Joyce JA, Freeman C, Meyer-Morse N, Parish CR, Hanahan D. A functional heparan sulfate mimetic implicates both heparanase and heparan sulfate in tumor angiogenesis and invasion in a mouse model of multistage cancer. Oncogene. 2005;24(25):4037–4051. doi: 10.1038/sj.onc.1208602. [DOI] [PubMed] [Google Scholar]
- 65.Parish CR, Freeman C, Brown KJ, Francis DJ, Cowden WB. Identification of sulfated oligosaccharide-based inhibitors of tumor growth and metastasis using novel in vitro assays for angiogenesis and heparanase activity. Cancer Res. 1999;59(14):3433–3441. [PubMed] [Google Scholar]
- 66.Nister M, et al. Expression of messenger RNAs for platelet-derived growth factor and transforming growth factor-alpha and their receptors in human malignant glioma cell lines. Cancer Res. 1988;48(14):3910–3918. [PubMed] [Google Scholar]
- 67.Carapancea M, et al. Dual targeting of IGF-1R and PDGFR inhibits proliferation in high-grade gliomas cells and induces radiosensitivity in JNK-1 expressing cells. J Neurooncol. 2007;85(3):245–254. doi: 10.1007/s11060-007-9417-0. [DOI] [PubMed] [Google Scholar]
- 68.Liu TJ, et al. Inhibition of both focal adhesion kinase and insulin-like growth factor-I receptor kinase suppresses glioma proliferation in vitro and in vivo. Mol Cancer Ther. 2007;6(4):1357–1367. doi: 10.1158/1535-7163.MCT-06-0476. [DOI] [PubMed] [Google Scholar]
- 69.Chakravarti A, Loeffler JS, Dyson NJ. Insulin-like growth factor receptor I mediates resistance to anti-epidermal growth factor receptor therapy in primary human glioblastoma cells through continued activation of phosphoinositide 3-kinase signaling. Cancer Res. 2002;62(1):200–207. [PubMed] [Google Scholar]
- 70.Salaita K, et al. Restriction of receptor movement alters cellular response: physical force sensing by EphA2. Science. 2010;327(5971):1380–1385. doi: 10.1126/science.1181729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Wykosky J, Gibo DM, Stanton C, Debinski W. EphA2 as a novel molecular marker and target in glioblastoma multiforme. Mol Cancer Res. 2005;3(10):541–551. doi: 10.1158/1541-7786.MCR-05-0056. [DOI] [PubMed] [Google Scholar]
- 72.Wang LF, et al. Increased expression of EphA2 correlates with adverse outcome in primary and recurrent glioblastoma multiforme patients. Oncol Rep. 2008;19(1):151–156. [PubMed] [Google Scholar]
- 73.Jackson EL, et al. PDGFR alpha-positive B cells are neural stem cells in the adult SVZ that form glioma-like growths in response to increased PDGF signaling. Neuron. 2006;51(2):187–199. doi: 10.1016/j.neuron.2006.06.012. [DOI] [PubMed] [Google Scholar]
- 74.Assanah M, Lochhead R, Ogden A, Bruce J, Goldman J, Canoll P. Glial progenitors in adult white matter are driven to form malignant gliomas by platelet-derived growth factor-expressing retroviruses. J Neurosci. 2006;26(25):6781–6790. doi: 10.1523/JNEUROSCI.0514-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Shih AH, Dai C, Hu X, Rosenblum MK, Koutcher JA, Holland EC. Dose-dependent effects of platelet-derived growth factor-B on glial tumorigenesis. Cancer Res. 2004;64(14):4783–4789. doi: 10.1158/0008-5472.CAN-03-3831. [DOI] [PubMed] [Google Scholar]
- 76.Johansson FK, et al. Identification of candidate cancer-causing genes in mouse brain tumors by retroviral tagging. Proc Natl Acad Sci U S A. 2004;101(31):11334–11337. doi: 10.1073/pnas.0402716101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Johansson FK, Goransson H, Westermark B. Expression analysis of genes involved in brain tumor progression driven by retroviral insertional mutagenesis in mice. Oncogene. 2005;24(24):3896–3905. doi: 10.1038/sj.onc.1208553. [DOI] [PubMed] [Google Scholar]
- 78.Abramsson A, et al. Defective N-sulfation of heparan sulfate proteoglycans limits PDGF-BB binding and pericyte recruitment in vascular development. Genes Dev. 2007;21(3):316–331. doi: 10.1101/gad.398207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Rolny C, Spillmann D, Lindahl U, Claesson-Welsh L. Heparin amplifies platelet-derived growth factor (PDGF)- BB-induced PDGF alpha -receptor but not PDGF beta -receptor tyrosine phosphorylation in heparan sulfate-deficient cells. Effects on signal transduction and biological responses. J Biol Chem. 2002;277(22):19315–19321. doi: 10.1074/jbc.M111805200. [DOI] [PubMed] [Google Scholar]
- 80.Ball SG, Shuttleworth CA, Kielty CM. Vascular endothelial growth factor can signal through platelet-derived growth factor receptors. J Cell Biol. 2007;177(3):489–500. doi: 10.1083/jcb.200608093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Xie J, et al. A role of PDGFRalpha in basal cell carcinoma proliferation. Proc Natl Acad Sci U S A. 2001;98(16):9255–9259. doi: 10.1073/pnas.151173398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Malmstrom J, Westergren-Thorsson G. Heparan sulfate upregulates platelet-derived growth factor receptors on human lung fibroblasts. Glycobiology. 1998;8(12):1149–1155. doi: 10.1093/glycob/8.12.1149. [DOI] [PubMed] [Google Scholar]
- 83.Warrington NM, et al. Spatiotemporal differences in CXCL12 expression and cyclic AMP underlie the unique pattern of optic glioma growth in neurofibromatosis type 1. Cancer Res. 2007;67(18):8588–8595. doi: 10.1158/0008-5472.CAN-06-2220. [DOI] [PubMed] [Google Scholar]
- 84.Welm BE, Dijkgraaf GJ, Bledau AS, Welm AL, Werb Z. Lentiviral transduction of mammary stem cells for analysis of gene function during development and cancer. Cell Stem Cell. 2008;2(1):90–102. doi: 10.1016/j.stem.2007.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Cancer Genome Atlas Research Network. Comprehensive genomic characterization defines human glioblastoma genes and core pathways. Nature. 2008;455(7216):1061–1068. doi: 10.1038/nature07385. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.