Summary
A key point of neural development is the commitment of progenitor cells to a specific neural fate. In all animals studied, proneural proteins — transcription factors of the basic helix-loop-helix (bHLH) family — are central to this process. The function of these factors is strongly influenced by the spatial and temporal context in which they are expressed. It is important to understand the molecular mechanisms by which developmental context interacts with and modifies the intrinsic functions and properties of the proneural proteins. Recent insights have been obtained in Drosophila and vertebrates from analysis of how bHLH proteins interact with other transcription factors to regulate target genes.
Introduction
Neuronal diversity is acquired by various spatial and temporal mechanisms that pattern the events of neural development. The defining event of neurogenesis itself is the switch from uncommitted, cycling progenitor cell to committed neural precursor cell, which has a relatively restricted cell division potential before it or its daughter cells differentiate as neurons or glia. The details of this step differ in different organisms, but a consistent feature is the involvement of basic helix-loop-helix (bHLH) transcription factors. In Drosophila, proneural bHLH factors are initially expressed in ectodermal cells, giving the cells competence to undergo neural commitment, and are then upregulated upon neural commitment, at which point they also trigger Notch-mediated lateral inhibition to suppress the competence of surrounding cells. In vertebrates, the proneural function is also closely associated with committing cycling progenitor cells to a neuronal fate, which involves activation of Notch signalling, inducing cell cycle exit, migration and terminal differentiation [1-4].
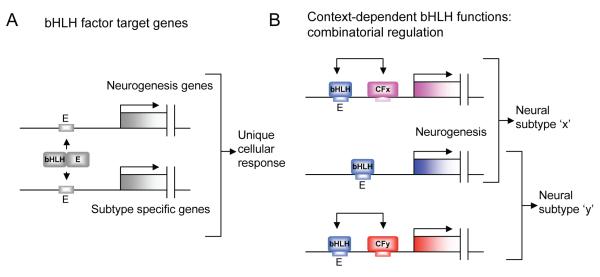
Proneural proteins are transcriptional activators that function as heterodimers with E proteins (Daughterless in Drosophila and typically E12 or E47 in vertebrates), which are generally more widely expressed and dimerise with multiple proneural factors. These bHLH heterodimers bind E box motifs (CANNTG) in the regulatory regions of their target genes (Fig. 1A). A number of observations are of major importance with regard to proneural bHLH proteins, their regulation of specific target genes, and how this relates to acquisition of neural diversity. Firstly, proneural proteins generally trigger neurogenesis, but different proneural proteins are required for different neuronal and/or glial cell types, which implies that each bHLH protein regulates both common (shared) target genes for neurogenesis and unique target genes for neuronal subtype characteristics (Fig. 1A). Thus, subtype specificity of bHLH factors underpins some of the cellular diversity in the nervous system [5-7]. Among other things, this has important implications for coaxing the production of specific neuronal subtypes from stem cells [8,9].
Figure 1.
(A) bHLH proteins regulate target gene sets by binding to E box motifs as heterodimers with an E protein (the latter generally not shown in subsequent figures). (B) A bHLH factor may regulate different subtype-specific targets in different places (or at different times) through combinatorial regulation with coexpressed transcription cofactors (CF). Arrows indicate protein interactions leading to synergistic regulation of the target.
Secondly, and somewhat paradoxically, an individual bHLH protein is required for several different neural cell types at different times or locations in development (see Table 1 for recent findings). Therefore, target gene specificity of a bHLH factor must be modulated by developmental context. A striking example is Drosophila Atonal, which is required for precursors of chordotonal proprioceptors in most of the body, olfactory receptors in the antenna, and R8 photoreceptors in the eye [5,10]. In each location, Atonal function cannot be substituted by other proneural bHLH genes [7]. bHLH proteins must therefore have different intrinsic properties that endow them with different target gene specificities, but these properties are modified strongly according to context. This review summarises recent research that begins to illuminate the molecular mechanisms by which context modifies the intrinsic functions and properties of neural bHLH factors.
Table 1.
Some major proneural factor functions in the nervous system
| Atonal | Proprioceptors (chordotonal organs) Olfactory receptors (sensilla coeloconica) Eye (R8 photoreceptor) Subset of brain neurons (differentiation) |
[5] |
| Achaete/Scute | External sense organs (tactile/chemosensory bristles) CNS neuroblast subset |
[5] |
| Amos | Olfactory receptors (sensilla basiconica and trichodea) Dbd neuron |
[5] |
| Math1 (Atoh1) |
Cerebellum (Granule Cells) Inner ear (sensory hair cells) Mechanoreceptors (Merkel cells) |
[52] |
| Dorsal spinal cord (commissural neurons) | [28] | |
| Ngn2 (Neurog2) |
Ventral neural tube (motorneurons) Forebrain (glutaminergic neurons) |
|
| Dentate gyrus | [53] | |
| Midbrain dopaminergic neurons | [54] | |
| Retinal ganglion cells | [6] | |
| Mash1 (Ascl1) |
Dorsal neural tube (commissural neurons) Neural crest (autonomic neurons) |
|
| Forebrain (GABAergic neurons) | [55] | |
| Forebrain (oligodendrocytes) | [56,57] | |
| Midbrain dopaminergic neurons | [31] |
Combinatorial regulation – cofactors provide spatial/temporal context
One explanation of context dependence of bHLH function is that certain subtype-specific target genes also require input from other regionally expressed transcription factors (referred to here as cofactors). Thus, context-dependent specificity results from different combinations of bHLH factors and cofactors whose expression overlaps in different places or times [11,12]. A long-standing theme is combinatorial control by bHLH and homeodomain (HD) proteins, and the mouse and Xenopus retinas provide good examples of this [13-15]. For instance, a combination of Mash1 and the HD factor Chx10 are required for bipolar cell fate [14]. Another well-studied example, recently extended by Sugimori et al. [16], comes from the mouse ventral spinal cord [17,18]. Here a combinatorial code of patterning factors (Olig2, Pax6, Nkx2.2) and proneural bHLH factors (Neurogenin (Ngn), Mash1) is postulated to produce neurons, astrocytes and oligodendrocytes in different locations or times [2,16].
In the Drosophila eye, genes of the Retinal Determination Gene Network (RDGN) (particularly sine oculis and the PAX6 homologues, eyeless and twin of eyeless) are prime candidates for providing the context for neurogenesis by Atonal [19]. It is not difficult to envisage that the RDGN not only activates Atonal expression for retinal neurogenesis [20,21], but also modulates Atonal target gene regulation in an eye-specific manner [22]. However, whilst interactions within the RDGN are well characterised, much less is known about how these patterning factors interact with Atonal. Analysis of newly identified eye-specific targets of these RDGN factors may be fruitful [23,24].
Enhancers provide the molecular context for specific target gene regulation
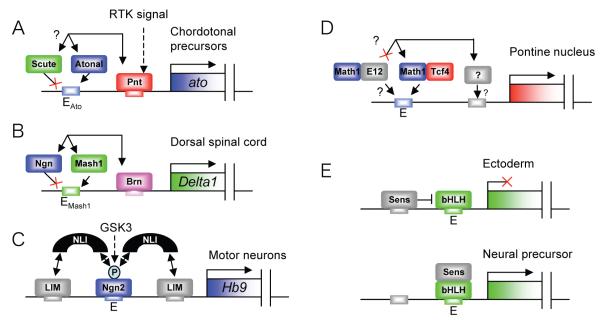
Cellular context is achieved by the co-expression of bHLH factors and cofactors. Combinatorial control is widely expected result from the co-occurrence of binding sites for the bHLH and cofactors in the enhancers of specific target genes, with different cofactors combining with a bHLH factor to regulate different targets (Fig. 1B). Thus, enhancers act in their well-known role as integrators of diverse regulatory inputs. However, few examples are currently known in detail. One is the ‘recruitment’ enhancer of atonal, which is regulated by the combination of Atonal (for autoregulation) and the ETS factor, Pointed [25] (Fig. 3A). The proteins bind cooperatively to adjacent binding sites in the enhancer. This combination of sites also occurs in the enhancer of another Atonal target — the dacapo gene, which encodes a cdk inhibitor [26]. Thus, a subset of Atonal targets only respond in the presence of Pointed, which itself is produced in response to receptor tyrosine kinase (RTK) signalling.
Figure 3.
Examples of specific target gene regulation by bHLH factors. (A) In Drosophila Atonal and Pointed bind cooperatively to adjacent sites in an atonal autoregulatory enhancer. Scute apparently cannot utilise the E box motif; it is not clear whether it can interact with Pnt. (B) Mouse Mash1 and Brn proteins bind cooperatively to adjacent sites in the DeltaM enhancer. Ngn2 apparently cannot utilise the E box motif even though it can interact with Brn proteins. (C) Alternative E protein dimerisation partners may regulate Math1 target gene specificity in the Pontine nucleus, putatively via specific interaction with an unknown cofactor. (D) Phosphorylation of Ngn2 is required for its interaction with NLI, thereby switching its ability to activate a subset of targets with Lim-HD factors. (E) Drosophila proneural factor activity is regulated by Sens acting as a binary switch. Sens can act as DNA-binding repressor or a non-DNA binding coactivator.
An analogous example in mouse is provided by combinatorial regulation of Delta1 by Mash1 and Brn factors, acting co-ordinately by binding cooperatively to adjacent E box and POU protein binding sites in the DeltaM enhancer [27] (Fig. 3B). The two binding sites form a characteristic motif that is also found in other likely downstream targets, including Delta3, Insm1 (Zn finger factor involved in differentiation) and Fbw7 (involved in cell cycle arrest and Notch degradation). In both these examples, the cofactor interaction is required only for a subset of targets, supporting a model in which bHLH factors interact with different cofactors to activate different subprograms of neurogenesis (Fig. 1B) [28].
Another example of combinatorial regulation is the cooperation between Ngn2/NeuroM and the LIM-HD factors, Lhx3 and Isl1, to specify motor neurons in the chick neural tube. Part of their joint function is to activate the Hb9 motor neuron-specifying factor [18]. This is achieved via two E boxes and two LIM-HD sites in the Hb9 motor neuron enhancer (Fig. 3C). Interestingly, in this case the sites are not directly adjacent, and instead a bridging cofactor, NLI, mediates the interaction between the DNA binding factors. This arrangement mirrors a Drosophila interaction between Scute and the GATA factor, Pannier, which is bridged by the NLI homologue, Chip [29].
The structure of target enhancers therefore provides the important molecular context for bHLH protein function. Collocation of binding sites within enhancers of targets determines whether a particular target gene responds to bHLH factors in a particular cellular context.
bHLH selectivity – DNA or protein interactions?
Whilst these are powerful examples of combinatorial control, an important but largely unanswered question is what provides the specificity of co-regulation by different bHLH proteins and different cofactors. Multiple bHLH factors and multiple potential coregulators may be present in the same cells, but targets respond to a specific combination. Is specificity for a particular bHLH factor driven by specificity of protein-protein interactions or DNA binding specificity (Fig. 2)? Unfortunately, in vivo E box occupancy by different bHLH factors (as measured by chromatin immunoprecipitation (ChIP)) may not clearly answer the question since occupancy may depend on protein interactions as much as on DNA binding affinity. In structure-function experiments, functional differences between bHLH proteins have been mapped to non-DNA contacting residues within the bHLH domain, whilst DNA-contacting residues tend to be highly conserved between different bHLH factors [7,10,30-33]. It is attractive to think therefore that these ‘specificity residues’ contact different cofactors thereby providing specificity of target gene regulation (Fig. 2A). However, it is also clear that different proneural bHLH factors do have different E box binding site preferences in vivo if not in vitro. For instance, Scute and Atonal-specific target genes have functionally distinct Scute- and Atonal-specific variant E box motifs [34]. Moreover, these E box motifs are differentially used by Scute and Atonal even in a non-neural cell culture system, suggesting that specialised cofactors may not always be required for bHLH specificity [35]. How can these observations be reconciled? Non-DNA-contacting ‘specificity’ residues in the bHLH domain may well cause conformation effects on DNA-contacting residues, thereby changing DNA interaction properties, perhaps in a manner induced or modified by cofactor interactions.
Figure 2.
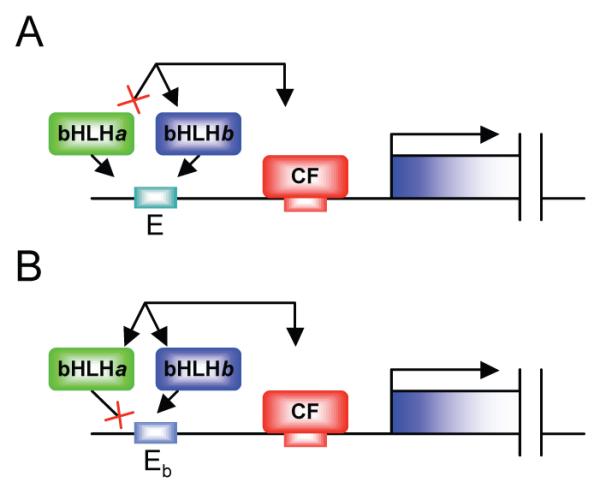
Mechanisms for specificity between bHLH factors. (A) Multiple bHLH factors can utilise an E box, but specific cofactor interactions determine that only bHLH factor ‘b’ results in target gene activation. (B) Multiple bHLH factors can interact with the cofactor, but specific DNA-protein interactions determine that only bHLH factor ‘b’ utilises that particular E box motif and results in target gene activation.
An elegant example of specifity in bHLH-cofactor interaction is provided by the DeltaM enhancer mentioned above. Whilst bHLH proteins Mash1 and Ngn2 are both generally capable of synergising with the Brn proteins, it appears that selective E box utilisation determines that only Mash1 synergises at the DeltaM enhancer [27] (Fig. 3B). A different explanation has been suggested for the mouse Hb9 promoter. Whilst NeuroM and Mash1 can both bind an E box in the Hb9 promoter (as measured by ChIP), only NeuroM can synergise with LIM-HD cofactors bound to the same promoter (via the NLI adaptor protein). Although not proven, it is suggested that this selective protein interaction provides the Hb9 promoter’s specificity [18] (Fig. 3C). Conceivably, expression of different adaptor proteins may be an important mechanism for determining target gene specificity in other contexts [36].
A simple model for bHLH function suggests that common (shared) neurogenesis targets are regulated via non-specific E boxes (Fig. 1B). Indirect evidence suggests this is true for the senseless gene, which has a single E box responding both to Scute and Atonal [37], and Prokineticin 2, with an E box that binds both Ngn1 and Mash1 [38]. However, other shared targets respond to different bHLH factors via separate subtype-specific enhancers. This includes Brd (regulated by two enhancers containing Atonal- and Scute-specific E boxes respectively)[34] and mouse Delta1 (two enhancers for Mash1 and Ngn1/2 respectively) [27]. Why should some shared targets be regulated in this way? Possibly it allows such targets to be regulated with different dynamics in different neural subtypes.
Mechanisms providing signalling context
Signalling context (the signals a cell is receiving) is clearly important for modulating bHLH activity in both space and time. The Atonal/Pointed interaction is one obvious way in which this is achieved [25]. Recently, the motor neuron Hb9 paradigm has been extended to show how bHLH specificity may be affected by signalling context via direct post-translational modification. Phosphorylation of Ngn2 has previously been shown to have several roles, including regulation of its stability [39]. Ma et al. [40] have now shown that serine phosphorylation by GSK3 is required for the subtype determination function of Ngn2, but apparently not its neurogenesis function. For the Hb9 motor neuron enhancer, this phosphorylation is necessary for Ngn2’s interaction with the NLI adaptor (Fig. 3C). Thus, a post-translational modification modulates the target gene specificity of a bHLH factor via controlling its capacity for protein-protein interactions. Tyrosine phosphorylation was previously shown to be required for Ngn2 function in migration and dendrite morphology of cortical pyramidal neurons [41], and it is possible that this modification also allows regulation of a specific subset of targets.
Expression of a coactivator protein may also underlie a temporal switch in activity rather than specificity. In Drosophila neurogenesis, proneural factors are thought to regulate only a few targets in neurally competent ectodermal cells (notably concerned with lateral inhibition) but many more upon commitment of the neural precursors. What promotes this switch in activity? A compelling series of papers shows that the Zn finger GPS factor, Senseless, is part of this switch: it can act as both a positive and negative modulator of proneural activity (Fig. 3D) [37,42-44]. There is a strong possibility that some aspects of this interaction are conserved [45,46]. It is not clearly known what switches the activity of Senseless, although a concentration-dependent switch is suggested [37]. Senseless provides temporal context for proneural bHLH activity rather than subtype specificity. However, recent evidence suggests that Senseless interaction may enhance the E box selectivity of different proneural factors, perhaps by strengthening bHLH recognition of different E box motifs [35].
Finally, the role of bHLH dimerisation partners should not be overlooked. In vertebrates there are several E protein partners [47], opening the possibility that choice of E protein may also provide a way of modulating proneural bHLH function. Flora et al. [47] found a specific requirement for Math1/Tcf4 heterodimers (as opposed to heterodimers of Math1 with E47, E12 or HEB) in the generation of the pontine nucleus neuronal structure (derived from the rhombic lip of the hindbrain [48]). They propose that this may reflect a unique ability of Math1/Tcf4 heterodimers to interact with tissue-specific cofactors [47] (Fig. 3), although choice of dimerisation partner might also affect DNA binding site preferences [49].
Conclusions
Although exciting progress is being made, we still have a very patchy understanding of the molecular mechanisms of bHLH specificity in relation to context. One message is that enhancers provide the all-important molecular context that enable different protein interactions to occur at different target genes even within the same cell [11]. A major bottleneck in progressing is a dearth of subtype-specific enhancers to be analysed in structure-function studies. Genome-wide target gene identification, bioinformatic and gene network approaches promise to add much more in the future [50,51].
References
- 1.Bertrand N, Castro DS, Guillemot F. Proneural genes and the specification of neural cell types. Nat Rev Neurosci. 2002;3:517–530. doi: 10.1038/nrn874. [DOI] [PubMed] [Google Scholar]
- 2.Guillemot F. Spatial and temporal specification of neural fates by transcription factor codes. Development. 2007;134:3771–3780. doi: 10.1242/dev.006379. [DOI] [PubMed] [Google Scholar]
- 3.Ge W, He F, Kim KJ, Blanchi B, Coskun V, Nguyen L, Wu X, Zhao J, Heng JI, Martinowich K, et al. Coupling of cell migration with neurogenesis by proneural bHLH factors. Proc Natl Acad Sci U S A. 2006;103:1319–1324. doi: 10.1073/pnas.0510419103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kageyama R, Ohtsuka T, Hatakeyama J, Ohsawa R. Roles of bHLH genes in neural stem cell differentiation. Exp Cell Res. 2005;306:343–348. doi: 10.1016/j.yexcr.2005.03.015. [DOI] [PubMed] [Google Scholar]
- 5.Grillenzoni N, de Vaux V, Meuwly J, Vuichard S, Jarman A, Holohan E, Gendre N, Stocker RF. Role of proneural genes in the formation of the larval olfactory organ of Drosophila. Dev Genes Evol. 2007;217:209–219. doi: 10.1007/s00427-007-0135-6. [DOI] [PubMed] [Google Scholar]
- 6.Matter-Sadzinski L, Puzianowska-Kuznicka M, Hernandez J, Ballivet M, Matter JM. A bHLH transcriptional network regulating the specification of retinal ganglion cells. Development. 2005;132:3907–3921. doi: 10.1242/dev.01960. [DOI] [PubMed] [Google Scholar]
- 7.Maung SM, Jarman AP. Functional distinctness of closely related transcription factors: a comparison of the Atonal and Amos proneural factors. Mech Dev. 2007;124:647–656. doi: 10.1016/j.mod.2007.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Berninger B, Guillemot F, Gotz M. Directing neurotransmitter identity of neurones derived from expanded adult neural stem cells. Eur J Neurosci. 2007;25:2581–2590. doi: 10.1111/j.1460-9568.2007.05509.x. [DOI] [PubMed] [Google Scholar]
- 9.Klein C, Butt SJB, Machold RP, Johnson JE, Fishell G. Cerebellum- and forebrain-derived stem cells possess intrinsic regional character. Development. 2005;132:4497–4508. doi: 10.1242/dev.02037. [DOI] [PubMed] [Google Scholar]
- 10.Quan X-J, Hassan BA. From skin to nerve: flies, vertebrates and the first helix. Cell. Mol. Life Sci. 2005;62:2036–2049. doi: 10.1007/s00018-005-5124-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Allan DW, Park D, St. Pierre SE, Taghert PH, Thor S. Regulators Acting in Combinatorial Codes Also Act Independently in Single Differentiating Neurons. Neuron. 2005;45:689–700. doi: 10.1016/j.neuron.2005.01.026. [DOI] [PubMed] [Google Scholar]
- 12.Baumgardt M, Miguel-Aliaga I, Karlsson D, Ekman H, Thor S. Specification of Neuronal Identities by Feedforward Combinatorial Coding. PLoS Biology. 2007;5:e37. doi: 10.1371/journal.pbio.0050037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ohsawa R, Kageyama R. Regulation of retinal cell fate specification by multiple transcription factors. Brain Research. 2008;1192:90–98. doi: 10.1016/j.brainres.2007.04.014. [DOI] [PubMed] [Google Scholar]
- 14.Hatakeyama J, Kageyama R. Retinal cell fate determination and bHLH factors. Semin Cell Dev Biol. 2004;15:83–89. doi: 10.1016/j.semcdb.2003.09.005. [DOI] [PubMed] [Google Scholar]
- 15.Wang JC, Harris WA. The role of combinational coding by homeodomain and bHLH transcription factors in retinal cell fate specification. Dev Biol. 2005;285:101–115. doi: 10.1016/j.ydbio.2005.05.041. [DOI] [PubMed] [Google Scholar]
- 16.Sugimori M, Nagao M, Bertrand N, Parras CM, Guillemot F, Nakafuku M. Combinatorial actions of patterning and HLH transcription factors in the spatiotemporal control of neurogenesis and gliogenesis in the developing spinal cord. Development. 2007;134:1617–1629. doi: 10.1242/dev.001255. [DOI] [PubMed] [Google Scholar]
- 17.Zhou Q, Anderson DJ. The bHLH transcription factors OLIG2 and OLIG1 couple neuronal and glial subtype specification. Cell. 2002;109:61–73. doi: 10.1016/s0092-8674(02)00677-3. [DOI] [PubMed] [Google Scholar]
- 18.Lee S-K, Pfaff SL. Synchronization of Neurogenesis and Motor Neuron Specification by Direct Coupling of bHLH and Homeodomain Transcription Factors. Neuron. 2003;38:731–745. doi: 10.1016/s0896-6273(03)00296-4. [DOI] [PubMed] [Google Scholar]
- 19.Silver SJ, Rebay I. Signaling circuitries in development: insights from the retinal determination gene network. Development. 2005;132:3–13. doi: 10.1242/dev.01539. [DOI] [PubMed] [Google Scholar]
- 20.Zhang T, Ranade S, Cai CQ, Clouser C, Pignoni F. Direct control of neurogenesis by selector factors in the fly eye: regulation of atonal by Ey and So. Development. 2006;133:4881–4889. doi: 10.1242/dev.02669. [DOI] [PubMed] [Google Scholar]
- 21.Tanaka-Matakatsu M, Du W. Direct control of the proneural gene atonal by retinal determination factors during Drosophila eye development. Dev Biol. 2008;313:787–801. doi: 10.1016/j.ydbio.2007.11.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Niwa N, Hiromi Y, Okabe M. A conserved developmental program for sensory organ formation in Drosophila melanogaster. Nature Genetics. 2004;36:293–297. doi: 10.1038/ng1308. [DOI] [PubMed] [Google Scholar]
- 23.Jemc J, Rebay I. Identification of transcriptional targets of the dual-function transcription factor/phosphatase eyes absent. Developmental Biology. 2007;310:416–429. doi: 10.1016/j.ydbio.2007.07.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ostrin EJ, Li Y, Hoffman K, Liu J, Wang K, Zhang L, Mardon G, Chen R. Genome-wide identification of direct targets of the Drosophila retinal determination protein Eyeless. Genome Res. 2006;16:466–476. doi: 10.1101/gr.4673006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.zur Lage PI, Powell LM, Prentice DRA, McLaughlin PM, Jarman AP. EGF receptor signalling triggers recruitment of Drosophila sense organ precursors by stimulating proneural gene autoregulation. Dev. Cell. 2004 doi: 10.1016/j.devcel.2004.09.015. [DOI] [PubMed] [Google Scholar]
- 26.Sukhanova MJ, Deb DK, Gordon GM, Matakatsu MT, Du W. Proneural basic helix-loop-helix proteins and epidermal growth factor receptor signaling coordinately regulate cell type specification and cdk inhibitor expression during development. Mol Cell Biol. 2007;27:2987–2996. doi: 10.1128/MCB.01685-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Castro DS, Skowronska-Krawczyk D, Armant O, Donaldson IJ, Parras C, Hunt C, Critchley JA, Nguyen L, Gossler A, Gottgens B, et al. Proneural bHLH and Brn proteins coregulate a neurogenic program through cooperative binding to a conserved DNA motif. Dev Cell. 2006;11:831–844. doi: 10.1016/j.devcel.2006.10.006. [DOI] [PubMed] [Google Scholar]
- 28.Saba R, Johnson JE, Saito T. Commissural neuron identity is specified by a homeodomain protein, Mbh1, that is directly downstream of Math1. Development. 2005;132:2147–2155. doi: 10.1242/dev.01781. [DOI] [PubMed] [Google Scholar]
- 29.Heitzler P, Vanolst L, Biryukova I, Ramain P. Enhancer-promoter communication mediated by Chip during Pannier-driven proneural patterning is regulated by Osa. Genes Dev. 2003;17:591–596. doi: 10.1101/gad.255703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Skowronska-Krawczyk D, Matter-Sadzinski L, Ballivet M, Matter J-M. The basic domain of ATH5 mediates neuron-specific promoter activity during retina development. Mol. Cell. Biol. 2005;25:10029–10039. doi: 10.1128/MCB.25.22.10029-10039.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Park CH, Kang JS, Kim JS, Chung S, Koh JY, Yoon EH, Jo AY, Chang MY, Koh HC, Hwang S, et al. Differential actions of the proneural genes encoding Mash1 and neurogenins in Nurr1-induced dopamine neuron differentiation. J Cell Sci. 2006;119:2310–2320. doi: 10.1242/jcs.02955. [DOI] [PubMed] [Google Scholar]
- 32.Nakada Y, Hunsaker TL, Henke RM, Johnson JE. Distinct domains within Mash1 and Math1 are required for function in neuronal differentiation versus neuronal cell-type specification. Development. 2004;131:1319–1330. doi: 10.1242/dev.01008. [DOI] [PubMed] [Google Scholar]
- 33.Quan X-J, Denayer T, Yan J, Jafar-Nejad H, Philippi A, Lichtarge O, Vieminckx K, Hassan BA. Evolution of neural precursor selection: functional divergence of proneural proteins. Development. 2004;131:1679–1689. doi: 10.1242/dev.01055. [DOI] [PubMed] [Google Scholar]
- 34.Powell LM, zur Lage PI, Prentice DRA, Senthinathan B, Jarman AP. The proneural proteins Atonal and Scute regulate neural target genes through different E-box binding sites. Mol. Cell. Biol. 2004;24:9517–9526. doi: 10.1128/MCB.24.21.9517-9526.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Powell LM, Deaton AM, Wear MA, Jarman AP. The specificity of Atonal and Scute bHLH factors: analysis of cognate E box binding sites and the influence of Senseless. Genes to Cells. 2008;13 doi: 10.1111/j.1365-2443.2008.01217.x. in the press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lim J, Lee OK, Hsu YC, Singh A, Choi KW. Drosophila TRAP230/240 are essential coactivators for Atonal in retinal neurogenesis. Dev Biol. 2007;308:322–330. doi: 10.1016/j.ydbio.2007.05.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Jafar-Nejad H, Acar M, Nolo R, Lacin H, Pan H, Parkhurst SM, Bellen HJ. Senseless acts as a binary switch during sensory organ precursor selection. Genes Dev. 2003;17:2966–2978. doi: 10.1101/gad.1122403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhang C, Ng KL, Li JD, He F, Anderson DJ, Sun YE, Zhou QY. Prokineticin 2 is a target gene of proneural basic helix-loop-helix factors for olfactory bulb neurogenesis. J Biol Chem. 2007;282:6917–6921. doi: 10.1074/jbc.C600290200. [DOI] [PubMed] [Google Scholar]
- 39.Vosper JM, Fiore-Heriche CS, Horan I, Wilson K, Wise H, Philpott A. Regulation of neurogenin stability by ubiquitin-mediated proteolysis. Biochem J. 2007;407:277–284. doi: 10.1042/BJ20070064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ma YC, Song MR, Park JP, Henry Ho HY, Hu L, Kurtev MV, Zieg J, Ma Q, Pfaff SL, Greenberg ME. Regulation of motor neuron specification by phosphorylation of neurogenin 2. Neuron. 2008;58:65–77. doi: 10.1016/j.neuron.2008.01.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hand R, Bortone D, Mattar P, Nguyen L, Heng JI, Guerrier S, Boutt E, Peters E, Barnes AP, Parras C, et al. Phosphorylation of Neurogenin2 specifies the migration properties and the dendritic morphology of pyramidal neurons in the neocortex. Neuron. 2005;48:45–62. doi: 10.1016/j.neuron.2005.08.032. [DOI] [PubMed] [Google Scholar]
- 42.Nolo R, Abbott LA, Bellen HJ. Senseless, a Zn finger transcription factor, is necessary and sufficient for sensory organ development in Drosophila. Cell. 2000;102:349–362. doi: 10.1016/s0092-8674(00)00040-4. [DOI] [PubMed] [Google Scholar]
- 43.Jafar-Nejad H, Tien AC, Acar M, Bellen HJ. Senseless and Daughterless confer neuronal identity to epithelial cells in the Drosophila wing margin. Development. 2006;133:1683–1692. doi: 10.1242/dev.02338. [DOI] [PubMed] [Google Scholar]
- 44.Acar M, Jafar-Nejad H, Giagtzoglou N, Yallampalli S, David G, He Y, Delidakis C, Bellen HJ. Senseless physically interacts with proneural proteins and functions as a transcriptional co-activator. Development. 2006;133:1979–1989. doi: 10.1242/dev.02372. [DOI] [PubMed] [Google Scholar]
- 45.Jafar-Nejad H, Bellen HJ. Gfi/Pag-3/Senseless Zinc Finger Proteins: a Unifying Theme? Mol. Cell. Biol. 2004;24:8803–8812. doi: 10.1128/MCB.24.20.8803-8812.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Shroyer NF, Wallis D, Venken KJT, Bellen HJ, Zoghbi HY. Gfi1 functions downstream of Math1 to control intestinal secretory cell subtype allocation and differentiation. Genes Dev. 2005;19:2412–2417. doi: 10.1101/gad.1353905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Flora A, Garcia JJ, Thaller C, Zoghbi HY. The E-protein Tcf4 interacts with Math1 to regulate differentiation of a specific subset of neuronal progenitors. Proceedings of the National Academy of Sciences. 2007;104:15382–15387. doi: 10.1073/pnas.0707456104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wang VY, Rose MF, Zoghbi HY. Math1 Expression Redefines the Rhombic Lip Derivatives and Reveals Novel Lineages within the Brainstem and Cerebellum. Neuron. 2005;48:31–43. doi: 10.1016/j.neuron.2005.08.024. [DOI] [PubMed] [Google Scholar]
- 49.Castanon I, Von_Stetina S, Kass J, Baylies MK. Dimerization partners determine the activity of the Twist bHLH protein during Drosophila mesoderm development. Development (Cambridge, England) 2001;128:3145–3159. doi: 10.1242/dev.128.16.3145. [DOI] [PubMed] [Google Scholar]
- 50.Gohlke JM, Armant O, Parham FM, Smith MV, Zimmer C, Castro DS, Nguyen L, Parker JS, Gradwohl G, Portier CJ, et al. Characterization of the proneural gene regulatory network during mouse telencephalon development. BMC Biol. 2008;6:15. doi: 10.1186/1741-7007-6-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Reeves N, Posakony JW. Genetic Programs Activated by Proneural Proteins in the Developing Drosophila PNS. Developmental Cell. 2005;8:413–425. doi: 10.1016/j.devcel.2005.01.020. [DOI] [PubMed] [Google Scholar]
- 52.Kawauchi D, Saito T. Mbh1 is involved in cerebellar granule cell differentiation downstream of a proneural gene, Math1. Neuroscience Research. 2007;58:S85–S85. [Google Scholar]
- 53.Galichet C, Guillemot F, Parras CM. Neurogenin 2 has an essential role in development of the dentate gyrus. Development. 2008;135:2031–2041. doi: 10.1242/dev.015115. [DOI] [PubMed] [Google Scholar]
- 54.Park CH, Kang JS, Yoon EH, Shim JW, Suh-Kim H, Lee SH. Proneural bHLH neurogenin 2 differentially regulates Nurr1-induced dopamine neuron differentiation in rat and mouse neural precursor cells in vitro. FEBS Lett. 2008;582:537–542. doi: 10.1016/j.febslet.2008.01.018. [DOI] [PubMed] [Google Scholar]
- 55.Poitras L, Ghanem N, Hatch G, Ekker M. The proneural determinant MASH1 regulates forebrain Dlx1/2 expression through the I12b intergenic enhancer. Development. 2007;134:1755–1765. doi: 10.1242/dev.02845. [DOI] [PubMed] [Google Scholar]
- 56.Parras CM, Hunt C, Sugimori M, Nakafuku M, Rowitch D, Guillemot F. The proneural gene Mash1 specifies an early population of telencephalic oligodendrocytes. J Neurosci. 2007;27:4233–4242. doi: 10.1523/JNEUROSCI.0126-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Petryniak MA, Potter GB, Rowitch DH, Rubenstein JL. Dlx1 and Dlx2 control neuronal versus oligodendroglial cell fate acquisition in the developing forebrain. Neuron. 2007;55:417–433. doi: 10.1016/j.neuron.2007.06.036. [DOI] [PMC free article] [PubMed] [Google Scholar]