Abstract
Background
Genetic alterations in human topoisomerase II alpha (TOP2A) are linked to cancer susceptibility. TOP2A decatenates chromosomes and thus is necessary for multiple aspects of cell division including DNA replication, chromosome condensation and segregation. Topoisomerase II alpha is also required for embryonic development in mammals, as mouse Top2a knockouts result in embryonic lethality as early as the 4-8 cell stage. The purpose of this study was to determine whether the extended developmental capability of zebrafish top2a mutants arises from maternal expression of top2a or compensation from its top2b paralogue.
Results
Here, we describe bloody minded (blm), a novel mutant of zebrafish top2a. In contrast to mouse Top2a nulls, zebrafish top2a mutants survive to larval stages (4-5 day post fertilization). Developmental analyses demonstrate abundant expression of maternal top2a but not top2b. Inhibition or poisoning of maternal topoisomerase II delays embryonic development by extending the cell cycle M-phase. Zygotic top2a and top2b are co-expressed in the zebrafish CNS, but endogenous or ectopic top2b RNA appear unable to prevent the blm phenotype.
Conclusions
We conclude that maternal top2a enables zebrafish development before the mid-zygotic transition (MZT) and that zebrafish top2a and top2b are not functionally redundant during development after activation of the zygotic genome.
Background
Topoisomerase (DNA) II alpha (TOP2A) is a nuclear protein which regulates DNA architecture during the mitotic phase of the cell cycle [1]. Studies in Xenopus have shown that its function in chromatin condensation is tightly coupled to prior DNA replication [2-4]. The expression of TOP2A is cell cycle regulated, reaching a peak in the G2/M phase [5]. Thus, up-regulated levels of TOP2A protein are found in proliferating cancer cells, and TOP2A is essential for the viability of these dividing cells. Consistent with its role in cell proliferation, genetic aberrations in TOP2A are linked to numerous human cancers [6]. To facilitate proper separation of chromatids and DNA replication, TOP2A generates transient double-stranded breaks in DNA [7-10]. Pharmacological targeting of TOP2A, which is extensively applied in cancer treatment, exploits this mechanism [11].
Vertebrate genomes contain two topoisomerases II paralogues. Human TOP2A and TOP2B have similar molecular masses (180 and 170 kDa, respectively) and share ~70% amino acid similarity, with the greatest divergence occurring in the C-terminal domain [12]. Despite structural similarities, these topoisomerases II isoforms have different expression patterns and functions [13]. In contrast to the expression of mammalian Top2a which peaks at G2/M, mammalian Top2b is expressed in differentiated tissues and its expression is not cell cycle regulated [14-17]. Several in vitro models have been utilised to study the loss-of-function effects associated with topoisomerase II genes. However, the critical role of Top2a genes for cell proliferation and survival necessitates conditional knockout models [18,19]. In vivo, genetic elimination of Top2a is dramatic, with mouse knockouts not developing beyond the 4-8 cell stage [20]. Underlying a distinct role, Top2b knockouts are not embryonic lethal. Instead Top2b is required during neuronal differentiation, for survival of some neural cells and neurite outgrowth [9,21,22]. Top2b null mice have defects in cerebral stratification and motor axons fail to contact skeletal muscles resulting in death soon after birth due to breathing impairment [23,24]. The topoisomerases II paralogues are also important as targets of anti-cancer drugs. ICRF-193, a catalytic inhibitor, blocks topoisomerase II-mediated ATP hydrolysis which is required to regenerate its active enzymatic form [25]. Etoposide, a topoisomerase II poison, inhibits the ability of topoisomerase II to re-ligate DNA molecules and therefore stabilises cleavable complexes of the enzyme leading to extensive fragmentation of DNA and cell death [26]. Topoisomerase II poisons are reported to mediate their cytotoxic effects through TOP2A, while double strand breaks and DNA rearrangements associated with secondary malignancies are due to TOP2B [27].
Two top2a mutant alleles have previously been documented in zebrafish: hi3635 generated by viral insertion [28] and can4 by ENU mutagenesis [29]. These mutants present with similar phenotypes including brain necrosis, abnormal tail curvature and death at 4-5 dpf. The can4 mutants have been studied more thoroughly and display reduced cell proliferation, mitotic spindle defects and increased DNA content. Recently, a zebrafish top2b mutant was reported with a phenotype distinct to top2a mutants, including defects in neurite targeting within the retinal inner plexiform layer and tectal neuropil [30]. These distinct phenotypes indicate separate functions of the zebrafish Top2a and Top2b isoforms.
Previous studies, however, do not explain the viability of zebrafish top2a mutants up to larval stages, in contrast to the early embryonic lethality of mouse Top2a knockouts. One explanation could relate to differential requirements of teleost and mammalian embryos for the timing of initiation of zygotic transcription, i.e. transcription from the embryo's own genome. The mid-blastula transition (MBT) refers to the moment during the blastula stage of embryonic development when expression of the zygotic genome starts, cell cycles lengthen, and cells acquire the ability to migrate [31]. The original term MBT (still used in relation to Xenopus) was expanded to include a phase of elimination of maternal transcripts and proteins starting before the activation of zygotic transcription [32]. This developmental event is referred as maternal-to-zygotic-transition (MZT) in recent studies [33]. In zebrafish, zygotic transcription increases gradually from the 10th (~2.75 hpf) to 13th cycle (~4.75 hpf) [34,35].
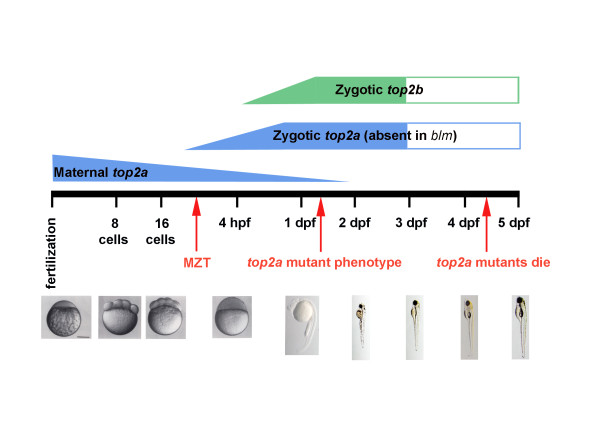
Here, we describe a novel zebrafish top2a mutant named "bloody minded" (blm) which was identified in an ENU mutagenesis screen. Mutant larvae can survive to ~5 dpf, although abnormal embryo morphology is discernable at ~27 hpf. We tested the hypotheses that the advanced development of zebrafish top2a mutants is due to either the presence of maternal Top2a or redundancy with the Top2b isoform. Our results are consistent with early embryonic development depending on maternal transcripts of zebrafish top2a. The absence of zygotic top2a is not fully compensated for by maternal, zygotic or ectopic top2b mRNA suggesting distinct functions of Top2a and Top2b in embryonic development.
Results
blm is a lethal recessive zebrafish mutant
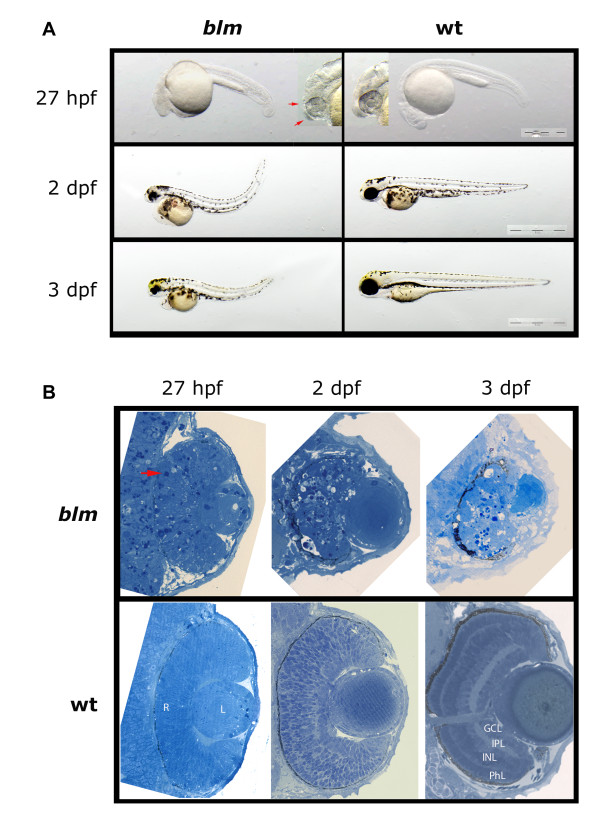
During F3 mutagenesis screens we recovered a recessive-lethal mutant named bloody-minded (blm). The blm phenotype is first visible at ~27 hpf when forebrain and midbrain atrophy is apparent and occasionally accompanied with CNS haemorrhage (Figure 1A). Increased cell death is prominent in the blm head, retina and lens at ~27 hpf (Figure 1B). The eyes fail to develop further so that at 2-3 dpf, blm eyes are significantly smaller than wildtype siblings and blm lenses protrude from the eye. At this stage, blm retinae fail to properly laminate and dying cells are visible. The body axes of blm mutants curves dorsally, cardiac oedema develops and blm mutants die at 4-5 dpf.
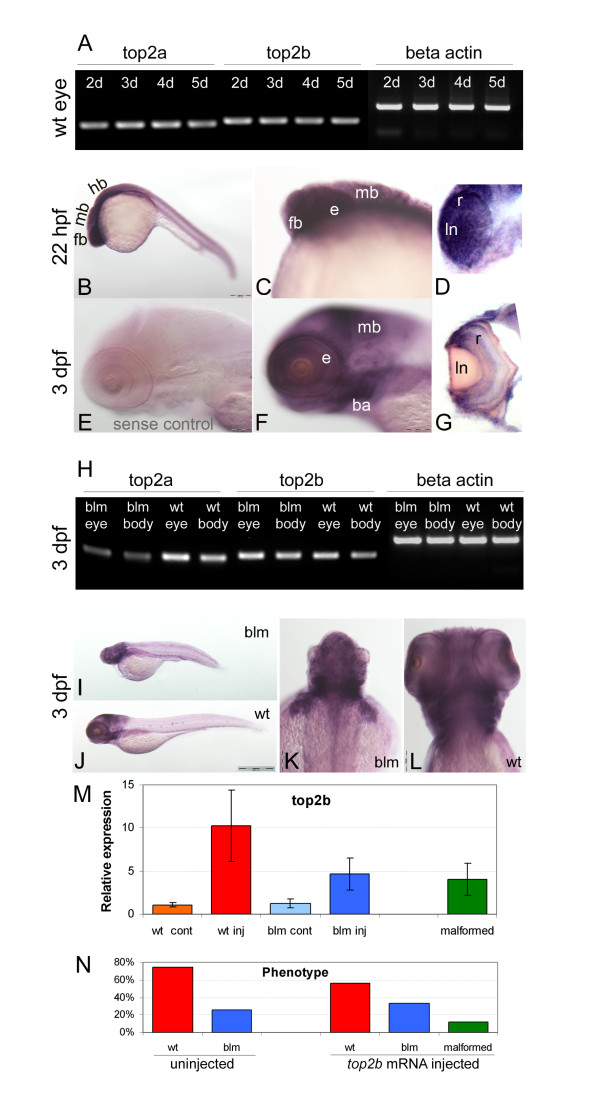
Figure 1.
Phenotype of the zebrafish bloody minded (blm) mutant. A) Mutants can be first recognized at ~27 hpf when signs of malformation in the head are visible (red arrows). At 2 and 3 dpf, eyes are much smaller and the tail is bent dorsally. B) Plastic sections through the eye reveal apoptotic cells in the retina (red arrow) and in the brain at 27 hpf. There is no proper lamination of the retina and necrosis proceeds. Abbreviations: hpf - hours post fertilisation, dpf - days post fertilisation, R - retina, L - lens, PhL - photoreceptor layer, INL - inner nuclear layer, IPL - inner plexiform layer, GCL - ganglion cells layer.
blm arises from a nonsense mutation in the top2a gene
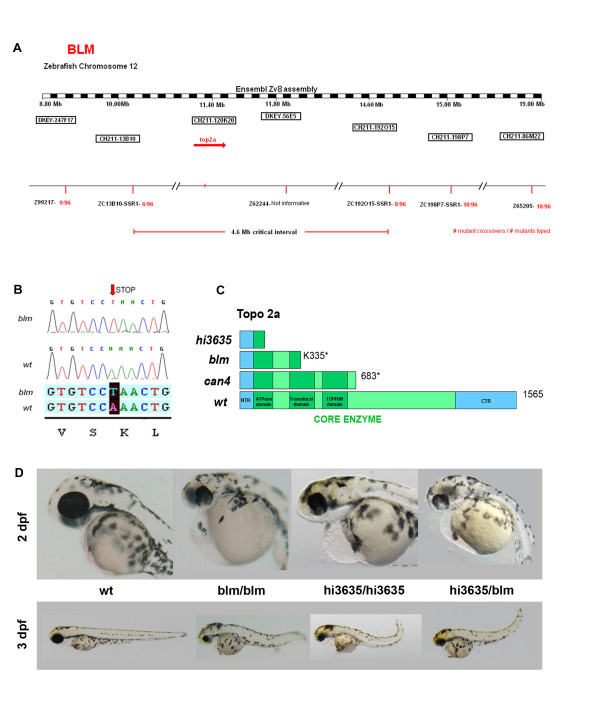
To determine the genetic locus of the blm mutation we performed bulk segregant analysis. Linkage to the blm phenotype was found on chromosome 12 with flanking Z-markers Z99217 and Z10806 (Figure 2A). Several new simple sequence repeat (SSR) markers were designed to narrow the critical interval of which zC13B10-SSR1 was most closely linked (6/96 recombinants). Analyses of the genetic maps in this interval revealed that a viral insertion mutant hi3635, with gross morphology similar to blm, co-segregated with the blm interval [28]. As hi3635 arises from an insertion in exon 1 of the gene encoding topoisomerase IIa (top2a), we chose top2a as a candidate gene for blm. Sequencing of whole larval cDNA from blm mutants reveals a point mutation (A→T) which introduces a premature stop codon at Lys residue 335 (K335X) (Figure 2B). The resulting truncated protein lacks the enzymatic domain (Figure 2C).
Figure 2.
blm arises from a mutation in the top2a gene. A) Mapping of blm reveals that the gene is located on chromosome 12. B) top2a was selected as a candidate gene and sequencing of mutant cDNA shows a point mutation (A→T) in Lys residue 335 resulting in a premature stop codon. C) A schematic of wildtype Top2a protein and the truncated proteins in hi3635, blm and can4 mutants D) Complementation assay performed to confirm that blm is top2a mutation. blm heterozygotes were crossed to hi3635 carriers and the resulting hybrid offspring present with blm phenotype.
To confirm that the blm phenotype arises from a mutation in the top2a gene we performed complementation assays (Figure 2D). Matings of blm carriers and hi3635 carriers fail to complement. Matings of blm+/- with hi3635+/- generated ~30% embryos with the blm phenotype, whereas outcrosses to wildtype fish did not. Thus, genomic and genetic data confirm that the blm mutant results from a nonsense mutation in the zebrafish top2a gene.
top2a mutants exhibit defects in cell cycle progression
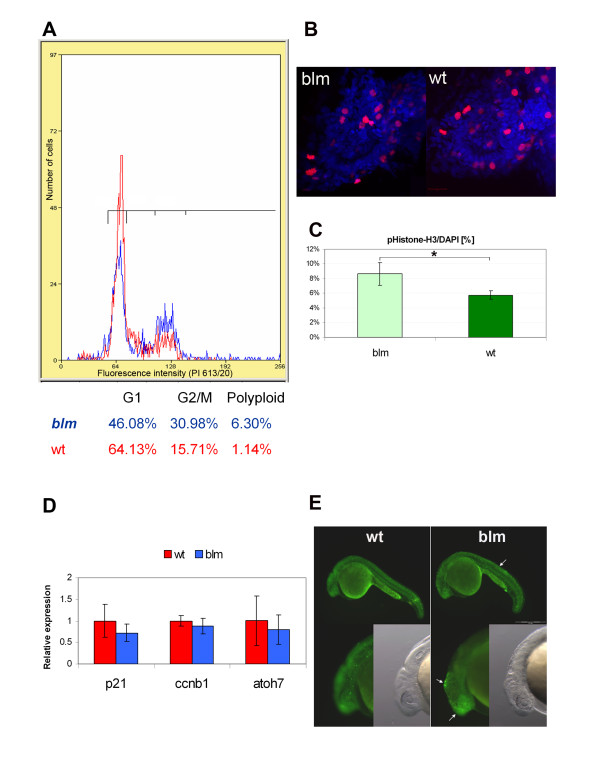
Topoisomerase II alpha genes are known to be critical for cell cycle progression. Therefore, we quantified, by flow cytometry, the distribution of blm cells within the major phases of the cell cycle. At 27 hpf, the proportion of dissociated blm cells in the G2/M phase (~31%) is approximately double that of wildtype siblings (~16%) consistent with defects in mitosis (Figure 3A). By immunohistochemistry, the total number of cells in the blm eye that express the G2/M marker phospho-histone H3 is lower than wildtype siblings (Figure 3B). However, consistent with the flow cytometry analyses (Figure 3A), the proportion of cells expressing the G2/M marker, normalized to the total number of nuclei, is significantly higher in blm mutants (8.7%) than in wildtype siblings (5.8%) (Figure 3C). At the transcript level, no significant difference is observed at 27 hpf for p21-like or ccnb1, G1 and G2 phase markers respectively, in blm mutants compared to siblings (Figure 3D). Equivalent levels of the post-mitotic retinal marker atoh7 were observed in blm and siblings indicating that retinogenesis had initiated in blm mutants (Figure 3D). With regards to cell death, a higher degree of apoptosis was observed in blm mutants at 24 hpf (Figure 3E). In summary, blm mutants exhibit an increased proportion of cells in G2/M phase, consistent with delayed cell cycle progression.
Figure 3.
top2a mutants exhibit defects in cell cycle progression. A) Flow cytometry analysis of larvae at 27 hpf (red plot - wt, blue - blm) reveals an increased fraction of cells in G2/M phase in blm mutants. B) Representative Z-series projections showing that the total number of mitotic cells (stained with PH3 antibody - red) is lower in the eye of 27 hpf blm mutants (n = 58) than in wild type larvae (n = 70). C) The percentage of mitotic cells (PH3 positive) relative to the total number of DAPI stained cell nuclei is significantly (p = 0.036) higher in blm mutants compared to wild type as assessed from n ≥ 23 sections through the head and spinal cord of the N = 5 mutant and wild type larvae. D) Real time PCR of cell cycle markers p21-like (G1 phase) and ccnb1 (G2/M phase) in wildtype and blm larvae at 27 hpf shows no significant difference in expression levels (p = 0.21 and 0.38, respectively). Expression of atoh7, a marker of retinogenesis, is also equivalent in eyes of blm and wildtype siblings. Presented data are average of 3 replicate experiments, each comprising pools of 16-35 larvae. Error bars represent the standard error of the mean (C-D). E) Acridine orange staining of apoptotic cells in wildtype and blm larvae at 24 hpf. Arrows point to regions of increased apoptosis in blm mutants.
Maternal top2a but not top2b is expressed pre-MZT
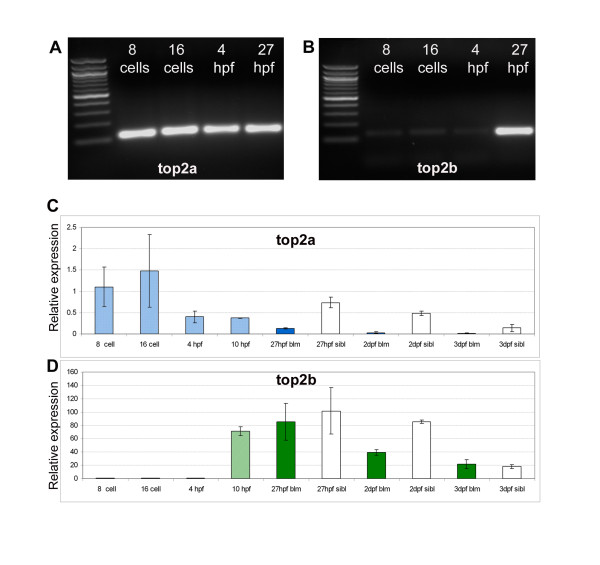
The fact that cell cycle progression is occurring in blm larvae contrasts dramatically with the essential requirement of Top2a for mammalian embryo development beyond the 4-8 cell stage. Therefore, we hypothesised that top2a-/- cells can proliferate in zebrafish blm mutants due to the presence of maternal top2a or functional redundancy with its top2b paralogue. The amino acid sequences of the zebrafish paralogues are 65% identical with the greatest divergence at the C-terminal end (Additional file 1). RT-PCR of RNA extracted from pre- (8 and 16 cell stages) and post-MZT (4 and 27 hpf) wildtype embryos demonstrates that maternal top2a transcript is abundant pre-MZT, whereas top2b transcript is not significantly expressed until after the onset of zygotic transcription (Figure 4A-B). Quantitative analysis by real-time PCR confirms these results (Figure 4C-D).
Figure 4.
top2a, but not top2b, is expressed pre-MZT. RT-PCR of wt embryos (A-B) and real time PCR of offspring from blm carriers (C-D) shows that maternal top2a (A, C), but not top2b (B, D), is present pre-MZT at 8- and 16-cell stages. top2a transcript levels decrease in blm siblings from 27 hpf to 3 dpf, and the expression in blm mutants is always significantly lower (C). The levels of top2b transcript also decrease in wildtype siblings from 27 hpf to 3 dpf, and although top2b levels in blm mutants and siblings is similar at 27 hpf, top2b is dramatically reduced in the mutants by 2 dpf (D). Error bars represent the standard error of the mean. At 8-cell, 16-cell, 4 hpf and 10 hpf stages mutant and wild type offspring are pooled. In graphs light blue (C) and light green bars (D) represent pooled wiltype and mutant embryos; dark blue/green bars represent blm mutants and white bars represent wildtype larvae.
Real-time PCR also demonstrates a reduced level of top2a transcript in blm embryos compared to wildtype siblings at 1-3 dpf (Figure 4C). Zebrafish top2b transcript is negligible before 4 hpf, but is abundantly expressed at 10 hpf, a stage after MZT and before blm phenotypes are detected (Figure 4D). At 27 hpf, top2b levels are similar in blm mutants and widtype siblings. In siblings, top2b levels remain high at 2 dpf. In contrast, top2b levels decrease by ~50% in blm mutants by 2 dpf, consistent with the morphological defects observed at this developmental stage. In summary, maternal top2a could account for the extended pre- and/or post-MZT development of blm mutants whereas zygotic top2b could only account for the extended post-MZT development.
No evidence of functional redundancy between top2a and top2b in vivo
RT-PCR analyses confirm that top2a and top2b are co-expressed in the eye at 2-5 dpf (Figure 5A). Wholemount in-situ hybridisation in 22 hpf wildtype larvae reveals predominant zygotic expression of top2b in the forebrain and eyes, organs that display severe morphological abnormalities from 27 hpf in blm mutants (Figure 5B-D). At 3 dpf, top2b is expressed in the forebrain, midbrain, branchial arches and eyes (Figure 5F-G). T-PCR confirms that top2b is expressed in the blm eyes and body at 3 dpf (Figure 5H). Although the size of the eyes and head is reduced, the spatial expression of top2b in blm mutants at 3 dpf is not affected (Figure 5I-L). Overall, this suggests that top2b has biological potential to rescue affected tissues in blm mutants but cannot do so due to functional divergences between the top2a and top2b paralogues. In agreement, ectopic expression of top2b mRNA in blm mutants shows no evidence of rescuing the blm phenotype (Figure 5M-N and Additional file 2). Larvae overexpressing top2b by ~3.8 fold still exhibit the blm phenotype (Figure 5M). Morphologically, the average eye diameter of blm larvae overexpressing top2b is not significantly different from uninjected blm larvae (Additional file 2C). Thus, overexpression of top2b in blm larvae at non-toxic levels shows no evidence of rescuing the blm phenotype. In summary, our data suggests that top2a and top2b are not functionally redundant during embryo development.
Figure 5.
Overlapping expression, but functional divergence of top2a and top2b paralogues in vivo. A) RT-PCR shows that both top2a and top2b are expressed in the eye at 2, 3, 4 and 5 dpf. B-G) Wholemount in situ hybridization reveals that top2b is expressed in the anterior of wildtype larvae including the forebrain, midbrain and eye (B-D) at 22 hpf and in the forebrain, midbrain, branchial arches and retina (F-G) at 3 dpf. E is the negative control sense probe; D and G are sections through the eye. H) RT-PCR at 3 dpf shows similar abundant expression of top2b in the eye and body of blm mutants and wildtype sibling. I-L) Wholemount in situ hybridization reveals a similar spatial expression pattern of top2b in blm (I, K) and wildtype larvae (J, L) at 3 dpf. M) top2b expression levels at 33 hpf following injection of top2b mRNA into 1-2 cell stage offspring of blm carriers. blm larvae overexpressing top2b RNA by 2.7-5.9 fold show no evidence of phenotypic rescue. N = 3 replicate experiments, n = 50 uninjected wild type (wt cont), n = 29 top2b RNA injected wild type (wt inj), n = 13 uninjected blm (blm cont), n = 17 top2b RNA injected blm (blm inj), n = 6 top2b RNA injected malformed. N) Phenotypes recorded at 33 hpf following injecting offspring of carriers of blm mutation with zebrafish top2b mRNA (N ≥ 3 replicate experiments, n = 117 uninjected wild type, n = 40 uninjected blm, n = 29 top2b RNA injected wild type, n = 17 top2b RNA injected blm, n = 6 top2b RNA injected malformed). Error bars represent the standard error of the mean. Abbreviations: ba - branchial arches, e - eye, fb - forebrain, hb - hindbrain, ln - lense, mb - midbrain, r - retina.
Chemical inhibition of Top2a delays development of pre-MZT embryos
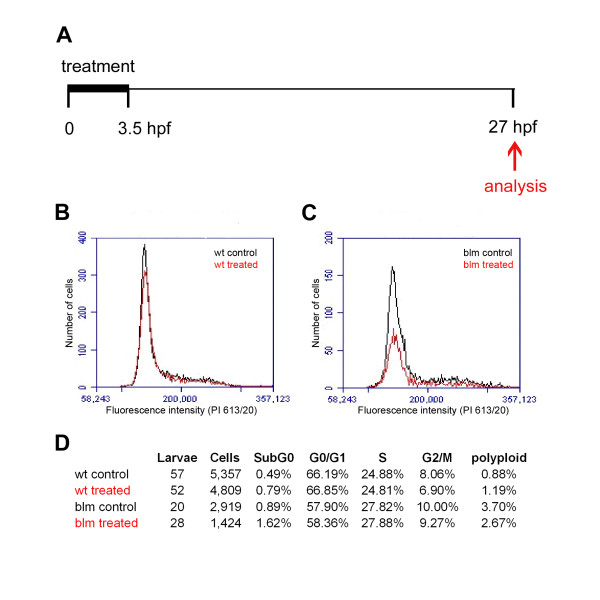
To evaluate if maternal topoisomerase II proteins present in zebrafish embryos are required for early development, we chemically inhibited their function. ICRF-193 is a chemical inhibitor of TOP2A and TOP2B. With the goal of targeting the drug to maternal Top2a, and avoiding effects on zygotic Top2b, 1-2 cell stage embryos from matings of blm carriers were treated with ICRF-193 for ~3.5 hours, the drug removed before MZT and the embryos allowed to develop to 27 hpf. No discernible morphological defects were observed in ICRF-193 treated embryos compared to controls (Additional file 3). In replicate experiments, the cell cycle profile of treated embryos was analysed at 27 hpf (Figure 6A). Compared to their DMSO-treated controls, transient inhibition does not dramatically affect the percentage of sibling- or blm-treated cells in G0/G1, S, G2/M or polyploid categories (Figure 6B-D). A slight increase in the subG0 population of treated embryos is observed. Thus, transient treatment of pre-MZT embryos with a topoisomerase inhibitor causes no obvious morphological or cell cycle defects at a later developmental stage.
Figure 6.
Chemical inhibition of Top2a pre-MZT does not affect cell cycle profile at 27 hpf. A) Schematic of the experimental procedure. 1-2 cell stage embryos were treated with 100 μM ICRF-193 for 3.5 hours when the drug was removed and the cell cycle profile analysed at 27 hpf. B-C) Flow cytometry analysis of cell cycle profiles of wt (B) and mutant (C) embryos. D) Table documenting the percentage of cells in cell cycle phases following ICRF-193 treatment.
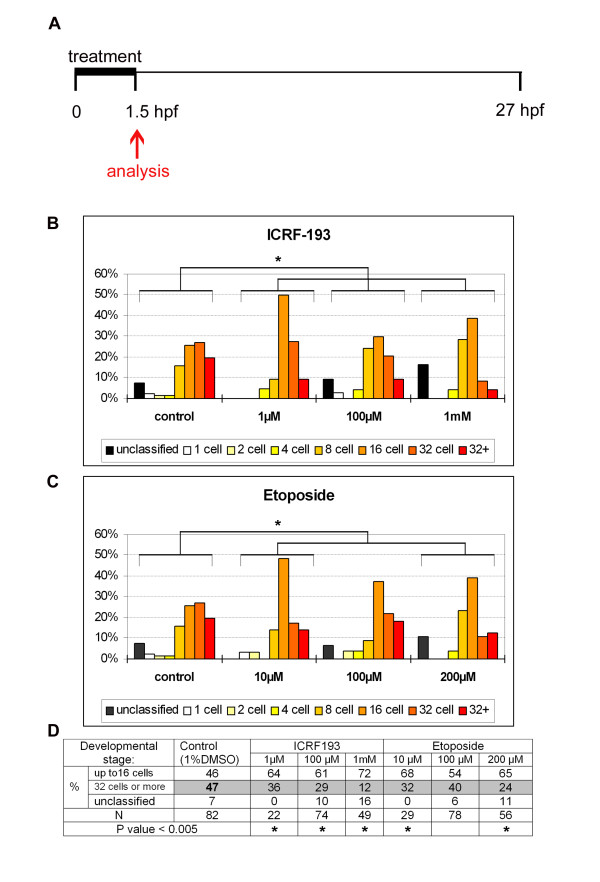
In an alternative approach (Figure 7A), focusing specifically on earlier cell divisions, zebrafish embryos at the 1-2 cell stage were treated for ~1.5 hours with a topoisomerase II inhibitor (ICRF-193) or poison (etoposide) and their developmental progression quantified according to established developmental staging series (Additional file 4). In embryo medium alone or in the presence of a drug not targeting topoisomerase II (L-AP4, a metabotropic glutamate receptor agonist), ~60-70% of embryos are staged at 16 cells (Additional file 5). In vehicle-treated controls (1% DMSO), performed concurrently with siblings, embryos are distributed equivalently across the 8-32+ cell stages (Figure 7B-C). In contrast, in ICRF-193 and etoposide-treated siblings there is an altered distribution peaking in the 8-16 cell stages (Figure 7B-C). These differences are statistically significant (p < 0.005) when compared to DMSO controls (Figure 7B-C). 1 mM ICRF-193 demonstrates a ~26% increase in the numbers of embryos ≤ 16 cell stage, and up to a ~35% reduction in the number of embryos > 16 cell stage (Figure 7D). The reduced number of cell divisions in over one quarter of the population is significant, considering the short treatment duration, the necessity for the drugs to penetrate the chorion, and the very high concentration of Top2a in zebrafish embryos (Figure 4), [29]. In summary, we conclude that maternal Top2a is required for the normal rate of embryonic cell division in developing zebrafish.
Figure 7.
Chemical inhibition of Top2a disrupts pre-MZT development of zebrafish embryos. A) Schematic of the experimental procedure. 1-2 cell stage embryos were treated with Top2a inhibitors for 1.5 hours and their developmental progression was analysed. B-C) Graphs representing the percentage of embryos at a range of developmental stages following treatment with ICRF-193 (B) or etoposide (C). Top2a inhibition changes the staging distribution of embryos which peak at 16-32 cell stage in vehicle controls (DMSO) to peaking at the 8-16 cell stage in drug treated populations. The "unclassified" category includes embryos that do not fit into the standard developmental morphologies, ones in which all cells do not have typical optical transparency, and ones that are malformed. D) Table grouping the percentage of embryos at ≤ 16 cell stage or ≥ 32 cell stage for each drug treatment. Top2a inhibition increases the number of embryos ≤ 16 cell stage by up to 26%, and decreases the number of embryos ≥ 32 cell stage by up to 35%. Asterisks represent statistically significant differences (p < 0.005) to DMSO controls.
Inhibition of Xenopus Top2a results in an extended M-phase in cycling extracts
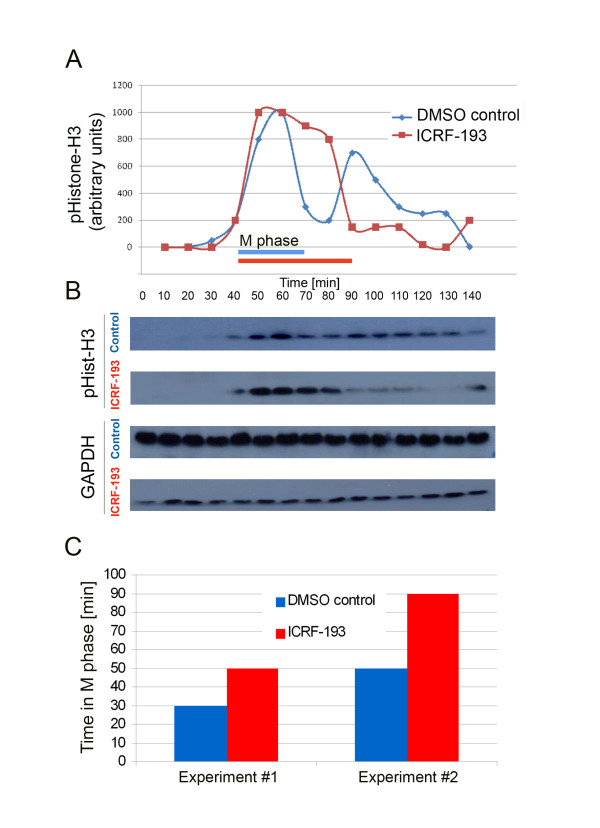
To further analyze the effect of Top2a inhibition on pre-MZT cell cycle progression we utilized cycling extracts prepared from activated Xenopus laevis eggs. These extracts recapitulate, in vitro, the cell cycle of the intact fertilized egg, with extracts oscillating between S phase and mitosis as in pre-MBT embryos [36]. In control extracts the first mitotic peak of phosphohistone H3 expression was observed after 60 minutes, with a subsequent mitotic peak observed at 90 minutes, thus recapitulating the 30 minute cycling interval observed in whole embryos (Figure 8A). Addition of ICRF-193 to extracts at time 0 resulted in a much longer M phase compared to control extracts (Figure 8A-B). Results from two different cycling extracts indicate a ~40% lengthened M phase following topoisomerase II inhibition (Figure 8C). Additionally, there was no evidence of a subsequent mitotic cycle in the following 50 minutes. In summary, we conclude that inhibiting maternal Top2a with ICRF-193 delays early embryonic cell cycle progression in M phase and may interfere with subsequent S phase entry.
Figure 8.
Inhibition of maternal Top2a in pre-MBT cycling extracts delays cell cycle progression by lengthening the M-phase. A) Cycling extracts from activated Xenopus eggs were treated with 20 μM Top2a inhibitor ICRF-193 diluted in 1% DMSO. To monitor cell cycle progression samples were taken every 10 minutes and phospho-histone H3 expression relative to GAPDH expression plotted. Blue and red bars indicate M phase in DMSO control and ICRF-193 treated extracts, respectively. B) Western blot analysis of phospho-Histone H3 and GAPDH expression levels. C) The increase in M phase length in two different cycling extracts treated with ICRF-193. M phase lengthened on average by 40% (SD = 3.14).
Discussion
Top2a mutagenic susceptibility
Vertebrate topoisomerase II alpha genes appear particularly susceptible to somatic and germline mutations. Amplifications of the human TOP2A locus is associated with somatic mutations in subsets of breast, bladder and gastric cancers [6]. Notably, three genetically-distinct zebrafish top2a mutant alleles have now been uncovered (hi3635, can4, blm) in independent mutagenesis screens, suggesting that the top2a locus is also particularly susceptible to germline DNA perturbations [28,29]. All zebrafish top2a mutants display equivalent gross morphological phenotypes: small eye and brain, CNS necrosis, abnormal tail curvature and death at 4-5 dpf. Although, no further characterisation of the hi3635 insertion mutant has been reported, can4 and blm top2a-/- alleles display similar cell cycle defects, including accumulation of mutant cells in G2/M and altered phosphohistone H3 expression (Figure 3), [29].
The blm phenotype
Our interpretation of the blm mutant phenotype is as follows (Figure 9). A nonsense mutation in the zebrafish top2a gene results in the expression of non-functional Top2a protein post MZT in blm mutants. The lower RNA expression levels may be due to lower transcription or increased decay of mutant top2a mRNA. The extensive embryonic and larval development of blm mutants arises from maternal expression of top2a, and not top2b which is negligible pre-MZT. Depletion of the maternal top2a transcript and Top2a protein results in the morphological phenotype of blm mutants at 27 hpf. Intracellular levels of top2a/Top2a diminish as daughter cells divide and as mRNA/protein degrades. For example, after only 10 divisions the level of top2a per daughter cell is expected to deplete by > 1000 fold relative to DNA content. Thus, tissues that are actively proliferating in later stages of development (e.g. eye) are preferentially affected. In those cells, Top2a depletion would likely perturb DNA decatenation during replication resulting in genomic instability and cell cycle checkpoint activation. The augmented levels of G2/M and apoptotic cells observed in blm mutants at ~27 hpf are consistent with activation of the known G2/M decatenation checkpoint present in adult cells leading to apoptosis.
Figure 9.
Model depicting how maternal top2a enables pre-MZT development of zebrafish embryos and post-MZT development of top2a mutants.
Overall our data lead us to conclusion that zebrafish Top2a and Top2b are not functionally redundant in vivo. The zebrafish paralogs are ~65% identical at the protein level; both containing N-terminal ATP-binding sites and a central domain tyrosine required for DNA cleavage and ligation (Additional file 1). Only the C-terminal domains, containing nuclear localization sequences and phosphorylation sites, are highly divergent [8]. Previous studies in human cell lines suggested that TOP2B can compensate for depleted TOP2A [37,38]. In contrast, abundant zebrafish top2b transcript at stages post-MZT is not sufficient to prevent the blm phenotype, despite extensive temporal expression in tissues that develop morphological abnormalities in blm mutants. The onset of the blm phenotype correlates with depletion of maternal top2a, at a stage when zygotic top2b is abundant, and we identify embryos overexpressing ectopic top2b that retain the blm phenotype. Furthermore, in contrast to top2a mutants, the recently described top2b mutant displays a distinct phenotype in post-mitotic cells and not in proliferating cells [30]. There is a potential compensatory role for maternal Top2b in pre-MZT stages that we cannot directly exclude. However, pre-MZT top2b expression is negligible [33] and we have not found evidence of maternal deposition of Top2b in the literature. Notably, the functional divergence we observe in vivo between zebrafish Top2a and Top2b is in agreement with other in vitro studies reporting that the unique cell cycle functions of the Top2a isoform are dependent on specific C-terminal sequences [5,12,38]. Overall, our in vivo studies support that top2a and top2b do not exhibit complete functional redundancy during zebrafish embryo development.
Role of maternal Top2a pre-MZT
Previous genetic approaches to analyse maternal top2a function using morpholino knockdowns have proven unsuccessful, probably because of the abundance of top2a/Top2a [29]. Therefore, to confirm that maternal top2a enables pre-MZT development, we pharmacologically targeted maternal Top2a protein using ICRF-193, a topoisomerase II catalytic inhibitor and etoposide, a topoisomerase II poison [11].
Our data supports the conclusion that maternal top2a is needed for the normal progression of pre-MZT embryos through developmental divisions. Both ICRF-193 and etoposide delay the progression of embryos through hierarchical developmental stages. This delay is reversible as washing out the drug results in the vast majority of zebrafish embryos proceeding beyond the 24 hpf, segmentation stage. It is likely that we are only partially inhibiting Top2a protein as complete inhibition is expected to arrest development earlier.
To explore the mechanism underlying the impaired developmental cell divisions in ICRF-193 treated embryos we utilised Xenopus cycling extracts which recapitulate in vitro the cell cycle of pre-MBT embryos [36]. Only topoisomerase II α is expressed in Xenopus eggs and embryos as demonstrated by the fact that depletion of Top2a from egg extracts abolishes topoisomerase II activity [39]. Our molecular analysis of Xenopus extracts treated with ICRF-193 indicates a lengthening of M phase. This is consistent with the previous morphological studies that observed anaphase stalling of pre-MZT zebrafish treated with etoposide [40].
We propose that the mechanism underlying delayed pre-MZT development and M phase lengthening following Top2 inhibition reflects activation of a cell cycle decatenation checkpoint. In the pre-MZT embryos lacking gap phases, Top2a inhibition results in a stoichiometric decrease in DNA decatenation during cell division activating a DNA decatenation checkpoint [40,41]. The M phase lengthening observed in ICRF-193 treated cycling extracts are consistent with defects in chromosome condensation and segregation most likely slowing the cycle and possibly activating a checkpoint in mitosis.
Inhibition of Top2a could be impacting on one or more phases of the cell division cycle and its effects in the simple early embryonic cell cycle and later somatic cycles where gap phases are present may be the result of different cell cycle processes being impeded. ICRF-193 is known to activate a G2 decatenation checkpoint in adult cells [42,43]. Interestingly, Top2a, but not Top2b, is required for activation of the decatenation checkpoint [43]. Catalytically inhibited Top2a exposes a C-terminal phospho-serine that recruits MDC1 to chromatin and activates the decatenation checkpoint [42]. As shown by our data, etoposide, which induces DNA damage in adult cells, phenocopies ICRF-193 in pre-MZT embryos. The likely explanation is the absence of Top2b and the absence of a DNA damage checkpoint in pre-MZT embryos [44]. This also suggests that etoposide alters Top2a topology to activate the decatenation checkpoint. In summary, we propose that inhibition of maternal Top2a activates a DNA decatenation checkpoint that delays mitosis and results in delayed embryonic development.
Phenotypic differences between mouse and zebrafish Top2a mutants
Why does maternal Top2a enable zebrafish top2a-/- mutants to progress to stages comprising of hundreds of thousands of cells, whilst mouse Top2a-/- knockouts fail to divide beyond the 4-8 cell stage? One possibility is that maternal Top2a is not expressed in mammalian embryos. This is not the case, as studies show that catalytically active Top2a is functional at the 1-cell stage in mouse embryos [45]. Alternatively, maternal topoisomerase II alpha may have differential stability in mice and zebrafish due to the timing of MZT which occurs between embryonic day 0.5 and 1.5 (2-cell stage) in mouse but between 2.75 and 4.75 hours after fertilization (from 1,024 cells stage) in zebrafish [34,35,46]. Although some maternal transcripts are still present in embryonic day 2.5 murine morulas [46], previous reports demonstrate that newly synthesised mouse Top2a at the 2-cell stage is essential for cell cycle progression [45].
Conclusions
The ability of zebrafish top2a-/- mutants to proceed through thousands of developmental cell divisions starkly contrasts with mammalian Top2a-/- knockouts which fail to divide beyond the 4-8 cell stage [20]. Our analyses reveal that pre-MZT, maternal top2a is sufficient to enable zebrafish top2a-/- mutants to progress through early development. Post-MZT, zebrafish top2b expression is insufficient to fully compensate for the absence of top2a in vivo and blm mutants die at ~5 dpf (Figure 9). Top2a inhibition or loss results in clear cell cycle defects in early embryonic cell cycles of both zebrafish and Xenopus, as well as in later somatic cell cycles where blm mutants show evidence of G2/M decatenation checkpoint activation.
Methods
All animal experiments were conducted under licences from the Department of Health and Children (B100/3641; B100/3003) and according to protocols approved by the University College Dublin Animal Research Ethics Committee (AREC-P-08-54; AREC-P-10-68).
Mutagenesis
Zebrafish males of AB strain were mutagenised with ENU and mated to wildtype females to generate F1 founder fish [47]. F1 founders were mated to wildtype fish to establish 370 F2 families. F3 offspring of random incrosses within 200 F2 families were screened for recessive mutations at 5 dpf. 25% of the offspring from family Y-008 carriers presented with blm phenotypes.
Light microscopy
Larvae at 27 hpf, 2 dpf and 3 dpf were euthanized with benzocaine and fixed in Sorensen phosphate buffer (pH 7.3) containing 4% PFA and 2.5% glutaraldehyde. Larvae were embedded in Epon resin and semi-thin (900 nm) sections stained with toluidine blue were photographed using a Leica DMLB microscope and a Leica DFC 480 camera.
Mapping
Carriers of blm (AB strain) mutants were map-crossed to wildtype Tubingen fish to generate hybrid carriers AB*/Tub. 96 blm and 96 normal siblings from map crosses were used for bulk segregation analysis. Linkage was found to chromosome 12 with the blm mutation flanked by Z-markers Z99217 and Z10806. Several new simple sequence repeat (SSR) markers were designed to further narrow the critical interval of which zC13B10-SSR1 was the closest (6/96 recombinants). Primers zC13B10-SSR1-F: CTCCAATCGAGAGTCCTCGT, zC13B10-SSR1-R: AGCTGAAGGCCTGCTGTAAA. The top2a gene was localised in the critical interval.
Sequencing
RNA from 3 dpf blm larvae was extracted using the Qiagen RNeasy Mini Kit and cDNA was synthesised using the Invitrogen Superscript III First Strand Synthesis for RT-PCR system. Six sets of primers were designed to amplify the whole reading frame of zebrafish top2a. The causative point mutation was sequenced using the following primers: forward 5'TGTTGCGCTACTGACTCGAC, reverse 5'TGGCCAGTTATGATGGATGA.
Complementation assay
Hi3635 carriers were obtained from ZIRC. Complementation matings of blm+/- ♀ × hi3635+/- ♂ and blm+/- ♂ × hi3635+/- ♀ were performed. Phenotypes were recorded at 2 and 3 dpf.
Flow cytometry
blm and wildtype sibling larvae were collected at 27 hpf. Embryos were dechorionated, rinsed in PBS and incubated for 10 minutes at 37°C in trypsin (1 mg/ml) with DNase I (1 U/200 μl) followed by mechanical disruption with a 25 G needle. Trypsin inhibitor was added to a final concentration of 1 mg/ml. After centrifugation, the cell pellet was fixed with 70% ethanol. Cells were rinsed with PBS, treated with RNase (10 μg/ml) and DNA stained with propidium iodide (40 μg/ml). DNA content was analysed using a Coulter EPICS XL-MCL or Accuri C6 flow cytometer.
Quantification of mitotic cells in the eye
Embryos at 27 hpf were fixed at 4°C overnight in 4% PFA, then dechorionated, dehydrated in methanol and stored at -20°C. Prior to immunostaining embryos were rehydrated in PBS, permeabilised for 5 min in proteinase K (20 μg/ml) and post-fixed in 4% PFA for 30 min. Larvae were treated with blocking solution (2% normal goat serum, 1% Triton X-100, 1% Tween-20 in PBS) for 60 min followed by incubation in the primary antibody: anti-phospho-histone H3 at 1:200 dilution (Upstate Biotech) for 20 hours. Primary antibody binding was detected with Cy3-conjugated goat anti-rabbit secondary antibody (dilution 1:200). Nuclei were counter stained with DAPI. Z-stack images of whole larval eyes (optical sections at 2 μm interval) were taken with Zeiss LMS 510 Meta laser confocal microscope (with 63 × objective). Images were analysed with Imaris v7.2.3 software. To quantify the proportion of mitotic cells, embryos at 27 hpf were fixed in PFA, cryoprotected and sectioned (12 μm thickness). Mitotic nuclei were labelled with anti-phospho-histone H3 antibody (Upstate Biotech) and counter stained with DAPI. Images of sections through the eyes, head and spinal cord (at least 3 per animal) were taken with Zeiss AxioImager M1 microscope (objective 100×) and all PH3 positive and DAPI labelled nuclei from the field (89.5 μm × 67.1 μm) were counted. The percentage of mitotic cells in 5 mutant and 5 wild type larvae was compared.
Acridine orange staining
To visualise apoptotic cells at 24 hpf, dechorinated wildtype and blm larvae were incubated with 5 μg/ml acridine orange (Sigma) for 30 minutes and washed in embryo medium. Larvae were anaesthetised with tricaine and imaged with an Olympus SZX2-ILLT microscope using fluorescence lamp and GFP filter.
RT-PCR and Q-RT-PCR
RNA was extracted using the Qiagen RNeasy Mini Kit from pools (15 to 70 individuals) of 8-cell, 16-cell, 4 hpf, 10 hpf, 27 hpf, 2 dpf and 3 dpf embryos or dissected eyes and rest of the body of 2, 3, 4 and 5 dpf larvae obtained from wildtype incrosses or blm carrier incrosses. cDNA was synthesised by reverse transcription after priming with random hexamers using the Invitrogen Superscript III system. Primers' sequences for cell cycle markers [41]: p21-likeF-CCGTAGACCATGAGGAGC; p21-likeR-GTCTCGTCCACTTCTTTCTTTC; ccnb1F-GAGTCACAGCAATAAACCAC; ccnb1R-AGGAAGGCTCAGACACAAC. Primers' sequences for top2a, top2b and βactin: top2aF-AACGAGACCATGCCTCACC; top2aR-CAAACCAGCCTCTTTCTTCG; top2bF-GCAGTTGGAGGAAACTCTGC; top2bR-AGCTTCACAGCCGCATCTAT; βactF-GAGAAGATCTGGCATCACAC; βactR-ATCAGGTAGTCTGTCAGGTC. Primers' sequences for atoh7 (ath5): atoh7F-CCGGAGAAGTTTGAGAGTGC and atoh7R- GCTCAGAGCCATCTGTAGGG. Real-Time PCR was performed with SYBR-Green chemistry. 18s rRNA amplification with TaqMan probe served as an internal reference. For the time course experiment an arbitrary value of 1 was assigned to the average expression at 8 cell stage and expression level at other time points was normalised to this denominator. Plots of quantitative RT-PCR are average from 2 or 3 replicas and error bars represent standard error of the mean.
Wholemount in situ hybridization
Full length top2b cDNA was amplified with AccuPrime Taq polymerase (Invitrogen) using forward and reverse primers binding to 5'UTR and 3'UTR (5UTRzfTop2b-TCTGGCCACACACAATAGAAA, 3UTRzfTop2b- CCAATCAGTTTTCTGGACCAA) and cDNA (made with random hexamers from total RNA of 1 dpf old larvae) as template. It was cloned into pGEM vector and used for synthesis of antisense and sense digoxigenin labelled probes with DIG in vitro transcription kit (Roche).
Larvae were fixed in 4% PFA (overnight at 4°C). 3 dpf specimens were treated for 20 mins with a bleach solution (30% H2O2, 50% formamide, 20% 20× SSC) to remove pigmentation and permeabilised with proteinase K (10 μg/ml, 22 minutes). Probe hybridisation was carried out at 62.5°C (overnight), followed by a series of formamide/SSC washes at the same temperature. Detection of hybridised probe was performed by incubation (overnight at 4°C) with AP-conjugated anti-digoxigenin Fab fragments (1:5,000; Roche) followed by colorimetric detection (NBT/BCIP, Roche). Whole larvae were imaged under 100% glycerol using an Olympus SZX2-ILLT stereo zoom microscope. 30 μm sections of stained larvae embedded in Tissue Tek (Sakura) were imaged using Zeiss Axioplan2 microscope equipped with AxioCam HRc camera.
Overexpression of top2b
The top2b coding region was amplified from cDNA of 1 dpf larvae using AccuPrime Taq polymerase (Invitrogen) and cloned into pGEM vector. This plasmid was used to synthesise full length top2b mRNA with the mMessage mMachine SP6 system (Albion Inc.). Offspring of blm carriers were microinjected at 1-2 cell stages with zebrafish top2b mRNA at 250 ng/μl (approximately 75 pg per embryo). Phenotypes were recorded at 33 hpf and eye diameter was measured with CellF software (Olympus Soft Imaging Solutions) after imaging with an Olympus SZX2-ILLT stereo zoom microscope. To verify overexpression of top2b, pools of embryos presenting with wildtype, blm or malformed phenotypes were collected at 33 hpf for RNA extraction. Q-RT-PCR was performed as described above (using primers top2bF-GCAGTTGGAGGAAACTCTGC and top2bR-AGCTTCACAGCCGCATCTAT). Expression of top2b mRNA in all samples was normalised to the expression level in uninjected wildtype controls.
Drug treatment
For transient treatment freshly laid eggs from blm carrier incrosses were incubated in embryo medium containing 100 μM ICRF-193 (Sigma) or 1% DMSO for 3.5 hours at 28°C and then the drug was replaced by fresh embryo medium. At 27 hpf larvae with mutant and wildtype phenotypes were separated and used for cell cycle analysis by flow cytometry.
For pre-MZT treatment, freshly laid wildtype AB eggs were placed in multi-well plates. Excess embryo medium was removed and replaced by a drug solution. Drugs were diluted in 1% DMSO in embryo medium at final concentrations of: 1 μM, 100 μM and 1 mM of ICRF-193 (Sigma) and 10 μM, 100 μM and 200 μM of etoposide (Ebewe). Embryos were incubated with drug for ~1.5 hours and then fixed with 4% PFA. Control embryos were incubated in 1% DMSO. The percentage of embryos reaching each development stage was calculated. Statistical analysis was performed using the Chi-squared test in GraphPad Prism.
Cell cycle extracts
Cycling extracts were prepared from unfertilized Xenopus laevis eggs activated using calcium ionophore A23187 (Sigma; C7522), as described in [36]. Extracts were treated with 20 μM ICRF-193 diluted in 1% DMSO or 1% DMSO alone at time zero. Samples were collected at 10 minute intervals and snap frozen in liquid nitrogen.
Western blotting
Protein samples were separated according to their apparent molecular mass under denaturing conditions on NuPAGE 4-12% Bis-Tris Gel, 1.0 mm × 17 well gels (Invitrogen), and transferred to PVDF membrane. After blocking in 5% Bovine Serum Albumin (BSA) in PBS-0.1% Tween-20, membranes were incubated with either anti-phospho-Histone H3 (Ser10) antibody from Millipore to detect mitosis (Cat. # 06-570) or GAPDH antibody from Cell Signaling (#3900), followed by incubation with horse-radish peroxidase-conjugated secondary antibodies. Proteins were visualized using Enhanced Chemiluminescence Western Blotting Substrate (Pierce; #32106). Densitometry was performed using the ImageJ program http://rsbweb.nih.gov/ij/.
Abbreviations
dpf: days post fertilization; hpf: hours post fertilization; MZT: mid-zygotic transition; TOP2A: topoisomerase 2 alpha; TOP2B: topoisomerase 2 beta.
Authors' contributions
BNK, YA, RC, FVE conceived and performed the mutagenesis experiment; BSR and SMcL characterized morphology; BSR, JRW and SMcL mapped and identified the mutation, BSR analyzed developmental expression pattern and cell cycle, BSR and OOL performed in situ hybridization, BSR, SMcL and JOS conceived and performed the drug treatment experiment and staging of embryos, CH and LOS conceived and performed cycling extract experiments, BSR and BNK drafted the manuscript. All authors read and approved the final manuscript.
Supplementary Material
Comparison of zebrafish Top2a and Top2b paralogues. A) Phylogenetic tree of Top2a and Top2b paralogues of 9 vertebrate species including zebrafish and other 4 teleost species. Protein sequences were retrieved from NCBI and Ensembl data bases. TOP2A: NP_001058.2 (human), NP_001003834.1 (zebrafish), NP_035753.2 (mouse), NP_001082502.1 (frog), NP_990122.1 (chicken), ENSTRUP00000041924 (fugu), ENSORLP00000005530 (medaka), ENSTNIP00000003411 (tetraodon), ENSGACP00000011260 (stickleback). TOP2B: NP_001059.2 (human), NP_001038656.1 (zebrafish), NP_033435.2 (mouse), XP_002932456.1 (frog), NP_990413.1 (chicken), ENSTRUP00000017341 (fugu), ENSORLP00000010828 (medaka), ENSTNIP00000021338 (tetraodon), ENSGACP00000010617 (stickleback). They were aligned by NCBI Multiple Alignment tool. Phylogenetic tree was generated with Seaview4 software (PhyML v3.0.1). Two distinct branches for Top2a and Top2b paralogues are apparent indicating that origin of two paralogues in zebrafish does not result from the genome duplication in teleosts. B) Alignment of zebrafish Top2a and Top2b protein sequences shows 65% identity (shaded residues) with the greatest divergence occurring at the C-terminal end. Asterisk indicates a serine residue in zebrafish Top2a which is associated with activation of the decatenation checkpoint in mammals.
Overexpression of top2b in blm embryos. Offspring of blm carriers were microinjected at 1-2 cell stages with zebrafish genomic top2b sequence (BAC clone CHORB736O22185Q) at 25 ng/μl (7.5 pg per embryo) or zebrafish top2b mRNA at 250 ng/μl (75 pg per embryo) and analysed at 33 hpf. A) Table of observed phenotypes upon injecting offspring of carriers of blm mutation with zebrafish top2b BAC clone or in vitro synthesised top2b RNA. B) Images of 33 hpf malformed larvae overexpressing top2b by ~4 fold following injecting with top2b mRNA. C) Ectopic expression of top2b mRNA does not rescue the small eye phenotype of blm embryos. Error bars represent the standard error of the mean.
Transient inhibition of Top2a pre-MZT. A) Dark field images showing the morphology of embryos at 27 hpf following treatment with ICRF-193 from 1-2 cell stage until 3.5 hpf B) Representative confocal images (projections of 30 slices taken at 0.5 μm intervals) of 3.5 hpf embryos stained with DAPI, which had been treated from 1-2 cell stage with 100 μM ICRF-193. Chromatin appears to be more compacted in the treated embryos but not extensive DNA damage was observed.
Staging of embryos treated with topoisomerase inhibitors. Top panel: images of normally developed embryos at 16, 32 and 64 cell stages. Bottom panels: example images of "not classified" or malformed embryos treated with 1% DMSO (vehicle control), ICRF-193 or etoposide. This group includes embryos with atypical shape: A) irregular shape, protruding animal pole, B) asymmetric animal pole and uneven cell size, C) irregular shape and opaque, D-E) undefined cell morphology and abnormal transparency, F) small, irregular-shaped animal pole, G) abnormal distribution of cells around yolk, H) undefined cell morphology with abnormal distribution around yolk, I) duplicated/split animal pole.
Effect of topoisomerase inhibitors on pre-MZT development of zebrafish embryos is specific. Treatment of 1-2 cell embryos with L-AP4 (metabotropic glutamate receptor agonist) does not change the distribution of developmental stages compared to untreated controls in embryo medium. Incubation in 1% DMSO (vehicle for topoisomerase inhibitors) increases the percentage of embryos in more advanced stages of development. A) Table and B) graphs representing the number and percentage, respectively of embryos at 8-32 cell developmental stages following incubation for 1 hour in embryo medium containing 10 μM, 200 μM or 1 mM L-AP4 (water dilutions), 1% DMSO or embryo medium alone.
Contributor Information
Beata Sapetto-Rebow, Email: beata.sapetto@ucd.ie.
Sarah C McLoughlin, Email: sarmc@mail.med.upenn.edu.
Lynne C O'Shea, Email: lynne.oshea@ucdconnect.ie.
Olivia O'Leary, Email: olivia.oleary1@gmail.com.
Jason R Willer, Email: j.willer@cellbio.duke.edu.
Yolanda Alvarez, Email: yolanda.alvarez@mail.com.
Ross Collery, Email: rcollery@mcw.edu.
Jacintha O'Sullivan, Email: osullij4@tcd.ie.
Freek Van Eeden, Email: f.j.vaneeden@sheffield.ac.uk.
Carmel Hensey, Email: carmel.hensey@ucd.ie.
Breandán N Kennedy, Email: brendan.kennedy@ucd.ie.
Acknowledgements
We thank members of BK lab for their involvement in the mutagenesis screen: Vincent Smyth, Maria Cederlund, Theresa Heffernan, Olaya Astudillo Fernandez, Fan Yang, Maria Morrisey, Antonino Glaviano, Victor Vendrell, Alison Reynolds, Lisa Shine. We thank Alfonso Blanco for assistance and advice with flow cytometry, Dimitri Scholz for assistance with confocal microscopy and advice on image analysis and Jun Yin for help with retrieving microarray data. We thank Michael R. Taylor for comments on the manuscript. This work was supported by Science Foundation Ireland grants: 04/IN3/B559, 06/RFP/BIM052, 07/SRC/B1156 (LOS).
References
- Roca J. Topoisomerase II: a fitted mechanism for the chromatin landscape. Nucleic Acids Res. 2009;37:721–30. doi: 10.1093/nar/gkn994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costanzo V, Shechter D, Lupardus PJ, Cimprich KA, Gottesman M, Gautier J. An ATR- and Cdc7-dependent DNA damage checkpoint that inhibits initiation of DNA replication. Mol Cell. 2003;11:203–13. doi: 10.1016/S1097-2765(02)00799-2. [DOI] [PubMed] [Google Scholar]
- Cuvier O, Hirano T. A role of topoisomerase II in linking DNA replication to chromosome condensation. J Cell Biol. 2003;160:645–55. doi: 10.1083/jcb.200209023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cuvier O, Stanojcic S, Lemaitre JM, Mechali M. A topoisomerase II-dependent mechanism for resetting replicons at the S-M-phase transition. Genes Dev. 2008;22:860–5. doi: 10.1101/gad.445108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adachi N, Kobayashi M, Koyama H. Cell cycle-dependent regulation of the mouse DNA topoisomerase IIalpha gene promoter. Biochem Biophys Res Commun. 1997;230:105–9. doi: 10.1006/bbrc.1996.5893. [DOI] [PubMed] [Google Scholar]
- Mano MS, Rosa DD, Azambuja E De, Ismael GF, Durbecq V. The 17q12-q21 amplicon: Her2 and topoisomerase-IIalpha and their importance to the biology of solid tumours. Cancer Treat Rev. 2007;33:64–77. doi: 10.1016/j.ctrv.2006.10.001. [DOI] [PubMed] [Google Scholar]
- McClendon AK, Rodriguez AC, Osheroff N. Human topoisomerase IIalpha rapidly relaxes positively supercoiled DNA: implications for enzyme action ahead of replication forks. J Biol Chem. 2005;280:39337–45. doi: 10.1074/jbc.M503320200. [DOI] [PubMed] [Google Scholar]
- McClendon AK, Osheroff N. DNA topoisomerase II, genotoxicity, and cancer. Mutat Res. 2007;623:83–97. doi: 10.1016/j.mrfmmm.2007.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nitiss JL. DNA topoisomerase II and its growing repertoire of biological functions. Nat Rev Cancer. 2009;9:327–37. doi: 10.1038/nrc2608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coelho PA, Queiroz-Machado J, Carmo AM, Moutinho-Pereira S, Maiato H, Sunkel CE. Dual role of topoisomerase II in centromere resolution and aurora B activity. PLoS Biol. 2008;6:e207. doi: 10.1371/journal.pbio.0060207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nitiss JL. Targeting DNA topoisomerase II in cancer chemotherapy. Nat Rev Cancer. 2009;9:338–50. doi: 10.1038/nrc2607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jenkins JR, Ayton P, Jones T, Davies SL, Simmons DL, Harris AL, Sheer D, Hickson ID. Isolation of cDNA clones encoding the beta isozyme of human DNA topoisomerase II and localisation of the gene to chromosome 3p24. Nucleic Acids Res. 1992;20:5587–92. doi: 10.1093/nar/20.21.5587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimura K, Saijo M, Ui M, Enomoto T. Growth state- and cell cycle-dependent fluctuation in the expression of two forms of DNA topoisomerase II and possible specific modification of the higher molecular weight form in the M phase. J Biol Chem. 1994;269:1173–6. [PubMed] [Google Scholar]
- Woessner RD, Mattern MR, Mirabelli CK, Johnson RK, Drake FH. Proliferation- and cell cycle-dependent differences in expression of the 170 kilodalton and 180 kilodalton forms of topoisomerase II in NIH-3T3 cells. Cell Growth Differ. 1991;2:209–14. [PubMed] [Google Scholar]
- Capranico G, Tinelli S, Austin CA, Fisher ML, Zunino F. Different patterns of gene expression of topoisomerase II isoforms in differentiated tissues during murine development. Biochim Biophys Acta. 1992;1132:43–8. doi: 10.1016/0167-4781(92)90050-a. [DOI] [PubMed] [Google Scholar]
- Tsutsui K, Tsutsui K, Hosoya O, Sano K, Tokunaga A. Immunohistochemical analyses of DNA topoisomerase II isoforms in developing rat cerebellum. J Comp Neurol. 2001;431:228–39. doi: 10.1002/1096-9861(20010305)431:2<228::AID-CNE1067>3.0.CO;2-M. [DOI] [PubMed] [Google Scholar]
- Lyu YL, Lin CP, Azarova AM, Cai L, Wang JC, Liu LF. Role of topoisomerase IIbeta in the expression of developmentally regulated genes. Mol Cell Biol. 2006;26:7929–41. doi: 10.1128/MCB.00617-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson M, Phua HH, Bennett SC, Spence JM, Farr CJ. Studying vertebrate topoisomerase 2 function using a conditional knockdown system in DT40 cells. Nucleic Acids Res. 2009;37:e98. doi: 10.1093/nar/gkp480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carpenter AJ, Porter AC. Construction, characterization, and complementation of a conditional-lethal DNA topoisomerase IIalpha mutant human cell line. Mol Biol Cell. 2004;15:5700–11. doi: 10.1091/mbc.E04-08-0732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akimitsu N, Adachi N, Hirai H, Hossain MS, Hamamoto H, Kobayashi M, Aratani Y, Koyama H, Sekimizu K. Enforced cytokinesis without complete nuclear division in embryonic cells depleting the activity of DNA topoisomerase IIalpha. Genes Cells. 2003;8:393–402. doi: 10.1046/j.1365-2443.2003.00643.x. [DOI] [PubMed] [Google Scholar]
- Tsutsui K, Tsutsui K, Sano K, Kikuchi A, Tokunaga A. Involvement of DNA topoisomerase IIbeta in neuronal differentiation. J Biol Chem. 2001;276:5769–78. doi: 10.1074/jbc.M008517200. [DOI] [PubMed] [Google Scholar]
- Nur EKA, Meiners S, Ahmed I, Azarova A, Lin CP, Lyu YL, Liu LF. Role of DNA topoisomerase IIbeta in neurite outgrowth. Brain Res. 2007;1154:50–60. doi: 10.1016/j.brainres.2007.04.029. [DOI] [PubMed] [Google Scholar]
- Lyu YL, Wang JC. Aberrant lamination in the cerebral cortex of mouse embryos lacking DNA topoisomerase IIbeta. Proc Natl Acad Sci USA. 2003;100:7123–8. doi: 10.1073/pnas.1232376100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang X, Li W, Prescott ED, Burden SJ, Wang JC. DNA topoisomerase IIbeta and neural development. Science. 2000;287:131–4. doi: 10.1126/science.287.5450.131. [DOI] [PubMed] [Google Scholar]
- Roca J, Ishida R, Berger JM, Andoh T, Wang JC. Antitumor bisdioxopiperazines inhibit yeast DNA topoisomerase II by trapping the enzyme in the form of a closed protein clamp. Proc Natl Acad Sci USA. 1994;91:1781–5. doi: 10.1073/pnas.91.5.1781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montecucco A, Biamonti G. Cellular response to etoposide treatment. Cancer Lett. 2007;252:9–18. doi: 10.1016/j.canlet.2006.11.005. [DOI] [PubMed] [Google Scholar]
- Azarova AM, Lyu YL, Lin CP, Tsai YC, Lau JY, Wang JC, Liu LF. Roles of DNA topoisomerase II isozymes in chemotherapy and secondary malignancies. Proc Natl Acad Sci USA. 2007;104:11014–9. doi: 10.1073/pnas.0704002104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amsterdam A, Nissen RM, Sun Z, Swindell EC, Farrington S, Hopkins N. Identification of 315 genes essential for early zebrafish development. Proc Natl Acad Sci USA. 2004;101:12792–7. doi: 10.1073/pnas.0403929101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dovey M, Patton EE, Bowman T, North T, Goessling W, Zhou Y, Zon LI. Topoisomerase II alpha is required for embryonic development and liver regeneration in zebrafish. Mol Cell Biol. 2009;29:3746–53. doi: 10.1128/MCB.01684-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nevin LM, Xiao T, Staub W, Baier H. Topoisomerase IIbeta is required for lamina-specific targeting of retinal ganglion cell axons and dendrites. Development. pp. 2457–65. [DOI] [PMC free article] [PubMed]
- Etkin LD. Regulation of the mid-blastula transition in amphibians. Dev Biol (N Y 1985) 1988;5:209–25. doi: 10.1007/978-1-4615-6817-9_7. [DOI] [PubMed] [Google Scholar]
- Tadros W, Lipshitz HD. The maternal-to-zygotic transition: a play in two acts. Development. 2009;136:3033–42. doi: 10.1242/dev.033183. [DOI] [PubMed] [Google Scholar]
- Aanes H, Winata CL, Lin CH, Chen JP, Srinivasan KG, Lee SG, Lim AY, Hajan HS, Collas P, Bourque G. et al. Zebrafish mRNA sequencing deciphers novelties in transcriptome dynamics during maternal to zygotic transition. Genome Res. 2011;21:1328–38. doi: 10.1101/gr.116012.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kane DA, Kimmel CB. The zebrafish midblastula transition. Development. 1993;119:447–56. doi: 10.1242/dev.119.2.447. [DOI] [PubMed] [Google Scholar]
- Pelegri F. Maternal factors in zebrafish development. Dev Dyn. 2003;228:535–54. doi: 10.1002/dvdy.10390. [DOI] [PubMed] [Google Scholar]
- Murray AW. Cell cycle extracts. Methods Cell Biol. 1991;36:581–605. [PubMed] [Google Scholar]
- Sakaguchi A, Kikuchi A. Functional compatibility between isoform alpha and beta of type II DNA topoisomerase. J Cell Sci. 2004;117:1047–54. doi: 10.1242/jcs.00977. [DOI] [PubMed] [Google Scholar]
- Linka RM, Porter AC, Volkov A, Mielke C, Boege F, Christensen MO. C-terminal regions of topoisomerase IIalpha and IIbeta determine isoform-specific functioning of the enzymes in vivo. Nucleic Acids Res. 2007;35:3810–22. doi: 10.1093/nar/gkm102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirano T, Mitchison TJ. Topoisomerase II does not play a scaffolding role in the organization of mitotic chromosomes assembled in Xenopus egg extracts. J Cell Biol. 1993;120:601–12. doi: 10.1083/jcb.120.3.601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikegami R, Rivera-Bennetts AK, Brooker DL, Yager TD. Effect of inhibitors of DNA replication on early zebrafish embryos: evidence for coordinate activation of multiple intrinsic cell-cycle checkpoints at the mid-blastula transition. Zygote. 1997;5:153–75. doi: 10.1017/s0967199400003828. [DOI] [PubMed] [Google Scholar]
- Duffy KT, McAleer MF, Davidson WR, Kari L, Kari C, Liu CG, Farber SA, Cheng KC, Mest JR, Wickstrom E. et al. Coordinate control of cell cycle regulatory genes in zebrafish development tested by cyclin D1 knockdown with morpholino phosphorodiamidates and hydroxyprolyl-phosphono peptide nucleic acids. Nucleic Acids Res. 2005;33:4914–21. doi: 10.1093/nar/gki799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo K, Yuan J, Chen J, Lou Z. Topoisomerase IIalpha controls the decatenation checkpoint. Nat Cell Biol. 2009;11:204–10. doi: 10.1038/ncb1828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bower JJ, Karaca GF, Zhou Y, Simpson DA, Cordeiro-Stone M, Kaufmann WK. Topoisomerase IIalpha maintains genomic stability through decatenation G(2) checkpoint signaling. Oncogene. pp. 4787–99. [DOI] [PMC free article] [PubMed]
- Ikegami R, Hunter P, Yager TD. Developmental activation of the capability to undergo checkpoint-induced apoptosis in the early zebrafish embryo. Dev Biol. 1999;209:409–33. doi: 10.1006/dbio.1999.9243. [DOI] [PubMed] [Google Scholar]
- Pierre J St, Wright DJ, Rowe TC, Wright SJ. DNA topoisomerase II is essential for preimplantation mouse development. Mol Reprod Dev. 2002;61:347–57. doi: 10.1002/mrd.10015. [DOI] [PubMed] [Google Scholar]
- Li L, Zheng P, Dean J. Maternal control of early mouse development. Development. 2010;137:859–70. doi: 10.1242/dev.039487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haffter P, Granato M, Brand M, Mullins MC, Hammerschmidt M, Kane DA, Odenthal J, van Eeden FJ, Jiang YJ, Heisenberg CP. et al. The identification of genes with unique and essential functions in the development of the zebrafish, Danio rerio. Development. 1996;123:1–36. doi: 10.1242/dev.123.1.1. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Comparison of zebrafish Top2a and Top2b paralogues. A) Phylogenetic tree of Top2a and Top2b paralogues of 9 vertebrate species including zebrafish and other 4 teleost species. Protein sequences were retrieved from NCBI and Ensembl data bases. TOP2A: NP_001058.2 (human), NP_001003834.1 (zebrafish), NP_035753.2 (mouse), NP_001082502.1 (frog), NP_990122.1 (chicken), ENSTRUP00000041924 (fugu), ENSORLP00000005530 (medaka), ENSTNIP00000003411 (tetraodon), ENSGACP00000011260 (stickleback). TOP2B: NP_001059.2 (human), NP_001038656.1 (zebrafish), NP_033435.2 (mouse), XP_002932456.1 (frog), NP_990413.1 (chicken), ENSTRUP00000017341 (fugu), ENSORLP00000010828 (medaka), ENSTNIP00000021338 (tetraodon), ENSGACP00000010617 (stickleback). They were aligned by NCBI Multiple Alignment tool. Phylogenetic tree was generated with Seaview4 software (PhyML v3.0.1). Two distinct branches for Top2a and Top2b paralogues are apparent indicating that origin of two paralogues in zebrafish does not result from the genome duplication in teleosts. B) Alignment of zebrafish Top2a and Top2b protein sequences shows 65% identity (shaded residues) with the greatest divergence occurring at the C-terminal end. Asterisk indicates a serine residue in zebrafish Top2a which is associated with activation of the decatenation checkpoint in mammals.
Overexpression of top2b in blm embryos. Offspring of blm carriers were microinjected at 1-2 cell stages with zebrafish genomic top2b sequence (BAC clone CHORB736O22185Q) at 25 ng/μl (7.5 pg per embryo) or zebrafish top2b mRNA at 250 ng/μl (75 pg per embryo) and analysed at 33 hpf. A) Table of observed phenotypes upon injecting offspring of carriers of blm mutation with zebrafish top2b BAC clone or in vitro synthesised top2b RNA. B) Images of 33 hpf malformed larvae overexpressing top2b by ~4 fold following injecting with top2b mRNA. C) Ectopic expression of top2b mRNA does not rescue the small eye phenotype of blm embryos. Error bars represent the standard error of the mean.
Transient inhibition of Top2a pre-MZT. A) Dark field images showing the morphology of embryos at 27 hpf following treatment with ICRF-193 from 1-2 cell stage until 3.5 hpf B) Representative confocal images (projections of 30 slices taken at 0.5 μm intervals) of 3.5 hpf embryos stained with DAPI, which had been treated from 1-2 cell stage with 100 μM ICRF-193. Chromatin appears to be more compacted in the treated embryos but not extensive DNA damage was observed.
Staging of embryos treated with topoisomerase inhibitors. Top panel: images of normally developed embryos at 16, 32 and 64 cell stages. Bottom panels: example images of "not classified" or malformed embryos treated with 1% DMSO (vehicle control), ICRF-193 or etoposide. This group includes embryos with atypical shape: A) irregular shape, protruding animal pole, B) asymmetric animal pole and uneven cell size, C) irregular shape and opaque, D-E) undefined cell morphology and abnormal transparency, F) small, irregular-shaped animal pole, G) abnormal distribution of cells around yolk, H) undefined cell morphology with abnormal distribution around yolk, I) duplicated/split animal pole.
Effect of topoisomerase inhibitors on pre-MZT development of zebrafish embryos is specific. Treatment of 1-2 cell embryos with L-AP4 (metabotropic glutamate receptor agonist) does not change the distribution of developmental stages compared to untreated controls in embryo medium. Incubation in 1% DMSO (vehicle for topoisomerase inhibitors) increases the percentage of embryos in more advanced stages of development. A) Table and B) graphs representing the number and percentage, respectively of embryos at 8-32 cell developmental stages following incubation for 1 hour in embryo medium containing 10 μM, 200 μM or 1 mM L-AP4 (water dilutions), 1% DMSO or embryo medium alone.