Abstract
Natural killer (NK) cells are circulating cytotoxic lymphocytes that exert potent and nonredundant antiviral activity and antitumoral activity in the mouse; however, their function in host defense in humans remains unclear. Here, we investigated 6 related patients with autosomal recessive growth retardation, adrenal insufficiency, and a selective NK cell deficiency characterized by a lack of the CD56dim NK subset. Using linkage analysis and fine mapping, we identified the disease-causing gene, MCM4, which encodes a component of the MCM2-7 helicase complex required for DNA replication. A splice-site mutation in the patients produced a frameshift, but the mutation was hypomorphic due to the creation of two new translation initiation methionine codons downstream of the premature termination codon. The patients’ fibroblasts exhibited genomic instability, which was rescued by expression of WT MCM4. These data indicate that the patients’ growth retardation and adrenal insufficiency likely reflect the ubiquitous but heterogeneous impact of the MCM4 mutation in various tissues. In addition, the specific loss of the NK CD56dim subset in patients was associated with a lower rate of NK CD56bright cell proliferation, and the maturation of NK CD56bright cells toward an NK CD56dim phenotype was tightly dependent on MCM4-dependent cell division. Thus, partial MCM4 deficiency results in a genetic syndrome of growth retardation with adrenal insufficiency and selective NK deficiency.
Introduction
Natural killer (NK) cells are circulating cytotoxic lymphocytes lacking antigen-specific T cell and B cell receptors (1, 2). They have been shown to exert potent and nonredundant antiviral activity and antitumoral activity in the mouse model (3–6). Their function in host defense in humans remains unclear, due to the lack of well-defined inherited disorders associated with a selective defect in NK cell development (7). Several children with a specific quantitative circulating NK cell defect but normal T cell counts have been reported (8–12). Some of these patients seem to be highly susceptible to viral infections, such as those caused by herpesviruses, in particular (8, 9). However, these cases were sporadic, and the defect in NK cells was documented after clinical viral diseases, raising the possibility that the viral infection might have contributed to the NK cell deficiency (13). We have previously described the first two forms of familial NK cell deficiency, in two unrelated families, defining a new Mendelian syndrome (OMIM 609981) (14, 15). In the first family, the eldest child died from cytomegalovirus (CMV) infection, whereas the second child was still healthy at the age of 13 years despite displaying a persistent lack of NK and NK T cells, with no diagnosed viral infection to date. However, this patient recently developed osteosarcoma (our unpublished observations). These two siblings also displayed severe intrauterine growth retardation, persistent neutropenia, and transient CD8+ T lymphopenia (14). Enhanced lymphocyte apoptosis has been shown to underlie this syndromic immunodeficiency (16). In 2006, we also reported 4 patients from a large nomadic Irish consanguineous kindred with NK cell deficiency and susceptibility to viral infections (15). The hematopoietic phenotype specifically affects the NK cell lineage. These patients also display adrenal insufficiency, as reported (17), and developmental abnormalities, including ante- and postnatal growth retardation and microcephaly in particular. One of the patients developed an Epstein-Barr virus–driven (EBV-driven) lymphoproliferative disorder, and two others developed severe respiratory illnesses that were probably of viral origin. We mapped this immunodeficiency to a single 12-Mb region on the centromeric region of chromosome 8 (15) and then tried to decipher the molecular genetic basis of this new syndrome combining selective NK cell deficiency, growth retardation, adrenal insufficiency, and predisposition to viral infections and, possibly, cancer (18).
Results
Identification of a mutation in MCM4.
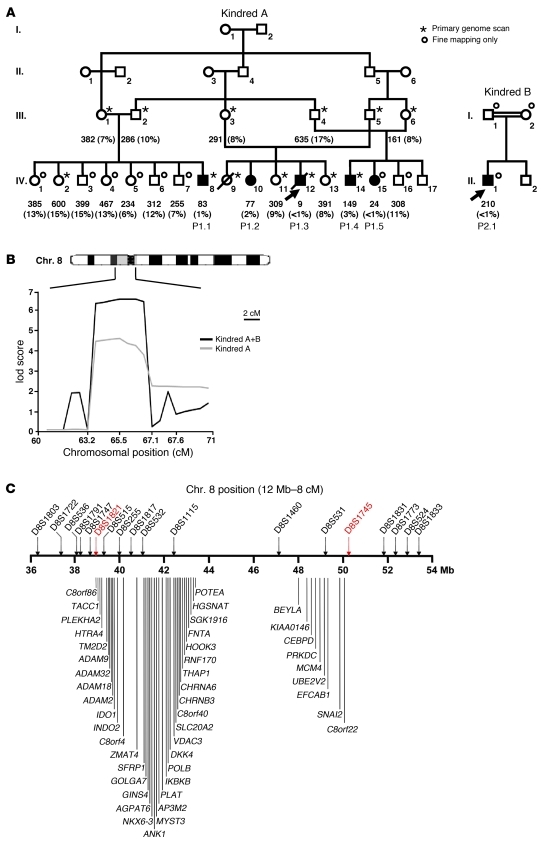
The interval previously linked to the familial defect mapped to the centromeric region of chromosome 8 (15). Two new members of this family were subsequently enrolled, including a patient with the same clinical phenotype, including a low NK cell count (<4%) (Figure 1A, kindred A). We also enrolled a new consanguineous nomadic Irish family, possibly related to the first family, including a child with growth retardation, adrenal insufficiency, a low NK cell count (<1%), and a history of recurrent infections with herpes simplex virus (HSV) and varicella zoster virus (VZV) (Figure 1A, kindred B). The growth retardation and adrenal insufficiency of all patients are detailed in Supplemental Table 1 (supplemental material available online with this article; doi: 10.1172/JCI61014DS1). Multipoint linkage analysis was performed with both families (a total of 6 patients and 18 healthy siblings), and the maximum lod score was obtained for the same region of chromosome 8, with a value increasing from 4.51 to 6.45 at marker D8S532 (see Supplemental Methods); the allelic profile was the same in both families, suggesting that the two families share the same genetic defect due to a founder effect. Fine mapping identified a narrower region of interest (8p11.23–q11.21) overlapping the centromeric region of chromosome 8 and extending from D8S1821 to D8S1745 (Figure 1B). None of the 45 genes encoding proteins in this region were obvious candidates. We therefore sequenced the coding region, the 5′ and 3′ UTR of all coding genes, and one miRNA from either genomic or complementary DNA (Figure 1C and see Supplemental Methods). All variations found in the patients are shown in Supplemental Figure 1A. We identified a homozygous substitution (A→G) in the acceptor splice site of intron 1 in minichromosome maintenance–deficient 4 (MCM4) (Figure 2A). This substitution, c.71-2A→G, shifts the splice site by 1 nucleotide and leads to the insertion of a single nucleotide into the cDNA, c.70_71insG (Figure 2B and Supplemental Figure 1B). This insertion creates a frameshift, with a premature stop codon at position 27 in the new protein, which is thus truncated, the normal protein being 863 amino acids long. The new stop codon is N-terminal to all known functional domains of MCM4 (Figure 2C). The same homozygous mutation was found in all 6 patients. The parents were heterozygous, and the healthy relatives were either heterozygous or homozygous WT. The mutation was not found in any database (NCBI, Ensembl, 1000 Genomes) or in any of the 1,003 individuals from 52 ethnic groups sequenced (the Human Genome Diversity Project–Centre d’Etude du Polymorphisme Humain [HGDP-CEPH] panel) (19), suggesting that it may be a rare pathogenic mutation rather than an irrelevant polymorphism. Finally, Hughes and colleagues identified the same mutation independently, through a whole-exome sequencing approach, in 8 other Irish patients with the same clinical phenotype (20).
Figure 1. Linkage analysis of NK cell deficiency in humans.
(A) Pedigrees of the two families. Generations are designated by Roman numerals I–IV. Patients with low counts of NK cells (P1.1, P1.2, P1.3, P1.4, P1.5, and P2.1) are represented by black symbols. The index case is indicated by an arrow. All other family members with normal NK cell counts are indicated by white symbols. A star indicates that the individual has been genotyped for all the microsatellites considered, whereas a circle indicates that genotyping has been carried out only for the 8p12–q12.2 region. Absolute numbers (per mm3 of whole blood) and percentages (% of lymphocytes) of NK cells are indicated for each individual, for the first whole-blood sample analyzed. (B) Multipoint linkage analysis of chromosome region 8p12–q12.2 by homozygosity mapping. Microsatellite positions are indicated in cM. In total, 9 microsatellites were genotyped between D8S1821 and D8S1745. The gray line represents kindred A lod score, and the black line represents the combined lod score for kindreds A and B. (C) Schematic representation of the candidate region (8p11.23–q11.21). The region of interest was delineated between microsatellites D8S1821 and D8S1745. This region is 12 Mb long and contains 1 microRNA and 45 predicted protein-coding genes.
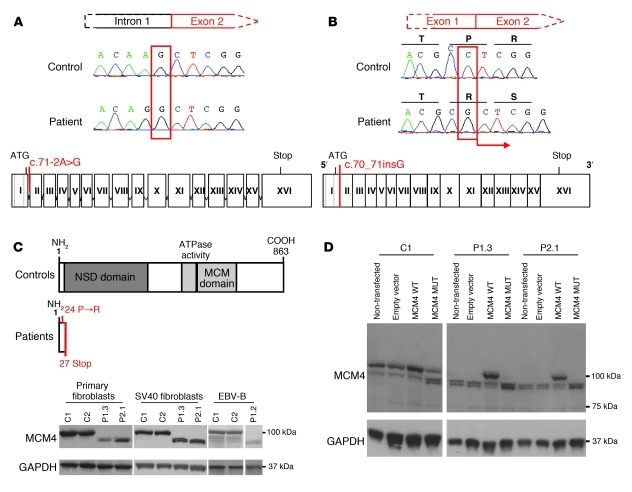
Figure 2. The MCM4 mutation is not associated with a loss of expression.
(A) Automated sequencing profiles showing the homozygous MCM4 c.70_71insG mutation in genomic DNA extracted from EBV-B cells of a patient and a WT control. Bottom: Schematic diagram of the structure of the MCM4 gene, consisting of 16 or 17 exons (Roman numerals), indicating the position of the mutation, which affects the acceptor splice site of intron 1. (B) Sequencing profile of a patient and a control indicating the insertion of a G nucleotide in the cDNA extracted from EBV-B cells. The position of the mutation is shown on a diagram of the MCM4 gene. The dotted lines represent the two mRNA transcripts produced from MCM4. The homozygous mutation leads to the insertion of an additional nucleotide between exons 1 and 2. (C) A schematic diagram of the MCM4 protein, which has an N-terminal serine/threonine-rich domain (dark gray) and a conserved MCM domain (light gray) including an ATP-binding site (black) toward its C terminus. The homozygous mutation results in a frameshift, creating a premature stop codon in exon 2. Bottom: Western blot analysis of MCM4 on total protein extracts from primary fibroblasts and SV40 fibroblasts from P1.3 and P2.1 and EBV-B cells from P1.2, controls. A polyclonal MCM4 antibody was used. (D) Complementation, by lentiviral particles, of SV40 fibroblasts from the controls and patients, with an empty pTRIP vector, an MCM4 WT vector, and the MCM4 c.71-2A→G mutation (MCM4 MUT). MCM4 was detected with a polyclonal antibody. The empty vector and non-transfected cells were used as a negative transfection control. In C and D, GAPDH was used as a loading control.
Impaired production of full-length MCM4 mRNA and protein.
Similar amounts of MCM4 mRNA were detected in EBV-transformed B cell lines (EBV-B cells) as well as primary and fibroblastic cell lines (SV40 fibroblasts) from patients and controls (Supplemental Figure 1C and see Supplemental Methods). This suggests that the insertion of a single nucleotide, due to the c.70_71insG mutation, did not destabilize the mRNA. By contrast, we detected no full-length MCM4 protein (~100 kDa) in patient 1.2 (P1.2) EBV-B cells as well as P1.3 and P2.1 primary and SV40 fibroblasts, by Western blotting with a polyclonal antibody recognizing the first 300 amino acids of MCM4, whereas this protein was detected in control cells (Figure 2C and Supplemental Figure 1D). Instead, we detected two other more rapidly migrating proteins of lower apparent molecular mass (~95 kDa and ~90 kDa) in the patients’ cells. These proteins were also detected in control cells, albeit in much smaller amounts than the full-length protein. Stable transfection with expression vectors carrying the WT MCM4 allele rescued normal MCM4 protein production (~100 kDa) in the SV40 fibroblasts from P1.3 and P2.1 (Figure 2D). Following the transfection of control cells with the mutated MCM4 cDNA, we detected the presence of the same two more rapidly migrating bands found in larger amounts in the patients’ cells (Figure 2D). Together, these results suggest that no WT MCM4 protein was produced in the patients’ cells and that the two more rapidly migrating bands were shorter forms of MCM4.
Reinitiation of MCM4 translation.
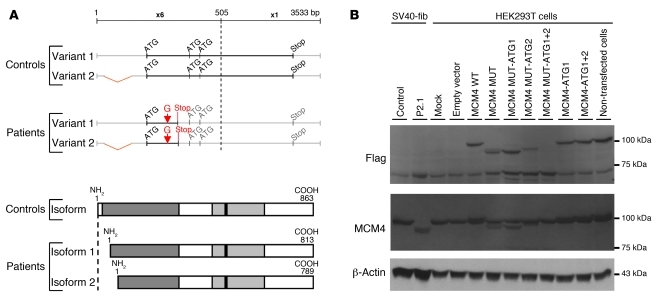
We then investigated the presence of MCM4 in SV40 fibroblasts from controls, P1.3 and P2.1, using two other antibodies, specific for the N-terminal and C-terminal domains of MCM4. The N-terminal antibody recognizes a peptide sequence N-terminal to the mutation. We detected no MCM4 protein (~100, ~95, and ~90 kDa) in whole-cell, cytoplasmic, and nuclear extracts from the patients’ SV40 fibroblasts. By contrast, in experiments with the C-terminal domain–specific antibody, the two smaller proteins (~95 and ~90 kDa) were detected in SV40 fibroblasts from the patients, but not in control cells (Supplemental Figure 2A). The two more rapidly migrating proteins (~95 and ~90 kDa) detected in the patients’ cells may be encoded by the mutant MCM4 gene and generated by the initiation of translation at two ATG codons in positions 51 and 75, giving rise to proteins with predicted sizes of 94 and 91 kDa, respectively (Figure 3A). We tested this hypothesis by transfecting HEK293T cells with various Flag-tagged expression vectors carrying WT and various mutant MCM4 alleles: the patients’ mutation (c.70_71insG) only (MUT), the patients’ mutation and the M51G substitution (MUT-ATG1), the patients’ mutation and the M75G substitution (MUT-ATG2), and all three mutations (MUT-ATG1+2). In Western blots probed with antibodies against Flag and MCM4, two proteins of ~94 and ~91 kDa were detected in MUT-transfected cells, whereas the ~94-kDa protein was not detected in the MUT-ATG1–transfected cells, and the ~91 kDa protein was not detected in the MUT-ATG2–transfected cells. Neither of these bands was detected in MUT-ATG1+2–transfected cells (Figure 3B). These findings imply that the c.70_71insG mutation results in the initiation of translation from two different ATG codons downstream from the mutation and the premature stop codon. Primary and SV40 fibroblasts from P1.3, P2.1, and controls were transfected with siRNAs against MCM4, to demonstrate that the two more rapidly migrating bands were indeed MCM4 isoforms (see Supplemental Methods). Transfection with 3 different siRNAs (si793, si1299, and si1325) against MCM4, but not with irrelevant siRNAs (EBNA or GL3), led to the disappearance of the full-length MCM4 protein from control cells and of the shorter isoforms from P2.1 fibroblasts, confirming that the two more rapidly migrating bands corresponded to isoforms of MCM4 generated by the initiation of translation downstream from the homozygous mutation in the patients’ cells (Supplemental Figure 2B and data not shown). These data suggest that the homozygous MCM4 mutation prevents the production of the N-terminal domain of MCM4, this protein domain being well conserved among vertebrate species (Supplemental Figure 3).
Figure 3. Characterization of the MCM4 isoforms detected in the cells of the patient tested.
Reinitiation of MCM4 protein translation. (A) Schematic diagram of the two potential reinitiation sites after the premature STOP codon and the corresponding isoforms of MCM4. Two ATG codons, at positions 51 and 75, are in the same open reading frame as the premature stop codon, leading to the production of two new isoforms, of 813 and 789 amino acids, respectively. The 5′ part of the MCM4 sequence is enlarged (×6) and shown at greater magnification than the 3′ part of the MCM4 sequence (×1). The two variants of MCM4 mRNA from the NCBI database are shown (NM 005914.3 and NM 182746.2). (B) MCM4 protein levels, in HEK293T cells, following transient transfection with a C-terminal Flag-tagged pCMV6 empty vector or pCMV6 MCM4 WT, pCMV6 MCM4 MUT, pCMV6 MCM4 MUT-ATG1, pCMV6 MCM4 MUT-ATG2, pCMV6 MCM4 MUT-ATG1+2, pCMV6 MCM4-ATG1, and pCMV6 MCM4-ATG1+2 vectors, were assessed by Western blotting of total protein extracts from each transfection with antibodies against Flag and against the MCM4 protein. Total protein extracts from non-transfected control SV40 fibroblast (SV40-Fib) cell lines from a control and patient P2.1 were used as a positive control. An antibody against β-actin was used as a loading control.
Normal MCM2-7 complex formation in patients’ cells.
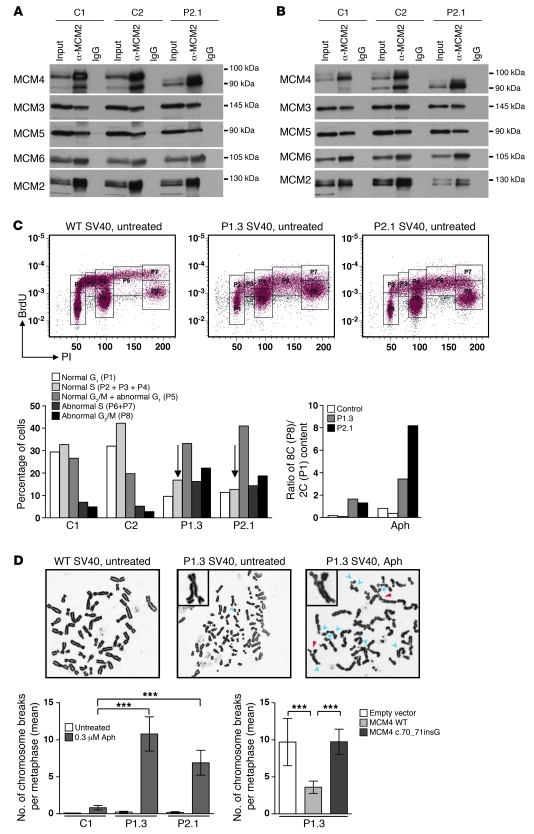
MCM4 forms a highly conserved hexameric complex with MCM2, MCM3, MCM5, MCM6, and MCM7; this complex is known as the minichromosome maintenance (MCM2-7) complex (21). The MCM2-7 complex is required for both the initiation and elongation phases of eukaryotic DNA replication (22, 23). The first step of DNA replication is the recruitment and loading of the inactive preformed hexameric MCM2-7 complex bound to CDT1 onto origins of replication by the origin recognition complex (ORC) and cell division cycle 6 (CDC6) to form a pre-replication complex (pre-RC). We assessed the effect of the mutation on MCM4 function by first examining the formation of the MCM2-7 complex. In mammalian nuclei, MCM heterocomplexes may take two different forms, one of which can be extracted with non-ionic detergents, the other being tightly bound to DNA and extractable by DNase I treatment (24). The chromatin-bound form is thought to be associated with pre-RCs. We therefore investigated the possible recruitment of the mutant MCM4 protein to the MCM2-7 complex and its loading onto chromatin. Detergent-soluble and DNase I–extracted fractions from cell extracts were subjected to immunoprecipitation with an anti-MCM2 antibody. Similar amounts of the MCM2, MCM3, MCM6, MCM5, and MCM4 (isoforms of 100 kDa in controls and ~94 kDa or ~91 kDa in patients) proteins were found in the cells from controls and patients, indicating that the two MCM4 isoforms in the patients were able to interact with the normal partners of MCM4, at least in SV40 fibroblasts (Figure 4A). The next step was to determine whether the MCM complex bound chromatin. The chromatin-bound fraction contained normal amounts of the MCM2-7 complex in both control cells and cells from patients (Figure 4B). Thus, the N-terminal domain of MCM4 was not essential for the formation of the MCM2-7 complex and its binding to DNA.
Figure 4. Functionality of the MCM4 isoforms detected in the cells of the patient.
(A) The Triton X–extractable fraction from patient and control SV40 fibroblasts was subjected to immunoprecipitation with a monoclonal antibody against MCM2. Protein extracts and immunoprecipitates were analyzed by immunoblotting with antibodies against MCM4, MCM3, MCM5, and MCM6. MCM2 was used as a loading control. (B) The chromatin-bound fraction was subjected to immunoprecipitation with a monoclonal antibody against MCM2. This procedure was carried out on the DNase I–extracted fractions of both control cells and cells from the patient. Protein extracts and immunoprecipitates were analyzed by immunoblotting with antibodies against MCM4, MCM3, MCM5, and MCM6. MCM2 was used as a loading control. Immunoprecipitation with IgG was used as a negative control. (C) Representative flow cytometry plots of the cell cycle of SV40 fibroblasts from controls and patients. Control (left), P1.3 (middle), and P2.1 (right) cell cycles in the absence of treatment. Transformation of cells with SV40 T antigen causes increased ploidy of all cells (59). P1 corresponds to normal G1 phase, P2+P3+P4 correspond to normal S phase, P5 corresponds to normal G2 phase plus abnormal G1 or failed mitosis, P6+P7 correspond to re-replication S phase, and P8 corresponds to 8C (P8) DNA content. Patients’ SV40 fibroblasts with or without 0.3 μM aphidicolin (Aph) treatment. (D) Representative chromosome spreads of chromosome breaks induced with or without aphidicolin. A WT metaphase chromosome without aberrations (left); a P1.3 metaphase with some aberrations indicated by blue arrowhead (middle); and a P1.3 metaphase after aphidicolin treatment, with chromosomal aberrations indicated by arrowheads (right). Blue arrowheads indicate chromatid breaks, and red arrowheads indicate chromosome exchanges. Bottom: Chromosome breaks (mean) per metaphase in P1.3 and P2.1 SV40 fibroblasts and in control SV40 fibroblasts. Complementation by lentiviral transduction with the WT MCM4 allele, the empty vector, or the c.70_71insG allele in P1.3 SV40 fibroblasts. Error bars indicate SEM. ***P < 0.0005, Student’s t test.
The MCM4 mutation causes a cell-cycle defect.
The second step in the initiation of DNA replication requires cyclin-dependent kinase–mediated (CDK-mediated) activation of the pre-RC to form the preinitiation complex (pre-IC), which in turn facilitates DNA unwinding and polymerization (25–28). This separation into two steps — pre-RC loading and activation — is crucial for the coordination of DNA replication and the prevention of re-replication (29). We addressed the impact of the mutated MCM4 allele on DNA replication itself by investigating the cell cycle and DNA content in SV40 fibroblasts, by propidium iodide (PI) and BrdU labeling with (Supplemental Figure 2C) and without aphidicolin (an inhibitor of DNA replication) pretreatment (Figure 4C). In the absence of aphidicolin treatment, the proportion of cells in the G1 and S phases was lower in the patients than in the controls, whereas the proportion of cells in G2/M phase was higher (Figure 4C). The DNA content of the patients’ SV40 fibroblasts was also abnormal, with an 8C/2C ratio much higher than that in control SV40 fibroblasts (Figure 4C), indicating disruption of the coordination of DNA replication. Treatment with aphidicolin amplified this phenotype (Figure 4C and Supplemental Figure 2C). Thus, MCM4 mutation affects DNA replication by disrupting the normal control of the prevention of re-replication and having an impact on the mitotic phase.
The MCM4 mutation causes genomic instability, which is complemented by expression of the WT allele of MCM4.
Complete and accurate DNA replication is essential for the maintenance of genetic integrity in all organisms. When replication is compromised, the DNA becomes more prone to breakage. Genomic instability was assessed in the presence and absence of aphidicolin treatment in SV40 fibroblasts from patients and controls, through assessment of chromatid breaks and chromosome exchanges. The mean number of breaks per metaphase differed significantly between patients and controls, with values of 0.82 obtained for control cells and of 9.25 and 10.79 for cells from P1.3 and P2.1, respectively (Figure 4D and Supplemental Table 2). Primary fibroblasts from P2.1 also contained radial chromosomes after aphidicolin treatment (Supplementary Figure 2D and Supplemental Table 2). Genomic instability was specific to aphidicolin treatment in the patients’ fibroblasts, as we observed no increase in the frequency of DNA breakage after treatment with a crosslinking agent, diepoxybutane (DEB) (data not shown). Thus, the mutant MCM4 protein promoted repair inefficiently at sites of replication stress caused by aphidicolin. We speculate that these sites correspond to the fragile sites that can be enhanced by aphidicolin treatment. The patients’ lymphocytes were more susceptible to DNA breakage following treatment with DEB, mitomycin C (30) and nitrogen mustard (ref. 31 and Supplemental Table 3), although breakage rates remained much lower than those in cells from patients with Fanconi anemia (32). We expressed the WT MCM4 or the patients’ MCM4 allele in patient fibroblasts and assessed the breakage phenotype, to demonstrate that the observed genomic instability was due to the absence of the full-length MCM4 in the patients’ cells. Expression of the WT allele greatly decreased the number of DNA breaks per metaphase (from 9.7 to 3.6 breaks per metaphase for the WT allele), whereas expression of the mutant allele did not (Figure 4D and Supplemental Table 2). The genomic instability phenotype of the patients’ cells described here is similar to that of mice with a hypomorphic Chaos3 mutation in MCM4. The causal mutation in this model, F345I, is a well-conserved amino acid located close to the zinc finger motif, which is important for interactions between MCMs. Mice homozygous for the mutant Chaos3 allele of the Mcm4 gene are viable, whereas Mcm4 knockout is lethal before implantation, indicating that the Chaos3 mutant allele is hypomorphic (33). Embryonic fibroblasts with the homozygous Chaos3 mutation have been reported to be susceptible to chromosome breaks in the presence of aphidicolin (33).
Impact of MCM4 deficiency on NK cell terminal differentiation.
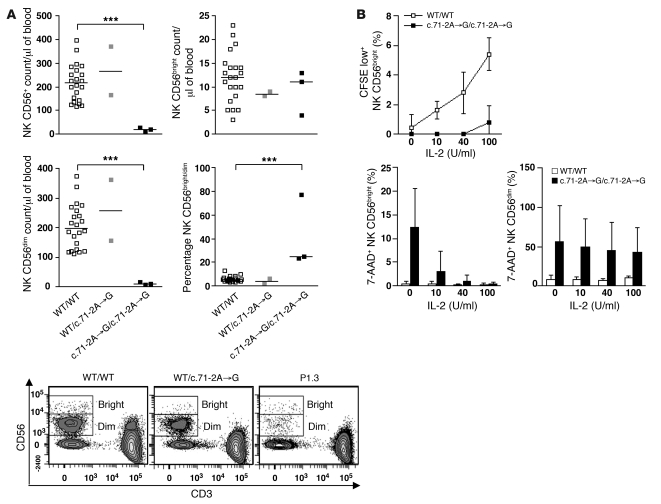
The immunological phenotype of the patients was characterized by a normal number of circulating cells of the myeloid and lymphoid subsets (Supplemental Figure 4, A and B, and Supplemental Figure 5A), with the exception of a selective decrease in the numbers of NK cells (Figure 5A) and a slightly smaller memory B cell subset (CD27+CD19+; Supplemental Figure 5A), despite ubiquitous MCM4 protein production (Supplemental Figure 4C). T cells proliferated normally in response to phytohemagglutinin (PHA) activation (Supplemental Figure 5B). The rate of apoptosis of PHA-activated T cell blasts cultured with or without IL-2 or IL-15 was similar in P1.2, P1.3, and controls (Supplemental Figure 5C). The proportion of lymphocytes was also affected in mice homozygous for the Chaos3 mutation. The percentages of T and B cells were lower and higher than normal, respectively, and, remarkably, a decrease in the proportion of NK cells was observed, supporting a role for MCM4 in NK cell development across species (Supplemental Figure 4D). However, the NK cell defect observed in this model was far too mild to reproduce the phenotype of patients, consistent with the lack of growth retardation in these hypomorphic Chaos3/Chaos3 mice. In humans, NK cells can be divided into two subsets on the basis of the surface density of CD56, an N-CAM isoform. In the linear differentiation model for human NK cells, CD56bright and CD56dim NK cells correspond to sequential steps in NK cell differentiation (34). CD56dim NK cells have a high surface density of the low-affinity Fc receptor CD16 (CD16hi) and account for 90% of peripheral blood NK cells. CD56bright NK cells are CD16lo/neg and account for less than 10% of circulating NK cells. CD56bright NK cells proliferate and produce IFN-γ in response to stimulation with cytokines, such as IL-2 and IL-18, whereas CD56dim NK cells are cytolytic and produce cytokines when they encounter target cells (35). The patients had very small numbers of NK cells, due to the selective loss of the CD56dim subset, resulting in an increase in the CD56bright/CD56dim cell ratio (Figure 5A). This finding was confirmed by the smaller percentage of CD16hi NK cells in the patients than in the controls (Supplemental Figure 6A and ref. 36). Thus, MCM4 deficiency was associated with a partial but effective blocking of the differentiation of NK cells into the CD56dim subset. Consistent with a role for MCM4 in the transition of CD56bright to CD56dim NK cells, MCM4 protein quantification showed that MCM4 was produced in slightly larger amounts in CD56bright cells than in CD56dim NK cells (Supplemental Figure 6B). CD56dim cells can be further subdivided into functional intermediates, based on a gradual increase in the surface expression of the sulfated carbohydrate CD57 and the progressive loss of surface CD94. The progression of CD56dim NK cells toward terminally differentiated CD57+CD94neg NK cells is accompanied by a loss of proliferative capacity and an increase in cytolytic activity (37, 38). Patients had smaller subsets of perforin+ NK cells, CD57+ NK cells, and CD94med/lo/neg NK cell subsets than controls, confirming the decrease in the size of all CD56dim subsets in patients (Supplemental Figure 6, A and C). By contrast, the CD3+perforin+ T cell subset was similar in patients and controls (Supplemental Figure 5D).
Figure 5. Homozygous MCM4 mutation and specific NK CD56dim deficiency.
(A) Quantitation, by flow cytometry, of peripheral total NK cells and of the CD56bright and CD56dim NK cell subsets in controls (n = 22), heterozygous subjects (n = 2), and homozygous patients (P1.1, P1.2, and P1.3). Horizontal bars represent medians. ***P < 0.0005, Student’s t test. Bottom: Representative flow cytometry plots of a homozygous WT sibling, a heterozygous sibling, and one patient. (B) PBMCs from 6 independent healthy controls and from one patient (P1.3) tested in 2 independent experiments were stained with CFSE and stimulated for 72 hours with various doses of IL-2. Apoptosis was assessed on NK subsets, by 7-AAD staining. Error bars indicate SD.
Proliferation and apoptosis of NK cells.
IL-2 and IL-15 promote NK cell proliferation and survival, respectively (39–43). We assessed whether the NK cell deficiency in patients was linked to a defect in NK cell proliferation and/or survival, as previously observed for T cells in another patient with a complete deficiency of NK and NK T cells as well as transitory CD8+ T lymphopenia (16) studied by single-cell flow cytometry analysis of intracytoplasmic CFSE dilution and 7-AAD staining. Unlike NK CD56bright cells from control individuals, the patients’ NK CD56bright cells did not proliferate in response to stimulation with IL-2 (Figure 5B) or IL-15 (Supplemental Figure 6D). We also observed excess spontaneous apoptosis in CD56bright NK cells in the absence of stimulation, this situation being reversed by treatment with exogenous IL-2 (Figure 5B) or IL-15 (Supplemental Figure 6D). Furthermore, some of the patients’ CD56dim NK cells displayed excess spontaneous apoptosis in vitro, and this was not corrected by the addition of exogenous IL-2 (Figure 5B) or IL-15 (Supplemental Figure 6D). Nevertheless, the patients’ NK CD56dim cells proliferated at rates similar to NK CD56dim cells from control individuals following stimulation with IL-2 or IL-15 (data not shown). These data suggest that the lack of NK cells was probably due to the accumulation of chromosomal aberrations during proliferation of the NK CD56bright subset, leading to the generation of only a few NK CD56dim cells. Thus, the full-length MCM4 protein is essential for NK cell differentiation, accounting for the selective lack of CD56dim NK cells in patients.
Discussion
We report here what we believe to be the first form of human MCM4 deficiency, in which a hypomorphic MCM4 allele is associated, in 6 Irish patients, with an autosomal recessive disease with growth retardation, adrenal insufficiency, NK deficiency, as well as predisposition to viral diseases and possibly also to cancer. An accompanying article (20) reports 8 other Irish patients with the same MCM4 genotype and the same clinical phenotype. The clinical phenotype of the patients, which is much more modest than that of mice with an embryonic lethal knockout of this gene (33), is accounted for by the hypomorphic nature of the mutant allele. The reinitiation of MCM4 translation results in the production of the two truncated isoforms, one lacking the first 50 amino acids and the other lacking the first 74 amino acids, in the patients’ cells. The N-terminal domain of the protein is well conserved in vertebrate species. The first 174 N-terminal amino acids of the yeast MCM4 protein form a serine/threonine-rich domain (NSD) including several protein kinase regulatory signals responsible for recruiting other DNA replication factors, leading to the formation of the pre-IC (26) required for progression through S phase (44–46). In budding yeast, the distal part of the NSD (residues 2–73), which has several CDK phosphorylation sites, supports Dbf4-Cdc7 protein kinase–independent (DDK-independent) growth, whereas the proximal part of this domain has an inhibitory role that is alleviated by DDK-mediated phosphorylation (44). It has also been shown, in HeLa cells, that phosphorylation of the serine/threonine residues of the NSD domain of MCM4 inhibits DNA replication (46). Downstream initiation from M51 or M75 eliminates 4 or 5 potential sites of phosphorylation, respectively, in the N-terminal domain of MCM4 in the patients’ cells. The regulatory function of the distal part of the MCM4 NSD is compromised in patients’ cells, as demonstrated by the lack of prevention of re-replication and the higher mitotic phase. The C-terminal residues of MCM4 form an MCM complex domain involved in MCM2-7 complex formation and loading onto DNA. In yeast, this domain has been found to suspend MCM helicase activity after the formation of sufficient single-stranded DNA for checkpoint activation during the cell cycle (47). We observed no defect in MCM complex formation or DNA loading in dermal fibroblasts from patients. However, we observed a high level of re-replication and DNA breakage in the patients’ SV40 fibroblasts, indicating that the NSD domain of human MCM4 is also important for the checkpoint control of DNA replication as well as maintenance of genome integrity.
The overall developmental phenotype of the patients is consistent and can be accounted for by a ubiquitous requirement of the MCM4-containing MCM2-7 complex for DNA replication and cell proliferation (22). Moreover, mice compound heterozygous for a hypomorphic Chaos3 allele and a null allele of Mcm4 fail to develop in utero, whereas homozygous Chaos3 mice are viable (33). This scenario is consistent with the clinical phenotype of patients homozygous for the MCM4 mutant allele, which is much less severe than that of mice with an Mcm4 gene knockout, which die in utero before implantation (33). In Caenorhabditis elegans, the homozygous lin-6 (Q88X) mutation in MCM4 causes defects in DNA replication, G2/M checkpoint arrest, and a decrease in cell proliferation rate (48). It has also recently been shown that mutations affecting pre-RC components, including ORC1, ORC4, ORC6, CDC6, and CDT1, cause Meier-Gorlin syndrome, which manifests as microcephalic primordial dwarfism (49–51). Growth retardation of a magnitude similar to that observed in patients with ORC4 mutations, in terms of height, weight, and head circumference, has been found in patients with MCM4 mutations, although these two disorders differ in terms of other features. For example, NK cell deficiency and adrenal insufficiency have been documented in patients bearing mutations in MCM4 but not in patients with Meier-Gorlin syndrome.
Mice carrying the Chaos3 hypomorphic allele of MCM4 also display chromosome instability in MEFs and develop mammary adenocarcinomas (33). Our findings indicate that the first 50 and 74 amino acids are not required for MCM complex formation and the loading of MCM onto chromatin, at least in dermal fibroblast cell lines. However, the higher rate of DNA breakage in patients’ leukocytes and dermal fibroblasts suggests that the N-terminal domain of MCM4 is involved in DNA replication and, specifically, in the maintenance of genome integrity during DNA replication. The impact of the N-terminal truncation of MCM4, however, seems to be cell context–dependent, at least in hematopoietic lineages, potentially accounting for the restricted developmental phenotype observed in the patients’ NK cells. Such DNA damage may result from inefficient pre-RC activation, as observed in cancer cells overproducing cyclin E–CDK2 kinase (52). Alternatively, the defect may be caused by a lack of checkpoint signaling via the MCM4 N-terminal domain. Either way, the genome is clearly unstable, and this may also account for the development of lymphoma in one of the patients. Predisposition to cancer is clearly associated with other chromosomal instability syndromes, such as ataxia telangiectasia, Bloom syndrome, RIDDLE syndrome, immunodeficiency centromeric instability facial anomalies syndrome, Fanconi anemia, LIG4 deficiency, and Nijmegen breakage syndrome. These syndromes are rarely associated with NK cell deficiency, and their association with selective NK deficiency has never been reported. Nevertheless, the NK cell deficit may have contributed to the development of EBV-driven lymphoma in one of the MCM4-deficient patients studied here.
Partial MCM4 deficiency is, to our knowledge, the first genetic etiology of a human disorder associated with selective NK cell deficiency to be described. In humans, the MCM4 deficit selectively affects the CD56dim subset of NK cells, which account for 90% of the mature circulating NK cells. Several reports have indicated that the NK CD56dim subset originates from the NK CD56bright subset and that the transition between these two subsets is associated with a decrease in the capacity of NK cells to proliferate (34, 53). Unlike control cells, the circulating NK cells in MCM4-deficient patients are mostly CD56bright and do not proliferate in response to IL-2 or IL-15. This suggests that the NK CD56bright subset cannot differentiate into the NK CD56dim subset in patients with MCM4 deficiency. The concomitant observation of a lack of proliferation of NK CD56bright cells and of the loss of the NK CD56dim subset in patients suggests that the final stage of NK differentiation requires the proliferation of NK CD56bright cells. Lutz et al. recently put forward this hypothesis, after comparing the rates of proliferation and apoptosis of NK CD56bright and NK CD56dim cells (54). Our analysis of the patients reported here suggests that this hypothesis may be correct and reveals that MCM4 is required for this process. Thus, the identification of this MCM4 deficiency sheds light not only on the genetic deficiency in these patients, but also on the mechanisms of NK cell differentiation, providing what we believe to be the first genetic evidence for the differentiation of CD56bright cells into CD56dim NK cells in humans.
Other lymphoid cells (T and B cells) have high rates of proliferation that are not affected in patients with MCM4 deficiency, the defect observed being specific to NK cells. Previous studies based on DNA labeling and cell cycle analysis in mice have shown that adult splenic NK cells proliferate more rapidly than total splenic T cells (55). Human peripheral blood NK cells have also been reported to proliferate more rapidly than T cells in healthy young and elderly women (54). The higher rate of proliferation in NK cells than in T cells may account for the higher sensitivity of NK cells in patients with MCM4 deficiency. This suggests that the CD56bright to CD56dim NK cell transition involves hyperproliferation in vivo. Other leukocytes, including T and B lymphocytes, may use a pathway not present in NK cells, and this may allow them to overcome the deletion of the N-terminal part of MCM4, this part of the protein being indispensable in NK cells. NK cells have potent and nonredundant antiviral and antitumoral activities in mice (4, 6, 7). NK cell deficiency in the MCM4-deficient patients may account for the development of viral diseases in the patients, possibly even including the EBV-lymphoproliferative disorder. However, impaired MCM4 production in other, not necessarily hematopoietic, cell types may have contributed to the infectious phenotype (56). Indeed, some viruses, such as EBV and CMV, inactivate the MCM complex by inhibiting MCM4 phosphorylation (57, 58). This process may also have contributed to the lymphoma observed in one of the patients. In any event, partial MCM4 deficiency defines an autosomal recessive syndrome with global growth retardation, adrenal insufficiency, lymphoma, viral infections, and a selective NK CD56dim deficiency.
Methods
Patients.
The detailed clinical features of this family are described in Supplemental Table 1. Briefly, in kindred A, 5 children had low NK cell counts in peripheral blood (<5%) and were born with severe intra- and extrauterine growth retardation and microcephaly, with mild or no facial dysmorphia. The index case, patient P1.3, was born in 1996 and displayed intra- and extrauterine growth retardation. At the age of 18 months, he presented hepatomegaly, splenomegaly, and lymphadenopathies. This may have reflected primary EBV infection, but remained unexplained. This patient had previously experienced recurrent childhood viral infections, with recurrent respiratory tract infections and lung disease consisting of bronchiectasis, fibrosis, and respiratory failure. At the age of 2 years and 9 months, he developed an EBV-driven lymphoproliferative disorder in the small bowel, and the resulting tumor was surgically removed. At the age of 5 years and 6 months, he developed clinical and biochemical features of adrenal insufficiency, leading to the introduction of replacement corticosteroid therapy (17). He died at the age of 14 years, from progressive respiratory failure. Similar clinical features were observed in the other 4 members of kindred A with NK cell deficiency. Three members of this family suffered from frequent lung infections: P1.2, born in 1991; P1.4, born in 1997; and P1.5, born in 2002. In two of these patients, P1.2 and P1.4, lung fibrosis and respiratory failure occurred, and P1.4 died at the age of 11 years, from progressive lung disease with features of inflammatory interstitial pneumonitis. Intra- and extrauterine growth retardation was observed in all 4 patients. Microcephaly was recorded in P1.1, P1.4, and P1.5. Adrenal insufficiency requiring corticosteroid replacement therapy developed in 3 patients, P1.2, P1.4, and P1.5, whereas P1.1, born in 1999, was less severely affected. P1.1 displays a mild failure to thrive but has suffered no documented episodes of infectious viral disease.
In kindred B, one patient, P2.1, had a low peripheral blood NK cell count (<1%). He also displayed intra- and extrauterine growth retardation and recurrent respiratory tract infections. He had recurrent infections with HSV and VZV. At the age of 8 years, he was diagnosed with adrenal insufficiency and underwent bone marrow transplantation due to a gradual deterioration of his health. In short-term cultures of lymphocytes with DNA crosslinkers, DNA breakage rates were always higher than controls in the members of kindred A (except P1.5, who was not tested) and in patient P2.1. Serological tests for EBV with IgG against EBNA were paradoxically negative for P1.3, whereas tests for IgM/IgA against VCA were positive. P1.1, P1.2, and P1.4 tested positive for IgG against EBNA and negative for IgG/IgM against VCA. P1.5 was not tested. Serological tests for HSV were positive in all 6 patients; those for VZV were positive in P1.1, P1.2, and P1.3, and those for CMV were positive in P1.4 but negative in all other patients.
Western blots.
Total protein was solubilized in extraction buffer (50 mM Tris HCl pH 7.4, 150 mM NaCl, 5 mM EDTA, and 1% Triton X-100 plus protease inhibitors plus phosphatase inhibitors). Cytoplasmic proteins were solubilized in cytoplasmic extraction buffer (10 mM HEPES pH 7.6, 10 mM KCl, 2 mM MgCl2, and 0.1 mM EDTA plus protease inhibitors plus phosphatase inhibitors). Nuclear proteins were solubilized in nuclear extraction buffer (50 mM HEPES pH 7.6, 50 mM KCl, 300 mM MgCl2, and 0.1 mM EDTA, 10% glycerol plus protease inhibitors plus phosphatase inhibitors). The extracted proteins were separated by electrophoresis in a Criterion XT Precast 10% Bis-Tris gel (Bio-Rad); molecular weight markers were included in the gel. Proteins were transferred onto membranes by iBlot Gel Transfer (Invitrogen), according to the manufacturer’s instructions. Nonspecific binding was blocked by incubation in 1× PBS, 5% BSA for 1 hour. The membrane was incubated overnight with primary antibody in 1× PBS, 1% BSA, 0.05% Tween 20 and then with peroxidase-conjugated secondary antibody (GE Healthcare) and ECL Western blotting substrate (Pierce). Anti–human MCM antibodies from Santa Cruz Biotechnology Inc. were used (MCM4 [clone H300], MCM2, MCM5). Anti-GAPDH and anti–β-actin antibodies (Santa Cruz Biotechnology Inc.) were used for normalization. An antibody specific for the N terminus of MCM4 (Poly6024) (BioLegend), an antibody specific for the C terminus of MCM4 (ab4459) (Abcam), and an anti-Flag monoclonal antibody (OriGene) were also used.
Transient and stable transfection.
The C-terminal Flag-tagged pCMV6 empty vector and the human MCM4 expression vector were purchased from Origene (RC206122). Other constructs were generated by direct mutagenesis with Phusion Taq from Finnzymes, according the manufacturer’s instructions: MCM4 MUT (Flag-CMV6 MCM4 with the c.70_71insG mutation; MCM4-ATG1 (Flag-CMV6 MCM4 with M51G mutation); MCM4 MUT-ATG1 (Flag-CMV6 MCM4 with the c.70_71insG and M51G mutations); MCM4-ATG2 (Flag-CMV6 MCM4 with the M75G mutation); MCM4 MUT-ATG2 (Flag-CMV6 MCM4 with the c.70_71insG and M75G mutations); MCM4 MUT-ATG1+2 (Flag-CMV6 MCM4 with the c.70_71insG, M51G, and M75G mutations); MCM4-ATG1+2 (Flag-CMV6 MCM4 with M51G and M75G mutations). We transiently transfected HEK293T cells with the various constructs, by the calcium phosphate method (kit from Invitrogen). The pTRIP plasmid for lentiviral vector transfection was obtained with Gateway cloning technology from Invitrogen, according to the manufacturer’s instructions. SV40 fibroblasts were transduced with lentiviral particles cultured in the presence of 2 μg/ml puromycin (InvivoGen).
Co-immunoprecipitation experiments.
Cells were lysed as previously described (24) to obtain the Triton X–extractable and DNase I–released MCM fractions. Briefly, patient and control cells were lysed in cold CSK buffer (10 mM PIPES [pH 6.8], 100 mM NaCl, 300 mM sucrose, 3 mM MgCl2, 0.1% Triton X, 1 mM ATP, protease inhibitors [Roche complete, Mini, EDTA-free]) for the Triton X–extractable fraction. The pellet was then digested with 1,000 U/ml DNase I for 30 minutes at 25°C, to extract the chromatin-bound MCM fraction. Both the Triton X–extractable and DNase I–released fractions were subjected to immunoprecipitation with the monoclonal anti-Mcm2 antibody (clone 2-2-40).
Cell cycle and BrdU incorporation analysis.
Cells were used to seed plates at a density of 1 × 105 cells/ml. For aphidicolin treatment, cells were incubated with 0.3 μM aphidicolin for 24 hours. The drug was removed by washing the cells with warm medium, and the cells were then pulse labeled with BrdU for 30 minutes, washed with PBS, and treated with trypsin. Cells were fixed with methanol and stained with anti-BrdU–FITC antibody (BD Biosciences) and propidium iodide, according to the manufacturer’s instructions. Flow cytometry analysis was carried out with the BD LSRII flow cytometry system and BD FACSDiva software.
Breakage analysis.
Primary or SV40 fibroblasts were treated with 0.3 μM aphidicolin for 24 hours, arrested by incubation with colcemid (0.167 μg/ml medium) for 2 hours, harvested, incubated for 10 minutes at 37°C in 0.075 M KCl, and fixed in freshly prepared methanol/glacial acidic acid (3:1 vol/vol). Cells were stored at 4°C. They were dispensed onto wet slides as required and air dried at 40°C for 60 minutes before staining with KaryoMAX Giemsa Gurr buffer (Invitrogen) for 3 minutes. The slides were rinsed with fresh Gurr buffer followed by distilled water, fully dried at 40°C for 60 minutes, and scanned with MetaSystems Metafer. For DEB treatment, the same procedure was followed, except that the cells were treated with 0.1 μg/ml DEB in medium for 72 hours before the addition of colcemid. Complementation of the breakage phenotype was analyzed in a blinded manner.
For lymphocyte DNA breakage experiments, blood cultures were exposed to 6 × 10–8 M MMC, 10–8 M HN2, or DEB at a concentration of 0.1 μg/ml. All cultures were harvested at 72 hours. Matched controls were analyzed in parallel. Colchicine was added to the cultures for a final hour before harvesting.
Phenotypic analysis of WT, Mcm4Chaos3/+, and Mcm4Chaos3/Chaos3 mice by flow cytometry.
Whole blood from anesthetized mice was collected and stained with anti-CD3, anti-CD19, and anti-NKp46 antibodies and analyzed by flow cytometry with a FACSCanto II cytometer (BD). NK cells were defined as CD3–NKp46+ cells within the lymphocyte gate.
Human T and B cell analysis by flow cytometry.
Immunologic analysis of the T and B cell compartments of whole-blood samples was performed by flow cytometry, with monoclonal antibodies against CD3, CD4, CD8, CD19, CD45RA, CD45RO, and CD27 (BD) and sorting by flow cytometry with a FACSCanto II cytometer (BD).
Human NK cell analysis and CD56bright/CD56dim sorting by flow cytometry.
Whole blood or PBMCs from healthy donors or patients were stained with fluorochrome-conjugated monoclonal antibodies against CD3, CD56, CD16, CD94, and CD57 and analyzed by flow cytometry with a FACSCanto II cytometer. All antibodies were supplied by BD Biosciences — Pharmingen. NK cells were defined as CD3–CD56+ cells in the lymphocyte gate. For the sorting of CD56bright and CD56dim NK cells, we first enriched whole-blood preparations in NK cells with the RosetteSep Human NK Cell Enrichment Cocktail (StemCell Technologies). The resulting mixture was subjected to gradient centrifugation on a Ficoll-Paque gradient, and the preparation enriched in NK cells was stained by incubation with anti-CD3 and anti-CD56 antibodies for 30 minutes at 4°C and sorted by flow cytometry, based on the surface density of CD56.
Cell stimulation and apoptosis analysis.
PBMCs were stained with CFSE (Invitrogen), plated in 96-well plates, and activated by incubation with various concentrations of IL-2 (Chiron Corp.) or IL-15 (R&D Systems). After 72 hours, PBMCs were stained by incubation for 30 minutes at 4°C with anti-CD3 and anti-CD56 mAbs and then with 7-AAD (BD).
PBMCs were plated in 96-well plates at a density of 106 cells/ml and activated by incubation with PHA (1:700). After 6 days of culture, PHA-activated T cells were plated in 96-well plates at a density of 106 cells/ml and activated by incubation with IL-2 or IL-15. DNA fragmentation was assessed in PHA-activated T cell blasts, by washing cells in 0.9% NaCl and then incubating them in a hypotonic solution containing propidium iodide (50 μg/ml; Sigma-Aldrich), 0.1% sodium citrate, and 1:100 Triton X-100. The staining patterns obtained were analyzed by flow cytometry (FACScan, BD Biosciences).
Statistics.
The mean values for the various measurements (such as the number of specific NK cells and mean levels of DNA breakage) were compared between controls and patients, using Student’s t tests implemented in R software ( http://www.r-project.org/; version 2.7.1). We compared the proportions of NK, T, and B cells observed in Chaos3/Chaos3 and WT/WT mice, using the nonparametric Wilcoxon test, as implemented in PROC NPAR1WAY of SAS Software version 9.1 (SAS Institute). For all analyses, P values less than 0.05 were considered significant.
Study approval.
Written informed consent was obtained from all family members. All experiments were approved by the Ethics Committee of St. James’ Hospital, Dublin, the IRB of Necker Hospital for Sick Children, and the IRB of Rockefeller University.
Supplementary Material
Acknowledgments
We thank family members for participating in this study. We thank all members of the two branches of the laboratory of Human Genetics of Infectious Diseases, including, in particular, Vanessa Sancho-Shimizu, Rebeca Pérez de Diego, Lazaro Lorenzo, Annabelle Cardon, Jacinta Bustamante, Guillaume Vogt, and Anne Puel for discussions. Laure Gineau was supported by a La Ligue Nationale Contre le Cancer (LNCC) grant and INSERM. This work was supported by the AXA Research Fund, the Action Concertée Incitative de Microbiologie, the Agence Nationale pour la Recherche, the St. Giles Foundation, the Rockefeller University Center for Clinical and Translational Science grant 5UL1RR024143, and the Rockefeller University. A. Smogorzewska is supported by the Burroughs Wellcome Fund Career Award for Medical Scientists and is a Rita Allen Foundation and Irma T. Hirschl scholar. B. Stillman is supported by National Cancer Institute grant CA13106. Jean-Laurent Casanova was an international scholar of the Howard Hughes Medical Institute until 2008.
Footnotes
Conflict of interest: The authors have declared that no conflict of interest exists.
Citation for this article: J Clin Invest. 2012;122(3):821–832. doi:10.1172/JCI61014.
See the related Commentary beginning on page 798.
References
- 1.Vivier E, et al. Innate or adaptive immunity? The example of natural killer cells. Science. 2011;331(6013):44–49. doi: 10.1126/science.1198687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Moretta L, Moretta A. Unravelling natural killer cell function: triggering and inhibitory human NK receptors. Embo J. 2004;23(2):255–259. doi: 10.1038/sj.emboj.7600019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.French AR, Yokoyama WM. Natural killer cells and viral infections. Curr Opin Immunol. 2003;15(1):45–51. doi: 10.1016/s095279150200002x. [DOI] [PubMed] [Google Scholar]
- 4.Orr MT, Lanier LL. Natural killer cell education and tolerance. Cell. 2010;142(6):847–856. doi: 10.1016/j.cell.2010.08.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cerwenka A, Lanier LL. Natural killer cells, viruses and cancer. Nat Rev Immunol. 2001;1(1):41–49. doi: 10.1038/35095564. [DOI] [PubMed] [Google Scholar]
- 6.Smyth MJ, Hayakawa Y, Takeda K, Yagita H. New aspects of natural-killer-cell surveillance and therapy of cancer. Nat Rev Cancer. 2002;2(11):850–861. doi: 10.1038/nrc928. [DOI] [PubMed] [Google Scholar]
- 7.Orange JS. Human natural killer cell deficiencies. Curr Opin Allergy Clin Immunol. 2006;6(6):399–409. doi: 10.1097/ACI.0b013e3280106b65. [DOI] [PubMed] [Google Scholar]
- 8.Biron CA, Byron KS, Sullivan JL. Severe herpesvirus infections in an adolescent without natural killer cells. N Engl J Med. 1989;320(26):1731–1735. doi: 10.1056/NEJM198906293202605. [DOI] [PubMed] [Google Scholar]
- 9.Etzioni A, Eidenschenk C, Katz R, Beck R, Casanova JL, Pollack S. Fatal varicella associated with selective natural killer cell deficiency. J Pediatr. 2005;146(3):423–425. doi: 10.1016/j.jpeds.2004.11.022. [DOI] [PubMed] [Google Scholar]
- 10.Ballas ZK, Turner JM, Turner DA, Goetzman EA, Kemp JD. A patient with simultaneous absence of “classical” natural killer cells (CD3-, CD16+, and NKH1+) and expansion of CD3+, CD4-, CD8-, NKH1+ subset. J Allergy Clin Immunol. 1990;85(2):453–459. doi: 10.1016/0091-6749(90)90155-w. [DOI] [PubMed] [Google Scholar]
- 11.Akiba H, Motoki Y, Satoh M, Iwatsuki K, Kaneko F. Recalcitrant trichophytic granuloma associated with NK-cell deficiency in a SLE patient treated with corticosteroid. Eur J Dermatol. 2001;11(1):58–62. [PubMed] [Google Scholar]
- 12.Ostenstad B, Giliani S, Mellbye OJ, Nilsen BR, Abrahamsen T. A boy with X-linked hyper-IgM syndrome and natural killer cell deficiency. Clin Exp Immunol. 1997;107(2):230–234. doi: 10.1111/j.1365-2249.1997.284-ce1174.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Vossen MT, et al. Absence of circulating natural killer and primed CD8+ cells in life-threatening varicella. J Infect Dis. 2005;191(2):198–206. doi: 10.1086/426866. [DOI] [PubMed] [Google Scholar]
- 14.Bernard F, et al. A novel developmental and immunodeficiency syndrome associated with intrauterine growth retardation and a lack of natural killer cells. Pediatrics. 2004;113(1 pt 1):136–141. doi: 10.1542/peds.113.1.136. [DOI] [PubMed] [Google Scholar]
- 15.Eidenschenk C, et al. A novel primary immunodeficiency with specific natural-killer cell deficiency maps to the centromeric region of chromosome 8. Am J Hum Genet. 2006;78(4):721–727. doi: 10.1086/503269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Eidenschenk C, et al. Familial NK cell deficiency associated with impaired IL-2- and IL-15-dependent survival of lymphocytes. J Immunol. 2006;177(12):8835–8843. doi: 10.4049/jimmunol.177.12.8835. [DOI] [PubMed] [Google Scholar]
- 17.O’Riordan SM, Lynch SA, Hindmarsh PC, Chan LF, Clark AJ, Costigan C. A novel variant of familial glucocorticoid deficiency prevalent among the Irish Traveler population. J Clin Endocrinol Metab. 2008;93(7):2896–2899. doi: 10.1210/jc.2008-0034. [DOI] [PubMed] [Google Scholar]
- 18.Casanova JL, Abel L. Primary immunodeficiencies: a field in its infancy. Science. 2007;317(5838):617–619. doi: 10.1126/science.1142963. [DOI] [PubMed] [Google Scholar]
- 19.Cann HM. CEPH maps. Curr Opin Genet Dev. 1992;2(3):393–399. doi: 10.1016/S0959-437X(05)80148-0. [DOI] [PubMed] [Google Scholar]
- 20.Hughes CR, et al. MCM4 mutation causes adrenal failure, short stature, and natural killer cell deficiency in humans. . J Clin Invest. doi: 10.1172/JCI60224. doi: 10.1172/JCI60224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Costa A, Onesti S. Structural biology of MCM helicases. Crit Rev Biochem Mol Biol. 2009;44(5):326–342. doi: 10.1080/10409230903186012. [DOI] [PubMed] [Google Scholar]
- 22.Bell SP, Dutta A. DNA replication in eukaryotic cells. Annu Rev Biochem. 2002;71:333–374. doi: 10.1146/annurev.biochem.71.110601.135425. [DOI] [PubMed] [Google Scholar]
- 23.Arias EE, Walter JC. Strength in numbers: preventing rereplication via multiple mechanisms in eukaryotic cells. Genes Dev. 2007;21(5):497–518. doi: 10.1101/gad.1508907. [DOI] [PubMed] [Google Scholar]
- 24.Fujita M, Kiyono T, Hayashi Y, Ishibashi M. In vivo interaction of human MCM heterohexameric complexes with chromatin. Possible involvement of ATP. J Biol Chem. 1997;272(16):10928–10935. doi: 10.1074/jbc.272.16.10928. [DOI] [PubMed] [Google Scholar]
- 25.Gambus A, et al. GINS maintains association of Cdc45 with MCM in replisome progression complexes at eukaryotic DNA replication forks. Nat Cell Biol. 2006;8(4):358–366. doi: 10.1038/ncb1382. [DOI] [PubMed] [Google Scholar]
- 26.Moyer SE, Lewis PW, Botchan MR. Isolation of the Cdc45/Mcm2-7/GINS (CMG) complex, a candidate for the eukaryotic DNA replication fork helicase. Proc Natl Acad Sci U S A. 2006;103(27):10236–10241. doi: 10.1073/pnas.0602400103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pacek M, Tutter AV, Kubota Y, Takisawa H, Walter JC. Localization of MCM2-7, Cdc45, and GINS to the site of DNA unwinding during eukaryotic DNA replication. Mol Cell. 2006;21(4):581–587. doi: 10.1016/j.molcel.2006.01.030. [DOI] [PubMed] [Google Scholar]
- 28.Forsburg SL. Eukaryotic MCM proteins: beyond replication initiation. Microbiol Mol Biol Rev. 2004;68(1):109–131. doi: 10.1128/MMBR.68.1.109-131.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Remus D, Beuron F, Tolun G, Griffith JD, Morris EP, Diffley JF. Concerted loading of Mcm2-7 double hexamers around DNA during DNA replication origin licensing. Cell. 2009;139(4):719–730. doi: 10.1016/j.cell.2009.10.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rosendorff J, Bernstein R. Fanconi’s anemia — chromosome breakage studies in homozygotes and heterozygotes. Cancer Genet Cytogenet. 1988;33(2):175–183. doi: 10.1016/0165-4608(88)90027-1. [DOI] [PubMed] [Google Scholar]
- 31.Deviren A, Yalman N, Hacihanefioglu S. Differential diagnosis of Fanconi anemia by nitrogen mustard and diepoxybutane. Ann Hematol. 2003;82(4):223–227. doi: 10.1007/s00277-003-0614-4. [DOI] [PubMed] [Google Scholar]
- 32.Auerbach AD. Diagnosis of fanconi anemia by diepoxybutane analysis. Curr Protoc Hum Genet. 2003;Chapter 8:Unit 8.7. doi: 10.1002/0471142905.hg0807s37. [DOI] [PubMed] [Google Scholar]
- 33.Shima N, et al. A viable allele of Mcm4 causes chromosome instability and mammary adenocarcinomas in mice. Nat Genet. 2007;39(1):93–98. doi: 10.1038/ng1936. [DOI] [PubMed] [Google Scholar]
- 34.Romagnani C, et al. CD56brightCD16– killer Ig-like receptor– NK cells display longer telomeres and acquire features of CD56dim NK cells upon activation. . J Immunol. 2007;178(8):4947–4955. doi: 10.4049/jimmunol.178.8.4947. [DOI] [PubMed] [Google Scholar]
- 35.Caligiuri MA. Human natural killer cells. Blood. 2008;112(3):461–469. doi: 10.1182/blood-2007-09-077438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Nagler A, Lanier LL, Cwirla S, Phillips JH. Comparative studies of human FcRIII-positive and negative natural killer cells. J Immunol. 1989;143(10):3183–3191. [PubMed] [Google Scholar]
- 37.Yu J, et al. CD94 surface density identifies a functional intermediary between the CD56bright and CD56dim human NK-cell subsets. Blood. 2010;115(2):274–281. doi: 10.1182/blood-2009-04-215491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lopez-Verges S, et al. CD57 defines a functionally distinct population of mature NK cells in the human CD56dimCD16+ NK-cell subset. Blood. 2010;116(19):3865–3874. doi: 10.1182/blood-2010-04-282301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kennedy MK, et al. Reversible defects in natural killer and memory CD8 T cell lineages in interleukin 15-deficient mice. J Exp Med. 2000;191(5):771–780. doi: 10.1084/jem.191.5.771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lodolce JP, et al. IL-15 receptor maintains lymphoid homeostasis by supporting lymphocyte homing and proliferation. Immunity. 1998;9(5):669–676. doi: 10.1016/S1074-7613(00)80664-0. [DOI] [PubMed] [Google Scholar]
- 41.Suzuki H, Duncan GS, Takimoto H, Mak TW. Abnormal development of intestinal intraepithelial lymphocytes and peripheral natural killer cells in mice lacking the IL-2 receptor beta chain. J Exp Med. 1997;185(3):499–505. doi: 10.1084/jem.185.3.499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ohteki T, Ho S, Suzuki H, Mak TW, Ohashi PS. Role for IL-15/IL-15 receptor beta-chain in natural killer 1.1+ T cell receptor-alpha beta+ cell development. J Immunol. 1997;159(12):5931–5935. [PubMed] [Google Scholar]
- 43.Fehniger TA, Cooper MA, Caligiuri MA. Interleukin-2 and interleukin-15: immunotherapy for cancer. Cytokine Growth Factor Rev. 2002;13(2):169–183. doi: 10.1016/S1359-6101(01)00021-1. [DOI] [PubMed] [Google Scholar]
- 44.Sheu YJ, Stillman B. The Dbf4-Cdc7 kinase promotes S phase by alleviating an inhibitory activity in Mcm4. Nature. 2010;463(7277):113–117. doi: 10.1038/nature08647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ishimi Y, Komamura-Kohno Y. Phosphorylation of Mcm4 at specific sites by cyclin-dependent kinase leads to loss of Mcm4,6,7 helicase activity. J Biol Chem. 2001;276(37):34428–34433. doi: 10.1074/jbc.M104480200. [DOI] [PubMed] [Google Scholar]
- 46.Ishimi Y, Komamura-Kohno Y, Kwon HJ, Yamada K, Nakanishi M. Identification of MCM4 as a target of the DNA replication block checkpoint system. J Biol Chem. 2003;278(27):24644–24650. doi: 10.1074/jbc.M213252200. [DOI] [PubMed] [Google Scholar]
- 47.Nitani N, Yadani C, Yabuuchi H, Masukata H, Nakagawa T. Mcm4 C-terminal domain of MCM helicase prevents excessive formation of single-stranded DNA at stalled replication forks. Proc Natl Acad Sci U S A. 2008;105(35):12973–12978. doi: 10.1073/pnas.0805307105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Korzelius J, et al. C. elegans MCM-4 is a general DNA replication and checkpoint component with an epidermis-specific requirement for growth and viability. Dev Biol. 2011;350(2):358–369. doi: 10.1016/j.ydbio.2010.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Bicknell LS, et al. Mutations in the pre-replication complex cause Meier-Gorlin syndrome. Nat Genet. 2011;43(4):356–359. doi: 10.1038/ng.775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Bicknell LS, et al. Mutations in ORC1, encoding the largest subunit of the origin recognition complex, cause microcephalic primordial dwarfism resembling Meier-Gorlin syndrome. Nat Genet. 2011;43(4):350–355. doi: 10.1038/ng.776. [DOI] [PubMed] [Google Scholar]
- 51.Guernsey DL, et al. Mutations in origin recognition complex gene ORC4 cause Meier-Gorlin syndrome. Nat Genet. 2011;43(4):360–364. doi: 10.1038/ng.777. [DOI] [PubMed] [Google Scholar]
- 52.Ekholm-Reed S, Mendez J, Tedesco D, Zetterberg A, Stillman B, Reed SI. Deregulation of cyclin E in human cells interferes with prereplication complex assembly. J Cell Biol. 2004;165(6):789–800. doi: 10.1083/jcb.200404092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Chan A, et al. CD56bright human NK cells differentiate into CD56dim cells: role of contact with peripheral fibroblasts. J Immunol. 2007;179(1):89–94. doi: 10.4049/jimmunol.179.1.89. [DOI] [PubMed] [Google Scholar]
- 54.Lutz CT, et al. Human NK cells proliferate and die in vivo more rapidly than T cells in healthy young and elderly adults. J Immunol. 2011;186(8):4590–4598. doi: 10.4049/jimmunol.1002732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Jamieson AM, Isnard P, Dorfman JR, Coles MC, Raulet DH. Turnover and proliferation of NK cells in steady state and lymphopenic conditions. J Immunol. 2004;172(2):864–870. doi: 10.4049/jimmunol.172.2.864. [DOI] [PubMed] [Google Scholar]
- 56.Alcais A, Abel L, Casanova JL. Human genetics of infectious diseases: between proof of principle and paradigm. J Clin Invest. 2009;119(9):2506–2514. doi: 10.1172/JCI38111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kudoh A, et al. Phosphorylation of MCM4 at sites inactivating DNA helicase activity of the MCM4-MCM6-MCM7 complex during Epstein-Barr virus productive replication. J Virol. 2006;80(20):10064–10072. doi: 10.1128/JVI.00678-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Qian Z, Leung-Pineda V, Xuan B, Piwnica-Worms H, Yu D. Human cytomegalovirus protein pUL117 targets the mini-chromosome maintenance complex and suppresses cellular DNA synthesis. PLoS Pathog. 2010;6(3):e1000814. doi: 10.1371/journal.ppat.1000814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Chang TH, Ray FA, Thompson DA, Schlegel R. Disregulation of mitotic checkpoints and regulatory proteins following acute expression of SV40 large T antigen in diploid human cells. Oncogene. 1997;14(20):2383–2393. doi: 10.1038/sj.onc.1201196. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.