Abstract
We are never alone. Humans coexist with diverse microbial species that live within and upon us—our so-called microbiota. It is now clear that this microbial community is essentially another organ that plays a fundamental role in human physiology and disease. Basic and translational research efforts have begun to focus on deciphering mechanisms of microbiome function—and learning how to manipulate it to benefit human health. In this Perspective, we discuss therapeutic opportunities in the human microbiome.
WHY FOCUS ON THE MICROBIOME?
Humans have coevolved with a vast number of microbes—our microbiota—that live within and upon us. Recent discoveries make clear that our microbiota is more like an organ than an accessory: These microbes are not just key contributors to human health but a fundamental component of human physiology. Several human ailments have been closely linked to (such as the inflammatory bowel diseases and obesity) or are thought to be influenced by (such as asthma and type I diabetes) the composition of the gut microbial community (1–6). Like a polymorphism in the human genome, a change in the human microbiome—the combined genomes of the bacterial species in our microbiota—can lead to a new phenotype that causes a disease or contributes to its progression. Unlike the human genome, the microbiome is a “flow reactor” of genes: Its composition is dynamic, and we can (in principle) exert control over the content and architecture of our microbial communities. In the coming years, the plasticity of the microbiome will be harnessed by a new category of therapeutics that target the microbiota and modulate microbe-microbe and microbe-host interactions (Fig. 1).
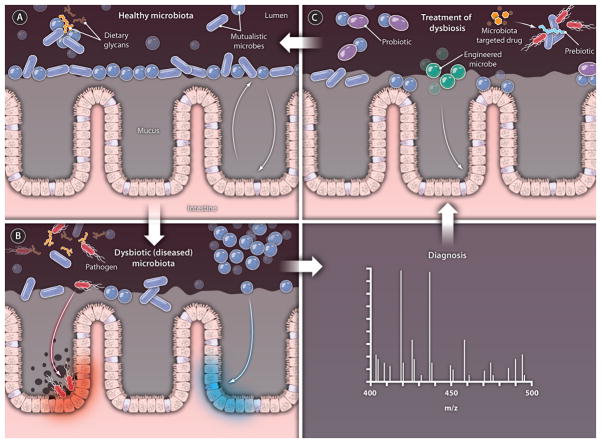
Fig. 1. Tipping the balance.
Many factors can cause a shift in the microbiota from (A) a healthy state to (B) a dysbiotic or diseased state. Detection of small molecules that serve as reporters of community membership and function can be used to diagnose the perturbed community and to determine the appropriate treatment (C). In the healthy state (A), a dense population of microbes inhabits the intestine. Resident microbes (tktk) compete for nutrients [such as dietary polysaccharides] (tktk), and an ecologically stable state is maintained through a complex and poorly understood network of host-host and host-microbe interactions. As shown in (B), diverse factors contribute to community disruption and disease, such as pathogen emergence, other alterations in the microbiota composition, and changes in microbiota function and interaction with the mucosa. Treatment of dysbiosis (C) can be accomplished with probiotics (tktk), prebiotics (tktk), microbes that confer gain-of-function, and microbiota-targeted drugs.
CREDIT: C. BICKEL/SCIENCE TRANSLATIONAL MEDICINE
Several early conclusions have emerged from studies that enumerate intra- and inter-person differences among microbiomes: (i) Typical human-associated communities harbor tens to hundreds of prominent species; (ii) communities from the same body site on different people are more similar than communities from different body sites on the same person; (iii) despite these similarities, when it comes to gut communities the number of microbial species that are unique to an individual far exceeds the number that is shared from person to person; and (iv) monozygotic twins or members of the same household (related or unrelated) have more closely related gut communities than people who are not related by genetic or environmental factors (2, 7). Little is known about how these patterns map to differences in the gene roster and how differences in the gene roster influence community function. However, the plasticity, relevance to human health, and individualized nature of the human microbiome ensure its incorporation into the coming era of personalized medicine. This Perspective focuses on three future goals of microbiome-focused therapeutics research: altering the composition of the community, endowing it with new functions, and using the microbiota to diagnose disease.
TWEAKING THE COMMUNITY COMPOSITION
Human-associated microbial communities are nonrandom assemblages of microbes adapted to specific body habitats. Microbiome-wide association studies (MWASs) in humans or animal models have revealed that substantial deviations from “normal” are observed in several disease states. In some cases, an etiological relationship between the microbiota and disease state has been established by conferring the pathological phenotype via microbiota transfer from diseased animals to healthy germ-free recipients (1, 8, 9). Alternatively, when microbiota alteration is a consequence rather than cause of a disease, such dysbiosis—microbial imbalance—may potentiate the disease or promote comorbidities. Several current research strategies directed at manipulating the microbiota in specific, predictable ways likely will spawn new interventions for rebalancing maladaptive communities.
Probiotics and prebiotics
To date, efforts to alter the composition of the gut community have been dominated by two therapeutic modalities that have not yet gained wide traction in the medical community: probiotics and prebiotics. Probiotics are defined as live microbes that confer a health benefit when consumed in adequate quantities. Many dietary supplements and foods that contain live microbes have been studied for their effects on diverse aspects of human health, ranging from colonic motility to skin allergy. Although the potential for probiotics to have diverse biological impact is great, the one widely substantiated benefit in humans is the improvement of symptoms associated with acute or chronic diarrhea (10, 11). Two related challenges stand in the way of the widespread adoption of probiotic therapies in the clinic: (i) Little is known about the effects these agents have on the gut community and host physiology. Less is known of the molecular mechanisms by which probiotics exert their effects, although there are some exceptions. The demonstration that Lactobacillus salivarius secretes a Listeria-killing bacteriocin—a toxin produced by one strain of bacteria to block the proliferation of a related competitor strain—that can protect mice from infection by this food-borne pathogen provides an example of how mechanisms that underlie probiotic activity can be elucidated and lends credibility to biological potential (12). More recently, probiotic Bifidobacterium species have been shown to protect the host from Escherichia coli infection by producing acetate (13). (ii) Probiotics, which are often classified as food or a dietary supplement, do not typically undergo a rigorous and unbiased evaluation, which can result in variable product quality and unsubstantiated claims. Establishing a regulatory framework in which probiotics are held to a set of appropriate standards may provide incentive for industry and academic researchers to explore the therapeutic mechanisms and potential of these agents, encourage understandably skeptical physicians to adopt probiotic therapies, and cull ineffective products. Thus, in the future, probiotics—like biologics and small-molecule drugs today—may be developed by pharmaceutical and biotechnology companies, approved by the FDA, and prescribed by physicians.
Another opportunity for the field is to expand the limited range of microbial genera currently included in probiotic preparations. Most probiotic organisms are adapted to live within fermented foods (such as yogurt) and are temporary or low-abundance members of the gut community. Strains of the two dominant intestinal phyla, Bacteroidetes and Firmicutes, may prove more effective as probiotics at conferring health benefits than the currently popular Lactobacillus species. Likewise, strains chosen for carrying out a specific function may be useful as agents for personalized therapy. For example, Oxalobacter formigines, a resident gut species that degrades dietary oxalates, is associated with protection from hyperoxaluria and its often accompanying calcium oxalate kidney stones (Fig. 2) (14).
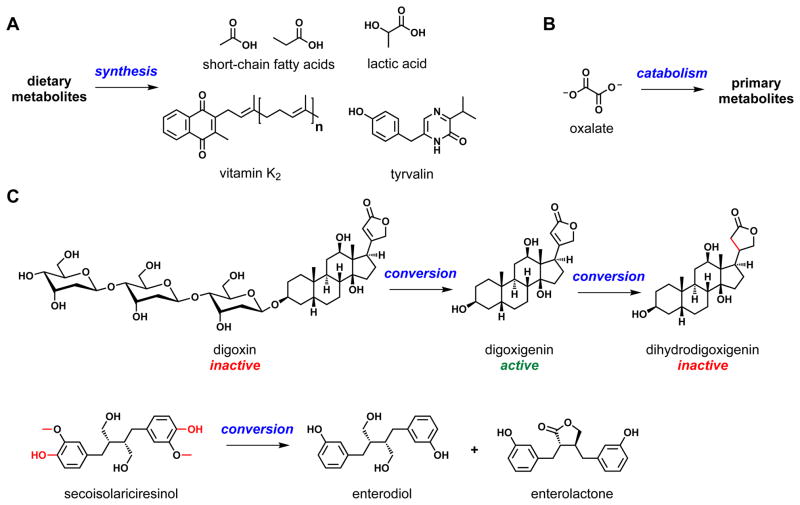
Fig. 2. Transformers.
Shown are three examples of chemical transformations catalyzed by human-associated bacteria. (A) Human microbiota use dietary metabolites as raw material for the synthesis of a variety of small molecules, including short-chain fatty acids, lactic acid, vitamin K2, and tyrvalin. (B) Oxalobacter formigines, a resident gut species that degrades dietary oxalates, is associated with protection from hyperoxaluria and calcium oxalate kidney stones. (C) Gut bacteria chemically convert drugs and dietary molecules such as digoxin and secoisolariciresinol (an antioxidant found in a variety of seeds), respectively, into metabolites with important differences in biological activity.
CREDIT: C. BICKEL/SCIENCE TRANSLATIONAL MEDICINE
Prebiotics are compounds administered to a microbial community to selectively stimulate growth of a specific subset of microbes. The study of prebiotic compounds has largely been confined to purified plant polysaccharides (for example, inulin) that resist host digestion and reach the distal gut. Enrichment of a specific prebiotic polysaccharide in one’s diet may permit preferential expansion of a microbial group that is well adapted to its use, but because patterns of prebiotic metabolism by resident gut microbiota are not well understood the outcomes of prebiotic administration can be unpredictable. As prebiotics have considerable therapeutic potential—and the human diet is, in essence, a mix of poorly understood prebiotics—the molecular effects of these molecules on the gut community should be explored in a way that promotes predictability by connecting prebiotic molecules to the microbiotal gene products that metabolize them. In certain instances, it will be necessary to also understand the bioactive small molecules that are produced as a function of prebiotic-stimulated secondary metabolism. Prebiotics may benefit from the co-administration of probiotics, and both prebiotic and probiotic therapies would benefit from knowing the incident microbial community (that is, the host “microbiotype”; see below).
Lastly, although the microbiota manipulation properties of probiotics and prebiotics have been studied largely in the context of the intestinal flora the concepts are applicable to other human-resident microbial communities. The skin creams and toothpastes of the future may contain live cultures or prebiotics that help treat atopic dermatitis (15) and prevent dental caries (16).
Small-molecule and biological drugs
A third strategy for altering community composition is to develop more conventional small-molecule or biological drugs targeted either to the microbiota or to microbiota-responsive host factors. Because the microbiota reside on exposed surfaces of the human body, they provide a wealth of previously unidentified drug targets that is easily accessed; indeed, gut bacteria routinely encounter orally administered drugs prior to absorption. This phenomenon is important for two reasons. First, promising lead compounds that target the microbiota are less likely to suffer from problems with absorption, metabolism, distribution, and excretion. Lipinski’s Rule of Five, which establishes chemical guidelines for determining drug-like properties, has influenced lead compound selection and development for the past decade (17). Lead compound structural requirements will likely be less stringent for compounds that target unique cell surface proteins of the microbiota and do not need to be absorbed by the host; however, compounds that target cytoplasmic proteins in the microbiota will have the same permeability challenges as antibiotics. Microbiota-targeted compounds engineered to be metabolized by bacteria or to avoid absorption by the host could have the advantage of reduced toxicity.
Second, the microbial biotransformation of certain drugs can change their function. For example, gut bacteria convert the prodrug digoxin to the active product digoxigenin by glycoside hydrolysis (18). In ~10% of patients, digoxigenin is reduced by the gut bacterium Eubacterium lentum to dihydrodigoxigenin, an inactive molecule, leading to as much as a twofold reduction in the serum concentrations of digoxigenin (Fig. 2). Co-administration of the antibiotic erythromycin decreases or eliminates the production of dihydrodigoxigenin in these subjects (19). Gut bacterial glycoside hydrolysis is responsible for converting the inactive glucuronide of SN-38, a cancer chemotherapeutic drug, to the active aglycone in the intestine, causing severe diarrhea. Wallace et al. recently showed that inhibiting the bacterial glycosidase responsible for this conversion prevents the regeneration of SN-38 from its glucuronide in the intestine, decreasing its toxicity (20). This result proves the principle that using microbiota-targeted drugs to achieve ends other than killing (21) or inhibiting virulence (22) could be a fruitful approach for altering human physiology by changing gut-community function.
Lastly, several recent studies have highlighted the targeting of pathogen-responsive host factors, such as the vacuolar adenosine triphosphatase (ATPase) (23), as a new strategy for combating infectious diseases (24). Host factors that respond to microbial mutualists—bugs that enhance the health of the host—may also become bona fide drug targets that when modulated could help shape the composition or function of human-associated microbial communities. For example, such a drug could target a host factor involved in a host-microbe interaction; modulation of this host factor could result in either the killing of undesirable microbes or the growth stimulation of desirable microbes.
Ecological challenges and opportunities
Treatments that alter community composition in a negative way are quite common; high-dose, broad-spectrum antibiotics can kill protective gut mutualists (25), leading to secondary infection by nefarious actors such as Clostridium difficile. Altering community composition in a positive way may not be quite as straightforward. Stable microbial ecosystems are often stubbornly resistant to changes in their composition (26), requiring robust interventions such as fecal bacteriotherapy to eliminate Clostridium difficile from the gut community (27). After milder perturbations, microbial communities can exhibit resilience—the property of returning to the pre-existing stable state—or they may settle in a distinct stable state (28). When such a state is reinforced through feedback from the environment (for example, chronic inflammation), the recalcitrance of the system may be formidable. Can principles of evolutionary dynamics be exploited to target key nodes in the population network? Will the landscape of host biology need to be simultaneously manipulated? Will therapies need to be tailored to an individual’s genotype and microbiotype? Careful ecological study of how natural and unnatural perturbations affect community composition may be the best guide to discovering successful approaches for predictable alteration of the human microbiota composition.
ENDOWING THE COMMUNITY WITH NEW FUNCTIONS
As the focus of human microbiome research shifts from “Who’s there?” to “What are they doing?” the molecular details of host-microbe and microbe-microbe interactions are beginning to emerge. These basic science efforts pave the way for translational research into mechanisms by which to genetically engineer the human microbiota to endow its members with new desirable functions that treat disease or promote health. For example, imagine taking a daily probiotic pill that contains four bacterial strains: The first one synthesizes and excretes a cholesterol-lowering drug and a cocktail of vitamins; the second one neutralizes carcinogenic free radicals and consumes dietary cholesterol and fat; the third one modulates your immune system to ward off infection and hold autoimmune disease in check; and the fourth one produces an appetite-suppressing compound, an antidepressant, and a cognitive enhancer. The field of microbiome research currently is dominated by microbiologists and immunologists. By embracing concepts and approaches from synthetic biology and systems biology, microbiome engineers will eventually be able to use microbial cells as a new platform to modulate human biology.
Microbial carting of therapeutic cargos
The delivery of drugs, therapeutic peptides, or vaccine antigens by microbes at mucosal surfaces holds tremendous promise for the treatment of diverse diseases. This concept was demonstrated presciently 10 years ago, when IL-10 delivery by an engineered strain of Lactococcus lactis prevented inflammatory bowel disease in a mouse model predisposed to develop this malady (29). Of particular interest is the production and delivery of peptides by microbes, because their access to the intestinal or nasal mucosa could potentially enable the delivery of biological drugs using a pill, cream, or spray rather than intravenous or intramuscular injection. In addition to heterologous expression of bioactive molecules and proteins, there are many examples of bacterial molecules that trigger host signaling pathways (for example, quorum-sensing molecules and ligands for toll-like receptors, which function in innate immunity). Undoubtedly, human-associated microbes produce a wealth of yet-to-be discovered molecules for the specific purpose of manipulating host biology. Co-opting such highly adapted signals to tune local or systemic immune responses could lead to a variety of applications, including decreasing susceptibility to pathogens, improving the efficacy of oral vaccination campaigns, and altering the trajectory of autoimmune conditions. Similarly, the gut microbiota may be a unique lever by which host energy balance can be tuned, either by exerting a direct influence on host metabolism or by changing the efficiency of microbiota-facilitated caloric harvest from the diet.
It may even be possible to engineer our microbiota to produce diffusible small molecules that enter our bloodstream, cross the blood-brain barrier, and exert neurological activities. One day, our microbiota may be engineered to produce stimulants, antidepressants, and satiety-inducing drugs, and if designed thoughtfully, microbial production could be modulated by dietary inputs dictated by the host or by microbial sensing of host biochemistry. Indeed, the intriguing possibility exists that some of our resident microbiota produce these compounds naturally and that their perception by the enteric and central nervous systems represents a fundamental form of microbe-host communication.
Other potential functions of the microbiota
In addition to engineering the microbiota to interact directly with the host in new ways, it may be possible to optimize microbiome functions with indirect effects on the host. Catabolism of cholesterol in the gut, production of ultraviolet light–absorbing molecules on the skin, and secretion of pathogen-killing antimicrobials in the urogenital or respiratory tracts all are plausible.
Two threshold questions await: First, will it be necessary to “reprogram” the gut community to stably embed a new member, or will it be simpler and safer to use a strain engineered to have a finite and defined residence time as a chassis for probiosis? Synthetic ecology will undoubtedly be a lively area of research in the coming years, and its pioneers will benefit from the tools and strategies of synthetic biology and the accumulated knowledge about microbial ecology.
Second, when and how will regulatory agencies and the general public come to accept the prospect of using genetically modified microorganisms to treat and prevent disease? Early efforts will likely involve engineered probiotics for treating serious diseases with poor standard-of-care treatments. In addition, the risk of unintended colonization may need to be mitigated by engineering strains that undergo a finite number of cell divisions or are auxotrophic for a co-administered metabolite. General acceptance is likely to take a long time; in the meantime, a key challenge will be to profile natural gut residents and probiotics to find strains that already perform some of these functions.
DIAGNOSING DISEASE AND DISEASE RISK
Exploiting new knowledge about connections between the microbiota and human health will be important not just to treat disease but also to diagnose it. The microbial components of feces, urine, sputum, and mucus have the two key features of a diagnostic sample: They are easily obtained and they hold a trove of molecular information relevant to disease.
Genes versus metabolites
Recent studies reveal that gut bacterial gene rosters are highly individualized; thus, rapid biological interpretation represents a substantial challenge. Given that small molecules—not genes—are the primary agents that mediate host-microbe and microbe-microbe interactions, small-molecule profiles from mass spectrometry or nuclear magnetic resonance spectroscopy may provide a more easily interpreted readout of collective community function. Metabolomic studies have revealed that the microbiota-derived metabolite p-cresol competes with acetaminophen for a sulfonation detoxification pathway (30). Differences among individuals in p-cresol abundance suggest that changes in community composition and function have implications for myriad processes of biological importance. Other important microbial metabolites such as short-chain fatty acids and hydrogen sulfide are likely harbingers of a new universe of small molecules that can be used to infer community function and predict biological effects on the host.
The microbiome in the era of personalized genomic medicine
The Human Microbiome Project of the U.S. National Institutes of Health and related international efforts are defining the varieties of a “normal” microbiome, studying how changes in the microbiota relate to disease and developing new technologies to enable community characterization. As new technologies allow the comprehensive evaluation of transcript, protein, and metabolite amounts in unpurified clinical samples such as fecal matter or skin swabs, the bottleneck will be in the data analysis: Can we identify how the abundance of key microbial gene products and metabolites correlate with disease state and disease risk? The torrent of high-dimensional data are creating important challenges and opportunities for bioinformaticians, statisticians, and computational biologists.
AN EVOLUTIONARY CONTEXT
Although human biology is largely the product of mutation and natural selection in the ancient past, the contemporary microbiome may deviate substantially from that of its ancestors as a result of “unnatural” modern influences. For example, antibiotic treatment can radically influence the human microbiota. And although the duration of antibiotic impact on the microbiota remains an open question, the loss of co-evolved commensal species that are targets of antibiotics in developed regions of the world, such as Helicobacter pylori, may have large health consequences such as altering predisposition to autoimmune disease (31). In addition, some bacterial-directed vaccines appear to alter the composition of microbiota (32–34). Combined with sharp changes in diet and the increasingly sanitary conditions in which many humans live, our current state of colonization may be dysbiotic or at least perturbed. The marked differences of the fecal microbiota of children from a rural village in Burkina Faso compared with Westernized children of northern Italy may provide a glimpse into the problems of defining a healthy baseline for the microbiota (35). Human-associated microbial communities thus are a moving target; the evolutionary plasticity that makes them so attractive therapeutically may translate into a considerable challenge in defining what is “normal” or “healthy.”
References and Notes
- 1.Turnbaugh PJ, Ley RE, Mahowald MA, Magrini V, Mardis ER, Gordon JI. An obesity-associated gut microbiome with increased capacity for energy harvest. Nature. 2006;444:1027–1031. doi: 10.1038/nature05414. [DOI] [PubMed] [Google Scholar]
- 2.Turnbaugh PJ, Hamady M, Yatsunenko T, Cantarel BL, Duncan A, Ley RE, Sogin ML, Jones WJ, Roe BA, Affourtit JP, Egholm M, Henrissat B, Heath AC, Knight R, Gordon JI. A core gut microbiome in obese and lean twins. Nature. 2009;457:480–484. doi: 10.1038/nature07540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ley RE, Turnbaugh PJ, Klein S, Gordon JI. Microbial ecology: Human gut microbes associated with obesity. Nature. 2006;444:1022–1023. doi: 10.1038/4441022a. [DOI] [PubMed] [Google Scholar]
- 4.Wen L, Ley RE, Volchkov PY, Stranges PB, Avanesyan L, Stonebraker AC, Hu C, Wong FS, Szot GL, Bluestone JA, Gordon JI, Chervonsky AV. Innate immunity and intestinal microbiota in the development of Type 1 diabetes. Nature. 2008;455:1109–1113. doi: 10.1038/nature07336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Penders J, Thijs C, van den Brandt PA, Kummeling I, Snijders B, Stelma F, Adams H, van Ree R, Stobberingh EE. Gut microbiota composition and development of atopic manifestations in infancy: The KOALA Birth Cohort Study. Gut. 2007;56:661–667. doi: 10.1136/gut.2006.100164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Frank DN, St Amand AL, Feldman RA, Boedeker EC, Harpaz N, Pace NR. Molecular-phylogenetic characterization of microbial community imbalances in human inflammatory bowel diseases. Proc Natl Acad Sci USA. 2007;104:13780–13785. doi: 10.1073/pnas.0706625104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dethlefsen L, McFall-Ngai M, Relman DA. An ecological and evolutionary perspective on human-microbe mutualism and disease. Nature. 2007;449:811–818. doi: 10.1038/nature06245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Garrett WS, Lord GM, Punit S, Lugo-Villarino G, Mazmanian SK, Ito S, Glickman JN, Glimcher LH. Communicable ulcerative colitis induced by T-bet deficiency in the innate immune system. Cell. 2007;131:33–45. doi: 10.1016/j.cell.2007.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Vijay-Kumar M, Aitken JD, Carvalho FA, Cullender TC, Mwangi S, Srinivasan S, Sitaraman SV, Knight R, Ley RE, Gewirtz AT. Metabolic syndrome and altered gut microbiota in mice lacking Toll-like receptor 5. Science. 2010;328:228–231. doi: 10.1126/science.1179721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Allen SJ, Martinez EG, Gregorio GV, Dans LF. Probiotics for treating acute infectious diarrhoea. Cochrane Database Sys Rev. 2010;11:CD003048. doi: 10.1002/14651858.CD003048.pub3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bernaola Aponte G, Bada Mancilla CA, Carreazo Pariasca NY, Rojas Galarza RA. Probiotics for treating persistent diarrhoea in children. Cochrane Database Sys Rev. 2010;11:CD007401. doi: 10.1002/14651858.CD007401.pub2. [DOI] [PubMed] [Google Scholar]
- 12.Corr SC, Li Y, Riedel CU, O’Toole PW, Hill C, Gahan CG. Bacteriocin production as a mechanism for the antiinfective activity of Lactobacillus salivarius UCC118. Proc Natl Acad Sci USA. 2007;104:7617–7621. doi: 10.1073/pnas.0700440104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fukuda S, Toh H, Hase K, Oshima K, Nakanishi Y, Yoshimura K, Tobe T, Clarke JM, Topping DL, Suzuki T, Taylor TD, Itoh K, Kikuchi J, Morita H, Hattori M, Ohno H. Bifidobacteria can protect from enteropathogenic infection through production of acetate. Nature. 2011;469:543–547. doi: 10.1038/nature09646. [DOI] [PubMed] [Google Scholar]
- 14.Kaufman DW, Kelly JP, Curhan GC, Anderson TE, Dretler SP, Preminger GM, Cave DR. Oxalobacter formigenes may reduce the risk of calcium oxalate kidney stones. J Am Soc Nephrol. 2008;19:1197–1203. doi: 10.1681/ASN.2007101058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Krutmann J. Pre- and probiotics for human skin. J Dermatol Sci. 2009;54:1–5. doi: 10.1016/j.jdermsci.2009.01.002. [DOI] [PubMed] [Google Scholar]
- 16.Hillman JD. Genetically modified Streptococcus mutans for the prevention of dental caries. Antonie van Leeuwenhoek. 2002;82:361–366. doi: 10.1023/A:1020695902160. [DOI] [PubMed] [Google Scholar]
- 17.Lipinski CA, Lombardo F, Dominy BW, Feeney PJ. Experimental and computational approaches to estimate solubility and permeability in drug discovery and development settings. Adv Drug Deliv Rev. 1997;23:3–25. doi: 10.1016/S0169-409X(96)00423-1. [DOI] [PubMed] [Google Scholar]
- 18.Goldin BR. Intestinal microflora: Metabolism of drugs and carcinogens. Ann Med. 1990;22:43–48. doi: 10.3109/07853899009147240. [DOI] [PubMed] [Google Scholar]
- 19.Lindenbaum J, Rund DG, Butler VP, Jr, Tse-Eng D, Saha JR. Inactivation of digoxin by the gut flora: reversal by antibiotic therapy. N Engl J Med. 1981;305:789–794. doi: 10.1056/NEJM198110013051403. [DOI] [PubMed] [Google Scholar]
- 20.Wallace BD, Wang H, Lane KT, Scott JE, Orans J, Koo JS, Venkatesh M, Jobin C, Yeh LA, Mani S, Redinbo MR. Alleviating cancer drug toxicity by inhibiting a bacterial enzyme. Science. 2010;330:831–835. doi: 10.1126/science.1191175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Clatworthy AE, Pierson E, Hung DT. Targeting virulence: A new paradigm for antimicrobial therapy. Nat Chem Biol. 2007;3:541–548. doi: 10.1038/nchembio.2007.24. [DOI] [PubMed] [Google Scholar]
- 22.Bartlett JG. Narrative review: The new epidemic of Clostridium difficile-associated enteric disease. Ann Intern Med. 2006;145:758–764. doi: 10.7326/0003-4819-145-10-200611210-00008. [DOI] [PubMed] [Google Scholar]
- 23.König R, Stertz S, Zhou Y, Inoue A, Hoffmann HH, Bhattacharyya S, Alamares JG, Tscherne DM, Ortigoza MB, Liang Y, Gao Q, Andrews SE, Bandyopadhyay S, De Jesus P, Tu BP, Pache L, Shih C, Orth A, Bonamy G, Miraglia L, Ideker T, García-Sastre A, Young JA, Palese P, Shaw ML, Chanda SK. Human host factors required for influenza virus replication. Nature. 2010;463:813–817. doi: 10.1038/nature08699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Allison SD, Martiny JB. Colloquium paper: Resistance, resilience, and redundancy in microbial communities. Proc Natl Acad Sci USA. 2008;105(Suppl 1):11512–11519. doi: 10.1073/pnas.0801925105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Dethlefsen L, Relman DA. Microbes and Health Sackler Colloquium: Incomplete recovery and individualized responses of the human distal gut microbiota to repeated antibiotic perturbation. Proceedings of the National Academy of Sciences of the United States of America. 2010 doi: 10.1073/pnas.1000087107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Camilli A, Bassler BL. Bacterial small-molecule signaling pathways. Science. 2006;311:1113–1116. doi: 10.1126/science.1121357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Khoruts A, Dicksved J, Jansson JK, Sadowsky MJ. Changes in the composition of the human fecal microbiome after bacteriotherapy for recurrent Clostridium difficile-associated diarrhea. J Clin Gastroenterol. 2010;44:354–360. doi: 10.1097/MCG.0b013e3181c87e02. [DOI] [PubMed] [Google Scholar]
- 28.Dethlefsen L, Huse S, Sogin ML, Relman DA. The pervasive effects of an antibiotic on the human gut microbiota, as revealed by deep 16S rRNA sequencing. PLoS Biol. 2008;6:e280. doi: 10.1371/journal.pbio.0060280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Steidler L, Hans W, Schotte L, Neirynck S, Obermeier F, Falk W, Fiers W, Remaut E. Treatment of murine colitis by Lactococcus lactis secreting interleukin-10. Science. 2000;289:1352–1355. doi: 10.1126/science.289.5483.1352. [DOI] [PubMed] [Google Scholar]
- 30.Clayton TA, Baker D, Lindon JC, Everett JR, Nicholson JK. Pharmacometabonomic identification of a significant host-microbiome metabolic interaction affecting human drug metabolism. Proc Natl Acad Sci USA. 2009;106:14728–14733. doi: 10.1073/pnas.0904489106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Blaser MJ, Falkow S. What are the consequences of the disappearing human microbiota? Nat Rev Microbiol. 2009;7:887–894. doi: 10.1038/nrmicro2245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Regev-Yochay G, Dagan R, Raz M, Carmeli Y, Shainberg B, Derazne E, Rahav G, Rubinstein E. Association between carriage of Streptococcus pneumoniae and Staphylococcus aureus in children. JAMA. 2004;292:716–720. doi: 10.1001/jama.292.6.716. [DOI] [PubMed] [Google Scholar]
- 33.Bogaert D, van Belkum A, Sluijter M, Luijendijk A, de Groot R, Rümke HC, Verbrugh HA, Hermans PW. Colonisation by Streptococcus pneumoniae and Staphylococcus aureus in healthy children. Lancet. 2004;363:1871–1872. doi: 10.1016/S0140-6736(04)16357-5. [DOI] [PubMed] [Google Scholar]
- 34.Croucher NJ, Harris SR, Fraser C, Quail MA, Burton J, van der Linden M, McGee L, von Gottberg A, Song JH, Ko KS, Pichon B, Baker S, Parry CM, Lambertsen LM, Shahinas D, Pillai DR, Mitchell TJ, Dougan G, Tomasz A, Klugman KP, Parkhill J, Hanage WP, Bentley SD. Rapid pneumococcal evolution in response to clinical interventions. Science. 2011;331:430–434. doi: 10.1126/science.1198545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.De Filippo C, Cavalieri D, Di Paola M, Ramazzotti M, Poullet JB, Massart S, Collini S, Pieraccini G, Lionetti P. Impact of diet in shaping gut microbiota revealed by a comparative study in children from Europe and rural Africa. Proc Natl Acad Sci USA. 2010;107:14691–14696. doi: 10.1073/pnas.1005963107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.M.A.F. is an advisor for PureTech Ventures/Enlight Biosciences; J.L.S. is on the scientific advisory board of Ganeden Biotech and Metametrix. Citation: Sonnenburg JL, Fischbach MA. Community health care: Therapeutic opportunities in the human microbiome. Sci Transl Med. 2011;3:78ps12. doi: 10.1126/scitranslmed.3001626.