Abstract
Myxoid round cell liposarcoma (MRCLS) is a common liposarcoma subtype characterized by a translocation that results in the fusion protein TLS:CHOP as well as by mixed adipocytic histopathology. Both the etiology of MRCLS and the mechanism of action of TLS:CHOP remain poorly understood. It was previously shown that ET-743, an antitumor compound with an unclear mechanism of action, is highly effective in patients with MRCLS. To identify the cellular origin of MRCLS, we engineered a mouse model in which TLS:CHOP was expressed under the control of a mesodermally restricted promoter (Prx1) in a p53-depleted background. This model resembled MRCLS histologically as well as functionally in terms of its specific adipocytic differentiation–based response to ET-743. Specifically, endogenous mesenchymal stem cells (MSCs) expressing TLS:CHOP developed into MRCLS in vivo. Gene expression and microRNA analysis of these MSCs showed that they were committed to adipocytic differentiation, but unable to terminally differentiate. We also explored the method of action of ET-743. ET-743 downregulated TLS:CHOP expression, which correlated with CEBPα expression and adipocytic differentiation. Furthermore, PPARγ agonists enhanced the differentiation process initiated by ET-743. Our work highlights how clinical observations can lead to the generation of a mouse model that recapitulates human disease and may be used to develop rational treatment combinations, such as ET-743 plus PPARγ agonists, for the treatment of MRCLS.
Introduction
Despite the fact that the characteristic translocation of myxoid round cell liposarcoma (MRCLS) that results in the fusion protein TLS:CHOP was characterized almost 20 years ago (1, 2), the etiology of MRCLS — in terms of both cell of origin and the mechanism of action by which the translocation product promotes tumorigenesis — remains elusive. Although a preadipocytic precursor remains the most obvious contender, direct proof for the latter is lacking. Neither transgenic mice expressing TLS:CHOP under the adipocytic specific promoter AP2 (3) nor adipose-derived mesenchymal stem cells (MSCs) engineered to express TLS:CHOP (4) develop liposarcomas. On the contrary, transgenic mice expressing TLS:CHOP under the ubiquitous E1Fα promoter (5) and bone marrow–derived MSCs ex vivo expanded and engineered to express TLS:CHOP (6) give rise to MRCLS. Furthermore, previous work has suggested that TLS:CHOP’s tumorigenicity is related to suppression of PPARγ activity. The PPARγ pathway is active in early adipocytic differentiation (i.e., before AP2 expression; ref. 3), which supports the conjecture that TLS:CHOP exerts its oncogenic effects early in mesenchymal development. On the other hand, our previous work assigning differentiation time points to liposarcomas suggested that MRCLS corresponds to mid-to-late stages of MSC adipocytic differentiation (7). To complicate things further, several reports indicate that the introduction of TLS:CHOP into nonadipocytic cells (e.g., NIH3T3, ref. 8; HT1080, ref. 9; p53null adipose-derived MSCs, ref. 4) does not enhance tumorigenesis, but rather redirects the tumors to phenocopy MRCLS. p53 abnormalities have been historically the most cited secondary genetic events in MRCLS. p53 immunoreactivity has been reported to range from 6.7% to 100%, whereas gene alterations range from 2.8% to 28.5%, with a greater preponderance in the round cell component (10–13) and prognosticating a poor outcome (10). However, the mechanistic relationships between TLS:CHOP and secondary genetic events remain mainly unexamined.
Yet despite the relative lack of understanding of both MRCLS etiology and TLS:CHOP biology, a compound isolated from the sea squirt, Ecteinascidia turbinata (ET-743; also known as ecteinascidin, yondelis, and trabectedin), is highly effective in patients with MRCLS and other sarcomas (14). However, ET-743 has an unclear mechanism of action (15). It has been reported that ET-743 binds the minor groove of DNA and induces the formation of DNA adducts. It is less efficient in cells deficient in transcription-coupled nucleotide excision repair, but more potent in cells deficient in homologous recombination. ET-743 may sterically hinder transcription factor binding, inhibit the in vitro production of the proinflammatory mediators, and modulate the transcriptional activity of specific sarcoma-related fusion genes, as seen not only in MRCLS (15), but also in Ewing’s sarcomas (16, 17). However, only in patients with MRCLS, rare but dramatic in vivo tumor cellular differentiation has been observed (14, 18).
This article begins with a description of our single-institution experience of 42 sarcoma patients treated with ET-743. Our clinical observations, specifically focusing on the dramatic, rapid, and unusual response of one patient with a diagnosis of MRCLS, was the starting point for the translational studies described herein, in which we sought to better delineate the cell of origin of MRCLS as well as the mechanism of action of ET-743. Specifically, we generated a biologically faithful mouse model of MRCLS by genetically engineering transgenic mice expressing TLS:CHOP in a mesodermally restricted pattern in a p53-depleted background. We then used these mice, and supporting in vitro models, to show that ET-743 rapidly differentiated the formed tumor in vivo along the adipocytic lineage, an effect that was enhanced by PPARγ agonists. Finally, using gene expression and microRNA RT-PCR–based profiling, we provided evidence to suggest that TLS:CHOP prematurely commits progenitor cells to an inappropriate lineage, resulting in the initiation of a differentiation program that cannot be terminally executed in that inappropriate state.
Results
ET-743 promotes adipocytic differentiation in a patient with MRCLS.
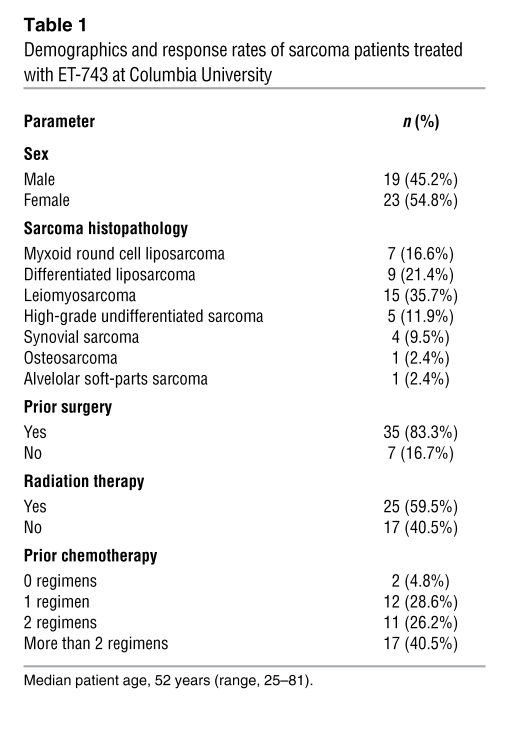
As part of the expanded access and compassionate-use program of ET-743 in the United States, we treated 42 patients with a pathologically confirmed diagnosis of sarcoma with ET-743 between October 2006 and August 2009 (see Table 1, Supplemental Tables 1 and 2, and Supplemental Text; supplemental material available online with this article; doi: 10.1172/JCI60015DS1). As shown in Figure 1A, 1 patient with a diagnosis of abdominal MRCLS (referred to herein as MRCLS patient 1) was alive 3 years after completing 12 cycles of treatment of ET-743 for locally recurrent, inoperable disease. MRCLS patient 1’s abbreviated history (see Supplemental Text for details) is as follows: upon presenting to us and after pathologically confirming recurrence (Figure 1B), the patient underwent baseline imaging (Figure 1C) and started on ET-743 at a dose of 1.5 mg/m2. Upon first reevaluation at 6 weeks (i.e., after 2 3-week cycles), the patient had lost 22 pounds and reported improved energy, improved breath capacity, lessened early satiety, and decreased urinary frequency. Although per RECIST criteria, the dimensions of the previously described mass were unchanged; the density of the mass had changed remarkably, from a density and pattern characteristic of soft tissue to that of air or fat (Figure 1C). Although “progressive decrease of contrast enhancement without tumour shrinkage” has been previously reported (14), this radiologic pattern is highly dramatic, especially compared with other radiologic patterns of patients who achieve prolonged stable disease with ET-743 (Supplemental Figure 1). To better understand the radiologic change, we performed a second biopsy of the mass and found the pathology to be consistent with atypical adipocytes (Figure 1B). MRCLS patient 1 received 10 subsequent cycles of ET-743 without any further radiologic changes. After completing 12 cycles, surgical options to remove the large adipocytic mass were discussed; however, the overwhelming majority opinion of our internal multidisciplinary team, as well as several external sarcoma oncologists and surgeons, was to observe. Although the patient was alive at the conclusion of the present study (i.e., 2 years later), the most recent images confirmed recurrence: the patient’s adipocytic mass was never resorbed and appeared to be reacquiring soft-tissue density (Figure 1C). MRCLS patient 1 is currently being rechallenged with ET-743.
Table 1 .
Demographics and response rates of sarcoma patients treated with ET-743 at Columbia University
Figure 1. ET-743–mediated adipocytic differentiation in MRCLS patient 1.
(A) Survival curve of ET-743–treated patients by major subtype. LMS, leiomyosarcoma; HGUPS, high-grade undifferentiated pleomorphic sarcoma; DDLS, dedifferentiated liposarcoma; Other, synovial sarcoma, osteosarcoma, or alveolar soft parts sarcoma (see Table 1). P = 0.0572, χ2 distribution, 4 degrees of freedom. (B) Histopathology of MRCLS patient 1 before (week 1) and after 2 rounds of (week 6) ET-743 treatment. Original magnification, ×100. (C) CT imaging of MRCLS patient 1 before and after ET-743, as well as 2 years after discontinuing ET-743 (week 158). (D and E) IHC of p53 in MRCLS patient 1 before (D) and after (E) ET-743 treatment. Mutational analysis revealed a TP53 mutation (exon 7, codon 225, Val>Ala), in this patient. (F–K) IHC of the 6 other MRCLS patients in our series, none of which displayed a mutation (Supplemental Table 2). Original magnification, ×200.
Although MRCLS patient 1 had a dramatic differentiation-based response, the other 6 MRCLS patients in this series did not. Since all MRCLS patients were confirmed via split-probe clinical diagnostic testing to detect expressed TLS:CHOP, we sought to assess whether p53 status accounts for the difference in response. Tumor tissue was obtained from the 7 MRCLS cases, paraffin embedded, and stained for p53. Of the 7 MRCLS patients, only tumor tissue from MRCLS patient 1 stained positive for p53 (Figure 1, D–K). Additionally, genomic DNA was extracted from frozen tumor tissues from primary surgeries, and mutational analysis was performed for p53 exons 5–9. In agreement with the immunohistochemistry (IHC), a TP53 mutation (exon 7, codon 225, Val>Ala) was detected only in tumor genomic DNA from MRCLS patient 1 (Supplemental Table 3). Although we cannot rule out the possibility that p53 mutations are present in the nonanalyzed portions of TP53, the mutational and IHC analyses together suggest that TP53 was aberrant only in this patient. Of note, this specific TP53 mutation in MRCLS was previously reported (13).
Modeling MRCLS in a mouse.
ET-743 has been previously reported to promote adipocytic differentiation in MRCLS (14, 18) in both in vitro assays and treated patients, although our results with MRCLS patient 1 represents an extreme example of ET-743’s clinical differentiation capacity. However, in these reports, in vitro differentiation was underwhelming compared with selective differentiation in patients (14, 18). Because substantial problems with mouse models of this disease remain, as discussed above, to our knowledge the in vivo differentiation results observed sporadically in patients have never been methodically modeled in transgenic MRCLS mouse models. Thus, as a first step in studying the differentiation capacity of ET-743 in vivo (either alone or in combination with other adipocytic differentiation–promoting drugs) we generated transgenic mice expressing TLS:CHOP (type II) under the mesoderm-specific promoter Prx1 enhancer element and flanked 5′ by an additional insulator element and 3′ with the SV40 polyadenylation signal (ref. 19 and Supplemental Figure 2A). Prx1-TLS:CHOP transgenic founders (referred to herein as pTC mice) were confirmed by Southern blotting (Supplemental Figure 2B). Expression analysis via RT-PCR showed that TLS:CHOP was detected mainly in appendicular skeletal tissues (i.e., muscle and fat of thigh; Supplemental Figure 2C). pTC mice were viable, had normal life spans, and did not form tumors.
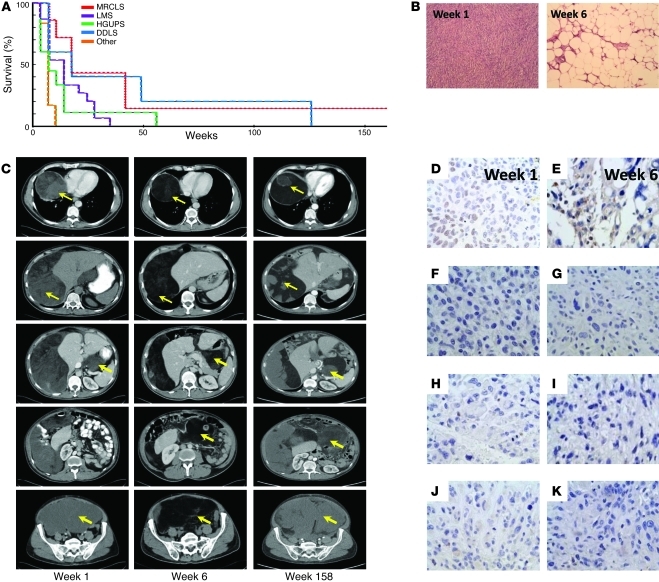
Since p53 mutations are common in MRCLS (10, 11) and, in our series, were specific to MRCLS patient 1, we crossed pTC mice to mice with constitutive deletion of p53 alleles (referred to herein as p53null mice). Progeny of that were pTCp53+/– or pTCp53–/– (referred to as pTCp53het and pTCp53null mice, respectively) were observed for tumor formation compared with parental p53het, p53null, and pTC mice (n = 25 per group). All p53-depleted mice formed tumors and were euthanatized (p53null, average 12 weeks; pTCp53null, average 14 weeks; p53het, average 64 weeks; pTCp53het, average 70 weeks; Figure 2A); the differences in overall tumor occurrence/survival curves were statistically significant (P = 9.5 × 10–52, χ2 distribution, 4 degrees of freedom). p53null mice developed predominantly lymphomas (92%), 66% of which were in a non-Prx1 distribution. pTCp53null mice developed a fairly equal representation of lymphomas and sarcomas (55% and 45%, respectively), with 71% of the sarcomas occurring in an established Prx1 distribution (i.e., appendicular skeleton). p53het mice also developed a fairly equal representation of lymphomas and sarcomas (48% and 52%, respectively); however, only 46% of the sarcomas occurred in an established Prx1 distribution. Finally, pTCp53het mice developed predominantly sarcomas (74%), 76% of which occurred in an established Prx1 distribution (Figure 2A). If TLS:CHOP only changed the phenotype of tumors induced as a result of p53 depletion, the distribution of lymphomas to sarcomas should not change; thus, we conclude that TLS:CHOP expression synergizes with p53 in the formation of MRCLS.
Figure 2. pTCp53null mice give rise to sarcomas.
(A) Survival curve of p53null, pTCp53null, p53het, pTCp53het, and pTC mice (n = 25 per group). Inset pie charts show histopathology occurrence and distribution. Sarc:Prx1-D, sarcoma with Prx1 distribution; Sarc:Non-Prx1-D, sarcoma with non-Prx1 distribution; Lymp:Prx1-D, lymphoma with Prx1 distribution; Lymp:Non-Prx1-D, lymphoma with non-Prx1 distribution. (B) Histopathology (H&E) of sarcomas from p53null, pTCp53null, p53het, and pTCp53het mice. Original magnification, ×100. (C) Gross demonstration of pTCp53null mouse forming a sarcoma and IHC expression of TLS:CHOP. Original magnification, ×200.
Histologically, sarcomas from p53null mice resembled sarcomas from p53het mice, and sarcomas from pTCp53null mice resembled sarcomas from pTCp53het mice. pTCp53het and pTCp53null mice also had a less cellular appearance than their p53 counterparts (Figure 2B). Sarcomas from pTCp53het and pTCp53null mice also occurred more distally than their p53 counterparts and were positive for TLS:CHOP expression, as determined by RT-PCR (Supplemental Figure 3A) and IHC (Figure 2C).
ET-743 differentiation of MRCLS in a transgenic model aided by PPARγ agonists.
Having generated a TLS:CHOP-dependent transgenic mouse model of MRCLS, we sought to test the differentiation potency of ET-743 in an in vivo system, both alone and in combination with the PPARγ agonist rosiglitazone (RSG), which has previously been reported to promote non-MRCLS differentiation (20–22). pTCp53null mice with Prx1 distribution of tumors underwent fine needle aspiration (FNA) and cytological determination of whether the tumor forming was a sarcoma or lymphoma. pTCp53null mice FNA-diagnosed with lymphomas were sacrificed, and those FNA-diagnosed with sarcomas were sequentially assigned to 1 of 4 groups: no treatment, ET-743 treatment (0.05 mg/kg via tail vein every 4 days); RSG treatment (25 mg/kg via lavage daily), or concurrent ET-743 and RSG treatment. All mice were treated until palpable tumor reached 1 cm in greatest dimension. A similar FNA-based approach was used to identify p53null mice developing sarcomas for corresponding controls. 20 mice were assigned to each cohort and followed for survival and/or tumor burden, as defined by tumor greater than 1 cm in longest dimension and/or animal distress. Notably, we purposely chose low ends of the reported spectrum of in vivo usage of these drugs to allow for concurrent usage without exacerbating toxicity (23–25).
We observed 4 survival patterns (P = 6.5 × 10–35, χ2 distribution, 3 degrees of freedom; Figure 3A). The lowest survival rate was observed for untreated and RSG-treated pTCp53null mice (median, 80 days), leading us to conclude that, as observed above, pTCp53null mice developing sarcomas have shorter survival than p53null mice with sarcomas, and RSG treatment does not improve outcomes in either case. Treated and untreated p53null mice developing sarcomas had equal survival rates (median, 99 days), which confirmed that treatment with either ET-743 or RSG in the absence of TLS:CHOP had no effect. Sarcoma-developing pTCp53null mice treated with ET-743 had increased survival (median, 100 days), which suggests that ET-743 has specific efficacy in this model of MRCLS. Finally, sarcoma-developing pTCp53null mice treated with both ET-743 and RSG had the longest survival of all groups (median, 134 days), which suggests that RSG potentiates ET-743 efficacy — despite having no single agent activity, as shown above. Notably, the cause of death in all pTCp53null mice treated with ET-743 with RSG were secondary lymphomas appearing in a non-Prx1 distribution. pTCp53null mice treated with ET-743 alone died from a combination of primary (FNA-determined) sarcoma progression or secondary lymphomas (approximately 1:1 ratio). All untreated and RSG-treated pTCp53null mice and all treated and untreated p53null controls died from the primary (FNA-determined) sarcomas.
Figure 3. RSG enhances ET-743–mediated adipogenesis of sarcomas in pTCp53null mice.
(A) Survival curve of p53null and pTCp53null mice receiving no treatment (NT), ET-743 (0.05 mg/kg via tail vein every 4 days), RSG (25 mg/kg via lavage daily), or both ET-743 and RSG concurrently (n = 20 per group). Treatment was initiated shortly after pathologic confirmation (tumor approximately 0.25–0.5 cm in greatest dimension) and continued until tumor was approximately 1 cm in greatest dimension or until mice showed signs of distress. On average, treatment lasted for 4–8 weeks, depending on the extent of response. All 4 survival curves for p53null mice were completely superimposable. Original magnification, ×100. (B) Quantitative RT-PCR analysis (average of 3 replicate experiments) of TLS:CHOP in p53null and pTCp53null mice, assessed at sacrifice. Values are shown relative to untreated pTCp53null group. (C) Transcriptional run-on assay from short-term single-cell suspensions (in vitro 24-hour cultures) of sarcomas from p53null and pTCp53null mice (see Methods) after in vivo treatment as in A, reflecting transcriptional activity of TLS:CHOP (from the Prx1 promoter) as well as PPARγ, CEBPα, and GADPH (from their respective endogenous promoters). (D and E) RT-PCR (average of 3 replicate experiments) of the indicated adipocytic differentiation gene set in bone marrow–derived MSCs of 8-week-old WT, p53null, and pTCp53null mice (D) or in pTCp53null mice treated as in A for 4 weeks (E). Values are shown relative to respective WT or untreated control.
Tumors were pathologically analyzed and confirmed for TLS:CHOP transgene expression via RT-PCR (Supplemental Figure 3A). All treated and untreated p53null and RSG-treated pTCp53null mice developing sarcomas were histologically identical to the sarcomas of the same groups in Figure 2B. However, sarcoma-developing pTCp53null mice treated with ET-743 showed dramatic adipocytic differentiation, which was further enhanced by concurrent treatment with RSG (Figure 3A). We subsequently performed pilot experiments using cohorts of 5 mice treated with ET-743 (5 mg/kg), RSG (150 mg/kg), or both concurrently. Substantial toxicity and early mouse death was observed, without a notable increase in either differentiation or cytotoxic effect. We also performed sequential combinations (ET-743 for 2 weeks before starting RSG, as well as RSG for 2 weeks before starting ET-743) of cohorts of 5 mice; as above, we did not observe a marked improvement in either differentiation or tumor cytotoxicity. However, we cannot rule out the possibility that some intermediate dose or an alternative dosing schedule may result in different outcomes.
As a first step in elucidating the mechanism of differentiation induction after ET-743 treatment without and with RSG, we assayed TLS:CHOP expression in the recovered tumors via quantitative RT-PCR. Compared with untreated and RSG-treated sarcoma-developing pTCp53null mice, sarcoma-developing pTCp53null mice treated with ET-743 alone had 40% less TLS:CHOP expression, and concurrent RSG treatment resulted in 65% less TLS:CHOP expression (Figure 3B). Because complete elimination of TLS:CHOP should approximate quantitative RT-PCR values obtained from p53null mice, which do not express TLS:CHOP, we concluded that ET-743–mediated adipocytic differentiation correlates with TLS:CHOP downregulation, and the effect is further enhanced in the presence of RSG. TLS:CHOP downregulation has been previously reported in 1 of 2 MRCLS cell lines examined after short-term high-dose in vitro treatment with ET-743 (18). Our demonstration of ET-743–mediated TLS:CHOP downregulation in vivo may reflect the prolonged treatment schedule used, which may also explain the dramatic differentiation potential induced by ET-743 we observed in vivo, as opposed to the relatively mild potential previously observed in vitro.
To exclude the possibility that TLS:CHOP mRNA downregulation reflects adipogenic differentiation–mediated downregulation of the Prx1 promoter, we performed transcriptional run-on assays on single-cell suspensions of cells isolated from the tumors of mice treated as above. Transcriptional run-on assays reflect a gene’s transcriptional activity, rather than steady-state levels (see Methods for details). As seen in Figure 3C, cells from sarcomas forming in p53null mice did not show any detectable expression of synthesis of PPARγ, CEBPα, or TLS:CHOP, whereas GADPH showed relatively stable transcriptional activity in the presence of treatment with ET-743, RSG, or the combination. On the contrary, in cells from sarcomas of pTCp53null mice, TLS:CHOP transcriptional activity was detectable in untreated cells, whereas treatment with ET-743, RSG, or both concurrently did not change TLS:CHOP activity (identical to results with GADPH control). However, treatment with RSG did result in detectable upregulation in PPARγ transcription, and combination treatment with RSG and ET-743 resulted in further upregulation of both PPARγ and CEBPα, but once again without a detectable change in TLS:CHOP transcription. However, the obvious decline in TLS:CHOP RNA, as measured by RT-PCR (Figure 3B), leads us to suggest that ET-743 regulates TLS:CHOP posttranscriptionally and independently of the Prx1 promoter. Downregulation of TLS:CHOP at the protein level was also observed using isolated MSCs from p53null mice in vitro that were infected with a CMV promoter–based lentivector expressing TLS:CHOP and treated with ET-743 (Supplemental Text and Supplemental Figure 4), which further validated our conclusion that ET-743 regulation of TLS:CHOP is promoter/transcriptionally independent.
It is currently unclear whether TLS:CHOP inhibits differentiation of adipogenic committed precursors or commits mesenchymal precursors to an adipogenic differentiation program that is incapable of being completed. Given our previous demonstration that MRCLS corresponds to moderately differentiated MSCs differentiating along the adipocytic lineage (7), we sought to determine the baseline adipocytic differentiation of 52 genes in mouse TLS:CHOP-expressing tumors, using a high-throughput quantitative RT-PCR platform (Figure 3D and Supplemental Table 4). MSCs were extracted and RNA isolated from either WT, p53null, or pTCp53null mice (Supplemental Figure 5) at approximately 8 weeks of age. Based on the 79 adipocytic gene differentiation set previously described by us (7) for differentiating human MSCs, we here focused specifically on genes with direct mouse homologs (56 of 79) and for which quantitative RT-PCR primers/conditions could be worked out (52 of 56). Of the 52 adipocytic genes examined herein, 17 (33%) had significantly higher levels in pTCp53 tumors than in bone marrow–derived MSCs from WT mice or in p53 tumors; only 1 gene, KRT18, showed repeatedly lower levels in pTCp53 mice. This observation was in agreement with the overall pattern of baseline adipocytic differentiation (and thus loss of epithelial markers) seen in even the untreated tumors of pTCp53 mice. Thus, as we observed previously in human MRCLS (7), expression of TLS:CHOP in our mouse model at baseline promoted adipocytic differentiation.
We next examined the effect of treating mice with ET-743 and RSG using the above-described adipocytic gene expression panel (Figure 3E). Mice were left untreated or treated as described above (n = 10 per group) for 4 weeks from initial tumor observation and then sacrificed, after which tumor RNA was recovered and analyzed via RT-PCR for relative expression. Treatment with ET-743 directly increased the expression pattern of some adipocytic genes dramatically (e.g., COL11A1); for others, partial increases were observed that were enhanced in the presence of RSG (e.g., FABP4). However, no gene expression increased with RSG alone. These data support the in vivo response data (Figure 3A); together, they suggest that RSG as a single agent cannot initiate an adipocytic program in MRCLS, but can accelerate an adipocytic differentiation program initiated by ET-743. This is also consistent with the data in Figure 3B, which demonstrated that only ET-743 initiated downregulation of TLS:CHOP, and RSG further enhanced the effect.
ET-743 induces CEBPα alone, and ET-743 in conjunction with RSG induces PPARγ.
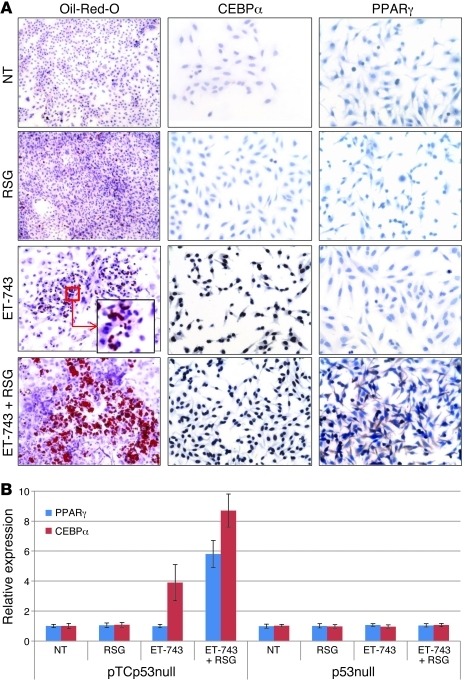
The above data suggested that TLS:CHOP both promotes adipocytic differentiation (as TLS:CHOP-expressing sarcomas were histologically more fatty and had more and higher expression of adipocytic genes) and simultaneously suppresses it (as treatment with ET-743 and PPARγ decreased TLS:CHOP and allowed for further adipocytic development). To better understand the effect of TLS:CHOP in giving rise to sarcomas, we isolated sarcoma precursors (i.e., MSCs; refs. 6, 26–28) from the bone marrow of sarcoma-developing pTCp53null mice (with confirmed TLS:CHOP expression; see Supplemental Figure 5) and further cultured them in the presence of adipocytic differentiation medium (ADM), either alone or in combination with RSG (1 μM), ET-743 (1 nM), or both ET-743 and RSG. Neither ADM nor ADM plus RSG induced any noticeable adipocytic differentiation, as assayed via Oil Red O staining for fat formation. However, ADM plus ET-743 induced some degree of adipocytic differentiation, an effect that was greatly enhanced when RSG was added to the combination of ADM and ET-743 (Figure 4A). IHC staining for PPARγ and CEBPα showed that ADM plus ET-743 induced only CEBPα, but not PPARγ, whereas ADM plus RSG did not induce CEBPα or PPARγ. However, the combination of ADM, ET-743, and PPARγ in these sarcoma precursors was able to induce both CEBPα and PPARγ (Figure 4A). Notably, PPARγ showed a mixed cytoplasmic/nuclear pattern, characteristic of an activated PPARγ pathway (29). Taken together, these data suggest that RSG is able to induce PPARγ expression only in the presence of ET-743–induced CEBPα.
Figure 4. ET-743 alone induces CEBPα, and ET-743 in conjunction with RSG induces PPARγ.
(A) MSCs from pTCp53null mice were treated with ET-743 or RSG in addition to ADM for 21 days. Drugs were changed with medium every 3 days. At the end of 21 days, cells were either stained with Oil Red O (to visualize lipid deposition) or analyzed via IHC for CEBPα and PPARγ expression. Original magnification, ×100 (Oil Red O); ×200 (IHC); ×1,000 (inset). (B) MSCs from pTCp53null and p53null mice were transfected with reporter assays for CEBPα and PPARγ after 21 days of differentiation as in A. Values denote average of 3 replicate experiments (see Methods).
To validate these IHC observations, we transfected MSCs from pTCp53null mice separately with transcriptional reporter vectors for CEBPα and PPARγ and treated them as described above. Treatment with ADM plus ET-743 caused CEBPα induction, whereas combination treatment with ADM, ET-743, and RSG resulted in induction of PPARγ and CEBPα, in agreement with the IHC data. Of note, MSCs from p53null mice showed no changes in these genes after identical treatment (Figure 4B).
MicroRNA patterns in TLS:CHOP tumors relate to stage of differentiation.
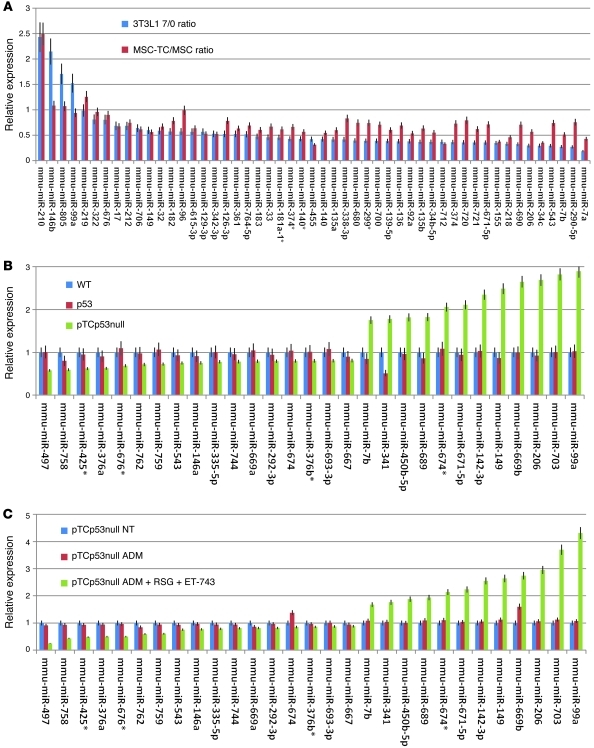
Since we previously showed that gene expression of MRCLS tumors corresponds to moderate degrees of adipocytic differentiation (7), we next used microRNAs in order to establish the degree of adipocytic differentiation, in a manner we believe to be novel. We first examined whether a previously identified set of microRNAs differentially expressed in 3T3L1 fibroblasts undergoing adipocytic differentiation (30) trended similarly in MSCs from pTCp53 compared with p53null mice. We identified 52 of 386 microRNAs previously assayed as being differentially expressed (P < 0.05) as a function of adipocytic differentiation. We then used a microRNA PCR array (see Methods) to assess whether those microRNAs trended similarly in the TLS:CHOP-expressing tumors. Expression data are presented as ratios of before treatment/after treatment values; thus, values greater than 1 indicate microRNAs that increased as a function of 3T3L1 differentiation or in the presence of TLS:CHOP (compared with MSCs), whereas values less than 1 reflect microRNAs that decreased during differentiation or in the presence of TLS:CHOP. Furthermore, if differentiation trended with TLS:CHOP expression, both values would be either above or below 1. Of the 52 microRNAs, 47 (90.3%) that decreased or increased with adipocytic differentiation in 3T3L1 cells were similarly modulated in MSCs when TLS:CHOP was expressed (Figure 5A). Only miR99a did not sustain a trend with TLS:CHOP expression, and the difference in miR146b, mir805, miR219, and miR96 were not statistically sound to claim correlation. This finding suggests that TLS:CHOP promotes adipocytic differentiation.
Figure 5. MicroRNA patterns in TLS:CHOP tumors relate to differentiation stage.
(A) MicroRNA expression, shown as the ratio of expression in 3T3L1 cells after 7 days relative to before differentiation (3T3L1 7/0 ratio; blue) compared with the ratio of expression in pTCp53 relative to WT MSCs (MSC-TC/MSC ratio; red). (B and C) microRNA expression of an adipocyte-specific differentiation microRNA panel in WT bone marrow–derived MSCs and p53null and pTCp53null sarcomas (B) or in pTCp53null mice left untreated or treated for 22 days with ADM or with the combination of ADM, RSG, and ET-743 (C), expressed relative to respective WT or untreated control. Values in A–C denote average of 3 replicate experiments (see Methods).
To more specifically assess the degree of adipocytic differentiation as measured by microRNA expression patterns of tumors from pTCp53null mice, we differentiated WT bone marrow–derived MSCs along the adipocytic lineage (26) and isolated total cellular RNA prior to (day 0), midway through (day 11), and at the end of (day 22) the differentiation protocol. Global microRNA expression was assessed using a mouse microRNA PCR array (see Methods), and 26 differentially expressed microRNAs (P < 0.05, ANOVA) were found to trend either directly or inversely with adipocytic differentiation (Supplemental Figure 6), as we previously described for gene expression differentiation (7). We then used this panel of 26 microRNAs to similarly assess their relative expression in MSCs from WT, p53null, and pTCp53null mice. Only miR341 was differentially expressed when MSCs from WT and p53null mice were compared; however, MSCs from pTCp53null mice showed substantial decreases or increases in the corresponding microRNAs compared with MSCs from either WT or p53null mice (Figure 5B). Furthermore, the latter increases or decreases corresponded perfectly to the patterns of those microRNAs seen during adipocytic differentiation (Supplemental Figure 6).
We next treated MSCs from pTCp53null mice either with ADM or with the combination of ADM, ET-743, and RSG for 22 days and assayed for expression levels of the 26-microRNA adipocytic differentiation set. In agreement with previous data showing that ex vivo exogenous introduction of TLS:CHOP into MSCs blocks adipocytic differentiation (6), MSCs from pTCp53null mice were also blocked (Supplemental Figure 7). However, treatment of pTCp53null mouse MSCs with the combination of ADM, ET-743, and RSG overcame this block and allowed for appropriate modulation (i.e., either increasing or decreasing microRNAs in the direction observed for adipocytic differentiation) of the 26 adipocyte-specific microRNAs (Figure 5C).
MSCs expressing TLS:CHOP are potential MRCLS precursors.
To assess whether MSCs from pTCp53null mice are precursors of MRCLS, we inoculated 1 × 106 ex vivo–expanded MSCs from pTCp53null and p53null mice subcutaneously into NOD-SCID-Gamma (NSG) mice. We also inoculated MSCs from pTC mice, but these (just as the parental mice) did not form tumors. MSCs from pTCp53null mice formed tumors rapidly and histopathologically resembled MRCLS, with TLS:CHOP detectable via IHC (Figure 6A). Xenografted mice were left untreated or treated with RSG, ET-743, or both ET-743 and RSG as described above, starting when a tumor nodule was palpable (i.e., 2–3 mm in diameter; n = 10 per group; Figure 6B). In line with our observations in the transgenic mouse models, only tumors forming from pTCp53 mouse–derived MSCs grew more slowly in the presence of ET-743, an effect that was greatly enhanced by the addition of RSG (Figure 6B). Histopathology of treated pTCp53null tumors showed adipocytic differentiation induction that was characteristic of transgenic animals (Figure 6C). Taken together, these data indicated that MSCs from pTCp53null mice can phenotypically and functionally (in terms of adipocytic differentiation response) recapitulate tumors observed in their transgenic counterparts, which suggests that MSCs from pTSp53null mice may be the progenitors of MRCLS.
Figure 6. MSCs expressing TLS:CHOP are potential precursors of MRCLS.
(A) Gross (left), H&E (middle), and IHC (right) positivity for TLS:CHOP of MSCs from pTCp53null mice injected subcutaneously into NSG mice. Arrow denotes developing tumor. Original magnification, ×200. (B) NSG mice were xenografted with MSCs from either pTCp53null or p53null mice, divided into cohorts of 10, and treated as in Figure 3 beginning when a tumor nodule was palpable (2–3 mm diameter; assigned as day 1). (C) Histopathology (H&E) of xenografts of MSCs from pTCp53null mice after treatment with RSG and/or ET-743. Original magnification, ×100.
Discussion
Here, we built on our clinical observations of the rapid and complete terminal adipocytic differentiation of one patient’s MRCLS after a brief treatment with ET-743 and made several insights into MRCLS etiology, the means by which ET-743 promotes differentiation specifically in MRCLS, and the enhancement of ET-743’s differentiation effect by PPARγ agonists. We engineered a mouse model of MRCLS in which the characteristic MRCLS translocation product, TLS:CHOP, was expressed under the control of a mesodermally restricted promoter, Prx1, in a p53-depleted background. This model was demonstrated to resemble MRCLS not only histologically, but also functionally, in terms of its specific adipocytic differentiation–based response to ET-743. We also showed that MSCs expressing TLS:CHOP developed into MRCLS in vivo, which suggests that MSCs harboring TLS:CHOP translocations are the precursors of MRCLS. Finally, we demonstrated, via both gene expression and microRNA profiling, that TLS:CHOP-expressing MSCs were simultaneously committed to adipocytic differentiation and blocked from executing the terminal differentiation program. Using the above model, we both confirmed the work done in MRCLS and extended it to the transgenic mouse setting, showing that ET-743 downregulated TLS:CHOP expression, that TLS:CHOP downregulation correlated with CEBPα expression and adipocytic differentiation, and that once the adipocytic differentiation process was reinitiated by ET-743, it could be subsequently accelerated by PPARγ.
The underlying mechanism of ET-743 is poorly understood (15), and the specific differentiation effect of ET-743 in MRCLS is likely the fortuitous coincidence of several biological occurrences. First, MRCLS is characterized by a fusion protein, TLS:CHOP, that inhibits adipocytic differentiation–promoting transcriptional programs (3, 18, 31). Second, ET-743 promotes TLS:CHOP dissociation from chromatin (18), and, as our data suggest, even posttranscriptional downregulation of TLS:CHOP. The specific mechanism by which ET-743 promotes posttranscriptional regulation of TLS:CHOP is currently unclear. One possibility, purely based on the initial characterization of TLS as both a DNA- and an RNA-binding protein (32), is that displacement of TLS:CHOP from DNA via ET-743 leads to TLS:CHOP binding of RNA (including possibly TLS:CHOP mRNA), which may affect RNA stability — especially in a p53-deficient background, because p53 also has dual DNA- and RNA-binding properties. Another possibility is that the differentiation and changes in microRNAs that result from TLS:CHOP displacement by ET-743 result in a specific microRNA that posttranscriptionally downregulates TLS:CHOP. Specifically, searching for microRNAs (miRbase database; http://www.mirbase.org) that may target the 200-bp area flanking the TLS:CHOP (type II) breakpoint identifies several hundred microRNAs, at least 1 of which was upregulated during ET-743–mediated differentiation of MRCLS (mmu-miR-674*; Figure 5C). Whether this microRNA or others can target TLS:CHOP specifically remains to be formally explored. However, it is interesting to highlight that an appreciation of the specificity of these points (a) explains the our observation that ET-743 did not show similar responses in other types of liposarcomas that do not express TLS:CHOP; (b) explains the lack of PPARγ agonist efficacy in MRCLS in the absence of ET-743, given that PPARγ agonists could not initiate TLS:CHOP downregulation in our studies; and (c) underlies the rationale for the combination of ET-743 presented here, in which ET-743 downregulation of TLS:CHOP and reinitiation of an adipocytic program was greatly enhanced by PPARγ agonists.
Our work highlights how this mouse model functionally and genetically recapitulating human MRCLS may be used effectively to test therapeutic combinations. Despite the success of ET-743 here and elsewhere (Figure 1 and refs. 14, 33), it is important to consider that such successes are rare. MRCLS patient 1 is the only patient of our set of 42, and the only MRCLS patient of 7, that is currently alive. PPARγ agonists have been previously explored in a phase II liposarcoma-specific trial without significant success (34), despite extremely promising early clinical (34) and preclinical (20) data. Although the hepatic toxicity observed with PPARγ agonists may have limited their clinical efficacy, our demonstration here that low doses in combination with ET-743 were sufficient to promote differentiation may overcome this limitation. However, because ET-743 also has notable liver toxicity, dose-exploratory and safety trials for the combination would still need to be done, even in the specific MRCLS population. It is possible that the transgenic mouse model we have generated only recapitulates a small percentage of patients: those deficient in TLS:CHOP and p53, specifically those that would respond dramatically to ET-743 without necessitating treatment with PPARγ agonists. In fact, a detailed examination of receptor tyrosine kinases and their downstream signaling pathways in MRCLS showed that RET and MET activation seems to be critical for MRCLS progression, with AKT activation being predominant in the round cell variant (35). The latter finding underlies the genetic heterogeneity of even specific cancer subtypes and suggests a potential need for generation of other, equally biologically faithful mouse models. Because a previously described MRCLS mouse model under the ubiquitous E1Fα promoter does not require TP53 depletion (5), it would be worth examining whether TLS:CHOP expression in nonprimary mesodermal progenitors and the resultant tumors are equally sensitive to ET-743 with or without RSG.
Will p53 predict differentiation-based responsiveness in MRCLS patients, as suggested by the model? Sarcoma cell lines with p53 mutations have previously shown greater cytotoxic ET-743 sensitivity than their WT counterparts (36, 37). However, retrospective clinical data linking p53 mutations in MRCLS patients to response, especially differentiation-based response, is currently lacking. Furthermore, the ET-743–based response observed for MRLCS seems to be particularly limited to TLS:CHOP type II MRCLS (14, 18, 24, 33), and the mechanism for the preferential response of type II versus other MRCLS types remains unclear. Our data suggesting that TLS:CHOP is posttranscriptionally regulated by ET-743 may set the stage to examine whether ET-743 similarly regulates other TLS-CHOP types and whether the difference in that regulation underlies the preferential response in TLS:CHOP type II MRCLS. In light of the posttranscriptional mechanism of action suggested by our present findings, examination of microRNA patterns in the various human MRCLS subtypes may be helpful in translating these mouse-based studies into a prognostic or even therapeutic modality.
Because MRCLS patient 1, and other similar patients, were not treated with PPARγ agonists, we cannot fully explain their specific responses as opposed to those of other MRCLS patients — other than to suggest, as above, that p53 deficiency may account for it. Yet we hope that the combination of ET-743 and a PPARγ agonist in MRCLS may make all patients respond as MRCLS patient 1 did. Furthermore, we suggest that the combination may result in more durable — if not complete — cures, as none of our pTCp53null mice treated with the combination died from sarcoma progression. This observation raises the possibility that had MRCLS patient 1 been treated with ET-743 and a PPARγ agonist, he may not have recurred. Although more work needs to be done, including the generation of a model in which TLS:CHOP is expressed and p53 is deleted specifically within the mesodermal lineage, our present work and the models generated herein suggest strongly that ET-743 and a PPARγ agonist would make a rational therapeutic combination in MRCLS.
Methods
Generation of transgenic animals, husbandry and genotyping, and drug treatment.
Transgenic mice were generated at the Transgenic Mouse Facility at the Irving Cancer Research Center (ICRC), Columbia University Cancer Center. The TLS:CHOP type II (gift of J.C. Tercero, PharmaMar, Madrid, Spain) was subcloned into an EcoRI site downstream of the Prx1 promoter (38), provided by C. Tabin (Harvard University, Cambridge, Massachusetts, USA). The Prx1 enhancer element is flanked 5′ with an insulator element derived from the chicken β-globin domain to protect against position effects at the site of transgene integration and 3′ by an SV40 polyadenylation signal (19). The construct was microinjected into fertilized eggs from a B6×B6 cross. Transgenic mice were initially identified via Southern analysis using a PCR generated using the following primers: Prx1 forward, TAGATCGTAGAGAGCC; TLS:CHOP reverse, ACTGAGTTCCATAGCCTG (~900 bp; Supplemental Figure 2). Founder mice were bred to WT B6 mice to generate subsequent generations. The founder line expressing the highest level of TLS:CHOP was chosen for further study. Subsequent generations were screened via PCR. pTC mice were subsequently crossed to p53+/– mice (Jackson Labs) and p53–/– mice (generated via p53+/– crossing). Mice were kept in pathogen-free housing at the ICRC animal facility. For genotyping, DNA was extracted from 5-mm mouse tails clipped from 2-week-old animals. Briefly, tails were incubated overnight in a mixture of Cell Lysis Solution (Qiagen) and proteinase K (Roche); DNA was precipitated using 100% ethanol dissolved in ultrapure water and assessed for concentration.
ET-743 (provided by PharmaMar) was prepared as a 1-mg/ml stock solution in absolute ethanol (stored at –20°C), further diluted with phosphate buffer (pH 4.0) immediately before administration, and given at 0.05 mg/kg via tail vein every 4 days. RSG (25 mg/kg; Cayman Chemicals) was given via lavage daily.
IHC and Western blot.
Tumor tissue samples were fixed in 10% buffered formalin for 24 hours, then embedded in paraffin blocks for further IHC assays. Sections were deparaffinized and then incubated in PBS containing 10% normal horse for at least 30 minutes. Primary mouse monoclonal antibodies to p53 (CalBiochem), CHOP (Santa Cruz), CEBPα (Abnova), and PPARγ (Abcam) were used. Sections were washed with PBS, after which slides were incubated with secondary anti-mouse antibodies (Vector Laboratories) at 1:500 dilution for 30 minutes, were washed with PBS, and incubated with avidin-biotin peroxidase complexes (Vector laboratories) at 1:25 dilution for 30 minutes. The peroxidase reaction was visualized by incubation in DAB (Sigma-Aldrich): 5 mg in 100 ml 0.5% Tris buffer with peroxide. Slides were counterstained in hematoxylin and mounted.
Isolation of bone marrow–derived progenitor cells.
WT, p53null, and pTCp53null mice were sacrificed at 8–12 weeks of age by cervical dislocation and cleaned thoroughly with ethanol. The lower limbs were dissected, and all connective tissue were carefully removed to expose the femur and tibia. Bone marrow was flushed with a 25-gauge needle into serum media (MEM-α with 20% FBS, 100 U/ml penicillin, and 100 μg/ml streptomycin; Invitrogen). A single-cell suspension was made by repeated aspiration and flushing of the bone marrow cells through the 25-gauge needle. Cells were allowed to attach to the surface of a cell culture plate for 72 hours, after which the media was replaced with fresh serum media. Cells were further expanded after growth for 14 days.
Reporter assays.
To assay for CEBPα and PPARγ expression, Cignal PPAR Reporter (luc) Kit: CCS-3026L and Cignal C/EBP Reporter (luc) Kit: CCS-001L (both SAS Biosciences), respectively, were used according to the manufacturer’s instructions. Specifically, MSCs from pTCp53null and p53null mice (isolated as described above) were grown to 75% confluence in 96-well culture plates. A transfection complex containing the SureFeECT transfection reagent and the reporter assay was then added to each well. Both a negative control (a GFP reporter construct in which GFP expression was controlled by a minimal promoter) and a positive control (a constitutively expressing GFP construct) were included. After 16 hours of transfection medium was changed to growth medium. After 24 hours of transfection, cells were treated with ET-743 and RSG, either alone or in combination. Dual Luciferase assay (Promega) was performed 18 hours after treatment; promoter activity values were expressed as arbitrary units using a Renilla reporter for internal normalization. All experiments including controls were done in triplicate, and the whole experiment was performed 3 separate times using different batches of MSCs.
microRNA analysis.
Mouse miRNome microRNA PCR Array (SAS Biosciences) was used per the manufacturer’s instructions. Specifically, total RNA was extracted from the tissue using the QIAzol Lysis Reagent, and the miRNeasy Mini Kit (Qiagen) was subsequently used to isolate small RNAs from samples. 100 ng of starting material small RNA were then reverse transcribed into first strand cDNA, the template for the PCR, using the RT2 microRNA First Strand Kit. The template was then combined with RT2 SYBR Green qPCR Mastermix and aliquoted into each well of The Mouse miRNome Complete (384-well) RTβ microRNA PCR Array profiles, in which each well contains a specific primer for each of the 579 most abundantly expressed and characterized microRNA sequences in the mouse microRNA genome as well as a universal primer. Quantitative PCR was performed according to the manufacturer’s instructions using the Stratagene Mx3005P Unit. Data were analyzed using the Stratagene MxPro Software Package. Each experiment was performed in triplicate.
Quantitative RT-PCR.
High-throughput quantitative RT-PCR was performed following the protocols and reagents of Mouse Adipogenesis PCR Array Kit (SAS Biosciences) modified to use the primer sets for the designated genes (Supplemental Table 4). For TLS:CHOP, total RNA was extracted from the indicated cell lines using an RNA extraction kit (Qiagen) according to the manufacturer’s instructions. 1 μg RNA was transcribed into cDNA using Super-Script III First Strand Synthesis System for RT-PCR (Invitrogen). RT-PCR reactions containing 500 ng cDNA, QuantiTect SYBR Green PCR mix (Qiagen), and corresponding primers were performed on a Stratagene MX3000P QPCR System with ROX reference dye (Stratagene). Primers for detection of TLS:CHOP fusion expression were as follows: forward, CAGCCAGCAGCCTAGCTATG; reverse, TGTCCCGAAGGAGAAAGGCAATG (10). Expression levels of the aforementioned genes were normalized to expression levels of the β-actin control gene for each sample. All experiments were performed twice, each time in triplicate. 1-tailed Student’s t test was used to assess the significance of differences in expression between indicated treatments. Mouse GAPDH was used as a normalizer: forward primer, 5′-CGGAGTCAACGGATTTGGTCGTAT-3′; reverse primer, 5′-AGCCTTCTCCATGGTGGTGAAGAC-3′.
Transcriptional nuclear run-on assays.
Assessment of transcriptional rates were performed using nuclear run-on assays following the protocol described in detail by Smale (39), with the following modifications. Sarcomas forming in either p53null or pTCp53null mice were subjected to 4 weeks of: no treatment, ET-743 (0.05 mg/kg via tail vein every 4 days), RSG (25 mg/kg via lavage daily), or both ET-743 and RSG concurrently. The last dose of drug was given on the day mice were sacrificed. 5 tumors per cohort were collected, and single-cell suspensions were generated by mincing with a scalpel, 0.25% trypsin digestion, and filtration through sterile stainless mesh. Cells were counted, and 108 cells were taken for further analysis. Cells were pelleted and chilled (resulting in polymerase pausing on actively transcribed genes), and nuclear extracts were isolated following lysing of the cytoplasm with NP40 lysis buffer. 150 μC of 3,000 Ci/mmol (10 mCi/ml) [α-32P]UTP was added to each tube along with an elongation buffer, resulting in elongation of radioactively labeled, actively transcribing genes. The entire procedure, from in vivo drug exposure to radioactive labeling of actively transcribing genes, was less than 12 hours. RNA was further purified as described previously (39) and hybridized to nitrocellulose membranes immobilized in a slot-blot with dsDNA containing probes for PPARγ (1 probe per gel). Gels were aligned on film and exposed. For the PPARγ probe, PCR product (primers TCTGAGTCTGTATGGAGTGACAT and GGCAATCCGTCTTCATCCACA, ~300 bp) was cloned from differentiated mouse MSCs along the adipocytic lineage corresponding to the 5′ end, cloned into topoTA vector, and linearized. For the CEBPα probe, PCR product (primers CAGTTGGGGCACTGGGTGGGC and GCACCGCGATGTTGTTGCGT, ~800 bp) was cloned from differentiated mouse MSCs along the adipocytic lineage corresponding to the 5′ end, cloned into topoTA vector, and linearized. For the GADPH probe, PCR product (primers TCGTCCCGTAGACAAAATGG and TTGAGGTCAATGAAGGGGTC, ~150 bp) were cloned from differentiated mouse MSCs along the adipocytic lineage corresponding to the 5′ end, cloned into topoTA vector, and linearized. For the TLS:CHOP probe, primers were as described above for RT-PCR, with the exception that the product was isolated, cloned into topoTA vector, and linearized.
Generating xenografts.
Cohorts of NOD/SCIDIL2Rg mice (Jackson Labs) were subcutaneously injected in the 2 flanks (1 × 106 cells/injection, 4 injections/mouse). Experiments were deemed concluded when the xenografts of untreated control recipients reached the largest cross-sectional diameter of 10–12 mm (or a tumor cross-sectional area of 100 mm2). Mice were randomized to no treatment control, or treatments indicated in Results. Each experimental group consisted of 5 animals. The entire experiment as depicted herein was performed 3 times; thus, 15 animals were included per arm in the final analysis shown in Figure 6. Tumor size, measured individually twice per week with microcalipers, was calculated as a cross-sectional area defined as length × width. Individual relative tumor size was calculated as the ratio of tumor size at initiation of therapy to that on the given day. Statistical analyses were performed using 1-way ANOVA for comparison of tumor growth in different treatment groups.
In vitro adipocytic differentiation.
Cells were seeded in 6-well plates at a density of 1 × 105 cells; at approximately 80% confluence, growth media was replaced with osteogenic medium or ADM (0.5 mM dexamethasone, 0.5 mM IMBX, and 50 μM indomethacin in complete culture media, changed biweekly for 21 or 22 days as indicated in the text). Cells were stained with Oil O Red. Where indicated, ADM was supplemented with ET-743 (1 nM) and/or RSG (1 μM).
Genotype analysis of patient biopsies.
We treated 42 patients as part of the phase III trial “A multicenter, open-label single-arm study of yondelis (Trabectedin, ET-743) for subjects with locally advanced or metastatic soft tissue sarcoma who have relapsed or are refractory to standard of care treatment” (ClinicalTrials.gov ID NCT00210665; Johnson and Johnson protocol no. ET743-SAR-3002). ET-743 (trabectedin, i.v. formulation) was provided by Johnson and Johnson Pharmaceutical Research and Development. See Table 1, Supplemental Tables 1 and 2, and Supplemental Text for details. Genomic DNA was extracted from biopsied tissue using standard proteinase K digestion and phenol/chloroform extraction methods. Single-strand conformation polymorphism and direct sequencing p53 (exons 5–9) were carried out using primer sequences and PCR conditions as previously described (40).
Histopathology of mouse tumors.
All mice were sacrificed within 24 hours of final treatment, and tumors were surgically excised, fixed in 10% formalin, embedded in paraffin, and stained with H&E as previously described (41).
Statistics.
Survival studies were performed with 20 animals per treatment group. We used χ2 analysis to compare the multiple groups. All data were analyzed on a Dell computer using the Prism 3.0 software package (GraphPad Software Inc.). The Bonferroni correction was applied to all multiple comparisons. All other data (e.g., microRNA, mRNA, and RT-PCR) are presented as mean ± SEM.
Study approval.
Clinical trial NCT00210665 and analysis of patient biopsy and tissue were approved by the IRB of Columbia University (nos. AAAB5863 and AAAA5213, respectively). Experiments using mice were approved by the IUCAC of Columbia University (no. AAAD9669). Patient consent was obtained prior to trial enrollment or biopsy.
Note added in proof.
After this manuscript was accepted for publication, MRCLS patient 1 underwent surgical resection of a 50-lb mass consisting of tissue that was pathologically classified as both MRCLS and normal fat.
Supplementary Material
Acknowledgments
The authors thank H. Ginsberg (Columbia University) for helpful discussions, C. Tercero for the TLS:CHOP vector, C. Tabin for the Prx1 promoter, and S. Siddiqi and F. Dela Cruz (Columbia University) for critical reading and review of the manuscript and data.
Footnotes
Conflict of interest: The authors have declared that no conflict of interest exists.
Citation for this article: J Clin Invest. 2012;122(3):886–898. doi:10.1172/JCI60015.
References
- 1.Crozat A, Aman P, Mandahl N, Ron D. Fusion of CHOP to a novel RNA-binding protein in human myxoid liposarcoma. Nature. 1993;363(6430):640–644. doi: 10.1038/363640a0. [DOI] [PubMed] [Google Scholar]
- 2.Rabbitts TH, Forster A, Larson R, Nathan P. Fusion of the dominant negative transcription regulator CHOP with a novel gene FUS by translocation t(12;16) in malignant liposarcoma. Nat Genet. 1993;4(2):175–180. doi: 10.1038/ng0693-175. [DOI] [PubMed] [Google Scholar]
- 3.Perez-Mancera PA, Vicente-Duenas C, Gonzalez-Herrero I, Sanchez-Martin M, Flores-Corral T, Sanchez-Garcia I. Fat-specific FUS-DDIT3-transgenic mice establish PPARgamma inactivation is required to liposarcoma development. Carcinogenesis. 2007;28(10):2069–2073. doi: 10.1093/carcin/bgm107. [DOI] [PubMed] [Google Scholar]
- 4.Rodriguez R, et al. FUS-CHOP fusion protein expression coupled to p53 deficiency induces liposarcoma in mouse but not in human adipose-derived mesenchymal stem/stromal cells. Stem Cells. 2011;29(2):179–192. doi: 10.1002/stem.571. [DOI] [PubMed] [Google Scholar]
- 5.Perez-Losada J, et al. Liposarcoma initiated by FUS/TLS-CHOP: the FUS/TLS domain plays a critical role in the pathogenesis of liposarcoma. Oncogene. 2000;19(52):6015–6022. doi: 10.1038/sj.onc.1204018. [DOI] [PubMed] [Google Scholar]
- 6.Riggi N, et al. Expression of the FUS-CHOP fusion protein in primary mesenchymal progenitor cells gives rise to a model of myxoid liposarcoma. Cancer Res. 2006;66(14):7016–7023. doi: 10.1158/0008-5472.CAN-05-3979. [DOI] [PubMed] [Google Scholar]
- 7.Matushansky I, et al. A developmental model of sarcomagenesis defines a differentiation-based classification for liposarcomas. Am J Pathol. 2008;172(4):1069–1080. doi: 10.2353/ajpath.2008.070284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Schwarzbach MH, et al. Comparable transforming capacities and differential gene expression patterns of variant FUS/CHOP fusion transcripts derived from soft tissue liposarcomas. Oncogene. 2004;23(40):6798–6805. doi: 10.1038/sj.onc.1207840. [DOI] [PubMed] [Google Scholar]
- 9.Engstrom K, et al. The myxoid/round cell liposarcoma fusion oncogene FUS-DDIT3 and the normal DDIT3 induce a liposarcoma phenotype in transfected human fibrosarcoma cells. Am J Pathol. 2006;168(5):1642–1653. doi: 10.2353/ajpath.2006.050872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Antonescu CR, et al. Prognostic impact of P53 status, TLS-CHOP fusion transcript structure, and histological grade in myxoid liposarcoma: a molecular and clinicopathologic study of 82 cases. Clin Cancer Res. 2001;7(12):3977–3987. [PubMed] [Google Scholar]
- 11.Dei Tos AP, et al. Molecular aberrations of the G1-S checkpoint in myxoid and round cell liposarcoma. Am J Pathol. 1997;151(6):1531–1539. [PMC free article] [PubMed] [Google Scholar]
- 12.Pilotti S, et al. Limited role of TP53 and TP53-related genes in myxoid liposarcoma. Tumori. 1998;84(5):571–577. doi: 10.1177/030089169808400512. [DOI] [PubMed] [Google Scholar]
- 13.Oda Y, et al. Frequent alteration of p16(INK4a)/p14(ARF) and p53 pathways in the round cell component of myxoid/round cell liposarcoma: p53 gene alterations and reduced p14(ARF) expression both correlate with poor prognosis. J Pathol. 2005;207(4):410–421. doi: 10.1002/path.1848. [DOI] [PubMed] [Google Scholar]
- 14.Grosso F, et al. Efficacy of trabectedin (ecteinascidin-743) in advanced pretreated myxoid liposarcomas: a retrospective study. Lancet Oncol. 2007;8(7):595–602. doi: 10.1016/S1470-2045(07)70175-4. [DOI] [PubMed] [Google Scholar]
- 15.D’Incalci M, Galmarini CM. A review of trabectedin (ET-743): a unique mechanism of action. Mol Cancer Ther. 2010;9(8):2157–2163. doi: 10.1158/1535-7163.MCT-10-0263. [DOI] [PubMed] [Google Scholar]
- 16.Manara MC, et al. The molecular mechanisms responsible for resistance to ET-743 (Trabectidin; Yondelis) in the Ewing’s sarcoma cell line, TC-71. Int J Oncol. 2005;27(6):1605–1616. [PubMed] [Google Scholar]
- 17.Grohar PJ, et al. Ecteinascidin 743 interferes with the activity of EWS-FLI1 in Ewing sarcoma cells. Neoplasia. 2011;13(2):145–153. doi: 10.1593/neo.101202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Forni C, et al. Trabectedin (ET-743) promotes differentiation in myxoid liposarcoma tumors. Mol Cancer Ther. 2009;8(2):449–457. doi: 10.1158/1535-7163.MCT-08-0848. [DOI] [PubMed] [Google Scholar]
- 19.Logan M, Martin JF, Nagy A, Lobe C, Olson EN, Tabin CJ. Expression of Cre Recombinase in the developing mouse limb bud driven by a Prxl enhancer. Genesis. 2002;33(2):77–80. doi: 10.1002/gene.10092. [DOI] [PubMed] [Google Scholar]
- 20.Tontonoz P, et al. Terminal differentiation of human liposarcoma cells induced by ligands for peroxisome proliferator-activated receptor gamma and the retinoid X receptor. Proc Natl Acad Sci U S A. 1997;94(1):237–241. doi: 10.1073/pnas.94.1.237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Demetri GD, et al. Induction of solid tumor differentiation by the peroxisome proliferator-activated receptor-gamma ligand troglitazone in patients with liposarcoma. Proc Natl Acad Sci U S A. 1999;96(7):3951–3956. doi: 10.1073/pnas.96.7.3951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mueller E, et al. Terminal differentiation of human breast cancer through PPAR gamma. Mol Cell. 1998;1(3):465–470. doi: 10.1016/S1097-2765(00)80047-7. [DOI] [PubMed] [Google Scholar]
- 23.Valoti G, et al. Ecteinascidin-743, a new marine natural product with potent antitumor activity on human ovarian carcinoma xenografts. Clin Cancer Res. 1998;4(8):1977–1983. [PubMed] [Google Scholar]
- 24.Frapolli R, et al. Novel models of myxoid liposarcoma xenografts mimicking the biological and pharmacologic features of human tumors. Clin Cancer Res. 2010;16(20):4958–4967. doi: 10.1158/1078-0432.CCR-10-0317. [DOI] [PubMed] [Google Scholar]
- 25.Girnun GD, et al. Regression of drug-resistant lung cancer by the combination of rosiglitazone and carboplatin. Clin Cancer Res. 2008;14(20):6478–6486. doi: 10.1158/1078-0432.CCR-08-1128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Matushansky I, et al. Derivation of sarcomas from mesenchymal stem cells via inactivation of the Wnt pathway. J Clin Invest. 2007;117(11):3248–3257. doi: 10.1172/JCI31377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tirode F, Laud-Duval K, Prieur A, Delorme B, Charbord P, Delattre O. Mesenchymal stem cell features of Ewing tumors. Cancer Cell. 2007;11(5):421–429. doi: 10.1016/j.ccr.2007.02.027. [DOI] [PubMed] [Google Scholar]
- 28.Riggi N, et al. EWS-FLI-1 expression triggers a Ewing’s sarcoma initiation program in primary human mesenchymal stem cells. Cancer Res. 2008;68(7):2176–2185. doi: 10.1158/0008-5472.CAN-07-1761. [DOI] [PubMed] [Google Scholar]
- 29.Burgermeister E, Seger R. MAPK kinases as nucleo-cytoplasmic shuttles for PPARgamma. Cell Cycle. 2007;6(13):1539–1548. doi: 10.4161/cc.6.13.4453. [DOI] [PubMed] [Google Scholar]
- 30.Mikkelsen TS, et al. Comparative epigenomic analysis of murine and human adipogenesis. Cell. 2010;143(1):156–169. doi: 10.1016/j.cell.2010.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Perez-Mancera PA, Bermejo-Rodriguez C, Sanchez-Martin M, Abollo-Jimenez F, Pintado B, Sanchez-Garcia I. FUS-DDIT3 prevents the development of adipocytic precursors in liposarcoma by repressing PPARgamma and C/EBPalpha and activating eIF4E. PLoS ONE. 2008;3(7):e2569. doi: 10.1371/journal.pone.0002569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nucleic Acids Res. 2002;30(19):4118–4126. doi: 10.1093/nar/gkf512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Grosso F, et al. Trabectedin in myxoid liposarcomas (MLS): a long-term analysis of a single-institution series. Ann Oncol. 2009;20(8):1439–1444. doi: 10.1093/annonc/mdp004. [DOI] [PubMed] [Google Scholar]
- 34.Debrock G, Vanhentenrijk V, Sciot R, Debiec-Rychter M, Oyen R, Van Oosterom A. A phase II trial with rosiglitazone in liposarcoma patients. . Br J Cancer. 2003;89(8):1409–1412. doi: 10.1038/sj.bjc.6601306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Negri T, et al. Functional mapping of receptor tyrosine kinases in myxoid liposarcoma. Clin Cancer Res. 2010;16(14):3581–3593. doi: 10.1158/1078-0432.CCR-09-2912. [DOI] [PubMed] [Google Scholar]
- 36.Moneo V, et al. Extreme sensitivity to Yondelis (Trabectedin, ET-743) in low passaged sarcoma cell lines correlates with mutated p53. J Cell Biochem. 2007;100(2):339–348. doi: 10.1002/jcb.21073. [DOI] [PubMed] [Google Scholar]
- 37.Martinez N, et al. Transcriptional signature of Ecteinascidin 743 (Yondelis, Trabectedin) in human sarcoma cells explanted from chemo-naive patients. Mol Cancer Ther. 2005;4(5):814–823. doi: 10.1158/1535-7163.MCT-04-0316. [DOI] [PubMed] [Google Scholar]
- 38.Martin JF, Olson EN. Identification of a prx1 limb enhancer. Genesis. 2000;26(4):225–229. doi: 10.1002/(SICI)1526-968X(200004)26:4<225::AID-GENE10>3.0.CO;2-F. [DOI] [PubMed] [Google Scholar]
- 39.Smale ST. Nuclear run-on assay. Cold Spring Harb Protoc. 2009;2009(11):pdb.prot5329. doi: 10.1101/pdb.prot5329. [DOI] [PubMed] [Google Scholar]
- 40.Oda Y, Sakamoto A, Satio T, Kawauchi S, Iwamoto Y, Tsuneyoshi M. Molecular abnormalities of p53, MDM2, and H-ras in synovial sarcoma. Mod Pathol. 2000;13(9):994–1004. doi: 10.1038/modpathol.3880180. [DOI] [PubMed] [Google Scholar]
- 41.Mills J, et al. Characterization and comparison of the properties of sarcoma cell lines in vitro and in vivo. Hum Cell. 2009;22(4):85–93. doi: 10.1111/j.1749-0774.2009.00073.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.