Abstract
Hybridization has been repeatedly put forward to explain the invasiveness of Rhododendron ponticum L. in the British Isles. The present study investigates the pattern of ecotypic differentiation and hybridization among native North American R. catawbiense and R. maximum, native R. ponticum from Georgia and Spain, and invasive R. ponticum from Ireland and aims to assess the contribution of hybridization for Rhododendron invasion in the British Isles. Six populations per taxon were analyzed with AFLP markers for genetic dissimilarity, subjected to germination and growth experiments, and tested for frost hardiness. We assessed variation in morphological and ecological characteristics to identify traits displaying evidence of hybridization, thus, promoting invasiveness. Molecular marker analyses revealed a clear distinction between North American R. catawbiense and R. maximum on the one hand, and all R. ponticum populations on the other hand, displaying a complete intermixture of native Spanish and invasive Irish populations. Multivariate analyses of traits revealed leaf length–width ratio, relative growth rates (RGRs) in leaf length, root biomass, and shoot–root ratio to significantly discriminate between the different taxa and unequivocally assigned invasive Irish R. ponticum to the Spanish phenotypes. While the Irish R. ponticum had similar growth traits as conspecific native R. ponticum provenances, germination and biomass allocation were more similar to North American R. catawbiense and R. maximum. Hybridization did not contribute to explaining invasiveness of R. ponticum in Ireland. The similarity in germination and biomass allocation of invasive Irish R. ponticum and North American species has evolved independently and can more probably be attributed to an independent shift within the Ponticum cluster in Ireland.
Keywords: AFLP, frost hardiness, germination, introgression, RGR, Rhododendron section Pontica
Introduction
Invading populations often experience evolutionary changes and many of these have been attributed to altered selection pressure in the new range (Mooney and Cleland 2001; Lee 2002; Stockwell et al. 2003; Lambrinos 2004; Maron et al. 2004; Barrett et al. 2008; Beckmann et al. 2009). Most frequently, such shifts have become evident in larger sizes and higher growth rates in invasive populations compared to native situations (see Bossdorf et al. 2005 for review). Common explanations for such patterns, for example, include the hypothesis of evolution of increased competitive ability (EICA) in the absence of enemy load in the new range (Blossey and Nötzold 1995; Müller–Schärer et al. 2004). As an alternative, but not necessarily exclusive suggestion, the evolution of increased growth has also been attributed to the absence of competition in a new range as was shown in a multispecies common garden experiment by Blumenthal and Hufbauer (2007). The authors conclude that the biotic release into environments with reduced competition, in particular, in high-resource environments, can favor the evolution of traits related to rapid growth and high net reproductive allocation. The evolution of more competitive phenotypes can be expressed in maximized growth and reproduction (Leger and Rice 2003) as well as in shifts in allocation patterns, for example, from decreased aboveground to increased belowground competition in the new range (Barney et al. 2009). While such differentiation in phenotypes can be quantified at the trait level in common garden attempts, more mechanistic explanations need to refer to hypotheses at the molecular level. The maintenance of high genetic diversity during invasions is a precondition for selection being able to act on (Hedrick 2005). Besides polyploidy, in particular, hybridization can augment genetic novelty and has been suggested to be an important driver promoting evolutionary change in the introduced range (Abbott 1992; Ellstrand and Schierenbeck 2000; Blum et al. 2007; Rieseberg et al. 2007; Prentis et al. 2008).
Invoking hybridization in invasion biology is attractive, since consequences of hybridization, such as fixed heterosis in new allopolyploids, can already have immediate impact on the invasive potential in early stages of plant expansion. Hybridization might also be important in later stages of establishment and spread if adaptive introgression and transgressive segregation aid in the colonization of new environments. An example for recent hybrid speciation is given by Senecio squalidus in the British Isles, a recombinant hybrid of S. aethnensis and S. chrysanthemifolius (James and Abbott 2005). Both parental species were introduced into botanical gardens in Britain in the 18th century, and until now, the hybrid descendant has become widespread throughout the country. Hybrids can exhibit traits that are novel or extreme relative to those of either parental line (Arnold and Hodges 1995) and can differ remarkably in growth rates, phenology, or traits of defense (Vilà et al. 2000). However, hybridization can also cause adaptive trait introgression, through which alleles are transferred from one species to another (Schweitzer et al. 2002). There are several examples that provide evidence for hybrid advantages in fitness-related traits, for example, mediated by increased lifetime fecundity or increased survivorship and higher fruit production for invasive hybrid-derived populations (Campbell et al. 2006; Ridley and Ellstrand 2009). Accordingly, information on demographic characteristics, in particular germination and survival, are important data to assess the species’ susceptibility to novel trait transgression (Hooftman et al. 2005). There is increasing evidence of intrataxon hybridization preceding the evolution of invasiveness (Schierenbeck and Ellstrand 2009). In particular, for ornamental plants or species of horticultural interest, hybridization between cultivars and native counterparts is increasingly drawing the researchers’ attention (Culley and Hardiman 2008; Ross and Auge 2008).
Many of the hypotheses raised above have also been consulted to explain Rhododendron ponticum L. invasion in the British Isles. Rhododendron ponticum is an Ericaceae shrub species that naturally occurs along the Black Sea coasts of Georgia (Caucasus) and Turkey, as well as in the southern part of the Iberian Peninsula, and was introduced to the British Isles in 1763 (Elton 1958) and used in gardens and estates as an ornamental plant (Dehnen–Schmutz and Williamson 2006). Many Rhododendron species from Asia, but also from North America have been introduced to Botanical Gardens in the United Kingdom, mainly for horticultural purposes, and much effort was put into breeding ambitions to make R. ponticum hardier by natural and artificial selection and by intended hybridization with related species (Dehnen–Schmutz and Williamson 2006). In addition, it was directly brought to habitats with suitable conditions, and naturalization in the wild seems to have occurred all over Britain until the mid of the 19th century (as summarized in Dehnen–Schmutz and Williamson 2006). Flowering starts early in the species’ life cycle, after 10–12 years (Cross 1975), which is a relatively short interval for a woody, long-lived plant and, thus, provides ample opportunity for contemporary evolution. In comparison to their native provenances, invasive Irish populations of R. ponticum exhibit both higher germination success and higher growth rates than native ones from Georgia and Spain (Erfmeier and Bruelheide 2005), and these traits have been identified as key factors for establishment and spread in the field (Erfmeier and Bruelheide 2004). Hybridization as a key factor underlying the evolution of invasiveness has repeatedly been suggested for explaining the colonization success of R. ponticum in the British Isles (Milne and Abbott 2000; Abbott et al. 2003; Rieseberg et al. 2007). Milne and Abbott (2000) used restriction fragment length polymorphisms of cpDNA and rDNA to study naturalized accessions of R. ponticum in the British Isles and detected a mainly Iberian provenance of these invasive occurrences with 99% of Iberian haplotypes. In addition, for both genetic and morphological markers, the authors found evidence of introgression from North American congeneric species of the Pontica subsection of the genus (Milne and Abbott 2000). In their study, introgression from North American R. catawbiense was remarkable, particularly, in eastern Scotland; therefore, the authors suggest that hybridization had induced cold tolerance that enabled southern Iberian R. ponticum provenances to spread into northern climates. However, in their study, the comparison of British and North American taxa included small sample sizes of the North American R. catawbiense and R. maximum. In addition, marker systems applied referred to plastid DNA that is maternally inherited and, thus, does not reflect effects of recombination. Thus, the question as to which mechanism has contributed to this differentiation encountered among the provenances remains a fundamental issue for explaining R. ponticum invasion. In particular, it is still unresolved whether hybridization might have contributed to this differentiation.
The present study aims to address the question of hybridization with North American R. catawbiense and/or R. maximum for explaining the invasive spread of R. ponticum in Ireland on the basis of both molecular marker profiles and quantitative traits of germination success and further ecological and morphological traits. We carried out a field sampling campaign including six populations each from European invasive and native R. ponticum in Ireland, Georgia, and Spain, respectively, and six populations each from North American R. catawbiense and R. maximum to gain seed and leaf material according to a common design and to test for similarities in genetic and quantitative markers. In particular, our aims were to (1) identify patterns of phenotypic trait variation and differentiation among populations within native and introduced Rhododendron taxa and (2) assess the relative contribution of hybridization to explaining the encountered patterns. Against this background, we discuss the evolutionary dimension of increased invasiveness of R. ponticum in Ireland.
Material and Methods
Study objects
All species and origins included in our study belong to the genus Rhododendron, section Pontica, within the Ericaceae family (Milne 2004). Areas of primary distribution of most Rhododendron species are SW China and the Himalayas. However R. ponticum distribution is confined to southwestern Eurasia, whereas R. maximum and R. catawbiense are native to southeastern North America. Natural occurrences of R. ponticum can be found mostly in forests on acid substrate of the eastern Balkan Peninsula, along the Black Sea Coast, and in riparian forests on the Iberian Peninsula, however, populations being in decline in that Iberian part of the native range (Mejías et al. 2007). In its invasive range, in the British Isles, R. ponticum has invaded forests, heathlands and bogs, and requires enormous control efforts and eradication attempts (Dehnen–Schmutz et al. 2004). Rhododendron catawbiense is a typical species of open woodlands and scrub at higher elevation in the Appalachian Mountains, USA. In contrast, R. maximum is preferably found in moist and wet forests in eastern North America. Although there is very little agreement between geographic location and phylogenetic position within the subsection Pontica (Milne 2004), there are nonetheless similarities among these species with respect to morphological traits. Leaf characteristics, for example, leaf apex and leaf base, are quite similar for R. ponticum and R. maximum. In contrast, R. ponticum and R. catawbiense are much more alike in terms of leaf coloring and glabrous twigs (Gleason and Cronquist 1963; Tutin et al. 1972; Clapham et al. 1987; Weakley 2000).
In accordance with the disjunctive distribution, R. ponticum in Turkey and in Georgia (Caucasus) is assigned to ssp. ponticum (Tutin et al. 1972; Davis 1978); whereas occurrences from southern Spain and Portugal are taxonomically addressed as ssp. baeticum (Boiss. & Reuter) Hand.-Mazz. (Tutin et al. 1972; Davis 1978; Clapham et al. 1987; Castroviejo et al. 1993). The taxonomic distinction between the two subspecies is clearly apparent in leaf shape differences: leaves of ssp. ponticum have a length of 12-18-(25) cm and are 2.5–3.5 times as long as wide, while leaves of ssp. baeticum are shorter with 6-12-(16) cm and have a larger length–width ratio of 3–5 than ssp. ponticum.
Material and sampling design
For the experiments and the molecular analyses, we used seed and leaf material from all three species; for R. ponticum, we sampled native subspecies from Georgia (ssp. ponticum) and from southern Spain (ssp. baeticum) as well as invasive provenances from Ireland. For each species and provenance (henceforth called taxa), six populations in the respective area of distribution were chosen randomly with the intention to cover maximum variation. We only included Rhododendron stands within forests at sites with a northern aspect and a slope of 10° to 20° to ensure comparability among the countries. Seeds from native R. ponticum were collected in Georgia (GEO) in August, from native R. ponticum in Spain (ESP) in October, from invasive R. ponticum in Ireland (IRE) in September, from native R. catawbiense (CAM) and R. maximum (MAM) in the Appalachian Mountains in North America in October, all in 2001. The exact locations of all 30 populations (five taxa with six populations each) sampled are provided in Table 1. Further details on the selection mode are described in Erfmeier and Bruelheide (2004).
Table 1.
Overview on populations sampled and location characteristics of the sites
| Taxon | Country | Population | Location | Elevation [m.a.s.l.] | Latitude | Longitude |
|---|---|---|---|---|---|---|
| GEO | Georgia | A | Banis-Khevi | 980 | 41°53′ | E 043°21′ |
| GEO | Georgia | B | Keda-Akutsa | 500 | 41°35′ | E 041°57′ |
| GEO | Georgia | C | Dandalo | 910 | 41°38′ | E 042°07′ |
| GEO | Georgia | D | Botanical Garden, Batumi | 85 | 41°41′ | E 041°43′ |
| GEO | Georgia | E | Djarnali | 175 | 41°33′ | E 041°36′ |
| GEO | Georgia | F | Mtirala | 960 | 41°39′ | E 041°47′ |
| ESP | Spain | G | Garganta de Puerto Oscuro | 790 | 36°30′ | W 005°37′ |
| ESP | Spain | H | Garganta de Passada Llana | 760 | 36°30′ | W 005°35′ |
| ESP | Spain | I | Arroyo del Montero | 660 | 36°29′ | W 005°35′ |
| ESP | Spain | K | Garganta de Enmedio | 445 | 36°32′ | W 005°38′ |
| ESP | Spain | L | Llanos del Juncal | 740 | 36°06′ | W 005°32′ |
| ESP | Spain | M | Rio de la Miel | 430 | 36°06′ | W 005°31′ |
| IRE | Ireland | N | National Park Killarney, Torc Mnts. | 60 | 52°00′ | W 009°30′ |
| IRE | Ireland | O | National Park Killarney, Ladies View | 35 | 51°58′ | W 009°35′ |
| IRE | Ireland | P | Glengariff | 35 | 51°45′ | W 009°33′ |
| IRE | Ireland | Q | Galtee Mnts. | 180 | 52°22′ | W 007°58′ |
| IRE | Ireland | R | Knockmealdown Mnts. | 220 | 52°15′ | W 007°57′ |
| IRE | Ireland | S | Greenan, Wicklow Mnts. | 120 | 52°55′ | W 006°18′ |
| MAM | USA | A | Two Chimneys, Great Smokey Mnts. National Park | 1075 | 35°38′ | W 083°28′ |
| MAM | USA | B | The Sinks, Great Smokey Mnts. National Park | 545 | 35°40′ | W 083°39′ |
| MAM | USA | C | Mingus Mill, Great Smokey Mnts. National Park | 605 | 35°30′ | W 083°19′ |
| MAM | USA | D | Saunakee Village Viewpoint, Blue Ridge Parkway National Park | 1365 | 35°25′ | W 083°02′ |
| MAM | USA | E | Linville Falls, Blue Ridge Parkway National Park | 1035 | 35°56′ | W 081°55′ |
| MAM | USA | F | Green Know, Blue Ridge Parkway National Park | 1390 | 35°42′ | W 082°14′ |
| CAM | USA | A | Two Chimneys, Great Smokey Mnts. National Park | 1120 | 35°38′ | W 083°28′ |
| CAM | USA | B | Mt. Sterling, Great Smokey Mnts. National Park | 1721 | 35°42′ | W 083°06′ |
| CAM | USA | C | Waterrock Knob, Blue Ridge Parkway National Park | 1815 | 35°27′ | W 083°08′ |
| CAM | USA | D | Richland Balsam Summit, Blue Ridge Parkway National Park | 1846 | 35°21′ | W 082°59′ |
| CAM | USA | E | Craggy Gardens, Blue Ridge Parkway National Park | 1725 | 35°41′ | W 082°22′ |
| CAM | USA | F | Grandfather Mountain, Blue Ridge Parkway | 1615 | 36°05′ | W 081°49′ |
GEO = Rhododendron ponticum, Georgia; ESP = R. ponticum, Spain; IRE = R. ponticum, Ireland; MAM = R. maximum, North America; CAM = R. catawbiense, North America.
Within each population, seeds were collected randomly from at least 20 fruiting individuals with a minimum distance of 5 m. Only ripe racemes were harvested. Afterwards released seeds were thoroughly mixed within each population's sample and stored in a dry, dark place at ambient temperatures of 10°C until further use. Accordingly, we collected cuttings in each population for frost hardiness studies on rerooted branches. For sampling and cultivation details, see Erfmeier and Bruelheide (2005). Leaf material for molecular genetic analyses was collected in each population according to a systematic sampling scheme on the basis of a 16 × 16 m grid with a mesh width of 4 m. Samples from the four central individuals each were included in the molecular analyses. Leaf material of all R. ponticum populations was collected in winter 1999/2000; R. maximum and R. catawbiense populations were sampled in autumn 2001. All leaves were dried and stored in silica gel prior to analysis.
Genetic analyses
From silica-dried leaves of a total of 120 individuals (i.e., four individuals per population with six populations per taxon), DNA was isolated with DNA Puregene cell and tissue Kit (Gentra Systems, Minneapolis, MN, USA) using 30–40 mg of dry leaf material. Genotyping was performed using the Amplified Fragment Length Polymorphism (AFLP) technique according to Vos et al. (1995) with modifications as described in Erfmeier and Bruelheide (2011). For each leaf sample, a total of three amplification procedures were run, including each DNA of two separate extractions. Fluorescently labelled PCR products were analyzed on an automated gel sequencer ABI PRISM ® 3100 (Applied Biosystems, Foster City, CA, USA) to infer sample specific fragment patterns. Fragments were analyzed with the software GEN SCANNER as described in Müller et al. (2005). The three replicates per individual were combined, compared, and translated into a 0-1 matrix. For recognition of presence and absence of peaks, in the three parallel analyses, a peak was considered as being present if the occurrence of peaks was provided in two or three of the replicates of this individual. All other peaks were regarded as error.
In order to identify traces of putative introgression, private and common markers were counted for all combinations of taxa. Assuming that hybrids should contain diagnostic alleles from their parent taxa, we counted diagnostic markers of the four potential parent taxa and tested for presence of these specific peaks in the Irish taxon.
Germination experiment
Seeds from each of the six populations per taxon were subjected to three different temperature regimes of 9/19°C, 16/26°C, and 23/33°C (night/day) with a thermo- and photoperiod of 16-h day length. Each 20 seeds were placed in petri dishes on 70 g of sterilized sand (105°C for 24 h), being covered with paper discs (Schleicher & Schuell GmbH, Dassel, Germany; diameter 90 mm), and kept constantly moist with deionized water. Petri dishes were regularly watered and sprayed with 50% ethanol solution twice a week to suppress mildew infection.
Each population was replicated three times at each temperature level, yielding 270 petri dishes being randomly positioned in controlled environment cabinets (Heraeus Vötsch, Vötsch Industrietechnik GmbH, Frommern, Germany). The environment cabinets were equipped with white light providing 120–150 µE/m2/s on average. Germination success was monitored every third day in the beginning and in total 10 times during the whole experimental period of 70 days. Seeds were considered to have germinated when the first radicle had emerged and were transferred at the cotyledon stage to pots (7 × 7 × 8 cm3) with 70%:30% sand–peat substrate for further cultivation.
Growth experiment
Seedlings for the growth experiment were acquired from the germination experiment and from additional seeds sown directly into pots. Seedlings from both cultivation attempts had about 3–4 weeks after germination for establishing in their pots before being exposed to growth treatments. In controlled greenhouse cabinets, the plants were subjected to a daily alternating temperature regime of either 9/19°C, 16/26°C, and 23/33°C and a thermo- and photoperiod of 8/16 (night/day) hours. Each cabinet was split into two layers of different light regimes: with the upper layer having a light treatment of 400 mE/m2/s and the layer underneath experiencing reduced light availability of 40 mE/m2/s. In addition, plants were subjected to two soil water level treatments of 25% and 15% (of dry weight). The combinations of these experimental settings resulted in a total of 12 experimental environments, to which all of the 30 populations available (each six populations by taxon) were assigned to. A total set of 360 individuals were randomly placed (within treatments) in the cabinets.
Water levels of the pots were assessed gravimetrically and readjusted every second day. At 3-week intervals, the pots were fertilized with a 0.25‰ NPK fertilizer (Flory 1, EUFLOR GmbH, München, Germany); after 6 weeks, fertilizer concentration was doubled for all pots to account for the seedling's increase in biomass. The seedling growth experiment ran for about 12 weeks. We studied variables of growth (increase in total plant height, number of leaves), variables of allocation (above- and belowground biomass, shoot–root ratio) and traits of leaf morphology (length and width of the largest leaf). Monitoring of size- and growth-related variables occurred once in the beginning and subsequently every 3 weeks. Biomass data were assessed at the end of the experiment by determining root and shoot dry weight. For variables of increase, we calculated relative growth rates (RGRs) following Hunt (1989).
Frost hardiness
Frost hardiness was determined on cuttings that had been harvested in autumn 2001 of all origins and taxa and that had been cultivated in the greenhouse at temperatures of 12/17°C (night/day) during the winter. For frost hardiness testing, we selected individuals from all populations and taxa at random and kept them for 7 d in a refrigerator of 4°C prior to the experiment for acclimation and hardening. From these individuals, we sampled leaves that still corresponded to plant material that had developed in the field. Exposure to frost treatments of these leaves started in April 2002.
Frost hardiness was quantified using the electrolyte leakage method according to Murray et al. (1989), which relates increasing tissue damage caused by frost to increasing rates of electrolyte loss. The experiment was carried out with 11 2 × 1 cm2 leaf rectangles from each individual, each one being assigned to one of 11 temperature levels applied, thus representing connected samples that allow for calculating nonlinear regressions across all temperature levels for every individual (Kathke and Bruelheide 2011). The five taxa with each six populations were represented by one individual each, yielding a total of 330 leaf samples. Leaf samples were subjected to freezing temperatures in a freezing chamber of –6°C, –9°C, –12°C, –15°C, –18°C, –21°C, –24°C, –27°C, and –30°C, respectively, or to a nonfreezing control temperature (+4°C). We chose a fine resolution of temperature treatments down to –30°C to reveal taxa differentiation more precisely and to grasp the full range of cold hardiness described for R. ponticum in literature (Sakai et al. 1986). Each temperature level lasted for 2 h with a cooling period of 1 h between levels. In addition, one leaf sample per individual was subjected to a liquid N2 (–196°C) treatment for 2 h to obtain maximum frost damage. After experimental frost exposure, leaf samples were put into tubes with 10-mL 3% iso-propanol solution. The electrical conductivity of the solution was measured (LF 2000, WTW) immediately after immersing the disk (C0) and subsequently repeated after 5 h, 19 h, 43 h, and 92 h (Ct). At the end of the measurement series, the solution with the leaf disk was boiled for 20 min, thus releasing all electrolytes from the cells, and final conductivity was measured as individual maximum reference (Cmax).
Each measurement was expressed as relative conductivity (RC) according to Murray et al. (1989) to account for variation in total electrolyte content. All RC values of one time series were fitted to a one-parameter nonlinear regression (proc nlin SAS 9.1, SAS Institute Inc., 2000):
The resulting parameter k indicates the rate of electrolyte leakage and can be used for further analyses as a measure of frost hardiness.
Statistical analysis
The presence/absence matrix of DNA fragments was analyzed with a cluster analysis based on unweighted pair group method with arithmetic average (UPGMA).
Data on growth, biomass, and on leaf morphology in the growth experiment were analyzed with general linear models (GLMs) for unbalanced data (type III sum of squares, proc glm). Prior to analysis, all growth and germination data were rank transformed as the majority of data lacked normal distribution (proc univariate; for appropriateness of rank transformation, see Brunner and Puri 2001; Quinn and Keough 2002). Maximum germination success was tested for effects of the taxon and the temperature applied, both considered fixed factors and with populations as random factor nested within taxon. Effects of taxon and temperature on germination velocity were tested in repeated measures analysis of variance (ANOVA; repeated statement in proc glm) with “time” as additional factor. Since our data did not satisfy the assumption of sphericity, numerator and denominator degrees of freedom were adjusted before determining significance levels according to Greenhouse–Geisser. In all growth analyses, we tested for main and interaction effects of taxon, temperature (both in germination and growth analyses), light and water (in growth analyses) as fixed factors and with populations as random factor nested in taxon (pop(tax)). Post-hoc tests were realized with Ryan–Einot–Gabriel–Welsh (REGWQ) multiple range tests.
For analysis of frost hardiness, we applied the same GLM to test for taxon, temperature, and their interaction effects on the rate of electrolyte leakage k. In addition, all variables were also compared using contrasts of the control temperature of +4°C versus all other temperatures of the freezing treatment separately by taxon (contrast and estimate statements, proc glm, SAS 9.1; SAS Institute Inc. 2000). The temperature at which 50% of the maximum k value occurred (LT50) was calculated by fitting a four-parameter nonlinear regression to the k values (Sigma Plot 9.0, Systat Software 2004) using the following equation:
with k the rate of electrolyte leakage; T the temperature to which the leaf sample was exposed; and a, b, c, and LT50 as regression parameters. LT50 was calculated for each taxon across all populations and temperature replicates.
All phenotypic data were summarized and analyzed by discriminant analysis in order to assign invasive and putative hybrid Irish phenotypes to one or more of the considered parental taxa. We performed a stepwise discriminant analysis (proc stepdisc) to select a subset of significant quantitative variables to discriminate between the four native Georgian, Spanish, and North American taxa. We applied a forward selection procedure, using all variables of growth, morphology, allocation, germination, and frost sensitivity, which consecutively entered the model according to the significance level of an F-test. Forward selection began with no variables in the model and stopped when no further variable could be added at the significance level of P = 0.05. Based on the final set of discriminating variables, we calculated the error probability of incorrect assignment for each of the four classified taxa by means of a discriminant procedure (proc discrim). In the following, the resulting discriminant function was applied to the Irish dataset to assign these invasive populations to either one of the European Georgian or Spanish R. ponticum or to the North American congeneric species R. catawbiense or R. maximum.
Illustration of taxon separation was performed with linear discriminant analysis (LDA) using R 2.10.1 based on the set of discriminating variables only. Multivariate observations were classified with lda (Mass package) and projected onto the first two linear discriminants.
Results
Genetic relationships between Rhododendron taxa
AFLP scores generated 478 polymorphic markers from three primer pairs across all 120 samples analyzed. All individuals had unique fragment combinations, thus, displayed discrete genotypes. Private alleles were rarely encountered: 4.4% of all alleles (i.e., 21 alleles) were present in all individuals, further 19.5% were shared by all populations (i.e., 93 alleles), and 50% were in common in all five taxa studied (i.e., 239 alleles; data not shown). A total of 24 alleles were found to be exclusive, that is, taxon-diagnostic, with 10 diagnostic alleles for R. catawbiense, six alleles each for R. maximum and Georgian R. ponticum, and each one diagnostic allele for Spanish and Irish R. ponticum (see Appendix S1). Inter-taxa comparisons between North American Rhododendron taxa with European R. ponticum taxa displayed only few, exclusively common alleles: while R. catawbiense exclusively shared one allele with Irish R. ponticum, the number of exclusively shared alleles between R. maximum and Irish R. ponticum was three alleles. For the North American taxa, the number of diagnostic markers shared with Georgian and Spanish R. ponticum was at a similar height, with five and three alleles, respectively, for R. catawbiense and with five and one alleles, respectively, for R. maximum.
The level of dissimilarity among individuals was low, ranging from 0.148 to 0.748. UPGMA analyses revealed a clear distinction of the three Rhododendron species (Fig. 1), with the major separation between a North American cluster including R. maximum and R. catawbiense and a European R. ponticum cluster. Within the North American cluster, both species were clearly distinguished with only one single R. maximum individual clustering among R. catawbiense. Within R. ponticum, the Georgian taxa clustered separately from the Iberian and Irish provenances. In contrast, individuals of the Spanish and the Irish provenances were completely intermixed and clustered together.
Figure 1.
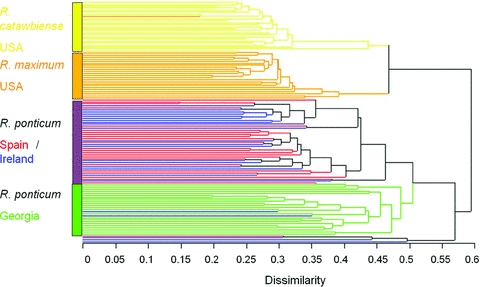
Cluster analyses based on unweighted pair group method with arithmetic average (UPGMA) of AFLP profiles of 24 genotypes each from Rhododendron catawbiense (North America), R. maximum (North America), invasive Irish R. ponticum, native Spanish R. ponticum, and native Georgian R. ponticum.
Germination characteristics
Maximum germination success differed clearly between the Rhododendron taxa tested after 10 weeks of temperature treatments (Table 2A; Fig. 2). We found the highest values for North American R. catawbiense and invasive Irish R. ponticum, which differed significantly from native Spanish R. ponticum. Maximum germination of native Georgian R. ponticum and North American R. maximum was intermediate and neither differed significantly from invasive Irish nor from native Spanish R. ponticum. Germination success was significantly higher at the intermediate temperature level compared to the lowest and the highest temperature levels.
Table 2.
GLM results for germination tests of Rhododendron taxa: (A) Maximum germination success after 10 weeks; Germination data were rank transformed prior to analysis. (B) Cumulative germination success expressed by repeated measures ANOVA with factor time. The mixed model was performed with populations as random factor nested within taxon and three replicates each per temperature level (n = 270). P GG adj = P values for Greenhouse–Geisser adjustment. Bold numbers indicate significant effects
| (A) | ||||||
|---|---|---|---|---|---|---|
| Source of variation | df | Type III SS | MS | F | P | |
| Taxon | 4 | 78136 | 19534 | 3.93 | 0.013 | |
| Error (pop(taxon)). | 25 | 124429 | 4974.3 | |||
| Temperature | 2 | 378100 | 189050 | 45.65 | <0.001 | |
| Temperature × taxon | 8 | 24615 | 3076.9 | 0.74 | 0.654 | |
| Pop(taxon) | 25 | 124365 | 4974.6 | 1.2 | 0.239 | |
| Error | 230 | 952522 | 4141.4 | |||
| (B) | ||||||
|---|---|---|---|---|---|---|
| Source of variation | df | Type III SS | MS | F | P | pGGadj |
| Time | 10 | 58595.2 | 5859.5 | 135.0 | <0.001 | <0.001 |
| Time × temperature | 20 | 30389.1 | 1519.5 | 35.0 | <0.001 | <0.001 |
| Time × taxon | 40 | 6118.3 | 153.0 | 3.5 | <0.001 | <0.001 |
| Time × temperature × taxon | 80 | 3188.6 | 39.9 | 0.9 | 0.681 | 0.543 |
| Time × pop(taxon) | 250 | 12809.7 | 51.2 | 1.2 | 0.034 | 0.203 |
| Error(time) | 2300 | 99813.1 | 43.4 | |||
| Greenhouse–Geisser–Epsilon | 0.185 | |||||
Figure 2.
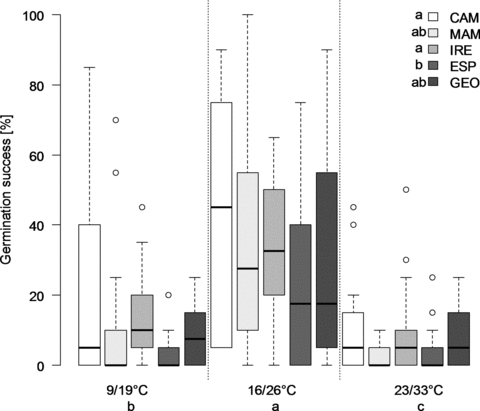
Germination success of Rhododendron taxa's seeds across three temperature regimes (n = 270). Medians, quartiles, minimum, and maximum refer to six populations with three replicates each per temperature (n = 18). Different letters indicate significant differences according to the REGWQ-test. CAM = North American R. catawbiense; MAM = North American R. maximum; IRE = invasive Irish R. ponticum; ESP = native Spanish R. ponticum; GEO = native Georgian R. ponticum. For statistical details referring to rank transformed data, see Table 2.
We found significant taxon effects for germination velocity, which was indicated by significant time × taxon interactions: seeds from R. catawbiense and Irish R. ponticum initially responded much faster to the temperature treatments than seeds from all other taxa (Table 2B). This pattern was consistent for all temperature levels studied, as displayed by nonsignificant time × taxon × temperature interaction effects.
Seedling characteristics
After 12 weeks of the experiment, all variables of growth and performance differed significantly between the five taxa (Table 3). Growth traits most often showed highest magnitude for the invasive Irish R. ponticum taxon, which had the largest relative growth rates in height, in number of leaves as well as in leaf length, and in width (Fig. 3A–C; Table 3). Relative growth rates in height and leaf width did not differ significantly among all R. ponticum taxa (Fig. 3A; Table 3), and for RGR in leaf length (Fig. 3C), both the Spanish and the Irish taxon displayed highest growth rates. Apart from RGR in number of leaves (Fig. 3B), we found a clear dissimilarity in all growth traits for invasive Irish R. ponticum and the American taxon R. catawbiense; the latter consistently displayed significantly lower growth rates than the invasive Irish congeneric R. ponticum.
Table 3.
GLM analysis for traits of (1) growth, (2) leaf morphology, and (3) biomass of R. ponticum seedlings after 12 weeks. All variables were rank transformed prior to analysis. Listed are effects of taxon, temperature treatments, light conditions, water conditions, and their interactions as fixed effects. ANOVA was performed with populations as random factor nested within taxon (n = 360)
| Source of variation | RGR Height | RGR no. of leaves | RGR leaf length | RGR leaf width | Leaf length | Leaf width | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| df | F | P | df | F | P | df | F | P | df | F | P | df | F | P | df | F | P | |
| Taxon | 4 | 7.48 | *** | 4 | 3.84 | * | 4 | 18 | *** | 4 | 8.73 | *** | 4 | 13.8 | *** | 4 | 6.75 | *** |
| Error | 25.8 | 25.8 | 25.7 | 25.9 | 25.6 | 25.6 | ||||||||||||
| Temperature | 2 | 0.47 | 2 | 15.98 | *** | 2 | 102 | *** | 2 | 86.3 | *** | 2 | 77.6 | *** | 2 | 69.8 | *** | |
| Taxon × temperature | 8 | 1.83 | 8 | 2.22 | 8 | 2.8 | ** | 8 | 2.13 | * | 8 | 0.96 | 8 | 1.15 | ||||
| Light | 1 | 987 | *** | 1 | 759.3 | *** | 1 | 759 | *** | 1 | 705 | *** | 1 | 737 | *** | 1 | 743 | *** |
| Taxon × light | 4 | 4.6 | ** | 4 | 0.75 | 4 | 0.2 | 4 | 0.43 | 4 | 2.23 | 4 | 2.36 | |||||
| Temperature × light | 2 | 0.37 | 2 | 5.39 | ** | 2 | 23.2 | *** | 2 | 11.1 | *** | 2 | 19.8 | *** | 2 | 18.5 | *** | |
| Taxon × temperature × light | 8 | 1.02 | 8 | 1.64 | 8 | 4.45 | *** | 8 | 3.23 | ** | 8 | 1.18 | 8 | 1.82 | ||||
| Water | 1 | 1.95 | 1 | 1.24 | 1 | 0.34 | 1 | 3.53 | 1 | 0.02 | 1 | 1.18 | ||||||
| Taxon × water | 4 | 0.54 | 4 | 0.97 | 4 | 0.8 | 4 | 0.38 | 4 | 0.6 | 4 | 0.23 | ||||||
| Temperature × water | 2 | 0.56 | 2 | 0.53 | 2 | 0.97 | 2 | 0.89 | 2 | 0.34 | 2 | 1.68 | ||||||
| Taxon × temperature × water | 8 | 0.76 | 8 | 0.76 | 8 | 1.76 | 8 | 1.48 | 8 | 0.77 | 8 | 1.03 | ||||||
| Light × water | 1 | 0.01 | 1 | 0.41 | 1 | 0.16 | 1 | 0.35 | 1 | 0.05 | 1 | 0.14 | ||||||
| Taxon × light × water | 4 | 0.25 | 4 | 0.16 | 4 | 2.06 | 4 | 3.57 | ** | 4 | 0.67 | 4 | 0.58 | |||||
| Temperature × light × water | 2 | 2.54 | 2 | 0.01 | 2 | 2 | 2 | 2.7 | 2 | 1.21 | 2 | 1.32 | ||||||
| Taxon × temperature × light × water | 8 | 0.48 | 8 | 0.37 | 8 | 1.27 | 8 | 1.03 | 8 | 0.42 | 8 | 0.55 | ||||||
| Pop(taxon) | 25 | 1.94 | ** | 25 | 1.91 | ** | 25 | 1.96 | ** | 25 | 1.64 | * | 25 | 2.43 | *** | 25 | 2.34 | *** |
| Error: MS(error) | 264 | 264 | 264 | 264 | 264 | 264 | ||||||||||||
| Source of variation | Leaf length–width ratio | SLA | Shoot biomass | Root biomass | Shoot–root ratio | |||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| df | F | P | df | F | P | df | F | P | df | F | P | df | F | P | ||||
| Taxon | 4 | 32.8 | *** | 4 | 9.61 | *** | 4 | 6.42 | ** | 4 | 3.45 | * | 4 | 10 | *** | |||
| Error | 25.6 | 27.9 | 25.8 | 25.9 | 27.3 | |||||||||||||
| Temperature | 2 | 37.3 | *** | 2 | 1.24 | 2 | 11.2 | *** | 2 | 8.87 | *** | 2 | 0.68 | |||||
| Taxon × temperature | 8 | 1.26 | 8 | 1 | 8 | 0.89 | 8 | 1.95 | 8 | 4.78 | *** | |||||||
| Light | 1 | 190 | *** | 1 | 120.5 | *** | 1 | 877 | *** | 1 | 721 | *** | 1 | 25.2 | *** | |||
| Taxon × light | 4 | 6.05 | *** | 4 | 3.76 | ** | 4 | 1.04 | 4 | 5.98 | *** | 4 | 11 | *** | ||||
| Temperature × light | 2 | 6.82 | ** | 2 | 19.17 | *** | 2 | 4.63 | * | 2 | 2.9 | 2 | 2.38 | |||||
| Taxon × temperature × light | 8 | 1.42 | 8 | 1.49 | 8 | 1.28 | 8 | 0.5 | 8 | 1.96 | ||||||||
| Water | 1 | 3.62 | 1 | 2.61 | 1 | 2.44 | 1 | 0.02 | 1 | 2.76 | ||||||||
| Taxon × water | 4 | 2.05 | 4 | 0.26 | 4 | 0.61 | 4 | 0.2 | 4 | 1.57 | ||||||||
| Temperature × water | 2 | 2.5 | 2 | 0.5 | 2 | 0.18 | 2 | 1.96 | 2 | 0.93 | ||||||||
| Taxon × temperature × water | 8 | 1.09 | 8 | 0.74 | 8 | 0.12 | 8 | 0.75 | 8 | 1.01 | ||||||||
| Light × water | 1 | 0 | 1 | 0.03 | 1 | 0 | 1 | 0.25 | 1 | 0.01 | ||||||||
| Taxon × light × water | 4 | 0.57 | 4 | 0.27 | 4 | 2.84 | * | 4 | 3.16 | * | 4 | 0.1 | ||||||
| Temperature × light × water | 2 | 1.48 | 2 | 2.2 | 2 | 0.95 | 2 | 0.18 | 2 | 1.06 | ||||||||
| Taxon × temperature × light × water | 8 | 1.27 | 8 | 1.28 | 8 | 0.42 | 8 | 1.09 | 8 | 0.79 | ||||||||
| Pop(taxon) | 25 | 2.42 | *** | 25 | 1.2 | 25 | 2.35 | *** | 25 | 2.5 | *** | 25 | 1.05 | |||||
| Error: MS(error) | 264 | 255 | 262 | 254 | 254 | |||||||||||||
Figure 3.
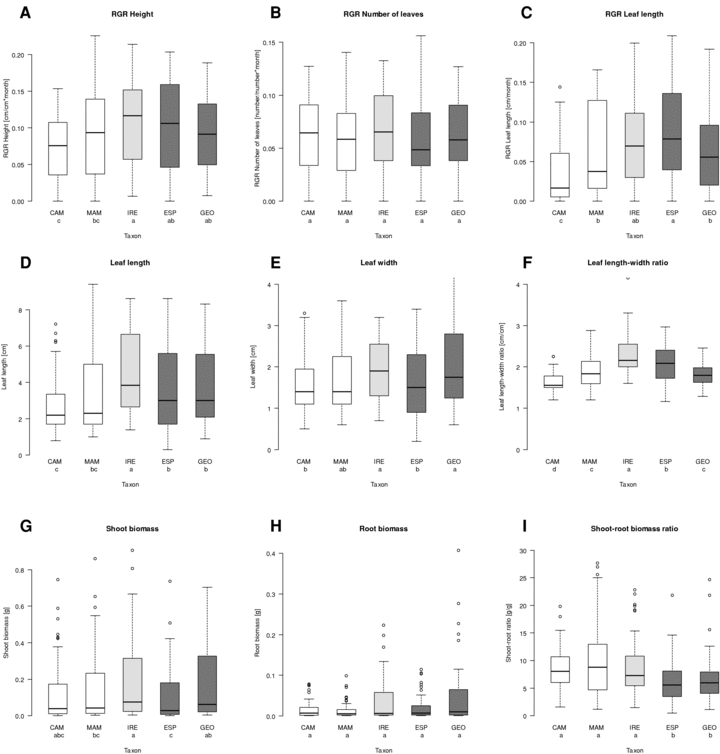
Taxon effects on traits of growth (a–c), leaf morphology (d–f), and biomass allocation (g–i) of R. ponticum seedlings across three temperature, two light, and two water regimes. (a) RGR in height, (b) RGR in number of leaves, (c) RGR in leaf length, (d) Leaf length, (e) Leaf width, (f) Leaf length-width ratio, (g) Shoot dry biomass, (h) Root dry biomass, (i) Shoot-root biomass ratio. Medians, quartiles, minimum, and maximum refer to six populations each as replicates across 12 treatments (n = 72). Different letters indicate significant differences according to the REGWQ-test. CAM = North American R. catawbiense; MAM = North American R. maximum; IRE = invasive Irish R. ponticum; ESP = native Spanish R. ponticum; GEO = native Georgian R. ponticum. For statistical details, see Table 3.
Leaf length and leaf-length-width ratio (Fig. 3D and F) were significantly larger for Irish R. ponticum than for all other taxa. Leaf width (Fig. 3E) and SLA (Table 3) did not differ between the invasive Irish taxon and native R. ponticum from Georgia and R. maximum. American R. catawbiense differed significantly in all leaf traits analyzed from invasive Irish R. ponticum (Table 3).
While the post-hoc test on root biomass revealed no significant differences between taxa (Fig. 3H), highest shoot biomass was encountered for invasive Irish and native Georgian R. ponticum in common with R. catawbiense (Fig. 3G); the Irish taxon displayed significantly higher aboveground biomass than Spanish R. ponticum and R. maximum. The shoot–root biomass ratio (Fig. 3I) was highest for the American Rhododendron taxa together with invasive Irish R. ponticum. All three taxa differed significantly from native R. ponticum taxa.
For most of the variables, we found some interaction effects indicating that the different taxa responded differently to different temperature and light environments, respectively (Table 3). RGR in number of leaves and shoot–root biomass ratio, for example, showed differential increase at warmer temperatures (see Appendix S2): in particular, North American R. catawbiense and R. maximum and the invasive Irish R. ponticum were able to increase growth and shoot–root ratios at warmer temperatures compared to native R. ponticum. Taxa × light interactions for RGR in height indicate that the invasive Irish R. ponticum and its native Spanish conspecific and R. maximum, in particular, seemed to profit from higher light conditions (Table 3; Appendix S2).
Frost hardiness
Tested across all taxa and compared to the control level, the electrolyte leakage rate k was significantly larger at all temperature levels of –18°C or below this value, suggesting general frost hardiness down to at least –15°C (see Appendix S3). The invasive Irish R. ponticum displayed the highest rates of frost damage and differed significantly from all other taxa (Fig. 4; Table 4). Damage was significantly different from the 4°C control level for Irish and Georgian samples at –18°C and for Spanish samples at –24°C, while for the North American taxa, we found no significant differences compared to the control. Frost hardiness expressed as LT50 values following nonlinear four-parameter regression was highest for North American R. catawbiense and R. maximum with LT50 values of –36.9°C and –31.4°C, respectively (Fig. 5). Among the R. ponticum origins, frost hardiness decreased from native Georgian (–24.0°C) to native Spanish (–16.3°C) and invasive Irish (−14.3°C) populations.
Figure 4.
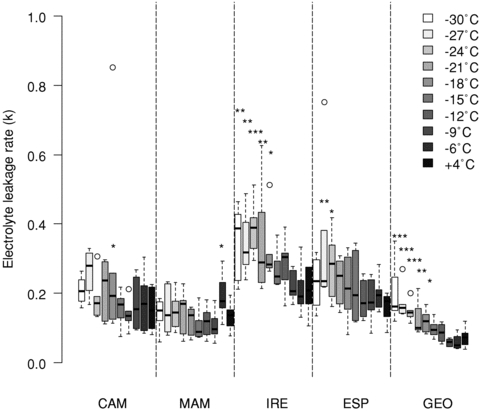
Electrolyte leakage rate k for the Rhododendron taxa at 10 different temperature levels. CAM = North American R. catawbiense; MAM = North American R. maximum; IRE = invasive Irish R. ponticum; ESP = native Spanish R. ponticum; GEO = native Georgian R. ponticum. Stars indicate significant differences according to contrasts between the control temperature of +4°C versus all other temperatures within each taxon. *P < 0.05, **P <0.01, ***P < 0.001.
Table 4.
Summary of GLM analysis for frost sensitivity (rate of electrolyte leakage k) of Rhododendron taxa. Listed are effects of taxon, temperature treatments, and their interactions as fixed effects. ANOVA was performed with populations as random factor nested within country (n = 330). Bold numbers indicate significant effects
| Source of variation | df | Type III SS | MS | F | P |
|---|---|---|---|---|---|
| Taxon | 4 | 1.237446 | 0.309361 | 20 | <0.001 |
| Error (pop(taxon)) | 25 | 0.386344 | 0.015454 | ||
| Temperature | 9 | 0.409223 | 0.045469 | 8.04 | <0.001 |
| Taxon × temperature | 36 | 0.267416 | 0.007428 | 1.31 | 0.121 |
| Pop (taxon) | 25 | 0.386344 | 0.015454 | 2.73 | <0.001 |
| Error | 225 | 1.272515 | 0.005656 |
Figure 5.
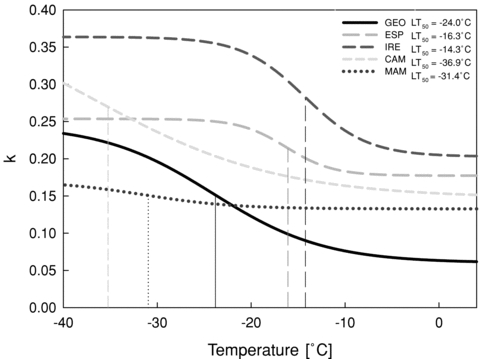
Estimation of the LT50 values for all Rhododendron taxa referring to k values and the corresponding temperature levels of the freezing treatment. Regression follows a four-parameter sigmoid function k = c+a/(1+exp(–(T–LT50)/b)), where T is the temperature to which the leaves were exposed and a, b, c, and LT50 are regression parameters. The regression was applied to pooled data per taxon (n = 60). CAM = North American R. catawbiense; MAM = North American R. maximum; IRE = invasive Irish R. ponticum; ESP = native Spanish R. ponticum; GEO = native Georgian R. ponticum. All regressions were significant (P < 0.001) with the exception of MAM. Vertical lines mark the LT50 values as position of the inflection point on the x-axis.
Assignment of invasive populations
Stepwise discriminant analysis revealed the variables RGR leaf length (F = 28.09, P < 0.001), leaf length–width ratio (F = 11.80, P < 0.001), root biomass (F = 9.14, P < 0.001), and shoot–root ratio (F = 4.74, P = 0.015) to discriminate significantly among the native Rhododendron taxa. The overall probability of misclassification was 8.3% (Table 5). Only two of the six native Georgian R. ponticum populations were misclassified as R. maximum; for all other populations of taxa, posterior probability of membership in the correct taxon was 100% (Table 5). Application of this discriminant function to the Irish dataset revealed 100% classification of the invasive Irish populations to the native Spanish congeneric ones (Table 5). Invasive Irish R. ponticum differed considerably from both North American taxa as well as from Georgian R. ponticum, and displayed closest relationship to the Spanish R. ponticum (Fig. 6).
Table 5.
Classification summary following linear discriminant analysis for calibration of the native taxa CAM, MAM, ESP and GEO, and the test taxon IRE. Number of observations and percent classified into taxon
| From tax | CAM | MAM | ESP | GEO | Total |
|---|---|---|---|---|---|
| CAM | 6 | 0 | 0 | 0 | 6 |
| 100% | 0 | 0 | 0 | 100% | |
| MAM | 0 | 6 | 0 | 0 | 6 |
| 0 | 100% | 0 | 0 | 100% | |
| ESP | 0 | 0 | 6 | 0 | 6 |
| 0 | 0 | 100% | 0 | 100% | |
| GEO | 0 | 2 | 0 | 4 | 6 |
| 0 | 33.3% | 0 | 66.7% | 100% | |
| Total | 6 | 8 | 6 | 4 | 24 |
| 25 | 33.3 | 25 | 16.7 | 1.0 | |
| Priors | 0.25 | 0.25 | 0.25 | 0.25 | |
| Error counts estimates for taxon | 0.083 |
| From tax | CAM | MAM | ESP | GEO | Total |
|---|---|---|---|---|---|
| IRE | 0 | 0 | 6 | 0 | 6 |
| 0 | 0 | 100% | 0 | 100% |
CAM = North American Rhododendron catawbiense; MAM = North American R. maximum; IRE = invasive Irish R. ponticum; ESP = native Spanish R. ponticum; GEO = native Georgian R. ponticum.
Figure 6.
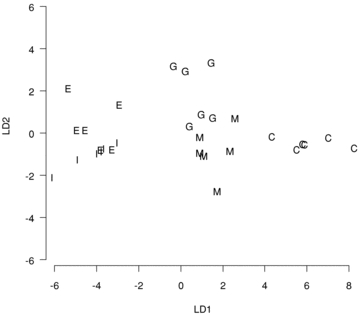
Ordination plot of the first and second linear discriminant functions of the six populations per Rhododendron taxon based on the discriminating variables RGR leaf length, leaf length–width ratio, root biomass, and shoot–root ratio. Proportion of explained variance for axis LD1 = 87.76%, LD2 = 6.72%. C = North American R. catawbiense; M = North American R. maximum; I = invasive Irish R. ponticum; E = native Spanish R. ponticum; G = native Georgian R. ponticum.
Discussion
In the present study, neither ecological nor morphological traits bore a general resemblance of Irish R. ponticum with North American R. catawbiense or R. maximum as reflected, in summary, in the discriminant analysis. Consistently, the AFLP data displayed a clear distinction between all R. ponticum taxa on the one hand and the two North American species on the other hand. In contrast, the present findings confirm the previously encountered high phenotypic similarity between Spanish and Irish R. ponticum (Erfmeier and Bruelheide 2005, 2010) as well as the Iberian provenance of invasive occurrences (Milne and Abbott 2000; Erfmeier and Bruelheide 2011) and, thus, suggest that mechanisms alternative to hybridization have to be considered.
Introgression and invasion
At the molecular level, we have to completely reject the hypothesis of introgression, since diagnostic marker analysis and cluster analysis displayed no evidence of gene transfer across species. Our present analyses suggest that hypotheses based on hybridization in invasive R. ponticum (Cox 1979; Milne and Abbott 2000) do not apply for Ireland. In particular, Milne and Abbott (2000) found very convincing support for this hypothesis based on cpDNA and rDNA analyses conducted on 260 accessions throughout the British Isles. In their analysis, 27 of their accessions had been introgressed with genetic material from R. catawbiense and two further accessions displayed introgression with R. maximum material. In their study, the sampled R. catawbiense material was much more frequently encountered within Scotland than in the rest of the British Isles. However, Milne and Abbott (2000) found no evidence of introgression in the 29 accessions sampled at two locations in Ireland, which is, thus, in agreement with our data for that particular region in Ireland. Milne and Abbott (2000) suggested that the higher amount of introgression encountered in eastern Scotland might reflect a higher required cold hardiness than in other regions of the British Isles. They concluded that the increased amount of introgression might be the result of either directional horticultural selection of hardy hybrids or of natural selection after naturalization favoring individuals with higher levels of introgression from R. catawbiense in cold regions. Increased frost hardiness has repeatedly been invoked as an example of introgression preceding invasiveness (Cox 1979; Ellstrand and Schierenbeck 2000). In their survey on cold hardiness in the genus Rhododendron, Sakai et al. (1986) described that the majority of the hardiest rhododendrons belongs to the Ponticum series. On the basis of visual assessment of leaf damage after experimental frost treatment and in accordance with our results, Sakai et al. (1986) determined lowest survival temperatures of –60°C for both R. catawbiense and R. maximum, thus, ranking about the hardiest species in the genus, whereas R. ponticum leaves survived down to –35°C, still showing considerable frost hardiness. However, in our study, data on frost hardiness, in full accordance with the genetic data, do not support the idea of frost gene introgression into Irish populations. Given the mild temperatures and the lack of frequent winter frost events in southern Ireland (see Erfmeier and Bruelheide 2010), the need of an increased frost resistance in that part of the range does not seem to have an adaptive value. Without regularly haunting frost events the effort of maintaining frost resistance does not make sense from an evolutionary point of view (Agrawal et al. 2004). In contrast, increased frost hardiness of invasive R. ponticum in the northern part of the British range as assumed by Milne and Abbott (2000) resulting from directional selection and, possibly, also including introgression, is still a probable scenario that calls for a concerted testing of both introgression by means of nuclear markers and frost hardiness by means of experimental determination on a large-scale sample of populations across the British Isles.
Trait similarities and divergences across taxa
Despite a lack of evidence of hybridization with R. catawbiense and R. maximum at the molecular level, we revealed higher similarities in some of the phenotypic responses between the invasive Irish R. ponticum and these North American species. In many cases, molecular markers do not necessarily reflect morphological traits or allozyme data (Hardig et al. 2000; Triest et al. 2000; Allendorf et al. 2001; Triest 2001). De Cock et al. (2003), for example, found phenotypical distinction of the hybrid taxon Salix rubens var. basfordiana from its parental taxon S. alba in morphological traits, although AFLP fingerprints failed to distinguish these groups. In particular, maximum germination as well as germination velocity of invasive Irish R. ponticum differed significantly from both conspecifics but resembled very much North American R. catawbiense. The most probable explanation for the similarities in germination features encountered in the present study is that similar but independent processes or chance effects have caused the observed genetic shifts. For invasive R. ponticum, a genetic shift in germination has been suggested before as adaptation to a more reliable environment in Ireland, lacking extreme drought or frost events during establishment in springtime, if compared to native sites in Georgia and Spain (Erfmeier and Bruelheide 2005). The relevance of differences in germination timing as trait of invasion was also demonstrated for other invasive plant species both in comparison of native and invasive species (Perglovà et al. 2009) as well as in comparisons of native and invasive populations (Kudoh et al. 2007; Hierro et al. 2009; Beckmann et al. 2011). These differences in germination traits have been shown to be heritable and to affect fitness (Leger et al. 2009), and also to be adaptive. For R. ponticum, a more precautious germination is of evolutionary advantage in cold regions of the native Caucasus range or under dry conditions on the Iberian Peninsula and might have been lost because of a relaxed selection pressure. Such a process can be enhanced by populations of hitherto separated origins that meet and mix (Lavergne and Molofsky 2007; Dlugosch and Parker 2008). In Irish R. ponticum populations, a high genetic diversity was shown to be maintained (Erfmeier and Bruelheide 2011), thus, these populations did not suffer from bottleneck effects but represent melting pot situations of increased genetic exchange. Verhoeven et al. (2011) pointed out that the benefit of admixtures is higher in the new range than at home sites because of a change in selection regime. In their review, the authors emphasize the importance of the shifted balance in costs and benefits of admixtures, and they consult a heterosis benefit as fitness boost in newly admixed populations contributing to an increased colonization success. Irrespective of inter-specific hybridization, such a heterosis effect based on admixed populations alone can account for the observed superior fitness of invasive populations (Keller and Taylor 2010; Verhoeven et al. 2011).
Relaxed selection regimes, probably supported by among population heterosis, would also explain the allocation and increased growth patterns found for Irish R. ponticum. The higher proportional investment in aboveground biomass is a beneficial strategy in a new, benign environment and allows for a more efficient occupancy of space, conferring a superior fitness in the face of novel habitats (Arnold and Hodges 1995). However, besides favorable abiotic conditions, the underlying cause of evolution of increased growth can also be an adaptation to open, noncompetitive environments. Blumenthal and Hubauer (2007) argue that such noncompetitive environments tend to select for traits such as rapid growth and high reproductive allocation in high resource environments. A release from competitive stress might also apply to Irish R. ponticum seedlings, indicated by an increased relative investment in shoot compared to root biomass. However, deciding about the relative importance of these explanations would require experimental testing of native and invasive R. ponticum origins in settings with different intensity levels of nutrient availability and competition.
Conclusions on invasiveness
In the present study, we found no evidence that introgressive hybridization was involved in the evolution of invasiveness in Irish populations. In contrast, the study confirms the invasiveness due to increased germination and effective aboveground growth of Irish R. ponticum, suggesting that the observed phenotypic differentiation between taxa must be attributed to other driving factors. Both molecular analyses and trait analyses of seedlings revealed a 100% congruence between Irish and Spanish R. ponticum populations. Given the common genetic basis of Irish R. ponticum with its Spanish ancestors and the only moderate differentiation between Irish and Spanish seedlings in growth characteristics, the Iberian genotypes might have a similar potential of becoming invasive, too, once appropriate environmental conditions are being provided.
Acknowledgments
The authors thank authorities and staff of the Georgian Academy of Sciences, Botanical Garden Batumi (Georgia), Los Alcornorcales Natural Park (Spain), Killarney and Wicklow National Parks (Ireland), and Blueridge Parkway National Park Headquarters (USA) for generous assistance in acquisition of plant and seed material. C. Meinen, M. Zopf, and U. Jandt provided valuable support in data collection. We acknowledge valuable comments by three anonymous reviewers on an earlier version of this manuscript. The project was supported by a grant of the German Research Foundation DFG (BR 1698/3).
Supporting Information
Additional Supporting Information may be found online on Wiley Online Library.
Appendix S1: Overview on diagnostic AFLP markers.
Appendix S2: Taxon effects of Rhododendron ponticum seedlings at three temperature levels.
Appendix S3: Overall temperature contrasts for frost hardiness (leakage rate k) of Rhododendron taxa.
Please note:Wiley-Blackwell is not responsible for the content or functionality of any supporting materials supplied by the authors. Any queries (other than missing material) should be directed to the corresponding author for the article.
References
- Abbott RJ, James JK, Milne RI, Gillies ACM. Plant introductions, hybridization and gene flow. Philos. Trans. R. Soc. Lond. B. 2003;358:1123–1132. doi: 10.1098/rstb.2003.1289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abbott RJ. Plant invasions, interspecific hybridization and the evolution of new plant taxa. Trends Ecol. Evol. 1992;7:401–405. doi: 10.1016/0169-5347(92)90020-C. [DOI] [PubMed] [Google Scholar]
- Agrawal AA, Conner JK, Stinchcombe JR. Evolution of plant resistance and tolerance to frost damage. Ecol. Lett. 2004;7:1199–1208. [Google Scholar]
- Allendorf FW, Leary RF, Spruell P, Wenburg JK. The problems with hybrids: setting conservation guidelines. Trends Ecol. Evol. 2001;16:613–622. [Google Scholar]
- Arnold ML, Hodges SA. Are natural hybrids fit or unfit relative to their parents? Trends Ecol. Evol. 1995;10:67–71.. doi: 10.1016/S0169-5347(00)88979-X. [DOI] [PubMed] [Google Scholar]
- Barney JN, Whitlow TH, DiTommaso A. Evolution of an invasive phenotype: shift to belowground dominance and enhanced competitive ability in the introduced range. Plant Ecol. 2009;202:275–284. [Google Scholar]
- Barrett SCH, Colautti R, Eckert CG. Plant reproductive systems and evolution during biological invasion. Mol. Ecol. 2008;17:373–383. doi: 10.1111/j.1365-294X.2007.03503.x. [DOI] [PubMed] [Google Scholar]
- Beckmann M, Erfmeier A, Bruelheide H. A comparison of native and invasive populations of three clonal plant species in Germany and New Zealand. J. Biogeogr. 2009;36:865–878. [Google Scholar]
- Beckmann M, Bruelheide H, Erfmeier A. Germination responses of three grassland species differ between native and invasive origins. Ecol. Res. 2011;26:763–771. [Google Scholar]
- Blossey B, Nötzold R. Evolution of increased competitive ability in invasive non-indigenous plants: a hypothesis. J. Ecol. 1995;83:887–889. [Google Scholar]
- Blum MJ, Bando KJ, Katz M, Strong DR. Geographic structure, genetic diversity and source tracking of Spartina alterniflora. J. Biogeogr. 2007;34:2055–2069. [Google Scholar]
- Blumenthal D, Hufbauer RA. Increased plant size in exotic populations: a common-garden test with 14 species. Ecology. 2007;88:2758–2765. doi: 10.1890/06-2115.1. [DOI] [PubMed] [Google Scholar]
- Bossdorf O, Auge H, Lafuma L, Rogers WE, Sieman E, Prati D. Phenotypic and genetic differentiation between native and introduced plant populations. Oecologia. 2005;144:1–11. doi: 10.1007/s00442-005-0070-z. [DOI] [PubMed] [Google Scholar]
- Brunner E, Puri ML. Nonparametric methods in factorial designs. Stat. Papers. 2001;42:1–52. [Google Scholar]
- Campbell LG, Snow AA, Ridley CE. Weed evolution after crop gene introgression: greater survival and fecundity of hybrids in a new environment. Ecol. Lett. 2006;9:198–1209. doi: 10.1111/j.1461-0248.2006.00974.x. [DOI] [PubMed] [Google Scholar]
- Castroviejo S, et al. Flora Iberica. Plantas vasculares de la península Ibérica y Islas Baleares. Vol. IV Cruciferae - Monotropaceae. Madrid, Spain: Real Jardín Botánico; 1993. [Google Scholar]
- Clapham AR, Tutin TG, Moore DM. Flora of the British isles. Cambridge, U.K: Cambridge Univ. Press; 1987. [Google Scholar]
- Cox PA. The larger species of Rhododendron. London: BT Batsford Ltd; 1979. [Google Scholar]
- Cross R. Biological Flora of the British Isles. Rhododendron ponticum L. J. Ecol. 1975;63:345–364. [Google Scholar]
- Culley TM, Hardiman NA. The role of intraspecific hybridization in the evolution of invasiveness: a case study of the ornamental pear tree Pyrus calleryana. Biol. Invasions. 2008;11:1107–1119. [Google Scholar]
- Davis PH. Flora of Turkey and the East Aegean Islands. Edinburgh, U.K: Edinburgh Univ. Press; 1978. [Google Scholar]
- Cock De, Lybeer B, Mijnsbrugge KVander, Zwaenepoel A, Peteghem PVan, Quataert P, Breyne P, Goetghebeur P, Slycken JVan. Diversity of the willow complex Salix alba–S. x rubens–S. fragilis. Silvae Genet. 2003;52:48–153. [Google Scholar]
- Dehnen-Schmutz K, Williamson M. Rhododendron ponticum in Britain and Ireland: social, economic and ecological factors in its successful invasion. Environ. Hist. 2006;12:325–350. [Google Scholar]
- Dehnen-Schmutz K, Perrings C, Williamson M. Controlling Rhododendron ponticum in the British Isles: an economic analysis. J. Environ. Manag. 2004;70:323–332. doi: 10.1016/j.jenvman.2003.12.009. [DOI] [PubMed] [Google Scholar]
- Dlugosch KM, Parker IM. Founding effects in species invasions: genetic variation, adaptive evolution, and the role of multiple introductions. Mol. Ecol. 2008;17:431–449. doi: 10.1111/j.1365-294X.2007.03538.x. [DOI] [PubMed] [Google Scholar]
- Ellstrand NC, Schierenbeck KA. Hybridization as a stimulus for the evolution of invasiveness in plants? Proc. Natl. Soc. U S A. 2000;97:7043–7050. doi: 10.1073/pnas.97.13.7043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elton CS. The ecology of invasions by animals and plants. London: Methuen; 1958. [Google Scholar]
- Erfmeier A, Bruelheide H. Comparison of native and invasive Rhododendron ponticum populations: growth, reproduction and morphology under field conditions. Flora. 2004;199:120–133. [Google Scholar]
- Erfmeier A, Bruelheide H. Invasive and native Rhododendron ponticum populations: is there evidence for genotypic differences in germination and growth? Ecography. 2005;28:417–428.. [Google Scholar]
- Erfmeier A, Bruelheide H. Invasibility or invasiveness? Effects of habitat, genotype, and their interaction on invasive Rhododendron ponticum populations. Biol. Invasions. 2010;12:657–676. [Google Scholar]
- Erfmeier A, Bruelheide H. Maintenance of high genetic diversity during invasion of Rhododendron ponticum. Int. J. Plant Sci. 2011;172:795–806. [Google Scholar]
- Gleason HA, Cronquist A. Manual of vascular plants of Northeastern United States and adjacent Canada. New York: D. VanNostrand Co., Inc.; 1963. [Google Scholar]
- Hardig TM, Brunsfeld SJ, Fritz RS, Morgan M, Orians CM. Morphological and molecular evidence for hybridization and introgression in a willow (Salix) hybrid zone. Mol. Ecol. 2000;9:9–24. doi: 10.1046/j.1365-294x.2000.00757.x. [DOI] [PubMed] [Google Scholar]
- Hedrick PW. Genetics of populations. Sudbury, MA: Jones and Bartlett Publishers; 2005. [Google Scholar]
- Hierro JL, Eren Ö, Khetsuriani L, Diaconu A, Török K, Montesinos D, Andonian K, Kikodze D, Janoian L, Villareal D, et al. Germination responses of an invasive species in native and non-native ranges. Oikos. 2009;118:529–538. [Google Scholar]
- Hooftmann DAP, Oostermeijer JGB, Jacobs MM, den Nijs HCM. Demographic vital rates determine the performance advantage of crop–wild hybrids in lettuce. J. Appl. Ecol. 2005;42:1086–1095. [Google Scholar]
- Hunt R. Basic growth analysis. London: Unwin Hyman; 1989. [Google Scholar]
- James JK, Abbott RJ. Recent, allopatric, homoploid hybrid speciation: the origin of Senecio squalidus (Asteraceae) in the British Isles from a hybrid zone on Mount Etna, Sicily. Evolution. 2005;59:2533–2547. [PubMed] [Google Scholar]
- Kathke S, Bruelheide H. Differences in frost hardiness of two Norway spruce morphotypes growing at Mt. Brocken, Germany. Flora. 2011;206:120–126. [Google Scholar]
- Keller SR, Taylor DR. Genomic admixture increases fitness during a biological invasion. J. Evol. Biol. 2010;23:1720–1731. doi: 10.1111/j.1420-9101.2010.02037.x. [DOI] [PubMed] [Google Scholar]
- Kudoh H, Nakayama M, Lihová J, Marhold K. Does invasion involve alternation of germination requirements? A comparative study between native and introduced strains of an annual Brassicaceae, Cardamine hirsuta. Ecol. Res. 2007;22:869–875. [Google Scholar]
- Lambrinos JG. How interactions between ecology and evolution influence contemporary invasion dynamics. Ecology. 2004;85:2061–2070. [Google Scholar]
- Lavergne S, Molofsky J. Increased genetic variation and evolutionary potential drive the success of an invasive grass. Proc. Natl. Acad. Sci. U S A. 2007;104:3883–3888. doi: 10.1073/pnas.0607324104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee CE. Evolutionary genetics of invasive species. Trends Ecol. Evol. 2002;17:386–391. [Google Scholar]
- Leger EA, Rice KJ. Invasive California poppies (Eschscholzia californica Cham.) grow larger than native individuals under reduced competition. Ecol. Lett. 2003;6:257–264. [Google Scholar]
- Leger EA, Espeland EK, Merrill KR, Meyer SE. Genetic variation and local adaptation at a cheatgrass (Bromus tectorum) invasion edge in western Nevada. Mol. Ecol. 2009;18:4366–4379. doi: 10.1111/j.1365-294X.2009.04357.x. [DOI] [PubMed] [Google Scholar]
- Maron JL, Vilà M, Bommarco R, Elmendorf S, Beardsley P. Rapid evolution of an invasive plant. Ecol. Monogr. 2004;74:261–280.. [Google Scholar]
- Mejías JA, Arroyo J, Marañon T. Ecology and biogeography of plant communities associated with the post Plio-Pleistocene relict Rhododendron ponticum subsp. baeticum in southern Spain. J. Biogeogr. 2007;34:456–472. [Google Scholar]
- Milne RI, Abbott RJ. Origin and evolution of invasive naturalized material of Rhododendron ponticum L. in the British Isles. Mol. Ecol. 2000;9:541–556. doi: 10.1046/j.1365-294x.2000.00906.x. [DOI] [PubMed] [Google Scholar]
- Milne RI. Phylogeny and biogeography of Rhododendron subsection Pontica, a group with a tertiary relict distribution. Mol. Phylogenet. Evol. 2004;33:389–401. doi: 10.1016/j.ympev.2004.06.009. [DOI] [PubMed] [Google Scholar]
- Mooney HA, Cleland EE. The evolutionary impact of invasive species. Proc. Natl. Acad. Sci. U S A. 2001;98:5446–5451. doi: 10.1073/pnas.091093398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murray MB, Cape JN, Fowler D. Quantification of frost damage in plant tissues by rates of electrolyte leakage. New Phytol. 1989;113:307–311. doi: 10.1111/j.1469-8137.1989.tb02408.x. [DOI] [PubMed] [Google Scholar]
- Müller J, Friedl T, Hepperle D, Lorenz M. Distinction between multiple isolates of Chlorella vulgaris (Chlorophyta, Trebouxiophyceae) and testing for conspecificity using amplified fragment length polymorphism and its rDNA sequences. J. Phycol. 2005;41:1236–1247. [Google Scholar]
- Müller-Schärer H, Schaffner U, Steinger T. Evolution in invasive plants: implications for biological control. Trends Ecol. Evol. 2004;19:417–422. doi: 10.1016/j.tree.2004.05.010. [DOI] [PubMed] [Google Scholar]
- Perglovà I, Pergl J, Skálová H, Moravková L, Jarošík V, Pyšek P. Differences in germination and seedling establishment of alien and native Impatiens species. Preslia. 2009;81:357–375. [Google Scholar]
- Prentis PJ, Wilson JRU, Dormontt EE, Richardson DM, Lowe AJ. Adaptive evolution in invasive species. Trends Plant Sci. 2008;13:288–294. doi: 10.1016/j.tplants.2008.03.004. [DOI] [PubMed] [Google Scholar]
- Quinn GP, Keough MJ. Experimental design and data analysis for biologists. Cambridge, U.K: Cambridge Univ. Press; 2002. [Google Scholar]
- Ridley CE, Ellstrand NC. Evolution of enhanced reproduction in the hybrid-derived invasive, California wild radish? Biol. Invasions. 2009;11:2251–2264. [Google Scholar]
- Rieseberg LH, Kim S-C, Randell RA, Whitney KD, Gross BL, Lexer C, Clay K. Hybridization and the colonization of novel habitats by annual sunflowers. Genetica. 2007;129:149–165. doi: 10.1007/s10709-006-9011-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ross CA, Auge H. Invasive Mahonia plants outgrow their native relatives. Plant Ecol. 2008;199:21–31. [Google Scholar]
- Sakai A, Fuchigami L, Weiser CJ. Cold hardiness in the genus Rhododendron. J. Am. Soc. Hortic. Sci. 1986;111:273–280. [Google Scholar]
- SAS Institute. SAS procedures guide. Cary, North Carolina, USA: 2000. [Google Scholar]
- Schierenbeck KA, Ellstrand NC. Hybridization and the evolution of invasiveness in plants and other organisms. Biol. Invasions. 2009;11:1093–1105. [Google Scholar]
- Schweitzer JA, Martinsen GD, Whitham TG. Cottonwood hybrids gain fitness traits of both parents: a mechanism for their long-term persistence? Am. J. Bot. 2002;89:981–990. doi: 10.3732/ajb.89.6.981. [DOI] [PubMed] [Google Scholar]
- Stockwell CA, Hendry AP, Kinnison MT. Contemporary evolution meets conservation biology. Trends Ecol. Evol. 2003;18:94–101. [Google Scholar]
- Triest L, Greef Bde, Bondt Rde, Slycken JVan. RAPD of controlled crosses and clones from the field suggests that hybrids are rare in the Salix alba-Salix fragilis complex. Heredity. 2000;84:555–563. doi: 10.1046/j.1365-2540.2000.00712.x. [DOI] [PubMed] [Google Scholar]
- Triest L. Hybridization in staminate and pistillate Salix alba and S. fragilis (Salicaceae): morphology versus RAPDs. Plant Syst. Evol. 2001;226:143–154. [Google Scholar]
- Tutin TG, Heywood JS, Burges NA, Moore DM, Valentine DH, Walters SM, Webb DA. Flora Europaea. 3: Diapensiaceae to Myoporaceae. Cambridge, U.K: Cambridge Univ. Press; 1972. [Google Scholar]
- Verhoeven KJF, Macel M, Wolfe LM, Biere A. Population admixture, biological invasions and the balance between local adaptation and inbreeding depression. Proc. R. Soc. B. 2011;278:2–8. doi: 10.1098/rspb.2010.1272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vilà M, Weber E, D'Antonio CM. Conservation implications of invasion by plant hybridization. Biol. Invasions. 2000;2:207–217. [Google Scholar]
- Vos P, Hogers R, Bleeker M, Reijans M, der Lee Tvan, Hornes M, Frijters A, Pot J, Peleman J, Kuiper M, Zabeau M. AFLP: a new technique for DNA fingerprinting. Nucleic Acids Res. 1995;23:4407–4414. doi: 10.1093/nar/23.21.4407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weakley . Flora of the Carolinas and Virginia. Working draft of 3 May 2000. Chapel Hill, NC: Univ. of North Carolina; 2000. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


