Abstract
Chromosomal instability (CIN) in tumors is characterized by chromosomal abnormalities and an altered gene expression signature; however, the mechanism of CIN is poorly understood. CCND1 (which encodes cyclin D1) is overexpressed in human malignancies and has been shown to play a direct role in transcriptional regulation. Here, we used genome-wide ChIP sequencing and found that the DNA-bound form of cyclin D1 occupied the regulatory region of genes governing chromosomal integrity and mitochondrial biogenesis. Adding cyclin D1 back to Ccnd1–/– mouse embryonic fibroblasts resulted in CIN gene regulatory region occupancy by the DNA-bound form of cyclin D1 and induction of CIN gene expression. Furthermore, increased chromosomal aberrations, aneuploidy, and centrosome abnormalities were observed in the cyclin D1–rescued cells by spectral karyotyping and immunofluorescence. To assess cyclin D1 effects in vivo, we generated transgenic mice with acute and continuous mammary gland–targeted cyclin D1 expression. These transgenic mice presented with increased tumor prevalence and signature CIN gene profiles. Additionally, interrogation of gene expression from 2,254 human breast tumors revealed that cyclin D1 expression correlated with CIN in luminal B breast cancer. These data suggest that cyclin D1 contributes to CIN and tumorigenesis by directly regulating a transcriptional program that governs chromosomal stability.
Introduction
Chromosomal instability (CIN) in tumors (1–3) is characterized by an elevated rate of gain or loss of whole chromosomes (i.e., aneuploidy) and/or as structural chromosomal aberrations (i.e., translocations, deletions, and duplications). Aneuploidy is one of the most striking differences between cancer and normal cells. The molecular mechanisms inducing CIN as well as the timing of CIN in tumor progression, invasion, and metastasis is poorly understood (4, 5). Cell cycle–associated factors have been implicated in CIN, including cyclin E (6). The relative enrichment of a molecular genetic signature of CIN-related genes has been used to quantitate a CIN score (7); this signature includes AURKB (a component of the chromosomal passenger complex [CPC]), TOP2A, CENPP, MLF1IP (a component of the CENPA-NAC kinetochore complex protein), ZW10 (a kinetochore-associated mitotic checkpoint protein), and CKAP2 (a mitotic spindle-associated protein) (3) as well as the retinoblastoma (pRb) protein. Supernumerary centrosomes increase the frequency of dual attachment of 1 sister kinetochore to 2 spindle poles. Cyclin E activity promotes centrosome duplication during S phase onset. Loss of pRb can also alter centrosome number and formation of micronuclei, leading to mis-segregation of chromosomes and aneuploidy (8). CIN occurs relatively early in tumor progression, whereas pRb loss occurs relatively late in the process of tumorigenesis, which raises the question of candidate mechanisms driving chromosomal aberrations in the early phase of tumor onset.
Cyclin D1 (CCND1) encodes the regulatory subunit of the holoenzyme that phosphorylates and inactivates pRb and the NRF1 proteins to regulate nuclear DNA synthesis and mitochondrial biogenesis (9–13). Quantitative single-cell analysis has shown that cyclin D1 levels oscillate during the cell cycle, increasing in a broad array of cell types during the G2 phase (14). Cyclin D1 expression is increased 3- to 8-fold in human breast, prostate, lung, and gastrointestinal malignancies (15–18). Furthermore, the cyclin D1 oncogene directly induces mammary gland tumors in mice (19). Cyclin D1 is required for oncogene-dependent growth, as genetic ablation of murine Ccnd1 impaired terminal alveolar breast bud development (20) and resulted in resistance to Ras- or ErbB2-induced mammary tumorigenesis and to APC-induced gastrointestinal tumorigenesis (21, 22). Over the last 2 decades, a substantial body of evidence has suggested cyclin D1 plays a direct role in transcriptional regulation (16). Cyclin D1 physically associates with, and regulates the transcriptional activity of, ERα (23) and more than 30 other transcription factors (TFs) (16). The histone acetyltransferases p300, p300/CREB-binding protein–associated factor (P/CAF), and AlB1 bind to cyclin D1 (24, 25). ChIP demonstrated cyclin D1 association within the local chromatin of target gene promoters that correlated with deacetylation of histone (H3), in particular at H3 lysine 9 (H3lys9). Deacetylation of H3lys9 was restored upon reintroduction of cyclin D1, which recruited HDAC1/HDAC3 (17, 22). Thus, cyclin D1 is recruited in the context of local chromatin to specific target genes (26, 27). Cyclin D1 recruitment to local chromatin was also associated with recruitment of p300 to regulate genes governing DNA damage repair signaling (26). Cyclin D1 was shown to regulate the activity of p300 independently of cyclin-dependent kinase (CDK) binding function. As p300 is regarded as a transcriptional cointegrator, cyclin D1 was proposed as a regulator of gene transcription through co-occupancy with p300 at target DNA-binding sites (26).
Recent studies demonstrated the occupancy of cyclin D1 in the context of local chromatin using ChIP-ChIP analysis on a –5.5 kb to +2.5 kb ChIP-ChIP microarray containing approximately 17,000 genes (28). In addition, cyclin D1 associated with the p300-related CREB-binding protein (CBP) in a proteomics screen and recruited CBP to the Notch1 gene to regulate its transcription. Here, we aimed to expand the interrogation of cyclin D1 TF binding sites to the entire genome and to include potential cyclin D1 interactions both within and outside the proximal 8 kb of a gene’s start site. We therefore performed ChIP of cyclin D1 followed by ChIP sequencing (ChIP-Seq) to map at high resolution the entire genomic region bound by cyclin D1. Functional pathway analysis of the gene regulatory elements bound by cyclin D1 uncovered enrichment for genes that govern chromosomal stability. Our data suggest that cyclin D1 contributes to CIN and tumorigenesis by directly regulating a transcriptional program that governs chromosomal stability.
Results
Defining genome-wide cyclin D1 binding sites.
In view of our prior findings that cyclin D1 occupies promoter regulatory regions in the context of local chromatin associated with the recruitment of p300 (17, 26, 29), we conducted genome-wide analysis of cyclin D1 genomic occupancy using ChIP-Seq analysis. In order to characterize genome-wide cyclin D1 binding sites, Ccnd1–/– mouse embryonic fibroblasts (MEFs) were transduced with a FLAG epitope–tagged expression vector encoding cyclin D1 (30). The exogenous levels of cyclin D1 in rescued cells were approximately 3-fold higher than basal levels (Supplemental Figure 1A; supplemental material available online with this article; doi: 10.1172/JCI60256DS1), consistent with the 3- to 8-fold increase in cyclin D1 levels observed in breast and other tumor types (18, 31). In order to characterize genome-wide cyclin D1 DNA binding sites in vivo, ChIP-Seq was conducted using Genpathway’s FactorPath discovery technology. We found 2,840 NCBI genes with intervals within 10 kb of the start site. A summary of the active regions and their proximity to NCBI-designated genes is given in Supplemental Table 1. Figure 1A shows the genome-wide distribution of 2,840 binding sites in relation to the transcriptional start site. Interestingly, peak values of active regions within the promoter were comparable to those 10 kb and beyond (Figure 1B), which suggests that cyclin D1 localizes to both promoter-proximal elements and very distant elements. Analysis of the tag density distribution at the promoter-proximal region identified the enrichment of cyclin D1 occupancy within approximately 500 bp of the transcriptional start site (Supplemental Figure 1, B and C).
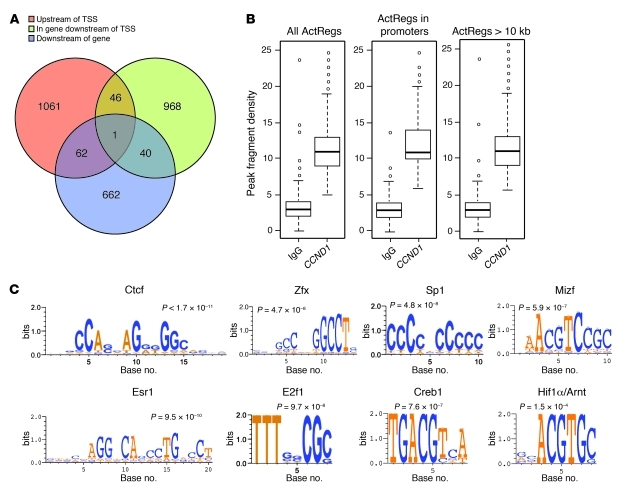
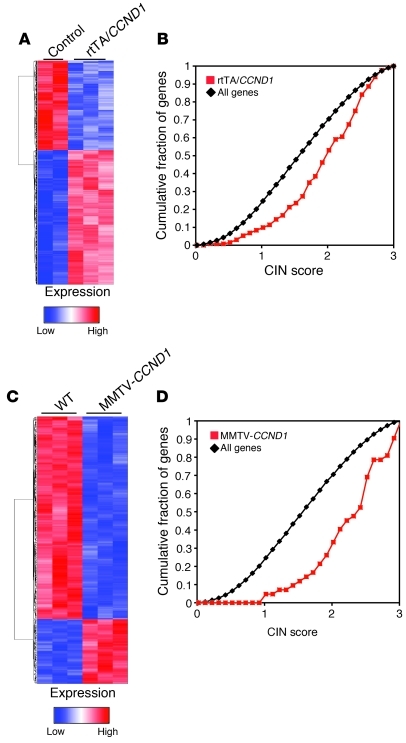
Figure 1. Identification of cyclin D1–bound sequences.
(A) Distribution of the 2,840 intervals with respect to neighboring genes. Intervals were categorized as being upstream of transcriptional start site (TSS), in gene and downstream of TSS, or downstream of the gene. Because some intervals had more than 1 associated gene, some were associated with more than 1 term. Upstream of TSS was defined as –10,000 to 0 bp. (B) Peak values in the anti-FLAG/CCND1 ChIP-Seq compared with IgG control. The peak values within promoter regions were similar to those 10 kb from the transcriptional start site. ActRegs, active regions. Bounds of the boxes denote SD; lines within boxes denote mean; whiskers denote interquartile range; symbols denote outliers. (C) Select example of conserved TF motifs enriched within the interval regions associated with cyclin D1. See Supplemental Table 2 for the full list of TFs.
In order to determine the TF binding sites enriched in the cyclin D1 peak interval sequences, we used the JASPAR Match server (32), employing a permutation test. Only those intervals within 2 kb of the transcription start site were processed. Figure 1C shows DNA recognition sequences and statistical significance for the top hit TFs Ctcf (a zinc finger protein; also known as CCCTC-binding factor), Zfx (a member of the krueppel C2H2-type zinc finger protein family), Sp1 (a TF belonging to the Sp/KLF family), Mizf (a zinc finger protein that has a key role in histone gene expression; also known as Hinfp), estrogen receptor R1 (Esr1; also known as ERα), E2f1 (a member of the E2F family that transcriptionally activates numerous genes involved in cell cycle regulation), Creb1 (a cAMP element–binding protein activated by cAMP usually through hormonal stimulation), and Hif1α/Arnt (which regulates key genes involved in hypoxic stress). A full list of TFs and their statistical significance is provided in Supplemental Table 2. The prevalence of TF motifs in the interval sequence was plotted for the top 20 TFs (Supplemental Figure 1D). We next asked whether a consensus sequence was present in the peak interval regions. We used the complete 3,222-peak interval data set and split the data into 4 groups based on proximity to nearest neighbor transcriptional start sites. In 3 of the groups, the motif was an exact copy of the CTCF invariant core sequence (Supplemental Figure 1E). We used luciferase reporter constructs containing multimeric copies of the consensus sequence alone to verify that cyclin D1 regulated the transcriptional activity of several members of the TF list (i.e., Myc, E2F, and Hif1α; Supplemental Figure 2). These data are consistent with a model in which cyclin D1 is recruited in the context of local chromatin to regulate gene transcription and occupies DNA elements that associate with TFs.
Cyclin D1 binds genes that regulate chromosomal stability.
An unbiased determination of the functional pathways using the annotation clustering feature of NIH Database for Annotation, Visualization, and Integrated Discovery (DAVID) demonstrated that cyclin D1 bound the regulatory region of the genes involved in RNA processing, mitochondrial function, and DNA organization and segregation (Figure 2A). Previous work by Bienvenu et al., using a ChIP promoter array that examined approximately 1% of the genome, demonstrated that cyclin D1 associates with approximately 900 genes in close proximity to the transcriptional start site (P < 1 × 10–4; ref. 26). Extension through ChIP-Seq to interrogate the additional components of the entire genome revealed considerable functional overlap, with additional functions identified through the global genomic analysis (Supplemental Figure 3).
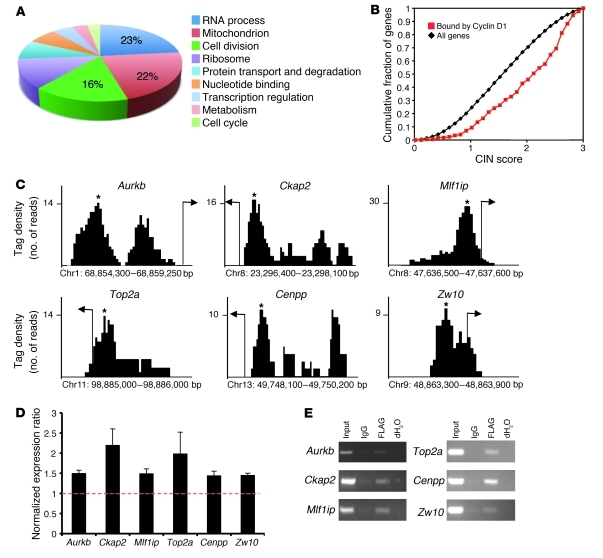
Figure 2. Cyclin D1 associates with genes involved in mitosis.
(A) Functional annotation clustering by DAVID of cyclin D1–associated genes, based on percent enrichment score of the top hits. (B) Cyclin D1–bound promoter regions (0 to –500 bp) were enriched in genes demonstrating an association with CIN (P < 0.0001). (C) Representative tag density profiles of cyclin D1–bound regions and their proximity to the transcriptional start site (arrow). Peak values for the intervals are denoted by asterisks. (D) Quantitative PCR on target mRNAs selected based on cyclin D1–associated genes. Shown are normalized expression ratios of Ccnd1–/– cells with MSCV-FLAG/CCND1 compared with MSCV-control (n = 4 separate cell lines; data are mean ± SEM). (E) ChIP analysis of Ccnd1–/– 3T3 cells transduced with MSCV-FLAG/CCND1 using anti-FLAG antibody. Primers were designed against the peak interval sequence.
Based on the functional annotation analysis, there were a large number of gene sets associated with cell division. We analyzed these sets further to extract the associated genes; a list of the genes associated with the Gene Ontology (GO) term cell division is provided in Supplemental Table 3. Most of the genes were involved in G2/M phase and cellular mitosis. Given the high number of genes that regulate mitosis, we determined whether the genomic regions that associate with cyclin D1 correlate with a CIN function. When genes were ranked based on CIN score (7), those regulatory regions occupied by cyclin D1 were significantly enriched (P < 0.0001; Figure 2B).
Most of the mitotic genes are involved in chromatin reorganization and chromosomal segregation during M phase. Representative tag density profiles for several members of the list are depicted in Figure 2C. The relative abundance of transcripts coding for proteins that regulate chromosomal segregation were increased around 1.5- to 2-fold by cyclin D1 expression, including Aurkb, Ckap2, Mlf1ip, and Zw10 (Figure 2D). We also performed quantitative RT-PCR on 2 other GO terms: protein catabolic process and RNA processing. The expression of the genes representative of those terms was also regulated by cyclin D1 (Supplemental Figure 4A). In addition, we verified by Western blot analysis that Aurkb, a key regulator of the mitotic checkpoint control, was increased in abundance by cyclin D1 (Supplemental Figure 4B). Cyclin D1 increased phosphorylation of H3S10, a target of Aurkb (33, 34). AURKB is overexpressed in human malignancies like prostate, colorectal, kidney, lung, and breast cancers (35), and its overexpression results in multinucleation and polyploidy in human cells. ChIP analysis using relevant and negative control primer sets (Figure 2E and Supplemental Figure 4C) confirmed the ChIP-Seq data, indicative of occupancy by cyclin D1 at the promoter regions of genes involved in regulation of chromosome segregation (i.e., Aurkb, Top2a, Cenpp, Mlf1ip, Zw10, and Ckap2). These results suggest that cyclin D1 contributes to CIN by transcriptional regulation of genes involved in mitosis.
Cyclin D1 promotes CIN.
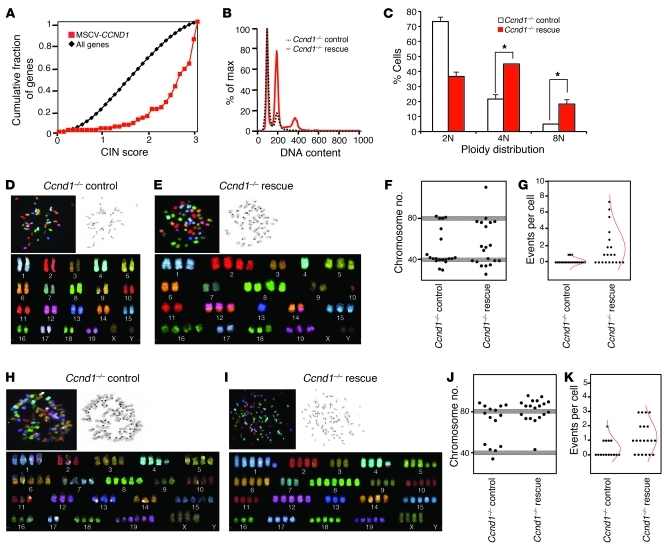
The cyclin D1–dependent enrichment of genes involved in chromosomal segregation and stability led us to determine the functional consequence using a cyclin D1 expression profile, fluorescence-activated cell sorting (FACS) analysis, and spectral karyotyping (SKY). We first used a previously published cyclin D1 expression profile to determine enrichment for the CIN profile. Induction of cyclin D1 expression in MEFs using a cyclin D1 retrovirus induced expression of CIN-associated genes (P < 0.0001; Figure 3A). We next assessed the effect of cyclin D1 on ploidy by reintroducing cyclin D1 into Ccnd1–/– cells (a process referred to herein as cyclin D1 rescue). The proportion of polyploid cells increased within 3 cellular divisions, increasing the relative proportion of 4N and 8N cells by 45% and 15%, respectively (Figure 3, B and C). Using SKY, we analyzed 20 metaphase spreads of cyclin D1–rescued versus control Ccnd1–/– cells (Figure 3, D and E, and Supplemental Figure 5, A and B). Graphical representation of the karyotype analysis of all 20 metaphases is shown in Figure 3F. Since mouse fibroblasts are prone to genomic instability when successively passaged in culture, a deviation of ±2 chromosomes at 2N and 4N is considered normal. By this criterion, 75% of the Ccnd1–/– MEF metaphases had a normal karyotype compared with 30% of the cyclin D1–rescued Ccnd1–/– MEFs (P < 0.001). Just as pronounced as the aneuploidy was the number of chromosomal aberrations observed in the cyclin D1–rescued Ccnd1–/– line. Defects identified by SKY are assigned as deletions, duplications, and translocations. There were significantly more translocations in the cyclin D1–rescued compared with control Ccnd1–/– MEFs (Figure 3G), although no significant differences in deletions and duplications were identified (Supplemental Figure 6, A and B). Most prevalent were the nonreciprocal translocations (NRTs) and reciprocal translocations, the latter of which was present in 7 of 20 metaphases analyzed. NRTs predominate in human carcinomas, as characterized by karyotype analysis, and contribute to carcinogenesis by carrying oncogenes at their breakpoints and also by distorting normal gene dosage (36). The number of NRTs in the cyclin D1–rescued line was 13 events, compared with 3 in the control. A full list of rearrangements is given in Supplemental Table 4. We also conducted SKY analysis of 3T3 control and cyclin D1–rescued Ccnd1–/– cells at passage 23 (P23) to determine whether the abnormal karyotype was present in late-passage cells (Figure 3, H and I, and Supplemental Figure 5, C and D). Although control Ccnd1–/– cells exhibited significantly higher polyploidy than did low-passage Ccnd1–/– MEFs, the rates were higher in cyclin D1–rescued Ccnd1–/– cells (P = 0.05; Figure 3J). In addition, there were substantially more reciprocal translocations and NRTs (Figure 3K), with little difference in deletions and duplication events between the 2 lines (Supplemental Figure 6, C and D). Taken together, these data suggest that acute rescue of Ccnd1–/– MEFs induces CIN with a high NRT rate.
Figure 3. Cyclin D1 rescue of Ccnd1–/– MEFs induces polyploidy and aneuploidy.
(A) The expression profile for cyclin D1–induced genes (63) was enriched for high CIN score (P < 0.0001). (B) PI staining demonstrated increased polyploidy in cyclin D1–rescued versus control Ccnd1–/– MEFs. (C) Quantitation of PI staining based on 3 separate cell lines (mean ± SEM). *P < 0.005. (D, E, H, and I) Representative metaphases from SKY of control and cyclin D1–rescued Ccnd1–/– MEFs at P6 (D and E) and 3T3 cells at P23 (H and I). Shown for each is an inverted DAPI image of the metaphase (top right), a raw spectral image of the metaphase (top left), and classification of the same metaphase (bottom). (F and J) Chromosomal number across metaphase spreads from control and cyclin D1–rescued Ccnd1–/– MEFs at P6 (F) and 3T3 cells at P23 (J), showing the total number of chromosomes for 20 mitotic spreads. Gray shading represents expected deviation from normal at 2N and 4N (± 2 chromosomes). P < 0.001, rescue vs. control, χ2 test of association. (G and K) Reciprocal translocations and NRTs in metaphase spreads from control and cyclin D1–rescued Ccnd1–/– MEFs at P6 (G) and 3T3 cells at P23 (K), shown as number of events per cell analyzed. The mean distribution is represented as a red curve.
Multipolar spindles and centrosome amplification predominate in cyclin D1–rescued Ccnd1–/– cells.
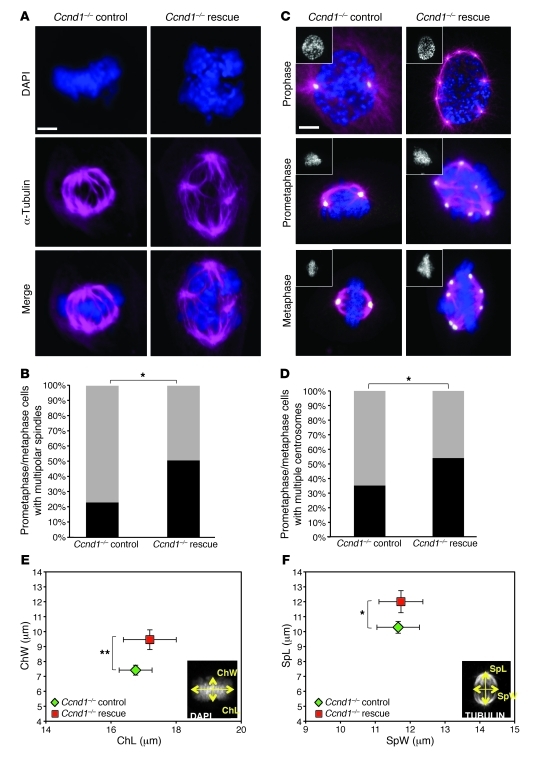
In order to screen for potential abnormalities in mitosis that could contribute to CIN, we performed immunofluorescence followed by high-resolution confocal imaging on 3T3 control and cyclin D1–rescued Ccnd1–/– cells using markers of mitotic spindles and centrosomes (α- and γ-tubulin, respectively). The number of cells with multipolar spindles increased 27% in cyclin D1–rescued relative to control Ccnd1–/– cells (P = 0.0289; Figure 4, A and B). Because multipolar spindles arise from abnormalities in centrosome number and distribution, we stained cells for γ-tubulin in conjunction with α-tubulin to quantitate the number of centrosomes. The percentage of prometaphase/metaphase cells with more than 2 centrosomes increased 19% in cyclin D1–rescued compared with control Ccnd1–/– cells (P = 0.0289; Figure 4, C and D). Spindle architecture was clearly abnormal in cyclin D1–rescued Ccnd1–/– cells, resulting in multipolar spindles that perpetuated from prophase to prometaphase and failed to coalesce at metaphase (Figure 4D). The alteration of spindle architecture was associated with the disruption of metaphase plate morphology, detected with DAPI staining (Figure 4E). Width and length of metaphase plates and spindles were measured in the same samples; consistent with the increase in spindle and centrosome abnormalities, plate width and spindle length were significantly increased in cyclin D1–rescued Ccnd1–/– cells (P = 0.0087 and P = 0.0486, respectively; Figure 4, E and F). Lagging chromosomes, anaphase bridges, and micronuclei were also observed in cyclin D1–rescued compared with control Ccnd1–/– cells (Supplemental Figure 7). These results demonstrated increased prevalence of centrosome amplification that contributed to mitotic spindle abnormalities.
Figure 4. Cyclin D1 induces centrosome amplification and mitotic spindle disorganization.
(A and C) Representative confocal maximum Z projections of mitotic cells (A; immunostained for α-tubulin [violet] and DAPI [blue]) and prophase, prometaphase, and metaphases (C; immunostained for α-tubulin [violet], γ-tubulin [yellow], and DAPI [blue and insets]) from control and cyclin D1–rescued Ccnd1–/– cells. Original magnification, ×60 NA1.4 oil objective, enlarged ×5 by digital zoom. Scale bars: 5 μm. (B and D) Frequency of mitotic cells with multiple polar spindles (B) and of prometaphase/metaphase cells with multiple centrosomes (γ-tubulin) and spindle disorganization (α-tubulin) (D). *P = 0.0289, χ2 analysis. Black bar, abnormal centrosome count (i.e., >2); gray bar, normal count (i.e., 2). (E and F) Spindle measurements on maximum Z projections of metaphase control and cyclin D1–rescued Ccnd1–/– cells. Data are mean ± SEM. Insets demonstrate metaphase plate (i.e., chromatin; Ch) width and length (measured using DAPI stain) and spindle (Sp) width and length (measured using tubulin stain). *P = 0.0486; **P = 0.0087.
Cyclin D1 promotes CIN expression profiles in vivo.
In order to directly assess the role of cyclin D1 in promoting CIN, we developed transgenic mouse mammary models to either acutely express cyclin D1 in the mammary gland using the tetracycline-inducible system or an MMTV-cyclin D1 system. For the tet-inducible transgenic mice (reverse tetracycline-controlled transactivator cross-mated with CCND1 transgenic [rtTA/CCND1]; Supplemental Figure 8A), RT-PCR analysis and Western blotting demonstrated that cyclin D1 expression levels were induced via tetracycline (Supplemental Figure 8, B and C). To examine the expression profiles induced by cyclin D1 in the mammary gland, we treated mice with tetracycline for 7 days, then performed microarray analysis to compare the cyclin D1 transgenic mice with rtTA-positive control mice that had undergone the same tetracycline regimen (Figure 5A). We then compared the genes that were most differentially regulated in the 2 sets (i.e., Fold>4 and log odds ratio of differential gene expression greater than 3 [B>3]) with the CIN signature gene set and found that the rtTA/CCDN1 gene profile was enriched for CIN (P < 0.001; Figure 5B). The MMTV-cyclin D1 transgenics (Supplemental Figure 8D) were confirmed by Northern blotting and Western blotting for FLAG-tagged cyclin D1 (Supplemental Figure 8E). Female MMTV-CCND1 and WT mice were monitored twice weekly for the development of palpable tumors. Those developing palpable tumors were sacrificed within a week of tumor detection. Kaplan-Meier survival and tumor-onset plots, and analyses with a log-rank test for curve comparisons, were performed between MMTV-CCND1 and WT lines (Supplemental Figure 8F). The first instances of tumor onset occurred at around 400 days in MMTV-CCND1 mice, whereas WT mice were tumor free at this age. At 760 days, the tumor-free fraction in the MMTV-CCND1 group was 42% compared with 85% in the WT group (P = 0.0018). The relative abundance of cyclin D1 was also assessed in normal mammary epithelial cells and found to be concordant with the increased level of cyclin D1 present in human breast cancer samples (Supplemental Figure 8G and ref. 31).
Figure 5. Heat maps displaying genes differentially regulated by cyclin D1 in transgenic mouse models.
(A) Genes differentially regulated between rtTA/CCND1 tumors (n = 3) and normal mouse mammary glands from rtTA control mice (n = 2), visualized by hierarchical clustering. (B) The most highly differentially regulated genes (Fold>2, B>3) were enriched for CIN in the rtTA/CCND1 profile (P < 0.0001). (C) Genes differentially regulated between MMTV-CCND1–induced tumors (n = 3) and normal mouse mammary glands (n = 2), visualized by hierarchical clustering. (D) The most highly differentially regulated genes (Fold>2, B>4) were enriched for CIN in the MMTV-CCND1 profile (P < 0.0001).
To determine the genes that are regulated by MMTV-CCND1, microarray analysis was performed on tumors obtained from age-matched mice and compared with mammary glands of WT (FVB) mice (Figure 5C). We then compared the genes that were most differentially regulated in the 2 sets (i.e., Fold>4 and B>3) with the CIN signature gene set and found the MMTV-CCND1 gene profile to be enriched for CIN (P < 0.0001; Figure 5D). Taken together, these data suggest that cyclin D1 induces enrichment of CIN score upon acute induction or with constitutive long-term expression in the mammary epithelium of mice.
High cyclin D1 expression correlates with CIN in luminal B breast cancer subtype.
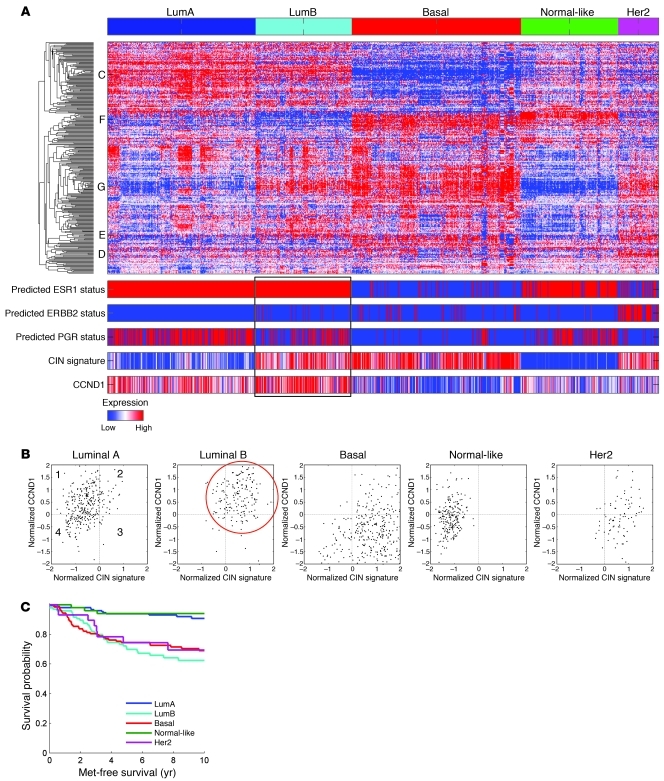
To analyze the association between CIN and cyclin D1 expression in the context of breast cancer, we aligned the expression of a 70-gene set with the highest CIN score against a collection of 2,254 breast cancer samples compiled from public microarray databases (37). We stratified the samples based on previously described breast cancer subtypes (38) and aligned them with the cyclin D1 expression profile across the dataset. A significant correlation among cyclin D1, CIN, and the luminal B subtype was identified (P < 1 × 10–10; Figure 6A). Scatter plots depicting CCND1 transcript level versus average CIN signature expression level revealed that the relationship between these expression levels was breast tumor subtype specific (Figure 6B). Kaplan-Meier plots showed differences in metastasis-free survival in this dataset (Figure 6C). We conclude that individuals with luminal B type breast cancer have elevated CIN expression profiles that correlate with high cyclin D1 expression.
Figure 6. Subtype classification of breast cancer microarray samples.
(A) Heatmap depicting samples from combined breast cancer microarray datasets that were assigned to the 5 breast cancer gene expression subtypes. The predicted ESR1, epidermal growth factor receptor (ERBB2), and progesterone receptor (PGR) statuses are shown together with CIN signature score and CCND1 expression level across the 5 subtypes. The luminal B subtype receptor status, CIN signature score, and cyclin D1 expression level is outlined. (B) CCND1 transcript level plotted versus average CIN signature expression level revealed that the relationship between high CIN score and high cyclin D1 expression was luminal B subtype specific (red circle). (C) Kaplan-Meier plot showing differences in metastasis-free status in this dataset (P = 6.4462 × 10–8).
Discussion
Our present findings indicate that cyclin D1 induces CIN. Cyclin D1 induced gene expression profiles characteristic of CIN in fibroblasts, in the mammary gland, and in cyclin D1–induced mammary tumors. Transient expression of cyclin D1 over 7 days in the mammary gland was sufficient to induce CIN gene expression. SKY analysis confirmed the induction of aneuploidy and polyploidy by cyclin D1 expression in Ccnd1–/– MEFs. Immunofluorescence demonstrated the occurrence of supernumerary centrosomes that formed multipolar spindles. A careful analysis of the relative abundance of the genes involved in CIN identified a cluster of genes regulating the G2/M checkpoint and mitosis. The relative abundance of these genes was increased by cyclin D1 expression, as confirmed by quantitative PCR. The finding that cyclin D1 induced CIN is of importance, as CIN is an early feature of tumorigenesis that may precede tumor suppressor loss (39, 40). Previous studies showed that cyclin E, but not cyclin D1, is capable of inducing CIN (6). However, cyclin D1 overexpression correlated with aneuploidy, supernumerary centrosomes, and spindle defects in mouse hepatocytes (41) and with aneuploidy and polyploidy in lymphoid tumors (42). In addition, cyclin D1 amplification correlated with centrosome amplification in bladder cancer (43). As cyclin D1 expression is increased in the early phases of tumorigenesis, cyclin D1 may be an important inducer of CIN in tumors.
Analysis of clinical samples with molecular genetic subtyping identified the correlation of the CIN signature with cyclin D1 overexpression and luminal subtype B breast cancer. The presence of CIN in this genetic subtype correlated with poor outcome. Previous studies examining the role of cyclin D1 in outcome have provided contradictory results, with some suggesting a positive correlation between cyclin D1 expression and outcome and others showing reduced survival (16, 44, 45). Cyclin D1 levels were induced in luminal A and B subtypes, but correlated with CIN in luminal subtype B. CIN is usually poorly tolerated by cells initiating cell death signaling. As luminal A and luminal B breast cancer subtypes have distinct molecular genetic profiles, there may be additional genetic changes in the luminal B tumors that allow the survival of cells with genomic instability. It may well be that the genetic subclassification, as conducted in the current studies, is important in determining the clinical significance of cyclin D1 overexpression. The recent identification of drugs targeting CIN (46, 47) may provide a rational basis for therapeutic substratification, supplementing with compounds targeting CIN in the luminal B subtype of breast cancer.
Here, we conducted a genome-wide analysis of cyclin D1 binding in the context of local chromatin using ChIP-Seq analysis. Our prior studies demonstrated the recruitment of cyclin D1 in the context of local chromatin to TF binding sites, which was associated with recruitment of SUV39H1 and HP1α and commensurate reduced acetylation and increased trimethylation of H3lys9 (17, 26). Subsequent studies by ChIP-ChIP covering –5.5 to +2.5 kb of a subset of promoters similarly identified cyclin D1 recruitment to a subset of target genes involved primarily in notch signaling and cellular proliferation (28).
How might cyclin D1 regulate gene expression in the context of local chromatin? Although intrinsic DNA sequence–specific binding of cyclin D1 has not been identified, cyclin D1 has been identified at sites of damaged DNA in the context of local chromatin (48, 49). Various TFs associate with cyclin D1 in IP–Western blot analysis, and the abundance of cyclin D1 can regulate the recruitment of TFs (22) and transcriptional coregulators (26, 29) in the context of local chromatin in ChIP assays. Given these findings, we had proposed that cyclin D1 is recruited either to DNA through sequence-specific binding proteins to regulate gene expression or to damaged DNA via Rad51 and the related repair complex, which thereby recruits BRCA proteins (29, 48). Cyclin D1 abundance determines the recruitment of cointegrator and chromatin remodeling proteins in ChIP assays, including p300/CBP (26, 29), SUV39H1, HP1α, and HDAC1/3 (17), and dictates acetylation and dimethylation of local histones (e.g., H3 and H4). The mechanisms permitting assembly of the cointegrator regulatory complex that are associated with cyclin D1 at a given cis element remain to be determined. Prior studies using cyclin D1 and p300 knockout mice showed that, in the case of genes governing the fidelity of DNA replication (e.g., MCM3, MCM4, and RfCH), their abundance was induced by cyclin D1 and reciprocally regulated by p300, consistent with previous findings that cyclin D1 inhibits p300 autoacetylation (26). Although the regulation of TFs and cointegrator activity was independent of the cdk-binding domain, the role of the cyclin D1 cdk-binding domain in regulating the CIN signature in vivo remains to be determined.
The current studies identified a distinct subset of cis elements occupied by cyclin D1, due in part to the distinct interrogation of the genome conducted herein. The current studies examined both noncoding and coding DNA and sites distal to the transcription start site and identified a proclivity for cyclin D1 to occupy the CTCF binding factor site. CTCF functions in chromatin reorganization and as an enhancer insulator (50). It is of interest that the cohesin complex — important in segregation of sister chromatids, which were altered in a cyclin D1–dependent manner — interacts with CTCF. The cohesin complexes are also found at a large fraction of CTCF sites in vivo (51, 52). Because CTCF is a chromatin reorganizer and has the potential to play a bidirectional role through the cohesin complex, it will be of interest to determine the relative importance of cyclin D1 in regulating CTCF-dependent global transcription.
Methods
Further information can be found in Supplemental Methods.
Cell culture, cell lines, and transgenic mice.
The MSCV-IRES-GFP retroviral vector and cyclin D1 WT constructs were previously described (53). Ccnd1+/+ and Ccnd1–/– primary MEF cultures were prepared as described previously (54). Cells were maintained in DMEM supplemented with 10% fetal bovine serum and 100 μg/ml each of penicillin and streptomycin.
Retrovirus production and infection.
Retroviral production and infection of Ccnd1–/– MEFs were described in detail previously (53). FACS-sorted (FACStar Plus; BD Biosciences) GFP+ cells were used for subsequent analysis.
ChIP-Seq analysis and TF enrichment.
Genpathway’s FactorPath method was carried out as previously described by Labhart et. al (55). Gene libraries were sequenced using Genome Analyzer II and aligned to the mouse genome using Eland (Illumina). TF enrichment in ChIP-Seq intervals was conducted using the Jasper server.
Western blotting and luciferase assays.
Whole cell lysates or homogenized tissue lysates (50 μg) were subjected to Western blotting as previously described (56). The following antibodies were used for Western blotting: guanine nucleotide dissociation inhibitor (GDI; ref. 57), cyclin D1 (catalog no. MS-210-P; NeoMarkers), FLAG-tagged M2 (catalog no. F1804; Sigma-Aldrich), β-tubulin (catalog no. T4026; Sigma-Aldrich), Aurkb (alias AIM-1, catalog no. 611082; BD Biosciences), phospho-H3S10 (catalog no. 06-570; Millipore), and GAPDH (catalog no. FL-335; Santa Cruz Biotechnology Inc.). Luciferase assays were conducted as described previously (58). A dose dependency for cyclin D1 was obtained using 50, 100, or 150 ng of plasmid DNA and 200 ng of reporter.
ChIP assay.
ChIP material was prepared in accordance with the Magna ChIP (Millipore) manufacturer’s guidelines. Briefly, 3-cm × 10-cm plates of actively growing late-passage MEFs (Ccnd1–/– MSCV-IRESD1) were fixed for 10 minutes with 37% paraformaldehyde (final concentration, 1%). Unreacted formaldehyde was quenched with 1 ml of 10× glycine. The 3 plates were washed twice with ice-cold PBS, and the pellets were harvested in 1 ml PBS with protease inhibitor cocktail and pooled together in a 15-ml tube in order to obtain 1.5 × 106 cells. DNA fragmentation of the pellets was achieved by sonication, 35 cycles of 20 seconds each at maximum speed using OMNI-Ruptor 4000 (OMNI International Inc). IP was performed with 10 μg FLAG-tagged M2 antibody (Sigma-Aldrich) and equivalent amount of mouse IgG as negative control. Washes and elution of the IP DNA were performed according to the Magna ChIP protocol (Millipore). PCR primers were designed based on the peak interval sequence associated with cyclin D1 (Supplemental Figure 5), and the PCR products were visualized by agarose gel electrophoresis.
PI staining.
1 × 106 randomly cycling cells were washed in PBS and fixed overnight in 70% ethanol. RNase A (10 mg/ml) was treated for 30 minutes at room temperature, washed, and stained with PI (20 μg/ml). PI staining was measured using a FACSCalibur flow cytometer (BD Biosciences).
Real-time PCR.
RNA quantitation was conducted in an Agilent 2100 bioanalyzer (Agilent Technologies), using Power SYBR Green (AB Biosciences) according to the manufacturer’s guidelines. Equal quantities of RNA were used for the reverse transcription reactions. Primers (Supplemental Figure 5) for all the genes were designed using GenScript’s bioinformatics tools (GenScript).
SKY.
SKY was carried out as described previously (59). Briefly, fluorescence color images of chromosomes stained by rhodamine, Texas Red, Cy5, FITC, and Cy5.5 were captured under a Nikon microscope equipped with a spectral cube and Interferometer module. SKY View software (version 1.62) was used to analyze chromosomal number and structural alterations of chromosomes, including simple balanced translocations, unbalanced translocations (i.e., NRTs), deletions, and duplications. At least 20 metaphases were analyzed per sample.
Microarray analysis.
Affymetrix Expression Console 1.1 or the R statistic console with limma package was used to compute Robust Multichip Average (RMA) expression values for the Mouse Gene 1.0 ST microarrays and Mouse 430A 2.0 microarrays. Microarray data have been deposited in GEO (accession no. GSE35076; http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE35076). The core set of probe set clusters was used with annotation version na30, dated December 2009. The dataset was imported into Matlab version R2010b (The Mathworks), and 1-way ANOVA was used to evaluate the significance of differential expression between biological conditions. Genes with a differential expression P value of 0.01 or less and an absolute fold change of 1.25 or more were clustered and visualized using a clustergram heatmap.
Analysis within public microarray datasets.
A breast cancer microarray dataset that was previously compiled from the public repositories Gene Expression Omnibus ( http://www.ncbi.nlm.nih.gov/geo/; ref. 60) and ArrayExpress ( http://www.ebi.ac.uk/arrayexpress/; ref. 61) was used to evaluate CIN and CCND1 transcript level expression in the context of clinical samples (37).
Immunofluorescence and confocal analysis.
Immunofluorescence was performed as previously described by Silkworth et al. (62).
Statistics.
To determine the number of ChIP-Seq peaks, a MACs algorithm was used (4.35% false discovery rate). Analysis of TF enrichment within the interval sequences produced by the ChIP-Seq data was computed using a permutation test. Enrichment for high CIN scoring genes between 2 sets was compared using Wilcoxon matched-pairs test. Kaplan-Meier plots were compared by log-rank test. Correlation between cyclin D1 expression and CIN was evaluated using χ2 test. For comparison between 2 independent groups, 2-tailed Student’s t test was used. A P value less than 0.05 was considered significant.
Study approval.
The Thomas Jefferson University Institutional Animal Care and Use Committee approved the mouse study protocols described herein.
Supplementary Material
Acknowledgments
This work was supported in part by Susan Komen Breast Cancer Foundation awards BCTR0504227 (to C. Wang) and PDF2000167 (to A. Arnold); by NIH grants R01CA70896, R01CA75503, and R01CA86072 (to R.G. Pestell), R01CA55909 (to A. Arnold), and R01CA12934 (to E.S. Knudsen); by the China Scholarship Council; by a Pennsylvania Department of Health grant (to C. Wang and R.G. Pestell); and by the Murray-Heilig Fund in Molecular Medicine (to A. Arnold). Work conducted at the Kimmel Cancer Center was supported by NIH Cancer Center Core grant P30CA56036 (to R.G. Pestell). The Pennsylvania Department of Health specifically disclaims responsibility for any analyses, interpretations, or conclusions.
Footnotes
Conflict of interest: The authors have declared that no conflict of interest exists.
Citation for this article: J Clin Invest. 2012;122(3):833–843. doi:10.1172/JCI60256.
References
- 1.Malumbres M, Barbacid M. Cell cycle, CDKs and cancer: a changing paradigm. Nat Rev Cancer. 2009;9(3):153–166. doi: 10.1038/nrc2602. [DOI] [PubMed] [Google Scholar]
- 2.Lengauer C, Kinzler KW, Vogelstein B. Genetic instabilities in human cancers. Nature. 1998;396(6712):643–649. doi: 10.1038/25292. [DOI] [PubMed] [Google Scholar]
- 3.Thompson SL, Bakhoum SF, Compton DA. Mechanisms of chromosomal instability. Curr Biol. 2010;20(6):R285–295. doi: 10.1016/j.cub.2010.01.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gollin SM. Mechanisms leading to chromosomal instability. Semin Cancer Biol. 2005;15(1):33–42. doi: 10.1016/j.semcancer.2004.09.004. [DOI] [PubMed] [Google Scholar]
- 5.Draviam VM, Xie S, Sorger PK. Chromosome segregation and genomic stability. Curr Opin Genet Dev. 2004;14(2):120–125. doi: 10.1016/j.gde.2004.02.007. [DOI] [PubMed] [Google Scholar]
- 6.Spruck CH, Won KA, Reed SI. Deregulated cyclin E induces chromosome instability. Nature. 1999;401(6750):297–300. doi: 10.1038/45836. [DOI] [PubMed] [Google Scholar]
- 7.Carter SL, Eklund AC, Kohane IS, Harris LN, Szallasi Z. A signature of chromosomal instability inferred from gene expression profiles predicts clinical outcome in multiple human cancers. Nat Genet. 2006;38(9):1043–1048. doi: 10.1038/ng1861. [DOI] [PubMed] [Google Scholar]
- 8.Mayhew CN, et al. RB loss abrogates cell cycle control and genome integrity to promote liver tumorigenesis. Gastroenterology. 2007;133(3):976–984. doi: 10.1053/j.gastro.2007.06.025. [DOI] [PubMed] [Google Scholar]
- 9.Weinberg RA. The retinoblastoma protein and cell cycle control. Cell. 1995;81(3):323–330. doi: 10.1016/0092-8674(95)90385-2. [DOI] [PubMed] [Google Scholar]
- 10.Kato J-Y, Matsushime H, Hiebert SW, Ewen ME, Sherr CJ. Direct binding of cyclin D to the retinoblastoma gene product (pRb) and pRb phosphorylation by the cyclin D-dependent kinase CDK4. Genes Dev. 1993;7(3):331–342. doi: 10.1101/gad.7.3.331. [DOI] [PubMed] [Google Scholar]
- 11.Ewen ME, Sluss HK, Sherr CJ, Matsushime H, Kato J-Y, Livingston DM. Functional interactions of the retinoblastoma protein with mammalian D-type cyclins. Cell. 1993;73(3):487–497. doi: 10.1016/0092-8674(93)90136-E. [DOI] [PubMed] [Google Scholar]
- 12.Sakamaki T, et al. Cyclin D1 determines mitochondrial function in vivo. Mol Cell Biol. 2006;26(14):5449–5469. doi: 10.1128/MCB.02074-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wang C, et al. Cyclin D1 repression of nuclear respiratory factor 1 integrates nuclear DNA synthesis and mitochondrial function. Proc Natl Acad Sci U S A. 2006;103(31):11567–11572. doi: 10.1073/pnas.0603363103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Stacey DW. Three observations that have changed our understanding of cyclin D1 and p27 in cell cycle control. Genes Cancer. 2010;1(12):1189–1199. doi: 10.1177/1947601911403475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pestell RG, Albanese C, Reutens AT, Segall JE, Lee RJ, Arnold A. The cyclins and cyclin-dependent kinase inhibitors in hormonal regulation of proliferation and differentiation. Endocr Rev. 1999;20(4):501–534. doi: 10.1210/er.20.4.501. [DOI] [PubMed] [Google Scholar]
- 16.Fu M, Wang C, Li Z, Sakamaki T, Pestell RG. Minireview: Cyclin D1: normal and abnormal functions. Endocrinology. 2004;145(12):5439–5447. doi: 10.1210/en.2004-0959. [DOI] [PubMed] [Google Scholar]
- 17.Fu M, et al. Cyclin D1 inhibits peroxisome proliferator-activated receptor gamma-mediated adipogenesis through histone deacetylase recruitment. J Biol Chem. 2005;280(17):16934–16941. doi: 10.1074/jbc.M500403200. [DOI] [PubMed] [Google Scholar]
- 18.Keyomarsi K, et al. Cyclin E and survival in patients with breast cancer. N Engl J Med. 2002;347(20):1566–1575. doi: 10.1056/NEJMoa021153. [DOI] [PubMed] [Google Scholar]
- 19.Wang TC, Cardiff RD, Zukerberg L, Lees E, Arnold A, Schmidt EV. Mammary hyperplasia and carcinoma in MMTV-cyclin D1 transgenic mice. Nature. 1994;369(6482):669–671. doi: 10.1038/369669a0. [DOI] [PubMed] [Google Scholar]
- 20.Sicinski P, et al. Cyclin D1 provides a link between development and oncogenesis in the retina and breast. Cell. 1995;82(4):621–630. doi: 10.1016/0092-8674(95)90034-9. [DOI] [PubMed] [Google Scholar]
- 21.Yu Q, Geng Y, Sicinski P. Specific protection against breast cancers by cyclin D1 ablation. Nature. 2001;411(6841):1017–1021. doi: 10.1038/35082500. [DOI] [PubMed] [Google Scholar]
- 22.Hulit J, et al. Cyclin D1 genetic heterozygosity regulates colonic epithelial cell differentiation and tumor number in ApcMin mice. Mol Cell Biol. 2004;24(17):7598–7611. doi: 10.1128/MCB.24.17.7598-7611.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zwijsen RM, Wientjens E, Klompmaker R, van der Sman J, Bernards R, Michalides RJ. CDK-independent activation of estrogen receptor by cyclin D1. Cell. 1997;88(3):405–415. doi: 10.1016/S0092-8674(00)81879-6. [DOI] [PubMed] [Google Scholar]
- 24.Zwijsen RM, Buckle RS, Hijmans EM, Loomans CJ, Bernards R. Ligand-independent recruitment of steroid receptor coactivators to estrogen receptor by cyclin D1. Genes Dev. 1998;12(22):3488–3498. doi: 10.1101/gad.12.22.3488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Reutens AT, et al. Cyclin D1 binds the androgen receptor and regulates hormone-dependent signaling in a p300/CBP-associated factor (P/CAF)-dependent manner. Mol Endocrinol. 2001;15(5):797–811. doi: 10.1210/me.15.5.797. [DOI] [PubMed] [Google Scholar]
- 26.Fu M, et al. Cyclin D1 represses p300 transactivation through a cyclin-dependent kinase-independent mechanism. J Biol Chem. 2005;280(33):29728–29742. doi: 10.1074/jbc.M503188200. [DOI] [PubMed] [Google Scholar]
- 27.Bienvenu F, Barre B, Giraud S, Avril S, Coqueret O. Transcriptional regulation by a DNA-associated form of cyclin D1. Mol Biol Cell. 2005;16(4):1850–1858. doi: 10.1091/mbc.E04-08-0654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bienvenu F, et al. Transcriptional role of cyclin D1 in development revealed by a genetic-proteomic screen. Nature. 2010;463(7279):374–378. doi: 10.1038/nature08684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wang C, et al. Cyclin D1 antagonizes BRCA1 repression of estrogen receptor alpha activity. Cancer Res. 2005;65(15):6557–6567. doi: 10.1158/0008-5472.CAN-05-0486. [DOI] [PubMed] [Google Scholar]
- 30.Zhou J, et al. Attenuation of Forkhead signaling by the retinal determination factor DACH1. Proc Natl Acad Sci U S A. 2010;107(15):6864–6869. doi: 10.1073/pnas.1002746107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yu Z, et al. A cyclin D1/microRNA 17/20 regulatory feedback loop in control of breast cancer cell proliferation. J Cell Biol. 2008;182(3):509–517. doi: 10.1083/jcb.200801079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sandelin A, Alkema W, Engstrom P, Wasserman WW, Lenhard B. JASPAR: an open-access database for eukaryotic transcription factor binding profiles. Nucleic Acids Res. 2004;32(database issue):D91–D94. doi: 10.1093/nar/gkh012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hsu JY, et al. Mitotic phosphorylation of histone H3 is governed by Ipl1/aurora kinase and Glc7/PP1 phosphatase in budding yeast and nematodes. Cell. 2000;102(3):279–291. doi: 10.1016/S0092-8674(00)00034-9. [DOI] [PubMed] [Google Scholar]
- 34.Giet R, Glover DM. Drosophila aurora B kinase is required for histone H3 phosphorylation and condensin recruitment during chromosome condensation and to organize the central spindle during cytokinesis. J Cell Biol. 2001;152(4):669–682. doi: 10.1083/jcb.152.4.669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tatsuka M, et al. Multinuclearity and increased ploidy caused by overexpression of the aurora- and Ipl1-like midbody-associated protein mitotic kinase in human cancer cells. Cancer Res. 1998;58(21):4811–4816. [PubMed] [Google Scholar]
- 36.Artandi SE, et al. Telomere dysfunction promotes non-reciprocal translocations and epithelial cancers in mice. Nature. 2000;406(6796):641–645. doi: 10.1038/35020592. [DOI] [PubMed] [Google Scholar]
- 37.Ertel A, et al. RB-pathway disruption in breast cancer: differential association with disease subtypes, disease-specific prognosis and therapeutic response. Cell Cycle. 2010;9(20):4153–4163. doi: 10.4161/cc.9.20.13454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hu Z, et al. The molecular portraits of breast tumors are conserved across microarray platforms. BMC Genomics. 2006;7:96. doi: 10.1186/1471-2164-7-96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Shih IM, Zhou W, Goodman SN, Lengauer C, Kinzler KW, Vogelstein B. Evidence that genetic instability occurs at an early stage of colorectal tumorigenesis. Cancer research. 2001;61(3):818–822. [PubMed] [Google Scholar]
- 40.Kops GJ, Weaver BA, Cleveland DW. On the road to cancer: aneuploidy and the mitotic checkpoint. Nat Rev Cancer. 2005;5(10):773–785. doi: 10.1038/nrc1714. [DOI] [PubMed] [Google Scholar]
- 41.Nelsen CJ, et al. Short term cyclin D1 overexpression induces centrosome amplification, mitotic spindle abnormalities, and aneuploidy. J Biol Chem. 2005;280(1):768–776. doi: 10.1074/jbc.M407105200. [DOI] [PubMed] [Google Scholar]
- 42.Aggarwal P, et al. Nuclear accumulation of cyclin D1 during S phase inhibits Cul4-dependent Cdt1 proteolysis and triggers p53-dependent DNA rereplication. Genes Dev. 2007;21(22):2908–2922. doi: 10.1101/gad.1586007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Del Rey J, et al. Centrosome clustering and cyclin D1 gene amplification in double minutes are common events in chromosomal unstable bladder tumors. BMC Cancer. 2010;10:280. doi: 10.1186/1471-2407-10-280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Jirstrom K, et al. Adverse effect of adjuvant tamoxifen in premenopausal breast cancer with cyclin D1 gene amplification. Cancer Res. 2005;65(17):8009–8016. doi: 10.1158/0008-5472.CAN-05-0746. [DOI] [PubMed] [Google Scholar]
- 45.Millar EK, et al. Cyclin D1b protein expression in breast cancer is independent of cyclin D1a and associated with poor disease outcome. Oncogene. 2009;28(15):1812–1820. doi: 10.1038/onc.2009.13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Manchado E, Malumbres M. Targeting aneuploidy for cancer therapy. Cell. 2011;144(4):465–466. doi: 10.1016/j.cell.2011.01.037. [DOI] [PubMed] [Google Scholar]
- 47.Tang YC, Williams BR, Siegel JJ, Amon A. Identification of aneuploidy-selective antiproliferation compounds. Cell. 2011;144(4):499–512. doi: 10.1016/j.cell.2011.01.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Li Z, et al. Alternative cyclin d1 splice forms differentially regulate the DNA damage response. Cancer Res. 2010;70(21):8802–8811. doi: 10.1158/0008-5472.CAN-10-0312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Jirawatnotai S, et al. A function for cyclin D1 in DNA repair uncovered by protein interactome analyses in human cancers. Nature. 2011;474(7350):230–234. doi: 10.1038/nature10155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Bell AC, West AG, Felsenfeld G. The protein CTCF is required for the enhancer blocking activity of vertebrate insulators. Cell. 1999;98(3):387–396. doi: 10.1016/S0092-8674(00)81967-4. [DOI] [PubMed] [Google Scholar]
- 51.Wendt KS, et al. Cohesin mediates transcriptional insulation by CCCTC-binding factor. Nature. 2008;451(7180):796–801. doi: 10.1038/nature06634. [DOI] [PubMed] [Google Scholar]
- 52.Rubio ED, et al. CTCF physically links cohesin to chromatin. Proc Natl Acad Sci U S A. 2008;105(24):8309–8314. doi: 10.1073/pnas.0801273105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Li Z, et al. Cyclin D1 regulates cellular migration through the inhibition of thrombospondin 1 and ROCK signaling. Mol Cell Biol. 2006;26(11):4240–4256. doi: 10.1128/MCB.02124-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wang C, et al. Cyclin D1 repression of peroxisome proliferator-activated receptor gamma expression and transactivation. Mol Cell Biol. 2003;23(17):6159–6173. doi: 10.1128/MCB.23.17.6159-6173.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Labhart P, et al. Identification of target genes in breast cancer cells directly regulated by the SRC-3/AIB1 coactivator. Proc Natl Acad Sci U S A. 2005;102(5):1339–1344. doi: 10.1073/pnas.0409578102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Bromberg JF, et al. Stat3 as an oncogene. Cell. 1999;98(3):295–303. doi: 10.1016/S0092-8674(00)81959-5. [DOI] [PubMed] [Google Scholar]
- 57.Lee RJ, et al. pp60(v-src) induction of cyclin D1 requires collaborative interactions between the extracellular signal-regulated kinase, p38, and Jun kinase pathways. A role for cAMP response element-binding protein and activating transcription factor-2 in pp60(v-src) signaling in breast cancer cells. J Biol Chem. 1999;274(11):7341–7350. doi: 10.1074/jbc.274.11.7341. [DOI] [PubMed] [Google Scholar]
- 58.Wu K, et al. Cell fate determination factor Dachshund reprograms breast cancer stem cell function. J Biol Chem. 2011;286(3):2132–2142. doi: 10.1074/jbc.M110.148395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Macville M, et al. Spectral karyotyping, a 24-colour FISH technique for the identification of chromosomal rearrangements. Histochem Cell Biol. 1997;108(4–5):299–305. doi: 10.1007/s004180050169. [DOI] [PubMed] [Google Scholar]
- 60.Barrett T, et al. NCBI GEO: mining tens of millions of expression profiles--database and tools update. Nucl Acids Res. 2007;35(database issue):D760–D765. doi: 10.1093/nar/gkl887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Brazma A, et al. ArrayExpress--a public repository for microarray gene expression data at the EBI. Nucleic Acids Res. 2003;31(1):68–71. doi: 10.1093/nar/gkg091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Silkworth WT, Nardi IK, Scholl LM, Cimini D. Multipolar spindle pole coalescence is a major source of kinetochore mis-attachment and chromosome mis-segregation in cancer cells. PLoS One. 2009;4(8):e6564. doi: 10.1371/journal.pone.0006564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Li Z, et al. Alternate Cyclin D1 mRNA Splicing Modulates p27KIP1 Binding and Cell Migration. J Biol Chem. 2008;283(11):7007–7015. doi: 10.1074/jbc.M706992200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.