Abstract
The question of how proneural bHLH transcription factors recognise and regulate their target genes is still relatively poorly understood. We previously showed that Scute and Atonal target genes have different E box motifs, suggesting that specific DNA interactions contribute to differences in their target gene specificity. Here we show that Scute and Atonal proteins (in combination with Daughterless) can activate reporter gene expression via their cognate E boxes in a non-neuronal cell culture system, suggesting that the proteins have strong intrinsic abilities to recognise different E box motifs in the absence of specialised cofactors. Functional comparison of E boxes from several target genes and site-directed mutagenesis of E box motifs suggests that specificity and activity require further sequence elements flanking both sides of the previously identified E box motifs. Moreover, the proneural cofactor, Senseless, can augment the function of Scute and Atonal on their cognate E boxes and therefore may contribute to proneural specificity.
INTRODUCTION
Basic-helix-loop-helix transcription factors are important for many different aspects of development in eukaryotic organisms, including neurogenesis, myogenesis, hematopoiesis and pancreatic development (2, 4, 7, 27) The Drosophila proneural proteins are Class II bHLH transcription factors essential for specifying the precursors of the various sense organs of the fly’s peripheral nervous system (3, 27). Different proneural proteins specify the precursors for different types of sense organs as demonstrated by loss and gain of function analyses. Members of the achaete-scute family, mainly Scute (Sc), specify the external sense organ precursors (5, 10), whereas Atonal (Ato) specifies the precursors for the fly’s stretch receptors (chordotonal organs), the R8 photoreceptors of the compound eye and a subset of the olfactory sensilla (17, 18). The remaining olfactory sensillla are specified by the Ato-related proneural protein, Amos (12).
For these related but distinct functions, proneural proteins are thought to activate directly both common (generic) and specific target genes. Examples of common target genes (those activated by both Sc and Ato) include senseless (sens) (1, 14, 25) and Bearded (Brd) (26, 33), both of which are genes needed for SOP selection from the proneural cluster (PNC). Subtype-specific target genes (those activated by only one proneural protein) are thought to have a role in sense organ subtype differentiation. Few direct target genes are known so far in this category. One example is TakR86C, an Ato target gene, which encodes a neuropeptide receptor expressed in a subset of embryonic chordotonal precursor cells (30). In addition, the ato and sc genes are themselves specific targets of their own gene products via autoregulatory enhancers (9, 37).
To activate target genes, the proneural proteins bind as heterodimers with the ubiquitously expressed Class I bHLH protein Daughterless (Da) to specific DNA sequences known as E boxes (CANNTG) in the target regulatory regions. By analogy with other bHLH factors (22) basic regions of the bHLH domains contact the residues of the E box core via the major groove of the double helix. Predicted DNA binding residues are conserved between Sc and Ato (8), and the basic regions of Amos and Ato are almost identical. While this is consistent with their regulation of common targets, the molecular mechanisms underlying the differences in target gene specificity of Sc and Ato proneural proteins are poorly known. Differences in functional specificity between Sc and Ato have been mapped to a number of bHLH domain residues (8). As these are predicted to face away from the DNA, it has been proposed that these variable residues make contacts with tissue-specific cofactors and that this underlies proneural protein specificity (8). However, we have shown that for Ato and Sc, differential utilisation of variant E-box DNA binding sites also makes a significant contribution to specificity of target gene activation (26). By comparing the E box binding sites for a number of target genes, we defined an extended Ato-specific variant E-box motif (EAto: awCAKGTGk) that differed from the previously defined Sc motif (ESc: gCAGSTGk) in its 5′ flanking bases and central bases (underlined). However, this is based on few sequences and it is not clear whether such motifs explain specific regulation of all Ato and Sc target genes. In vivo, these variants were shown to be important for specificity, but in a manner that is strongly influenced by developmental context, suggesting that cofactor interactions are also important (26). Some examples of the interplay of DNA and cofactor interactions come from studies of mouse proneural factor homologues. Whilst NeuroM and Mash1 can both bind the mouse HB9 promoter, only NeuroM can synergise with Isl1/Lhx3, and it is this selective protein interaction that provides the promoter’s specificity (20). Conversely, both Mash1 and Ngn2 are capable of synergising with the POU proteins, Brn1 and Brn2, but selective E box utilisation determines that only Mash1 synergises at the DeltaM enhancer (6).
Here, we have explored the influence of DNA binding site variation on Sc and Ato protein specificity in cell culture and in vivo reporter gene assays, and in vitro Surface Plasmon Resonance (SPR) analysis. In a number of cases, artificial enhancers consisting of concatemerised short (20 bp) E box sites can support specific reporter gene expression by their predicted cognate proneural protein in cell culture. In these cases it seems that 20 bp of DNA is sufficient for specificity without the need for additional specialised cofactors. There is, however, much variation in E box activity in cell culture despite similar E box motifs. This highlights the importance of other bases adjacent to the Sc and Ato E box motifs, and potentially the need for cofactors in vivo. Mutational analysis of E boxes confirms this. Additionally, we show that the proneural cofactor, Senseless (Sens) (14), can augment the function of Sc and Ato on their cognate E boxes and therefore may contribute to proneural specificity.
MATERIALS AND METHODS
Protein Purification
Protein purification was as described previously (26) from pRSET-Ato, pRSET-Da and pRSET-Sc expression plasmids, except the resin used was His-Select Cobalt Affinity Gel (Sigma). For SPR Experiments (Biacore) an additional ion-exchange chromatography step was included before refolding to improve the protein purity to greater than 95%.
Plasmid constructs
Protein expression constructs RactHAdh, RactH-Adh-Da and pAc-Sc were donated by Christos Delidakis (11), and pAc-Sens was donated by Hamed Jafar-Nejad (14). pAc-5.1-Ato was made as follows. The protein coding region of ato was amplified from pBS-84F#2 (18) primers: 5′-CGCGAATTCCCATACAGCAGCAGCAACATG-3′ and 5′-ATATCTAGAGCGCAGCAGATCCCCGAG-3′). The resulting PCR product was cloned in pAc5.1 (Invitrogen) using the underlined primer EcoRI and XbaI sites. pGL3-p (Promega) luciferase reporter constructs were made by transferring the appropriate concatemer sequences from pHStinger (ato-E1, sc-E1, TAKR86C-E2) or pBluescript (Brd-E1, Brd-E3, sens-E1, E(spl)mγ-E2, E(spl)mγ-E2mutA, E(spl)mγ-E2mutS, sc-E1 GG>AA, ato-E1 AA> GG). Concatemer constructs were made as described in (26) (see Table 1 for sequences). For the ato-E1 concatemer and its derivatives, a mutation was introduced to remove an adjacent Pointed protein binding site (37).
Table 1.
E boxes analysed in this study
| E box | Sequence | Origin | E box type | Ref. |
|---|---|---|---|---|
| ato-E1 | ACCATAACAGGTGGCACGGA | ato-RE. An ato autoregulatory enhancer | EAto | (26, 37) |
| sc-E1 | CGCGTGGCAGGTGTATTTAG | sc-SMC. A sc autoregulatory enhancer | ESc | (9) |
| Brd-E1 | TAGCGCGCAGGTGTTTCTCG | Sc-responsive enhancer from Brd gene | ESc | (33) |
| Brd-E3 | CCGACAACATGTGTTTAACG | Ato-responsive enhancer from Brd gene | EAto | (26, 33) |
| sens-E1 | TTCTAGGCAGGTGTGGCCCG | Pan-neural enhancer from sens gene | ESc | (14) |
| E(spl)mγ-E2 | GAAACGGCAGCTGTTCGCTC (GAGCGAACAGCTGCCGTTTC) |
Ato-responsive enhancer from E(spl)mγ gene |
EAto or ESc | (24) |
| TakR86C-E2 | GGGGTATCAGGTGTGCTGAAC | Ato-responsive enhancer from TakR86C gene |
EAto | (30) |
Six bp core E box is underlined and the region of match to the extended Ato and Sc E boxes consensus motifs is in bold. Note that E(spl)mγ-E2 matches either consensus depending on its orientation.
Cell culture and cotransfection
Drosophila S2 cells were grown at 27°C in Schneider’s insect medium (Sigma) plus 10% FBS (Invitrogen) and split on the day prior to cotransfection. Cells were used at 0.5×106 cells/ml for transfection with a mix of protein-expression (pAc5.1a. RactHAdh), luciferase reporter (pGL3-p) and control Renilla luciferase constructs (pRLCMV, Promega), using Effectene Transfection reagent (Qiagen) at a ratio of 25 μl Effectene per μg DNA. In each case total DNA per cotransfection was made up to 200 ng using ‘empty’ protein expression vector. Two different sets of conditions were used with respect to the amount of protein expression construct transfected, namely “High proneural” concentration using 20 ng of proneural protein and Da expression constructs per cotransfection and “Low proneural” where 1 ng of these expression constructs was used. The cells were incubated for 24 h at 27°C before harvesting.
Luciferase Assays
Transfected cells were pelleted at 8000 rpm for 6 min and resuspended in 200 μl Passive Lysis buffer (Promega). Dual Luciferase assays were carried out according to the standard protocol (Promega) in a Turner 20/20 or multiplate luminometer (Promega). Results are presented as the ratio of fold activation relative to a control cotransfection with ‘empty’ protein expression vector. Experiments were carried out in triplicate and results shown are plus and minus standard deviation. Additionally, similar results were obtained for at least 3 separate experiments for each expression and reporter construct combination. Standard Students T-tests were performed.
Germline transformation
The mutant concatemer sequence was cloned in pHStinger, to make (sc-E1 GG>AA)6-GFP and used in germline transformation as described previously (26) to make transgenic flies.
Surface Plasmon Resonance
Surface plasmon resonance (SPR) (23) experiments were carried out using a Biacore T100 instrument. HBS-EP+ buffer (10 mM HEPES (pH 7.4), 150 mM NaCl, 3 mM EDTA, 0.05% P20 surfactant) plus 0.1 mg/ml BSA was used for all experiments. All oligonucleotides for SPR were HPLC purified and from MWG. Biotinylated oligonucleotides (modified with 5′ Biotin TEG) were hybridised to complementary unmodified oligonucleotides. Biotinylated double-stranded DNA was immobilised on the sensor surface of flow cell 2 or 4 of a 4 flow cell Biacore SA (Streptavidin) Sensor chip as follows: Flow cells 1-4 were treated with 3 times 60 s injections of 50 mM NaOH, 1.5 M NaCl at a flow rate of 10 μl/min, with 60 s buffer washes in between. DNA was immobilised on the surface of flow cell 2 or 4 by injecting 25 nM DNA at 10 μl/min for 12 s. Flow cell 1 was used as “no DNA” control for flow cell 2 (and flow cell 3 for 4). The top strands for the double-stranded oligonucleotides (E-boxes underlined) immobilised on Streptavidin-modified chips were as follows: 40 bp 2 site oligonucleotides:
(ato-E1)2:
BiotinTEG-5′ ACCATAACAGGTGGCACGGAACCATAACAGGTGGCACGGA 3′
(sc-E1)2:
BiotinTEG-5′ CGCGTGGCAGGTGTATTTAGCGCGTGGCAGGTGTATTTAG 3′
36bp 1 site oligonucleotides:
ato-E1 Biotin TEG-5′ TGGTAGTAACCATAACAGGTGGCACGGAAG CCGCAC 3′ sc-E1 Biotin TEG-5′ CATGGCGACGCGTGGCAGGTGTATTTAGTCGAA 3′ These biotinylated oligonucleotides were hybridised to unmodified complementary strands before immobilisation to the SA chip as described above. Direct binding assays for these oligonucleotides were carried out, then unlabelled oligonucleotides were used in a competition assay as described in Teh et al. (35) to determine relative binding affinities for the following double stranded DNAs (top strand only shown):
sc-E1: 5′ CATGGCGACGCGTGGCAGGTGTATTTAGTCGAACGA 3′
ato-E1: 5′ TGGTAGTAACCATAACAGGTGGCACGGAAGCCGCAC 3′
E(spl)mγ-E2: 5′ CAAATCTAGAAACGGCAGCTGTTCGCTCTGCAAATT 3′
ato-E1M: 5′ TGGTAGTAACCATAAAAGGTTGCACGGAAGCCGCAC 3′
In each case the E box is underlined. The nucleotides mutated to ablate the E-box in ato-E1M are shown in bold.
RESULTS
ato-E1 and sc-E1 retain their specificity in a cell culture assay
We have used a Drosophila S2 cell culture (31) cotransfection assay with luciferase reporter constructs to explore the effect of E-box DNA sequence variation on transcriptional activation by Ato and Sc. Such a system has been used successfully in previous studies of bHLH protein interactions (11, 14, 32). Artificial enhancers were constructed consisting of several copies of short (20 bp) E-box-containing sequences driving a luciferase reporter gene (Table 1). Such concatemers of Ato-type or Sc-type E boxes have been shown to drive GFP-reporter expression in highly Ato- or Sc-specific patterns respectively in vivo (26).
The E boxes ato-E1 and sc-E1 are both derived from autoregulatory enhancers (Table 1). In the context of the ato femoral chordotonal enhancer, ato-E1 is needed for Ato autoregulation in a subset of chordotonal precursors (37), while sc-E1 in the context of the sc-SMC enhancer is needed for Sc autoregulation in all Sc-dependent SOPs (9). (ato-E1)7-GFP and (sc-E1)6-GFP concatemer reporter constructs give expression in only Ato- or Sc-dependent SOPs respectively (26). In the S2 cell culture assay, (ato-E1)7-luc was activated by both Ato/Da, and Sc/Da protein combinations, but the response was 2.5-fold greater with Ato/Da (Fig. 1A). Thus ato-E1 shows some specificity for its cognate proneural factor. Similarly, (sc-E1)6-luc showed specificity for Sc, and was activated about 4.2 fold more by Sc/Da than by Ato/Da (Fig.1A). Therefore, the specificity observed in vivo for these two concatemers can be partially reproduced in the S2 cell culture system. This supports the previous conclusion that the E boxes respond in vivo directly to their predicted cognate bHLH factors. It also suggests that the specificity of activation for these two E box sites can be attained outwith an in vivo cellular context and reinforces the importance of DNA sequence for specificity in transcriptional activation. Specificity is not as complete as in vivo however (26), suggesting additional factors, not expressed in S2 cells, may be important in vivo to improve specificity.
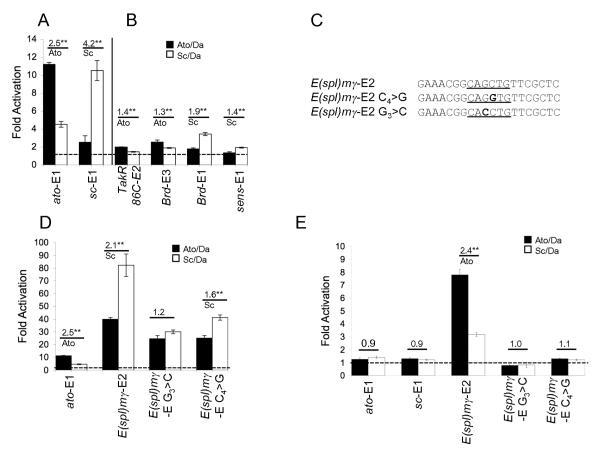
Figure 1. E-box concatemer luciferase constructs support Ato or Sc-specific activation in S2 cells.
Concatemer luciferase constructs as indicated were cotransfected into S2 cells with protein expression constructs for Da and Ato or Sc. (A) ato-E1 and sc-E1 show a preference for Ato and Sc respectively. (B) Other E boxes show weaker preferences for their presumed cognate proteins. (C) Sequence of the E(spl)mγ-E2 and mutated versions used in the concatemer reporter constructs in D and E. (D) E(spl)mγ-E2 supports very high levels of activation. Mutation of the central core reduces but does not abolish this. (E) E(spl)mγ-E2 is still activated at ‘low’ proneural transfection levels (see Materials and Methods). Mutating the core nucleotides now abolishes activity. The results are expressed as fold activation (± s.d.) relative to a control cotransfection using reporter construct and ‘empty’ protein expression vector. Fold activation of 1 (no activation above background) is indicated by a dashed line. The fold preference of each E box construct for Ato or Sc is indicated above the data pair, along with the statistical significance (** = p< 0.01; * = p<0.05).
Other E boxes also exhibit specificity
Having established that specific interactions can be reproduced in this assay, we then tested concatemers of other functional E-box sequences (Table 1). TakR86C-E2 and Brd-E3 are from Ato-responsive enhancers of the TakR86C and Brd genes respectively (26, 30) while Brd-E1 is from a Sc-responsive enhancer (26, 33). It is notable that these E boxes are activated less well in the S2 cell assay than ato-E1 and sc-E1 (Fig. 1B). However, some specificity is retained in each case. In vivo, a (TakR86C-E2)6-GFP concatemer was previously shown to give rise to expression in a subset of Ato-dependent precursors (26). This Ato specificity is partially retained by (TakR86C-E2)6-luc in the S2 cell assay. The Brd gene E boxes have not been tested in concatemeric reporter gene constructs in vivo. In the S2 cell assay, Brd-E3 shows Ato specificity as predicted, whilst Brd-E1 retains Sc specificity. Therefore, five E boxes show evidence of differentially responding to Ato or Sc proneural proteins in a manner predicted from their in vivo enhancer properties. Moreover, this specificity correlates well with the presence of an ESc or EAto sequence motif (Table 1).
Whilst these E boxes derive from Sc- or Ato-specific enhancers, there is evidence that some common target genes might be regulated by Ato and Sc via a single enhancer. Such a target gene is sens, which is thought to be regulated by Sc and Ato interacting with a single E box site, sens-E1 (14). Surprisingly, when this ‘pan-neural’ E box was tested in the S2 cell assay, little activation was achieved by either Sc or Ato (Fig. 1B). Although weak overall, activation was significantly stronger with Sc/Da than with Ato/Da. Interestingly, the sequence of sens-E1 conforms to the ESc motif (Table 1), suggesting that the S2 cell activity is reflecting the sequence of this E box rather than its proposed function in its native enhancer. However, activity is low despite this E box being a perfect match to the ESc motif (gCAGGTGt).
An E box with a ‘GC’ core activates very strongly
All the above E boxes have a G in the 4th position of the 6-bp core sequence (CANGTG – usually CAGGTG). However, a number of E boxes in proneural-responsive enhancers have a C in this position (CAGCTG). To examine the influence of this variant, we investigated an E box from the E(spl)mγ gene (E(spl)mγ-E2). The enhancer containing this E box appears to respond largely to Ato in vivo ((24) and unpublished observations). However, the palindromic core of this E box means that it matches the Sc consensus sequence motif in one orientation (gCAGCTGt) and almost matches the Ato consensus sequence motif in the other (aaCAGCTGc) (Table 1; Fig. 1C). Indeed, in the S2 cell assay E(spl)mγ-E2 is activated very strongly indeed by both Ato/Da and Sc/Da (Fig. 1D). Subsequently, the activation of (E(spl)mγ-E2)6-luc was tested at twenty-fold lower levels of cotransfected proneural protein construct. At this level of proneural factor, neither (ato-E1)7-luc nor (sc-E1)7-luc showed activation, whereas (E(spl)mγ-E2)6-luc still gave significant activation (Fig. 1E). Interestingly, at ‘high’ proneural concentrations, this E box showed some specificity for Sc, but at low proneural concentrations it showed specificity for Ato, reflecting better its predicted in vivo role.
The influence of the central two nucleotides of E(spl)mγ-E2 on both its strong activity and proneural specificity was tested by mutating the core GC. In one mutation, the E box was changed from gCAGCTGt to gCAGGTGt (C4>G; Fig. 1C), which still conforms completely to the Sc consensus binding site sequence. A second mutation was also examined: in this case gCAGCTGt was mutated to gCACCTGt (G3>C; Fig. 1C), which in reverse continues to match partially the Ato consensus binding sequence (aaCAGGTGc). Both mutated sequences show a loss of activity at low proneural concentration (Fig. 1E) and a drastic reduction in activation at high proneural concentration (Fig. 1D). Nevertheless, activity is still considerably higher than for other unmutated E boxes despite the fact that the 8-bp core of the sequence (gCAGGTGt) is exactly the same as that of sc-E1 and sens-E1 (Table 1). We conclude that a C in the fourth position of the E box core contributes to high E box activity, but other sequences outwith the E box consensus binding sequences must also contribute. In both mutations, no clear changes in specificity were observed.
Surface plasmon resonance analysis of interaction of Ato with cognate and non-cognate sites
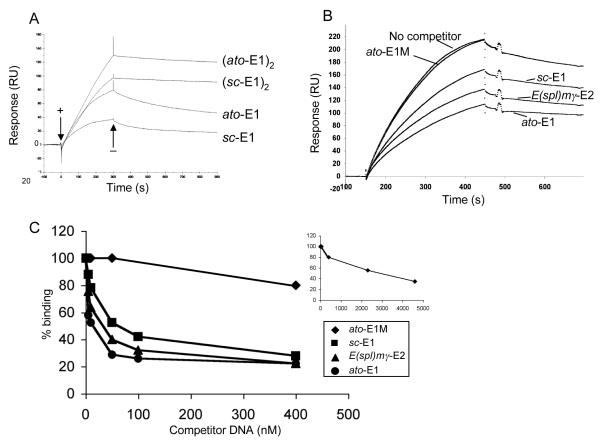
Previous in vitro binding experiments by gel retardation showed no difference in equilibrium binding affinities of Ato/Da or Sc/Da to ato-E1 or sc-E1 (26). To extend this analysis, an SPR assay (35) was used to explore further the relative affinities for Ato/Da interaction with cognate, non-cognate and mutated E-boxes. In SPR analysis, Ato/Da binding to immobilised ato-E1 or sc-E1 was characterised by a slow association step followed by a very slow dissociation (Fig. 2A lower two curves). Normalised curves (data not shown) indicated that the association phases were not markedly different in kinetics, but the dissociation from sc-E1 was faster than that from ato-E1, demonstrating a less stable complex. Subsequently, a competition assay was used to test the relative affinities for Ato/Da of various E box sequences in competition with immobilised ato-E1. In this assay, non-immobilised double-stranded competitor DNA was incubated with Ato/Da protein prior to SPR analysis. The reduction in the SPR signal for Ato/Da heterodimer binding to ato-E1 in the presence of 10 nM competitor (Fig. 2B) shows that ato-E1 itself competes most efficiently, with sc-E1 and E(spl)mγ-E2 competing less well. A mutated E box (ato-E1M) does not compete for binding at this concentration. A subsequent titration experiment determined the relative concentrations at which the four competitors competed 50% of the Ato/Da binding to ato-E1 (Fig. 2C). This again revealed ato-E1 to be the most efficient competitor (10 nM yields 50% competition), followed by E(spl)mγ-E2 (35 nM) and sc-E1 (50 nM). In comparison, ato-E1M competed extremely poorly (2.3 μM). Although the ato-E1 site showed the highest affinity for Ato/Da, it is apparent that there is no simple correlation of binding affinity with the magnitude or specificity of transcriptional activation in the S2 assay or in vivo. Notably, the highly active E(spl)mγ-E2 site does not show an unusually high affinity in vitro.
Figure 2. SPR measurements of proneural protein binding to E boxes.
(A) Sensorgrams for Ato/Da binding to immobilised DNAs containing single ato-E1 or sc-E1 sites (lower two curves) and tandem ato-E1 or sc-E1 sites (upper two curves), as indicated. A binding event is observed as a change in Response Units (RU) over time, based on the change in refractive index of the sensor chip induced by binding. Representative sensorgrams for 400 nM protein are shown. When Ato/Da is added (+) the association phases for the single site DNAs are biphasic and are followed by a slow dissociation when Ato/Da is removed from the flow-through solution (−). The dissociation from the tandem site DNAs is clearly much slower. (B,C) SPR competition assay to measure relative binding affinities of Ato/Da to different DNAs. (B) Sensorgrams showing the reduction of binding of Ato/Da to immobilised ato-E1 caused by the addition of competitor DNAs to the flow through solution. Representative sensorgrams are shown for 200 nM Ato/Da with 10 nM ato-E1, ato-E1M (mutated E box), sc-E1, E(spl)mγ-E2, or no competitor DNA. (C) Reduction of binding of Ato/Da to ato-E1 by titration with 0-400 nM competitor DNAs. The percent binding is shown relative to that obtained with no competitor DNA present. The inset shows the full titration for competition with the mutated E box DNA, ato-E1M.
Comparison of cooperativity of proneural protein binding to ato-E1 and sc-E1
Our analyses have shown that differences in E box behaviour in vivo and in the cell culture assay do not correlate with differences in protein-DNA interaction in vitro. In considering a possible molecular basis of in vivo specificity, we explored the role of cooperative binding to adjacent E box binding sites. Cooperative binding has been shown to be an important aspect of transcriptional activation, and the bHLH protein, MyoD, is known to bind cooperatively to adjacent E box binding sites in vitro (36). We speculated that cooperativity of proneural protein binding may be observed with cognate but not non-cognate E box sequences. To test this, SPR was used to analyse Ato/Da protein binding to immobilised DNAs containing either one E box (Fig. 2A, lower two curves) or two tandem E boxes (Fig. 2A, upper two curves) in the same configuration as used in the concatemer reporter gene constructs. For both ato-E1 and sc-E1, much slower Ato/Da dissociation from tandem sites was observed compared with one-site DNAs. This slower dissociation is indicative of cooperative interactions between Ato/Da heterodimers bound to adjacent sites. This shows for the first time that a proneural protein complex is capable of protein interactions that result in cooperative binding to E-box binding sites. Comparison of normalised (ato-E1)2 and (sc-E1)2 curves (results not shown), however, reveals no difference in the level of cooperativity seen with the cognate ((ato-E1)2) and non-cognate ((sc-E1)2) sites, and so differences in cooperative protein interaction would appear not to underlie E box specificity.
Effect of mutagenesis of concatemer E-box flanking sequence on specificity of activation in S2 cells
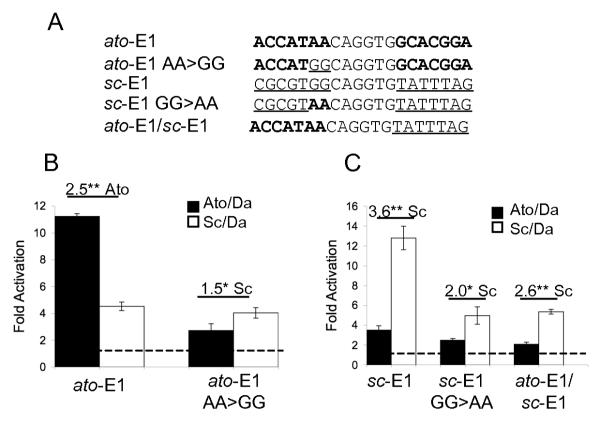
Although there is much variability between the E boxes tested, in the S2 cell assay E box specificity correlates quite well with the identity of the two 5′ nucleotides flanking the E box core, which previously defined distinct Ato- and Sc-specific E boxes (26). The importance of these flanking nucleotides was assessed by mutagenesis. Firstly, the 5′ flanking sequence of ato-E1 was mutated from AA to GG, thereby allowing the E box to conform to the Sc-specific consensus (ato-E1 AA>GG, Fig. 3A). In the S2 luciferase assay, ato-E1 AA>GG showed a small but significant specificity for activation by Sc (Fig. 3B). This largely results from a reduction in activation by Ato rather than an increase by Sc. Therefore, the flanking AA 5′ is necessary for Ato-specific activation of ato-E1; a flanking G, however, is not sufficient for full Sc-specific activation in the context of this concatemer construct.
Figure 3. ato-E1 and sc-E1 sites with altered flanking nucleotides support reduced activation in S2 cells.
(A) The mutations tested in S2 cell luciferase assay. Flanking nucleotides derived from ato-E1 (bold) or sc-E1 (underlined) are indicated. The 6-bp core E box is identical in all cases. (B) ato-E1 AA>GG drives much reduced activation and loses its specificity for Ato. (C) sc-E1 GG>AA has much reduced activation but retains some Sc specificity. Changing the whole 5′ flank of sc-E1 to the sequence 5′ of ato-E1 (ato-E1/sc-E1) yields a similar result.
The reciprocal mutation of sc-E1 yields a somewhat different result: mutation of the 5′ flanking GG to AA allows it to conform to the Ato-specific consensus sequence (sc-E1 GG>AA, Fig. 3A. Although this change reduces activation by Sc, it still remains higher than activation by Ato (Fig. 3C). There is, therefore, no reversal of specificity. The same result was observed when the whole 5′ flanking sequence of sc-E1 was altered to that of ato-E1 (Fig. 3C), thereby giving a hybrid concatemer with each repeat having a 5′ equivalent to ato-E1 and a 3′ equivalent to sc-E1 (ato-E1/sc-E1, Fig. 3A). Whilst this suggests that the 5′ G is essential for full Sc activation, it appears that unidentified sequences 3′ of the core E box also influence activity and specificity.
In vivo expression patterns for transgenic flies with (sc-E1 GG>AA)6-GFP
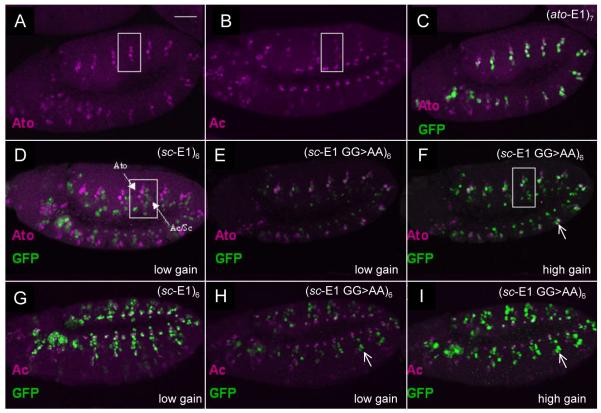
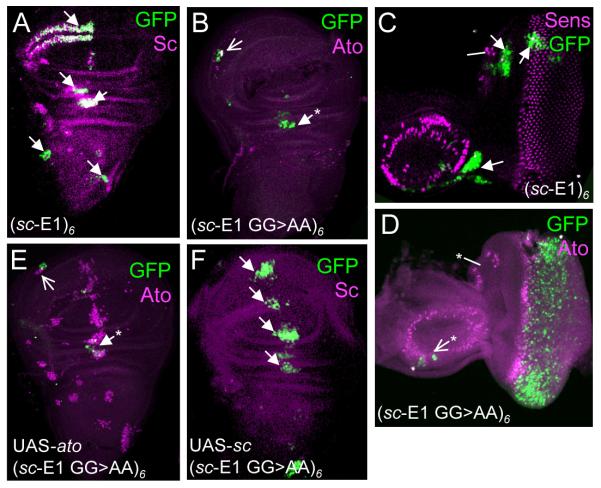
In S2 cells, mutating the sc-E1 concatemer to resemble an EAto motif (sc-E1 GG>AA) (Fig. 3C) did not result in regulation by Ato. To assess whether developmental context would improve specificity, the same concatemer construct was tested in vivo as a GFP reporter gene. In the embryo, unmutated (sc-E1)6-GFP gives strong expression in Sc-expressing SOPs and no expression in Ato-expressing SOPs (26) (Fig. 4D,G). This Sc-dependent expression was much reduced in the concatemer reporter with the sc-E1 GG>AA mutation (Fig. 4E,H), whereas some expression in Ato-dependent precursors is now apparent (Fig. 4F,I). Similarly in third instar larval imaginal discs, sc-E1 GG>AA supported reduced expression in Sc-dependent precursors in the wing, eye and leg imaginal discs compared to sc-E1 (Fig. 5A–D, and unpublished data). Additionally, a partial gain of expression was observed in Ato-expressing cells in the eye-antennal and wing discs (Fig. 5A–D), although no expression was observed in most Ato-expressing cells of the antenna or leg discs (data not shown). Thus, there appears to be a partial swap of specificity.
Figure 4. (sc-E1 GG>AA)6-GFP supports both Sc- and Ato-dependent expression in embryos.
(A) Expression of Ato in SOPs of stage 11 embryo. White box encloses a single abdominal segment. (B) Expression of Ac in SOPs of stage 11 embryo (Sc is similar). (C) (ato-E1)7-GFP showing overlap with Ato. (D) (sc-E1)6-GFP showing no overlap with Ato. Ato- and Sc-dependent SOPs are indicated by arrows. (E) Loss of GFP expression in (sc-E1 GG>AA)6-GFP embryos. Microscope gain settings for GFP the same as in D. (F) Increase of Ato-dependent GFP expression in (sc-E1 GG>AA)6-GFP embryos. Microscope gain higher to visualise weak GFP expression (G) Extensive overlap between (sc-E1)6-GFP and Achaete. (H) Loss of GFP expression in (sc-E1 GG>AA)6-GFP embryos. Microscope settings same as in G. (I) Loss of overlap between GFP and Achaete in (sc-E1 GG to AA)6-GFP embryos. Gain of GFP expression in Ato-dependent SOPs (circled). Scale bar is 50 μm. Colours are as indicated in the relevant panels.
Figure 5. (sc-E1 GG>AA)6-GFP supports both Sc- and Ato-dependent expression in imaginal discs.
(A) sc-E1 wing disc. Sites of GFP expression in Sc-dependent cells are arrowed. (B) sc-E1 GG>AA wing disc, showing reduced Sc-specific expression. Expression is present in the Ato-specific cells of the ventral radius (open arrow) and in some cells of the dorsal radius (asterisk) which are often observed in other Ato-dependent reporters (24). (C) sc-E1 eye-antennal disc showing areas of GFP corresponding to areas of Sc PNCs only (arrows). (D) sc-E1 GG>AA eye-antennal disc showing expression in Ato-specific photoreceptor precursors and a few Ato-dependent cells in the antennal portion (open arrow). Lack of overlap between Ato and GFP is likely to be due to the delay in GFP maturation (26). Interestingly, no expression is observed in the Ato-dependent ocellar precursors (asterisk). (E) sc-E1 GG>AA wing disc with ectopic Ato expression (109-68-Gal4; UAS-ato). No additional GFP expression arises from the additional Ato expression. (F) sc-E1 GG>AA wing disc with Sc overexpression (109-68-Gal4; UAS-sc) (earlier stage than A). The Sc overexpression drives strong extra GFP expression in the region of the third wing vein.
As a further test, the response of this reporter construct to proneural protein misexpression was assessed. UAS-ato or UAS-sc were misexpressed in imaginal disc proneural clusters using the driver line 109-68Gal4 (16). Interestingly, despite the change in specificity observed above, (sc-E1 GG>AA)6-GFP showed increased areas of expression only in response to UAS-sc (Fig. 5E,F and data not shown), even though UAS-ato can activate (ato-E1)7-GFP under similar conditions (26). In conclusion, the 5′ bases of the EAto and ESc motifs are important for proneural specificity in vivo, but this specificity is strongly dependent on developmental context.
Sens protein accentuates the specificity of Ato and Sc in the S2 cell assay
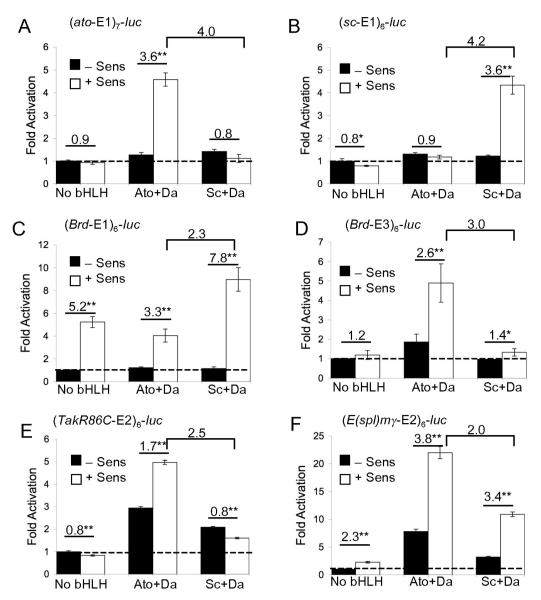
In the S2 cell assay, Ato and Sc have clear intrinsic specificities: in most cases each promotes significantly greater reporter gene activation via its cognate E boxes. However, specificity is not complete — the proteins can also activate non-cognate E boxes to some extent, whereas the same concatemer constructs (where tested) support more specific patterns of expression in vivo (26). One possibility is that interactions with other protein factors enhance proneural specificity and that these factors are not expressed in S2 cells. One such factor might be the Zn finger protein Sens, which has an important role in the selection of the single SOP from the PNC (1, 14, 25). Sens associates with Sc, Ac, Ato and Da in pulldown assays (1, 15) and it can enhance the activity of Ac in S2 cells in a non-DNA binding dependent manner (14). This suggests that Sens is a general proneural coactivator. Here, we address the questions of whether Sens influences the activity of all proneural factors, and whether it influences specificity. Transfection with Sens-expressing plasmid alone has little effect on transcriptional activation of any concatemer reporter construct except Brd-E1 (see below). However, compared with proneural proteins alone, cotransfection of Sens with proneural proteins resulted in higher levels of reporter activation with all combinations of protein and reporter construct tested (including Da alone and Amos, data not shown). Thus, Sens can indeed synergise with all proneural proteins tested in this assay, as predicted from pulldown experiments.
A different result was obtained at low levels of proneural protein. Under these conditions, there is normally very little activation of ato-E1 or sc-E1 reporter constructs by their cognate proteins (Fig. 6A,B). In the presence of Sens, however, a significant increase in activation was observed for the cognate proneural protein but not at all for the non-cognate protein. Therefore, Sens appears to enhance proneural activity in an E-box motif-specific manner.
Figure 6. Sens synergises with proneural proteins and increases specificity.
(A–F) Activation of concatemer constructs by Ato/Da or Sc/Da in the presence or absence of Sens. Annotation as for Fig. 1. Brackets denote the ratio of activation by Ato/Da and Sc/Da in the presence of Sens (specificity ratio). Proneural expression construct concentration is ‘low’ for all except (E).
Similar effects were observed for other concatemer reporter constructs. Thus, in the case of Brd-E3, Sens synergises with Ato/Da but not Sc/Da (Fig. 6D). For TakR86C-E2 there was no activation by any protein combination with Sens at low proneural concentrations, which reflects their lower ability in promoting transcriptional activation (see above). At high proneural concentration, Sens improved the Ato-specificity of TakR86C-E2 (Fig 6E). For Brd-E1 (Fig. 6C), Sens alone resulted in significant luciferase reporter activity, perhaps due to interaction with another bHLH factor present in the S2 cells. With the addition of proneural factors, this concatemer shows Sc specificity, and there is a small increase in the specificity ratio compared with high proneural concentrations in the absence of Sens (Fig. 1). The abnormally high activity of this construct in the absence of proneural factors may be obscuring their effect. As shown above, E(spl)mγ-E2 construct (Fig. 1E) supported high transcriptional activation even at low proneural concentration. In this case, Sens increased overall activity but did not alter the degree of specificity observed (Fig. 6F). In summary. specificity for cognate proteins was enhanced by Sens for four out of six E boxes tested. Hence, in many cases Sens enhances the specificity of both Sc and Ato for their cognate E boxes.
DISCUSSION
The proneural proteins exhibit very precise specificity in activation of different neurogenic programmes. Previously, we suggested that utilisation of different E-box motifs as binding sites may partly underlie this specificity (26). This was based on the finding that E boxes from Sc- and Ato-specific target genes conform to different consensus motifs. In this study we find further support for this. In a cell culture assay, artificial enhancers of concatemers of EAto or ESc sequences generally show specific activation by Ato or Sc proteins respectively. Nevertheless, our results also show that E box activity and specificity depends on complex features of the DNA surrounding the proneural-specific motifs both in cell culture and in vivo. The task of predicting by sequence analysis how proneural proteins regulate targets remains formidable.
Proneural specificity is exhibited in S2 cells
Transcription factor activity depends on a complex interplay of interactions with DNA and with other protein factors, including those bound to other sites within the enhancer. To concentrate on the role that proneural protein interaction with E-box binding sites plays in specificity, we analysed synthetic enhancers of concatemers of E-box-containing sequences in a cell culture reporter gene assay. Our previous study of Ato or Sc-specific enhancers relied on the analysis of expression patterns produced in transgenic flies carrying GFP reporter gene constructs (26). In that study, specific regulation by Sc or Ato was inferred indirectly from patterns of GFP expression. Here we show that much of this inferred specificity is also seen in a cell culture reporter gene assay, supporting the conclusion that Ato and Sc directly utilise different E box motifs. Thus, in general, the specificity of E box response (ratio of response to Sc and Ato) could be predicted from matches to ESc or EAto motifs identified previously (26). In most cases, this specificity also corresponded to the specificity of the native enhancer from which the E box was taken. An interesting exception is sens-E1: whilst this E box is proposed to respond to both Ato and Sc in vivo (14), it responds slightly better to Sc than to Ato in culture, which is more consistent with its ESc motif. It will be important to determine what other enhancer features allow such an E box to function as a common target of Ato and Sc in vivo.
Importantly, E box specificity is achieved outwith the appropriate cellular and developmental context of neurogenesis: S2 cells are embryonic, non-neural cells of likely haematopoietic origin (29, 31) and are not expected to contain neural-specific factors. Our results indicate therefore that proneural factors have intrinsic ability to utilise different E box motifs without the need for interactions with neural specific cofactors. The ESc and EAto motifs differ most notably in the bases flanking the 5′ end of the 6-bp core sequence (NG vs AW). There is evidence from the crystal structure of the MyoD bHLH domain-DNA complex that protein contacts are made with bases in this position (22), suggesting that similar direct contacts may influence E box utilisation by proneural proteins. The basic region amino acids making these contacts (R110, R117 and E118) are conserved in the proneural proteins, but in Ato the Rs are separated by three amino acids (LAA, equivalent to MyoD KAA) that are absent in Sc. This may influence how protein contacts are made with the flanking nucleotides. If so, this does not seem to be manifested as binding affinity differences in vitro. It is possible instead that differences in binding contacts may cause conformational effects that affect the activity of the proneural protein.
Determinants of E box function are complex
The above results point to the importance of distinct Ato and Sc E box motifs for proneural specificity. Several findings, however, demonstrate that these motifs must interact with other determinants of E box function. In the cell culture assay, swapping the flanking bases that distinguish these motifs caused a general reduction in E box activity rather than a change in specificity. For example when the flanking AA of ato-E1 was changed to GG (thereby matching perfectly the ESc consensus) the concatemer reporter construct was no longer able to respond well to either Sc or Ato. These results indicate that the consensus motifs alone are not sufficient for activity or specificity and that the surrounding DNA context (even within the short 20-bp sequences used) is important in the cell culture assay. Interestingly, in some instances specificity could be manipulated more successfully in vivo: (sc-E1 GG>AA)6-GFP transgenic flies showed GFP expression consistent with strongly reduced activation by Sc and a gain of activation in some specific locations by Ato. This suggests that in vivo the flanking G of the ESc motif is necessary but not sufficient for Sc regulation whereas the AA of the EAto motif is sufficient for Ato regulation in some contexts.
In addition to variation in specificity, the E boxes studied also show a wide range of functional efficiencies in the cell culture reporter assay. The very high activity of E(spl)mγ-E2 in this assay appears to be partly due to its central GC bases. The MyoD crystal structure shows a water-mediated hydrogen bond between E118 (also present in proneural proteins) and the central C in the E-box CAGCTG, which may strengthen this protein interaction (22). However, E(spl)mγ-E2 did not show an unusually high affinity for proneural proteins in vitro. Moreover, the central GC is not the sole explanation for the high activity of E(spl)mγ-E2: even when mutated to the more usual GG, E(spl)mγ-E2(C4>G) showed a relatively high level of activity. Strikingly, E(spl)mγ-E2(C4>G), sens-E1 and sc-E1 show large differences in activity in cell culture, even though they have identical motifs (gCAGGTGt).
Overall, our results above show that further sequences on both flanks of the ESc and EAto box motifs are also important for specificity and activity. One possibility is that the 20-bp DNA sequences used to construct the concatemers may include flanking sequences that interact with other protein factors to influence proneural specificity. Such adjacent sites have been identified for some mouse proneural E box binding sites (6, 20). Moreover, in its native enhancer, ato-E1 is adjacent to an Ets-domain transcription factor binding site (37) (although this site is mutated in the constructs used in this study). However, such cofactors would need to be expressed in S2 cells. Moreover, although the flanking sequences of the ato-E1 and sc-E1 sites are strongly conserved among Drosophila species (unpublished observations), we find no obvious shared sequence motifs in the 5′ and 3′ flanks of known Drosophila E boxes that might be cofactor binding sites. Whilst there is a potential POU factor binding sequence 5′ of the ato-E1 site, no members of the Drosophila POU family appear to be expressed during early neurogenesis (unpublished observations and data not shown). Alternatively, the further flanking bases may influence bHLH heterodimer interaction either through direct contacts or through indirect conformational effects. It is interesting that 3′ bases appear important as these may be predicted to affect Da interaction (37). It is notable that the Da homologue, E2A, has different half-site preferences when bound to Twi or MyoD (19).
Sens as a specificity cofactor for both Sc and Ato
The specificity of E-box concatemer constructs is generally more complete in vivo than in the S2 luciferase assay — notably proneural proteins can generally activate non-cognate E boxes to some extent in cell culture but not in vivo. One possibility is that the intrinsic specificity of proneural proteins must normally be enhanced by interaction with cofactors that are not present in S2 cells. In the cell culture assay, at high proneural levels we found that Sens enhanced proneural activity in a general manner. None of the constructs tested contain Sens binding motifs, so it is likely that enhancement occurs in a DNA-binding independent manner via protein-protein interactions as previously proposed (1, 14). At low proneural concentrations, however, the effect of Sens enhancement becomes selective. For many of the constructs tested, Sens only enhanced the activity of proneural proteins for concatemers consisting of their cognate E box. We suggest that proneural-Sens interaction may enhance the specificity of proneural-E box interaction. Thus, this is an interesting case in which proneural specificity can be influenced by a common cofactor, rather than requiring interaction with different subtype-specific cofactors. It remains to be determined whether Sens would enhance specificity on native enhancers as well as concatemer constructs. Moreover, it seems unlikely that Sens is a specificity factor for all proneural target genes. However, our results are consistent with Sens acting as a specificity cofactor in certain contexts – such as the proneural autoregulatory enhancers active in SOPs where there are high levels of Sens and proneural proteins present. Other non-DNA binding proneural protein interactors, such as Chip (28) and Kohtalo (21) may have a similar effect in other circumstances.
The effect of Sens could be explained by two models. Firstly, interaction of a proneural protein with a specific E-box motif may give rise to a specific conformation which results in an increased affinity for Sens protein. Alternatively, the Sens-proneural protein interaction may alter the proneural bHLH domain conformation thereby increasing its affinity for its cognate binding site (i.e. an induced fit model) (34). Indeed, variation in MyoD bHLH protein DNA sequence preferences have been previously observed to be due to conformational effects on basic region conformation arising due to binding partner differences (19) or amino acid composition of the basic region (13). In this view, proneural specificity relies on a combination of cognate DNA sequence recognition and protein-protein interactions. Important future work will be the identification of the amino acid residues of Ato and Sc necessary for their interaction with Sens and the determination of whether these influence DNA recognition.
ACKNOWLEDGEMENTS
This work was supported by grant 077266 from the Wellcome Trust and by the BBSRC. We thank Christos Delidakis and Hamed Jafar-Nejad for kindly providing plasmids, Sam Maung for technical assistance, Sandra Bruce and Malcolm Walkinshaw for help and advice with protein purification, and Sebastián Cachero, Ian Simpson and Petra zur Lage for manuscript comments. We also thank the Centre for Translational and Chemical Biology and the University of Edinburgh Protein Production Facility for assistance with characterisation.
REFERENCES
- 1.Acar M, Jafar-Nejad H, Giagtzoglou N, Yallampalli S, David G, He Y, Delidakis C, Bellen HJ. Senseless physically interacts with proneural proteins and functions as a transcriptional co-activator. Development. 2006;133:1979–89. doi: 10.1242/dev.02372. [DOI] [PubMed] [Google Scholar]
- 2.Berkes CA, Tapscott SJ. MyoD and the transcriptional control of myogenesis. Semin Cell Dev Biol. 2005;16:585–95. doi: 10.1016/j.semcdb.2005.07.006. [DOI] [PubMed] [Google Scholar]
- 3.Bertrand N, Castro DS, Guillemot F. Proneural genes and the specification of neural cell types. Nature Rev. Neuro. 2002;3:517–530. doi: 10.1038/nrn874. [DOI] [PubMed] [Google Scholar]
- 4.Bloor AJ, Sanchez MJ, Green AR, Gottgens B. The role of the stem cell leukemia (SCL) gene in hematopoietic and endothelial lineage specification. J Hematother Stem Cell Res. 2002;11:195–206. doi: 10.1089/152581602753658402. [DOI] [PubMed] [Google Scholar]
- 5.Campuzano S, Modolell J. Patterning of the Drosophila nervous system: the achaete-scute gene complex. Trends in Genetics. 1992;8:202–208. doi: 10.1016/0168-9525(92)90234-u. [DOI] [PubMed] [Google Scholar]
- 6.Castro DS, Skowronska-Krawczyk D, Armant O, Donaldson IJ, Parras C, Hunt C, Critchley JA, Nguyen L, Gossler A, Gottgens B, Matter JM, Guillemot F. Proneural bHLH and Brn proteins coregulate a neurogenic program through cooperative binding to a conserved DNA motif. Dev Cell. 2006;11:831–44. doi: 10.1016/j.devcel.2006.10.006. [DOI] [PubMed] [Google Scholar]
- 7.Chae JH, Stein GH, Lee JE. NeuroD: the predicted and the surprising. Mol Cells. 2004;18:271–88. [PubMed] [Google Scholar]
- 8.Chien C-T, Hsiao C-D, Jan LY, Jan YN. Neuronal type information encoded in the basic-helix-loop-helix domain of proneural genes. Proceedings of the National Academy of Sciences, USA. 1996;93:13239–13244. doi: 10.1073/pnas.93.23.13239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Culí J, Modolell J. Proneural gene self-stimulation in neural precursors: an essential mechanism for sense organ development that is regulated by Notch signalling. Genes Dev. 1998;12:2036–2047. doi: 10.1101/gad.12.13.2036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ghysen A, Dambly-Chaudière C. The specification of sensory neuron identity in Drosophila. BioEssays. 1993;15:202–208. doi: 10.1002/bies.950150502. [DOI] [PubMed] [Google Scholar]
- 11.Giagtzoglou N, Koumbanakis KA, Fullard J, Zarifi I, Delidakis C. Role of the Sc C terminus in transcriptional activation and E(spl) repressor recruitment. J Biol Chem. 2005;280:1299–1305. doi: 10.1074/jbc.M408949200. [DOI] [PubMed] [Google Scholar]
- 12.Goulding SE, zur Lage P, Jarman AP. amos, a proneural gene for Drosophila olfactory sense organs that is regulated by lozenge. Neuron. 2000;25:69–78. doi: 10.1016/s0896-6273(00)80872-7. [DOI] [PubMed] [Google Scholar]
- 13.Huang J, Weintraub H, Kedes L. Intramolecular regulation of MyoD activation domain conformation and function. Mol Cell Biol. 1998;18:5478–84. doi: 10.1128/mcb.18.9.5478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jafar-Nejad H, Acar M, Nolo R, Lacin H, Pan H, Parkhurst SM, Bellen HJ. Senseless acts as a binary switch during sensory organ precursor selection. Genes Dev. 2003;17:2966–78. doi: 10.1101/gad.1122403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jafar-Nejad H, Tien AC, Acar M, Bellen HJ. Senseless and Daughterless confer neuronal identity to epithelial cells in the Drosophila wing margin. Development. 2006;133:1683–92. doi: 10.1242/dev.02338. [DOI] [PubMed] [Google Scholar]
- 16.Jarman AP, Ahmed I. The specificity of proneural genes in determining Drosophila sense organ identity. Mech Dev. 1998;76:117–125. doi: 10.1016/s0925-4773(98)00116-6. [DOI] [PubMed] [Google Scholar]
- 17.Jarman AP, Grau Y, Jan LY, Jan YN. atonal is a proneural gene that directs chordotonal organ formation in the Drosophila peripheral nervous system. Cell. 1993;73:1307–1321. doi: 10.1016/0092-8674(93)90358-w. [DOI] [PubMed] [Google Scholar]
- 18.Jarman AP, Grell EH, Ackerman L, Jan LY, Jan YN. atonal is the proneural gene for Drosophila photoreceptors. Nature. 1994;369:398–400. doi: 10.1038/369398a0. [DOI] [PubMed] [Google Scholar]
- 19.Kophengnavong T, Michnowicz JE, Blackwell TK. Establishment of distinct MyoD, E2A, and twist DNA binding specificities by different basic region-DNA conformations. Molecular and Cellular Biology. 2000;20:261–72. doi: 10.1128/mcb.20.1.261-272.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lee S, Pfaff S. Synchronization of Neurogenesis and Motor Neuron Specification by Direct Coupling of bHLH and Homeodomain Transcription Factors. Neuron Neuron. 2003;Volume 38:731–745. doi: 10.1016/s0896-6273(03)00296-4. [DOI] [PubMed] [Google Scholar]
- 21.Lim J, Lee OK, Hsu YC, Singh A, Choi KW. Drosophila TRAP230/240 are essential coactivators for Atonal in retinal neurogenesis. Dev Biol. 2007;308:322–30. doi: 10.1016/j.ydbio.2007.05.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ma PCM, Rould MA, Weintraub H, Pabo CO. Crystal structure of MyoD bHLH domain-DNA complex: perspectives on DNA recognition and implications for transcriptional activation. Cell. 1994;77:451–459. doi: 10.1016/0092-8674(94)90159-7. [DOI] [PubMed] [Google Scholar]
- 23.Majka J, Speck C. Analysis of protein-DNA interactions using surface plasmon resonance. Adv Biochem Eng Biotechnol. 2007;104:13–36. [PubMed] [Google Scholar]
- 24.Nellesen DT, Lai EC, Posakony JW. Discrete enhancer elements mediate selective responsiveness of enhancer of split complex genes to common transcriptional activators. Dev Biol. 1999;213:33–53. doi: 10.1006/dbio.1999.9324. [DOI] [PubMed] [Google Scholar]
- 25.Nolo R, Abbott LA, Bellen HJ. Senseless, a Zn finger transcription factor, is necessary and sufficient for sensory organ development in Drosophila. Cell. 2000;102:349–62. doi: 10.1016/s0092-8674(00)00040-4. [DOI] [PubMed] [Google Scholar]
- 26.Powell LM, zur Lage PI, Prentice DRA, Senthinathan B, Jarman AP. The proneural proteins Atonal and Scute regulate neural target genes through different E-box binding sites. Mol. Cell. Biol. 2004;24:9517–9526. doi: 10.1128/MCB.24.21.9517-9526.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Quan X-J, Hassan BA. From skin to nerve: flies, vertebrates and the first helix. Cell. Mol. Life Sci. 2005;62:2036–2049. doi: 10.1007/s00018-005-5124-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ramain P, Khechumian R, Khechumian K, Arbogast N, Ackermann C, Heitzler P. Interactions between chip and the achaete/scute-daughterless heterodimers are required for pannier-driven proneural patterning. Mol Cell. 2000;6:781–90. doi: 10.1016/s1097-2765(05)00079-1. [DOI] [PubMed] [Google Scholar]
- 29.Ramet M, Manfruelli P, Pearson A, Mathey-Prevot B, Ezekowitz RA. Functional genomic analysis of phagocytosis and identification of a Drosophila receptor for E. coli. Nature. 2002;416:644–8. doi: 10.1038/nature735. [DOI] [PubMed] [Google Scholar]
- 30.Rosay P, Colas JF, Maroteaux L. Dual organisation of the Drosophila neuropeptide receptor NKD gene promoter. Mechanisms of Development. 1995;51:329–339. doi: 10.1016/0925-4773(95)00382-7. [DOI] [PubMed] [Google Scholar]
- 31.Schneider I. Cell lines derived from late embryonic stages of Drosophila melanogaster. J Embryol Exp Morphol. 1972;27:353–65. [PubMed] [Google Scholar]
- 32.Shirokawa JM, Courey AJ. A direct contact between the dorsal rel homology domain and Twist may mediate transcriptional synergy. Molecular and Cellular Biology. 1997;17:3345–55. doi: 10.1128/mcb.17.6.3345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Singson A, Leviten MW, Bang AG, Hua XH, Posakony JW. Direct downstream targets of proneural activators in the imaginal disc include genes involved in lateral inhibitory signaling. Genes and Development. 1994;8:2058–2071. doi: 10.1101/gad.8.17.2058. [DOI] [PubMed] [Google Scholar]
- 34.Spolar RS, Record MT., Jr. Coupling of local folding to site-specific binding of proteins to DNA. Science. 1994;263:777–84. doi: 10.1126/science.8303294. [DOI] [PubMed] [Google Scholar]
- 35.Teh HF, Peh WY, Su X, Thomsen JS. Characterization of protein--DNA interactions using surface plasmon resonance spectroscopy with various assay schemes. Biochemistry. 2007;46:2127–35. doi: 10.1021/bi061903t. [DOI] [PubMed] [Google Scholar]
- 36.Weintraub H, Davis R, Lockshon D, Lassar A. MyoD binds cooperatively to two sites in a target enhancer sequence: occupancy of two sites is required for activation. Proc Natl Acad Sci U S A. 1990;87:5623–7. doi: 10.1073/pnas.87.15.5623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.zur Lage PI, Powell LM, Prentice DRA, McLaughlin PM, Jarman AP. EGF receptor signalling triggers recruitment of Drosophila sense organ precursors by stimulating proneural gene autoregulation. Dev. Cell. 2004;5:687–696. doi: 10.1016/j.devcel.2004.09.015. [DOI] [PubMed] [Google Scholar]