Abstract
Individuals with Down syndrome (DS; also known as trisomy 21) have a markedly increased risk of leukemia in childhood but a decreased risk of solid tumors in adulthood. Acquired mutations in the transcription factor–encoding GATA1 gene are observed in nearly all individuals with DS who are born with transient myeloproliferative disorder (TMD), a clonal preleukemia, and/or who develop acute megakaryoblastic leukemia (AMKL). Individuals who do not have DS but bear germline GATA1 mutations analogous to those detected in individuals with TMD and DS-AMKL are not predisposed to leukemia. To better understand the functional contribution of trisomy 21 to leukemogenesis, we used mouse and human cell models of DS to reproduce the multistep pathogenesis of DS-AMKL and to identify chromosome 21 genes that promote megakaryoblastic leukemia in children with DS. Our results revealed that trisomy for only 33 orthologs of human chromosome 21 (Hsa21) genes was sufficient to cooperate with GATA1 mutations to initiate megakaryoblastic leukemia in vivo. Furthermore, through a functional screening of the trisomic genes, we demonstrated that DYRK1A, which encodes dual-specificity tyrosine-(Y)-phosphorylation–regulated kinase 1A, was a potent megakaryoblastic tumor–promoting gene that contributed to leukemogenesis through dysregulation of nuclear factor of activated T cells (NFAT) activation. Given that calcineurin/NFAT pathway inhibition has been implicated in the decreased tumor incidence in adults with DS, our results show that the same pathway can be both proleukemic in children and antitumorigenic in adults.
Introduction
Trisomy 21 is the most common cytogenetic abnormality observed at birth (about 1 out of 700 individuals) and one of the most recurrent aneuploidies seen in leukemia. As an acquired clonal chromosomal change, its incidence varies between 4.1% and 14.8% in hematological disorders and malignant lymphomas (1). Supporting the link between trisomy 21 and abnormal hematopoiesis, epidemiological studies have shown that individuals with Down syndrome (DS) have an increased frequency of leukemia but a lower incidence of solid tumors (2). Whereas recent studies implicated a subset of trisomic genes, including Erg, Ets2, Adamts1, and Dscr1, in tumor growth inhibition, in part through an altered angiogenesis (3–6), the role of trisomy 21, the initiating event in DS leukemogenesis, and the functional implication of specific genes at dosage imbalances that predispose to and/or participate in leukemogenesis remain unclear.
Children with DS are at an elevated risk of both acute megakaryoblastic leukemia (AMKL) and acute lymphoblastic leukemia (ALL) (7). Moreover, epidemiological studies showed that approximately 4%–5% of children with DS are born with transient myeloproliferative disorder (TMD), a clonal preleukemia characterized by an accumulation of immature megakaryoblasts in the fetal liver and peripheral blood (8, 9). Although TMD spontaneously disappears in most cases, TMD clones reemerge as AMKL in 20% of cases within 4 to 5 years (10). Extensive research has focused on identifying genetic abnormalities correlated with the transformation process. In addition to trisomy 21, acquired mutations of the transcription factor GATA1 have been observed in nearly all TMD and DS-AMKL cases (11–13). These GATA1 mutations, localized in exon 2, prematurely terminate translation of the full-length protein but allow for expression of a shorter isoform named GATA1s (11). Underscoring the requirement for trisomy 21 in DS-AMKL, humans without DS but with germline GATA1 mutations, analogous to those seen in TMD and DS-AMKL, have no predisposition to leukemia (14), and GATA1 mutations are not found in pediatric leukemia in the absence of trisomy 21. Other less frequent genetic abnormalities that lead to activation of myeloproliferative leukemia (MPL), JAK2, JAK3, and FLT3 have been identified in TMD/DS-AMKL samples (for review see ref. 7). Supporting a model of oncogenic cooperation, expression of these single genetic abnormalities, such as MPL W515L, JAK3 A572V, or JAK2 V617F, alters proliferation and differentiation properties of megakaryocytes but fails to reproduce a DS-AMKL–like phenotype in vivo (15–20).
Mouse models of DS and studies of human tissues show that trisomy 21 is sufficient to perturb hematopoiesis and enhance megakaryocyte development. For example, analysis of human trisomic fetal livers revealed a cell-autonomous enhanced proliferation of the megakaryocytic and erythroid compartments (21, 22). Furthermore, the Tc1, Ts65Dn, and Ts1Cje murine models for DS all display alterations of the erythro/megakaryocytic compartment, with Ts65Dn mice developing a progressive myeloproliferative disease (23–25), further demonstrating the preleukemic role of trisomy 21. Due to the relatively high number of trisomic genes in these partially trisomic animal models, identification of specific leukemia-predisposing genes has been challenging. While several groups have implicated trisomy of Erg with the myeloproliferative phenotype in Ts65Dn mice, its specific association with human DS-AMKL compared with that with non–DS-AMKL remains unclear (26–29). Of note, none of the DS mouse models develop a TMD or AMKL-like phenotype, even when mated to mice harboring Gata1 mutations analogous to those observed in human specimens (24, 25). These observations suggest that trisomy 21 and GATA1s expression are not sufficient to induce TMD or leukemia.
Here, to understand the functional implication of a partial trisomy 21, we used the Ts1Rhr murine model to recapitulate the progressive acquisition of genetic abnormalities seen in human DS specimens. We show that Ts1Rhr cooperates to some extent with Gata1s expression in fetal and adult hematopoiesis and that adding a third genetic event, expression of an activated allele of MPL that has been found in humans with DS-AMKL, induces megakaryoblastic leukemia in vivo. In addition, we demonstrate that dosage imbalance of the human chromosome 21 (Hsa21) gene DYRK1A, which encodes dual-specificity tyrosine-(Y)-phosphorylation–regulated kinase 1A, itself implicated in several DS developmental abnormalities (30) and in the decreased solid tumor incidence in adults with DS (3), contributes to the increased development of DS-AMKL in children. Since trisomy 21 is one of the most recurrent aneuploidies seen in hematological disorders, we believe that our findings will have implications beyond DS-AMKL, for malignancies such as hyperdiploid ALL and AML with acquired trisomy or tetrasomy of Hsa21.
Results
33 trisomic genes are sufficient to develop a progressive thrombocytosis.
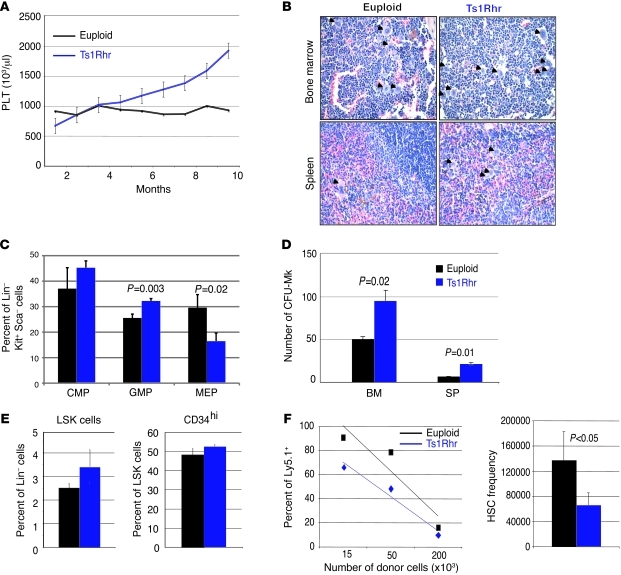
We began by studying hematopoiesis in the Ts1Rhr trisomic mouse model of DS, which is trisomic for 33 orthologous genes in the human DS critical region (DSCR) and spans 65%–70% of a minimal region recently associated with DS-TMD/AMKL (31, 32). Monthly analysis of complete blood counts revealed that Ts1Rhr mice have reduced red blood cell counts (Supplemental Figure 1A; supplemental material available online with this article; doi: 10.1172/JCI60455DS1) and develop a progressive thrombocytosis compared to their euploid littermates (Figure 1A). Moreover, these animals harbor an increased number of megakaryocytes, as demonstrated by histopathology and flow cytometry (Figure 1B, Supplemental Figure 1B, and data not shown). Ts1Rhr mice have an altered proportion of myeloid progenitors, characterized by a shift from megakaryocyte-erythroid progenitors (MEPs) toward granulocyte-monocyte progenitors (GMPs) (Figure 1C and Supplemental Figure 1C) and an increased proportion of CFU-GM colonies (Supplemental Figure 1D). Remarkably, the CFU-GM phenotype is reminiscent of that seen in both the Ts65Dn murine model of DS (23) and in human trisomic fetal liver cells (22). Moreover, bone marrow and spleen Ts1Rhr cells displayed an increased ability to form CFU-megakaryocyte (CFU-Mk) colonies in vitro, but not erythroid BFU-Es, compared with that in their wild-type littermates (Figure 1D and Supplemental Figure 1D). Eight- to twelve-week-old trisomic mice also had an increased proportion of Lin–Sca+Kit+ (LSK) cells (Figure 1E) but no significant variations in the LSK CD34hi subpopulations (containing short-term HSCs and multipotent progenitors) (Figure 1E). Through competitive transplant experiments in lethally irradiated mice, Ts1Rhr adult bone marrow cells appeared to be more functional than euploid ones (Figure 1F). In contrast to Ts1Cje mice and human samples, which display fetal hematopoietic abnormalities (21, 22, 24), no striking hematopoietic defects were seen in E13.5 Ts1Rhr embryos, apart from the significant increase of phenotypic fetal HSCs (Lin–Thy1.1loKit+Sca+Mac1+CD4–) (Supplemental Figure 1, E and F, and ref. 33).
Figure 1. Ts1Rhr mice develop a progressive myeloproliferative disorder associated with thrombocytosis.
(A) Monthly platelet (PLT) counts of euploid and Ts1Rhr mice. Mean ± SD. (B) H&E staining of bone marrow and spleen sections from old Ts1Rhr mice (>12 months) and wild-type mice (original magnification, ×400). The arrowheads point to megakaryocytes. (C) Histograms showing the percentages of myeloid progenitors of 12- to 14-month-old euploid and Ts1Rhr mice. CMP, Lin–c-kit+Sca–FcγRII/III–CD34+; GMP, Lin–c-kit+Sca–FcγRII/III+CD34+; MEP, Lin–c-kit+Sca–FcγRII/III–CD34–. Percentages of live cells are indicated. Mean ± SD. (D) Twelve- to fourteen-month-old Ts1Rhr bone marrow and spleen (SP) give rise to significantly more CFU-Mk colonies. Mean ± SD. (E) Histograms representing percentages of LSK populations and the percentage of CD34hi in the LSK population. Mean ± SD. (F) Functional HSC frequency in the Ts1Rhr bone marrow (1 out of 66,182 cells) compared with that in wild-type bone marrow (1 out of 137,284 cells), assessed by competitive transplants 8 weeks after transplantation. Mean ± SD.
Gata1s cooperates with Ts1Rhr in vivo.
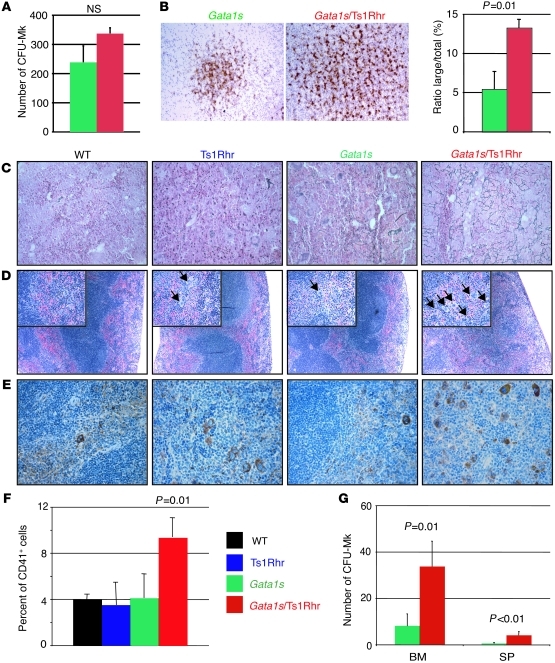
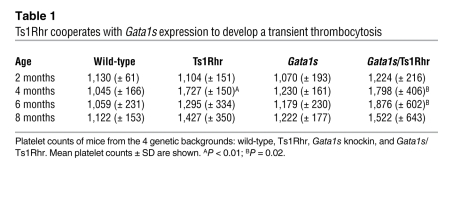
Having established that Ts1Rhr mice fail to develop leukemia, we next mated them with Gata1s knockin mice and assessed the oncogenic potential of these cooperating genetic events in vivo (15). We did not observe significant variations in proportions of the 4 genotypes at birth or significant enhancement of the fetal megakaryopoiesis, apart from the size of CFU-Mk colonies (Figure 2, A and B, Supplemental Figure 2, and data not shown). In line with previous reports, these observations confirm that, unlike human fetuses, murine trisomic fetal livers cells expressing Gata1s mutant protein do not develop a TMD-like phenotype (25). However, we observed that adult Gata1s/Ts1Rhr mice were mildly anemic and developed a transient thrombocytosis (Table 1 and Supplemental Figure 4A). Six-month-old double-transgenic mice displayed marrow fibrosis, with increased megakaryocytes present in clusters, splenomegaly, extensive extramedullary hematopoiesis, and an increased number of monocytes and megakaryocytes as well as CFU-GM and CFU-Mk colonies in the bone marrow and/or the spleen (Figure 2, C–G, Supplemental Figure 3, and Supplemental Figure 4, B–F). Compound Gata1s/Ts1Rhr mice show that Gata1 mutations synergize with partial trisomy to enhance the fetal megakaryocytic phenotype associated with Gata1s expression and to perturb adult hematopoiesis, emphasizing that dosage imbalance of these 33 specific genes is specifically correlated with abnormal megakaryopoiesis. Nevertheless, these 2 events are not sufficient to lead to leukemia in vivo.
Figure 2. Genetic interaction between trisomy for 33 orthologs of Hsa21 and the Gata1 mutation in vivo.
(A) CFU-Mk colony numbers from E13.5 fetal liver cells as well as (B) representative pictures (original magnification, ×100) of the CFU-Mk colonies (n = 3–4 per group) and a histogram comparing the large colonies/total colonies ratio. Mean ± SD. (C) Representative reticulin stains of bone marrow from 6-month-old mice (original magnification, ×400). (D and E) Representative spleen section images of (D) H&E (original magnification, ×100; ×400 [insets]) and (E) von Willebrand factor immunostaining (original magnification, ×400) of 6-month-old mice. The arrows point to megakaryocytes. (F) Histogram plots showing the proportion of CD41+ splenocytes at 6 months, as determined by flow cytometry. Mean percentages ± SD (n = 2–4 per group). (G) CFU-Mk colony number from bone marrow and spleen cells from the Gata1s and Gata1s/Ts1Rhr genetic backgrounds. Mean ± SD (n = 4–5 per group).
Table 1 .
Ts1Rhr cooperates with Gata1s expression to develop a transient thrombocytosis
Trisomy is functionally implicated in DS-AMKL establishment.
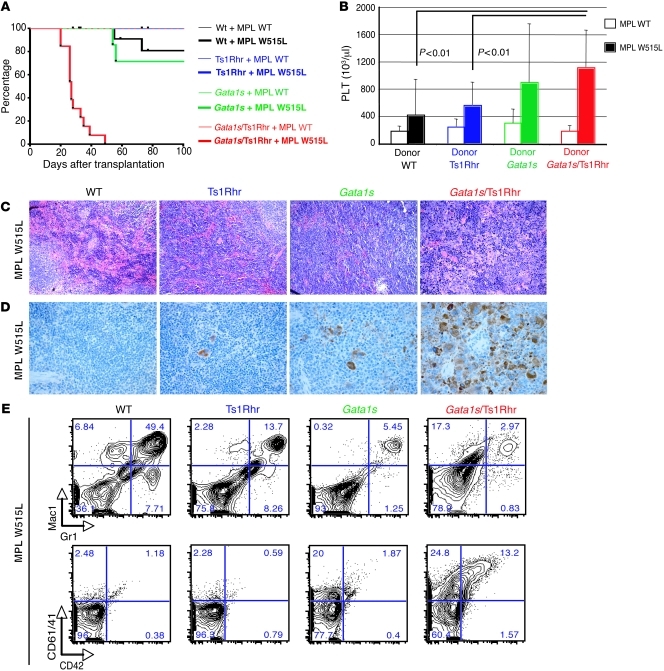
We next asked what the functional impact of trisomy 21 in a more complex disorder is. Given that human DS-AMKL has at least 3 known genetic abnormalities, we attempted to reproduce the leukemia in mice by adding a third oncogenic event to the Gata1s/Ts1Rhr background. Since MPL and JAK2/3 mutations have been identified in human AMKL specimens (18, 34, 35), we overexpressed these constitutively active mutant proteins in bone marrow cells from wild-type, Gata1s knockin, Ts1Rhr, or Gata1s/Ts1Rhr mice by retroviral transduction. To assess the specific impact of a trisomic cellular context on leukemia establishment, we used 6- to 8-month-old bone marrow donor cells, since no apparent phenotype was observed among the 4 different backgrounds. We discovered that 3 genetic events — trisomy for 33 orthologs of Hsa21 genes, Gata1 mutation, and expression of MPLW515L — are sufficient to cause a rapid and fatal leukemia in recipient mice (median survival, 27 days) (Figure 3A). The disease in the triple mutant mice was characterized by the presence of peripheral thrombocytosis and a profound bone marrow fibrosis (Figure 3B and Supplemental Figure 5B). Whereas there was no apparent change in spleen size (Supplemental Figure 5C), Gata1s/Ts1Rhr/W515L recipient mice have an effacement of the splenic architecture, due to infiltration by densely fibrotic tumor nodules comprised of megakaryoblasts and immature megakaryocytes (Figure 3, C and D, and Supplemental Figure 5D). Flow cytometric analyses of splenic cells confirmed the presence of the megakaryoblastic phenotype seen 4 weeks after transplantation, compared with a variable neutrophilic disease in the single or double mutant mice (Figure 3E and data not shown). As determined by Southern blot analyses, the megakaryoblastic phenotype we observed in the spleen of moribund recipient mice is an oligoclonal disorder (Supplemental Figure 6). Interestingly, spleen sections from moribund mice revealed that Gata1s/Ts1Rhr/W515L mice exhibit less infiltration by mature megakaryocytes than mice transplanted with wild-type, Ts1Rhr, or Gata1s bone marrow cells overexpressing MPL W515L (Supplemental Figure 5E). Liver sections from the mice with all 3 genetic abnormalities also demonstrated a substantial megakaryocytic infiltration (Supplemental Figure 5F). Moreover, we observed that MPL W515L overexpression results in an increased hematocrit in Ts1Rhr bone marrow cells, associated with an increased Ter119-positive population, and induces anemia and thrombocytosis when coupled with Gata1s (Supplemental Figure 5A, Figure 3B, and data not shown). Due to the failure associated with the rapid and profound marrow and spleen fibrosis, we failed to transplant this DS megakaryoblastic leukemia (DS-MkL) in secondary recipients. We separately overexpressed JAK3 A572V, another mutation associated with DS-AMKL, in the 4 different backgrounds and found that it cooperates with Gata1s and Ts1Rhr to lead to a fatal hematopoietic disorder in vivo (data not shown). However, as seen in previous bone marrow transplantation studies (36), expression of JAK3 A572V caused a hematolymphoid disorder characterized by a proliferation of CD8+ T cells and megakaryocytes (Supplemental Figure 7). Taken together, these data demonstrate that 3 genetic events are sufficient to lead to a DS-MkL. To date, we believe that this is the first murine model of megakaryoblastic leukemia involving trisomy 21, narrowing down the list of Hsa21 leukemia predisposing/promoting genes to 33 candidates and providing us with an in vivo platform to identify novel dysregulated targets/pathways associated with abnormal megakaryopoiesis.
Figure 3. Three oncogenic events, including a partial trisomy 21, cooperate to promote DS-MkL in vivo.
(A) Survival curves of mice transplanted with different combinations of oncogenic events (n = 6–13 per group). (B) Platelet counts of recipient mice 4 weeks after transplantation. Mean ± SD (n = 4–12 per group). (C) H&E-stained spleen sections of MPL W515L–overexpressing transplanted mice at 4 weeks after transplant (original magnification, ×400). (D) von Willebrand factor immunostaining of MPL W515L–overexpressing recipient mice 4 weeks after transplant, showing the complete megakaryocytic infiltration of the spleen only in the triple mutant mice (original magnification, ×400). (E) Representative flow cytometry plots reveal that triple mutants display marked megakaryocytic expansion in the spleen, while double or single mutants show neutrophilia. Percentages of live cells are indicated.
Functional screening of the trisomic genes implicated in DS-AMKL.
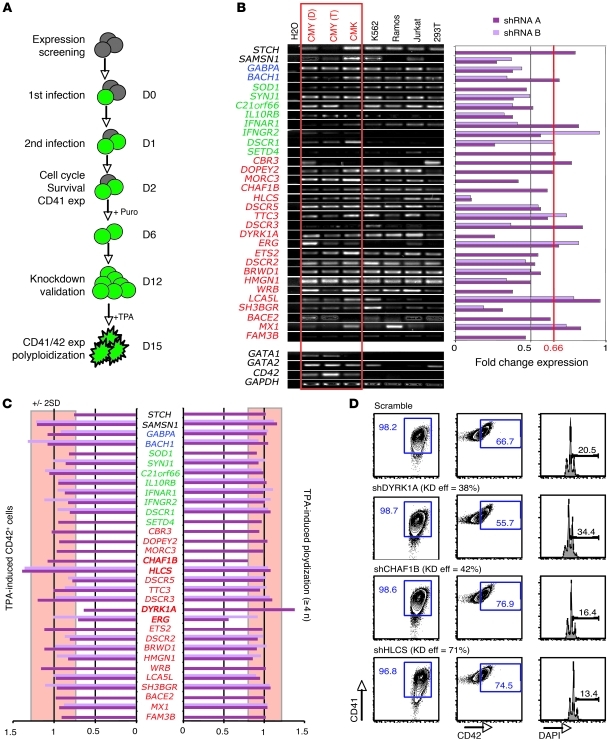
To gain insights into the specific Hsa21 genes that promote DS-AMKL, we designed an shRNA-based screening assay to assess the effects of reducing expression of individual genes on cell cycle, survival, CD41 expression, and enforced differentiation of human megakaryoblastic leukemia cells (Figure 4A). Although this strategy does not address the role of trisomy 21 outside the megakaryocyte lineage, the fact that Ts1Rhr mice and human fetuses with trisomy 21 show prominent expansion of megakaryocytes relative to that of other lineages (21) suggests that careful analysis of the role of Hsa21 genes in megakaryocytes is warranted. In addition to the 33 human orthologs of the Ts1Rhr trisomic genes, we selected other potential candidate genes based on human segmental trisomy studies and gene set enrichment analysis (GSEA) from available DS-AMKL gene expression profiles (26, 32, 37). We found that 32 out of the 50 selected genes were expressed in human DS-AMKL cell lines CMY and CMK (Figure 4B and data not shown). To analyze and select candidate genes, we normalized the raw value observed for each shRNA on the scramble control, calculated the average and SDs from all normalized data, and excluded every variation contained in ±2 SD for statistical and significant purposes. Under this stringent selection, we did not find significant variations in survival, cell cycle, or endogenous expression of CD41 by partial knock down of those 32 expressed genes (Figure 4B and data not shown). However, we found that knock down of 4 genes that are included in the DSCR — ERG, DYRK1A, CHAF1B, and HLCS — led to significant differences (>2 or <2 SDs from the mean) in TPA-induced differentiation and polyploidization of both CMY and CMK human DS-AMKL cell lines (Figure 4, C and D, and Supplemental Figure 8). Whereas DYRK1A and CHAF1B knock down showed a significant effect, with a modest knockdown efficiency (38% and 41%, respectively, in CMK), the functional implication of HLCS through dosage imbalance remains unclear (71% knockdown efficiency).
Figure 4. ERG, DYRK1A, CHAF1B, and HLCS are leading candidate DS leukemia-promoting oncogenes.
(A) Schematic representation of the strategy used to assess the functional implication of trisomic genes in human DS-AMKL cell lines. (B) RT-PCR of the DSCR and nearby genes selected for the functional screening in various cell lines, including DS-AMKL lines CMK and CMY (left panel). Red type indicates Ts1Rhr mice; green type indicates Ts1Cje mice; blue type indicates Ts65Dn mice; and black type indicates Tc1 mice. D, DMSO treated; T, TPA treated. Knockdown efficiency of the selected genes in the CMY cell lines (right panel). We hypothesize that a knockdown efficiency of at least 33% (0.66 threshold) artificially recapitulates the disomy of euploid cells. (C) Plots of normalized values of CD42 expression and DNA content of shRNA-infected CMY cells after treatment for 3 days with TPA. Changes outside of 2 SDs from the mean (red box) were considered significant. (D) Representative flow cytometry plots, showing effect of the DYRK1A, CHAF1B, and HLCS knock down during TPA-induced megakaryocytic differentiation of CMK cells. Percentages of live cells are indicated. Knockdown efficiency (KD eff) is shown. exp, expression; puro, puromycin selection.
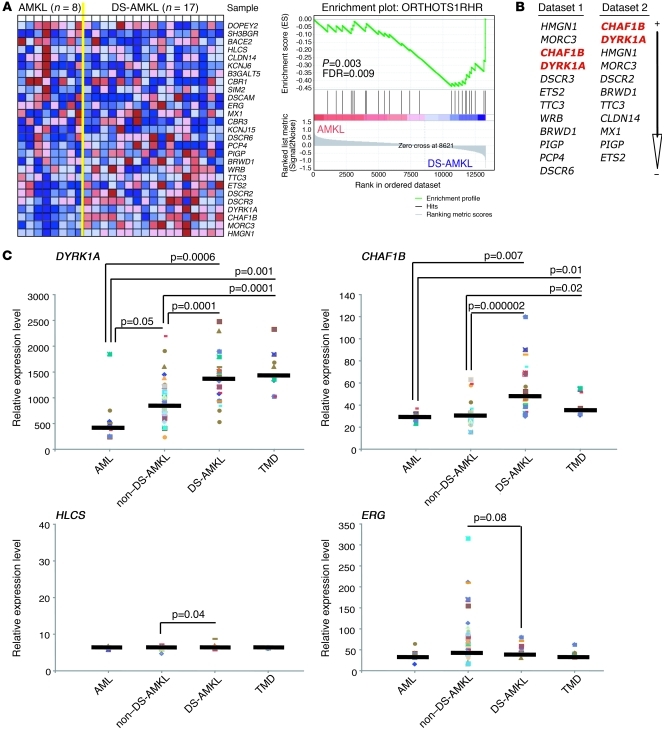
Since DS-AMKL encompasses only one subtype of megakaryocytic leukemia, we wondered to what extent the trisomic genes contributed to other forms of AMKL. Since the specific region has been specifically linked to DS leukemogenesis in murine (Ts1Rhr) and human specimens (32), we looked for a gene expression signature of the DSCR genes in DS-AMKL, assuming that those genes are not altered through other genetic abnormalities in non–DS-AMKL specimens. GSEA of genes contained in the human DSCR revealed that this entire region is moderately enriched in DS-AMKL compared with that for non–DS-AMKL (Figure 5A). However, DYRK1A and CHAF1B are among the top-ranked genes that are specifically enriched in the DS-AMKL subgroup, whereas HLCS and ERG are more widely expressed in all types of human AMKL (Figure 5B). Of note, HMGN1 and MORC3 are also enriched in DS-AMKL from 2 independent data sets, but shRNAs targeting either of those genes had no effect on CMY cell differentiation or polyploidization (Figure 4C). Careful analysis of DYRK1A and CHAF1B expression levels in different leukemic samples revealed that both are significantly overexpressed in DS specimens (including DS-AMKL and TMD) compared with those in non–DS-AMKL and/or pediatric AML (Figure 5C). HLCS is moderately overexpressed in DS-AMKL. Interestingly, ERG appeared to be more enriched in non–DS-AMKL than in DS specimens (Figure 5C). Finally, we observed an increased expression of DYRK1A, CHAF1B, and HLCS in megakaryocytes derived from human trisomic fetal livers (Supplemental Figure 9A).
Figure 5. DYRK1A and CHAF1B are overexpressed in DS-TMD and DS-AMKL.
(A) GSEA of the 33 trisomic human orthologs contained in Ts1Rhr mice derived from an available gene expression profile data set (62), comparing their relative expression in non–DS-AMKL compared with that in DS-AMKL and their relative enrichment. (B) Ranked list of chromosome 21 genes of the DSCR enriched in DS-AMKL compared with those in non–DS-AMKL from both data set 1 (shown in A) and data set 2 (26). (C) DYRK1A (probe set 209033_s_at), CHAF1B (probe set 204775_s_at), HLCS (probe set 209399_s_at), and ERG (probe set 211626_s_at) relative expression in pediatric AML (n = 9), non–DS-AMKL (n = 43), DS-AMKL (n = 20), and TMD (n = 8) (GC_RMA normalized probe values extracted from ref. 26). Gene expression in each sample (individual colored symbols), medians (horizontal bars), and P values (t test) are shown.
DYRK1A is a megakaryoblastic tumor-promoting gene that cooperates with Gata1s.
Since ERG is a known oncogene and has been extensively studied in human and animal abnormal megakaryopoiesis (29, 38), we focused our studies here on DYRK1A (a serine/threonine kinase), CHAF1B (a chromatin assembling factor), and HLCS (an enzyme that catalyzes biotin binding to carboxylases and histones), whose functions in hematopoiesis have not been yet reported. We first assessed expression of Dyrk1a, Chaf1b, and Hlcs in our murine model of DS-AMKL. Although it appears that they were all moderately overexpressed in megakaryocytes derived from Ts1Rhr and Gata1s/Ts1Rhr bone marrow cells, only Dyrk1a was significantly enriched in moribund Gata1s/Ts1Rhr/MPL W515L recipient mice (Supplemental Figure 9, B and C, and Figure 6A). In ex vivo assays using murine bone marrow cells, Dyrk1a overexpression induced robust expansion of low ploidy CD41-positive megakaryocytes (Figure 6, B and C), whereas CHAF1B or HLCS overexpression produced a less potent induction of megakaryopoiesis. Of note, the megakaryoblastic effect of Dyrk1a was enhanced in Gata1s mutant progenitors, arguing for functional cooperation between these proteins in bone marrow (Figure 6, C and D). Although Dyrk1a overexpression increased CD41-positive megakaryocytes in wild-type fetal liver cultures, we did not observe an enhancement in Gata1s FL cells, likely due to the profound enhanced megakaryopoiesis in mice of this genotype (data not shown). To test whether DYRK1A kinase activity is required for the megakaryoblastic phenotype, we overexpressed a catalytically inactive mutant, Dyrk1a-K179R (39), in wild-type or Gata1s mutant bone marrow cells. Dyrk1a-K179R failed to support a substantial expansion of CD41+/CD42+ cells in either the wild-type or Gata1s mutant background (Figure 6D).
Figure 6. Dyrk1a is a prominent megakaryocytic tumor-promoting gene.
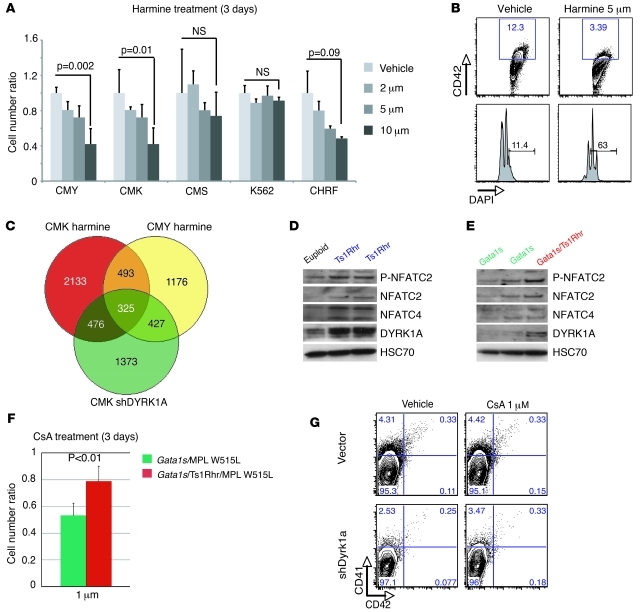
(A) Fold change gene expression values of Dyrk1a, Chaf1b, and Hlcs, as assessed by real-time PCR, in CD41-positive cells isolated from spleens of Gata1s/Ts1Rhr/MPL W515L recipient mice, compared with those in Gata1s/MPL W515L CD41-selected spleen cells 4 weeks posttransplantation. Mean ± SD. (B) Representative flow cytometry plots depicting the proportion of CD41+ cells derived from cultures of bone marrow progenitors infected with Dyrk1a, CHAF1B, or HLCS encoding viruses or control vector. Percentages of live cells are indicated. (C) Overexpression of Dyrk1a leads to reduced polyploidization of megakaryocytes. Mean ± SD (n = 3–5 per group). *P < 0.004, **P < 0.0008 compared with control infected. (D) Fold change increase in percentage of CD41+ and CD42+ cells after expression of wild-type or kinase-inactive alleles of Dyrk1a in wild-type or Gata1s bone marrow progenitors. Mean percentages ± SD (n = 2–4 per group). (E) Representative flow cytometry plots of Dyrk1a shRNA and control infected progenitors cells cultured under megakaryocytic conditions. Bone marrow cells were derived from Ts1Rhr and Gata1s/Ts1Rhr mice. Percentages of live cells are indicated. (F) Treatment of double (Gata1s/MPL W515L) and triple (Gata1s/Ts1Rhr/MPL W515L) mutant cells with harmine reveals that trisomic cells are more sensitive to DYRK1A inhibition in vitro. Mean ± SD (n = 3–4 per group).
To determine whether the elevated megakaryoblastic proliferation driven by GATA1s and trisomy requires elevated expression of Dyrk1a, we first attempted to mate Ts1Rhr mice with Dyrk1a knockout mice (40) but could not obtain Dyrk1a disomic offspring to analyze hematopoiesis and reproduce our multistep pathogenesis, probably due to the breeding deficiencies associated with Dyrk1a+/– mice. Next, we cultured Ts1Rhr and Gata1s/Ts1Rhr bone marrow cells with either a Dyrk1a-targeting shRNA or harmine, a small-molecule inhibitor of DYRK1A kinase activity (41). Knockdown of Dyrk1a reduced the proportions of megakaryocytes expanded from both genotypes (Figure 6E). In parallel, we established murine cell lines by overexpression of MPL W515L in Gata1s and Gata1s/Ts1Rhr progenitor cells, which partly reproduced the surface markers expression phenotype observed in our triple mutant mice (Supplemental Figure 10C). Growth of the trisomic Gata1s/Ts1Rhr/MPL W515L megakaryoblastic cell lines was sensitive to harmine inhibition, while euploid Gata1s/MPL W515L cells were not (Figure 6F). Furthermore, proliferation of human DS-AMKL cell lines was more sensitive to harmine treatment than non-DS human cells (Figure 7A). In addition, we verified that 5 μM harmine treatment recapitulated the phenotype observed with DYRK1A knockdown during TPA-induced megakaryocytic differentiation (Figure 4, C and D, and Figure 7B). Taken together, harmine inhibition and Dyrk1a knockdown experiments both confirm that DYRK1A is required for excessive expansion of trisomic megakaryoblasts.
Figure 7. DYRK1A dosage imbalance is correlated with NFAT pathway dysregulation in human and murine primary cells.
(A) Ratio of live cells treated with serial dilution of the Harmine inhibitor at 3 days (n = 3 per group) normalized on untreated cells. Mean ± SD. (B) Representative FACS plots, showing the effect of 5 μM harmine on a 3-day TPA-induced megakaryocytic differentiation of CMY. (C) Venn diagram showing the number of common genes dysregulated between treated or infected CMK and CMY cells during TPA-induced megakaryocytic differentiation. (D and E) Representative Western blots of DYRK1A, NFATC2, phospho-NFATC2 (P-NFATC2), and NFATC4 expression (D) in CD41-enriched spleen cells from euploid mice compared with that in Ts1Rhr mice (n = 2) and (E) in Gata1s mice (n = 2) compared with that in Gata1s/Ts1Rhr mice. (F) Gata1s/Ts1Rhr/MPL W515L triple mutant cells are less sensitive to the NFAT/calcineurin inhibitor cyclosporine A (CsA) than the non-trisomic Gata1s/MPL W515L cells in liquid culture. Mean ± SD (n = 5 per group). (G) Flow cytometry analysis of CD41 and CD42 populations derived from Ts1Rhr bone marrow progenitors infected with control or DYRK1A shRNA and treated for 3 days with cyclosporine A or vehicle. Percentages of live cells are indicated.
DYRK1A dosage imbalance alters the calcineurin/nuclear factor of activated T cells pathway in DS-AMKL.
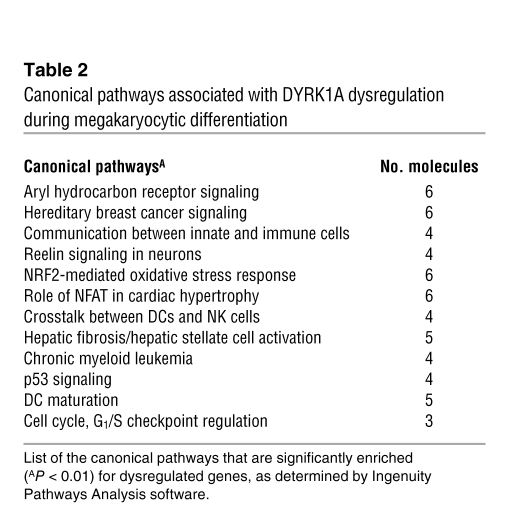
DYRK1A regulates multiple cellular processes through the phosphorylation of several substrates, including nuclear factor of activated T cells (NFAT) (30, 42). To identify targets of DYRK1A in megakaryoblasts, we performed global expression analysis of harmine-treated and DYRK1A shRNA–infected human DS-AMKL cell lines during TPA-induced differentiation. We identified 325 genes whose expression was commonly dysregulated (Figure 7C). Ingenuity pathway analysis revealed that the reduced activity of DYRK1A was associated with expression changes in 2 known DYRK1A target pathways, NFAT and TP53 (Table 2). Since NFAT factors have been implicated in megakaryopoiesis (43, 44) and because dosage imbalance of Dyrk1a has been functionally correlated with common disorders of DS through NFAT pathway alteration (40, 45, 46), the calcineurin/NFAT signaling pathway is an enticing candidate pathway for development of DS-AMKL.
Table 2 .
Canonical pathways associated with DYRK1A dysregulation during megakaryocytic differentiation
Phosphorylation of NFAT by DYRK1A leads to nuclear export of activated NFAT factors and the subsequent inhibition of their transcriptional activity. To begin dissecting the DYRK1A-regulated NFAT activity in megakaryocytes, we first correlated the increased expression levels of DYRK1A to NFATC2 and NFATC4 in megakaryocytes derived from Ts1Rhr and Gata1s/Ts1Rhr mice as compared with those from euploid littermates (Figure 7, D and E). To our surprise, it appears that trisomic cells had an increased expression of NFATC2 and NFATC4. However, we correlated increased phosphorylation of NFATC2 transcription factors to the DYRK1A overexpression in Ts1Rhr cells, consistent with reduced NFAT signaling in DS. The activity of DYRK1A on NFAT transcription factors is opposed by the phosphatase calcineurin, which promotes the dephosphorylation/activation of the NFAT factors and their subsequent nuclear translocation. Thus, we predicted that treatment of cells with cyclosporine A, an inhibitor of calcineurin, would give the opposite phenotype as that of harmine treatment. To ensure the functional correlation between Dyrk1a overexpression and NFAT signaling inhibition, we cultured Gata1s/MPL W515L and Gata1s/Ts1Rhr/MPL W515L cells with 1 μM cyclosporine A and found that euploid cells were significantly more sensitive to growth inhibition than trisomic cells (Figure 7F). Finally, we investigated the effect of cyclosporine A on derivation of megakaryocytes from Ts1Rhr mice and show that Dyrk1a knockdown trisomic cells were more sensitive than the control infected bone marrow cells (Figure 7G). These results are consistent with the model that DYRK1A modulates megakaryoblastic expansion through the inhibition of the calcineurin/NFAT pathway in DS-AMKL.
Discussion
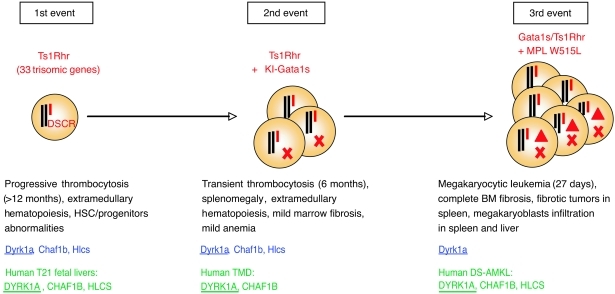
Here we show using a mouse model of 3 oncogenic events that trisomy of 33 gene orthologs of Hsa21, a GATA1 mutation, and a MPL mutation are sufficient to induce DS-MkL in vivo (Figure 8). Moreover, we identified DYRK1A as a potent megakaryoblastic tumor-promoting gene in the DSCR. DYRK1A has a role in embryonic stem cell fate regulation in DS (47), and dosage imbalance of the murine Dyrk1a gene is functionally correlated with heart and neurological disorders, similar to those seen in patients (40, 45, 46). Recently, Dyrk1a, together with another chromosome 21 ortholog, Dscr1, were shown to markedly diminish angiogenesis, resulting in a suppression of tumor growth (3), as observed in patients with DS. Although the DSCR1 gene does not appear to be overexpressed in DS-AMKL, and its knockdown did not reveal significant variations in our screening, it is remarkable to note that the same genes at dosage imbalance can be tumor suppressors or tumor promoters in a context-dependent manner. Based on our studies, we conclude that the differential effects of alterations in pathways downstream of these genes likely account for the discrepancy between solid tumor inhibition and leukemia predisposition in DS (3–6).
Figure 8. Schematic representation of the multistep pathogenesis model of murine DS-MkL and phenotypes associated with the presence of the partial trisomy.
Trisomic genes associated with each step in murine (blue) and human samples (green) are indicated below. Note that DYRK1A (underlined) is the only trisomic gene overexpressed at each step in both species. Triangles represent the third hit (MPL W515L in this study). KI, knockin; T21, trisomy 21, X, GATA1 mutation.
We and others have previously shown that the Hsa21 ETS protein ERG is a megakaryocytic oncogene that cooperates with GATA1 mutations to enhance megakaryopoiesis (29, 38, 48). Interestingly, our study shows that the ERG oncogene is not enriched in DS-associated leukemia compared with that in other pediatric AML specimens (Figure 6B). More surprisingly, ERG appears to be overexpressed in non–DS-AMKL. Of note, previous studies have shown that ERG is not specifically overexpressed in trisomic fetal livers compared with that in euploid ones (21). The same observation has been reported for another known oncogene of the chromosome 21, RUNX1 (21, 26), although, unlike Erg, Runx1 trisomy does not seem to be implicated in the myeloproliferative disorder developed by the Ts65Dn mice (23, 27). Those observations might reflect a higher ERG overexpression in non–DS-AMKL driven by other genetic abnormalities, whereas a low, increased ERG expression in DS leukemia could be sufficient to cooperate with other trisomic genes, as demonstrated in our study, such as DYRK1A, CHAF1B and/or HLCS. Since ERG overexpression has been associated with various types of cancer (49) and recently correlated with promotion and maintenance of T acute lymphoblastic leukemia (50, 51), additional experiments will be required to ensure that ERG trisomy is directly implicated in DS-AMKL as opposed to oncogenesis in general.
Our screening strategy identified 4 Hsa21 genes as candidates to promote excessive megakaryopoiesis. Although we believe that these genes represent strong candidates for the megakaryocytic phenotype, trisomy of these genes or others may also be responsible for promoting B cell acute lymphocytic leukemia and for altering hematopoietic stem and progenitor cell expansion and/or survival in such a way as to promote tumorigenesis. Future studies to uncover Hsa21 genes that contribute to other aspects of DS leukemogenesis are needed.
We identified Dyrk1a as a prominent megakaryoblastic tumor-promoting gene of the human DSCR. Although we were not able to genetically engineer the Ts1Rhr/Dyrk1a+/+/– background by using Dyrk1a knockout mice (40), several lines of evidence support a direct and specific role of DYRK1A dosage imbalance in leukemic predisposition in DS. First, overexpression of Dyrk1a cooperates with GATA1s expression to markedly increase proliferation of megakaryoblasts/immature megakaryocytes. Second, Dyrk1a knockdown decreases the megakaryocytic expansion seen in trisomic mice. Third, inhibition of DYRK1A activity with a small-molecule kinase inhibitor affects megakaryoblastic proliferation from trisomic progenitors to a much greater extent than that from euploid cells. Finally, DYRK1A expression is markedly and specifically upregulated in both human and murine DS-AMKL cells. This latest observation correlates with a recent study showing that DYRK1A is upregulated in trisomic fetal livers and is predicted to be a new potential biomarker for screening DS fetuses. Interestingly, it has been shown that the transcription factor E2F1 increases DYRK1A expression by enhancing promoter activity (52). Added to the recent observation that GATA1s protein failed to repress E2F target genes (53), this might lead to a further increased Dyrk1a expression and potentially explain the cooperation we observed in the Gata1s background and account for the DYRK1A enrichment observed in TMD and DS-AMKL compared with that in non–DS-AMKL and pediatric AML.
The DYRK kinase family is part of the CMGC group, which also includes mitogen-activated protein kinases (MAPKs), cyclin-dependant kinases (CDKs), glycogen synthase kinases, and CDK-like kinases (42). Unlike, MAPKs regulated by upstream protein kinases, catalytic activation of DYRK proteins occurs by autophosphorylation, which appears to occur by an intramolecular reaction immediately after translation (54). Since DYRK kinases always appear to be in a catalytically active state, their function is thought be controlled by changes in expression levels. Thus DYRK1A dosage imbalance is predicted to directly alter the regulation of its targets. Here we show that increased DYRK1A dosage in trisomic megakaryocytes is linked to an inhibition of NFAT factors activation. Interestingly, whereas the calcineurin/NFAT pathway has been shown to favor lymphoid leukemia progression (55), its inhibition has been associated with a megakaryocytic accumulation, through the overexpression of the immunophilin FKBP51 in idiopathic myelofibrosis (56). Since the calcineurin/NFAT pathway has been shown to be active in megakaryocytes (43, 44, 57), those results emphasize its functional implication in this lineage. Interestingly, our data also show that murine trisomic cells have an increased expression of NFATC2 and NFATC4 transcription factors, leading that the hypothesis that calcineurin/NFAT signaling may be altered through multiple processes in our murine model. Of note, NFATC4 also appears to be overexpressed in human DS-AMKL compared with that in non–DS-AMKL (data not shown). We also observed a modest enrichment of the calcineurin pathway in non–DS-AMKL compared with that in DS-AMKL but failed to find a NFAT signature using these gene expression analyses, probably due to the fact that most of the known NFAT target genes have been identified in cardiac and T cells. Additional experiments will be required to establish the functional implication of this pathway in human DS-AMKL and understand how NFAT factors expression is regulated during normal and pathologic megakaryopoiesis.
Although our murine model reproduced most of the human DS-AMKL pathogenesis, we only observed slight variations in the fetal liver, which does not account for the TMD phenotype commonly seen in humans with DS. While this discrepancy may reflect species-specific differences, there are several other possible explanations. For example, the difference may be due to the absence of trisomic genes, miRNAs (such as miR125b-2) (58), or regulatory elements that act during fetal hematopoiesis. Interestingly, Carmichael et al. described clonogenicity and HSC function impairment in the Ts1Cje murine model that we did not observe in the Ts1Rhr background (24). While this discrepancy could be related to experimental design, it may also emphasize the possible contribution of trisomic genes outside of the DSCR to this hematopoietic phenotype. Only a few trisomic genes in the Ts1Cje mice, but not in the Ts1Rhr mice, have a known function in fetal hematopoiesis. Among them, however, Runx1 appears to be a relevant candidate, because its dosage imbalance has been associated with abnormal HSC function during fetal hematopoiesis and, more recently, with the increased c-KIT and GATA2 expression in human DS fetal livers (59, 60). Other than Runx1, genes to consider include Dscr1, Sod1, and Son. Alternatively, the failure to fully recapitulate the human fetal phenotype may be due to a requirement for additional genetic abnormalities that do not spontaneously appear in mice. In line with these hypotheses, neither DYRK1A nor CHAF1B overexpression appears to specifically cooperate with fetal GATA1s expression, unlike our observations in adult progenitors, arguing for a time- and/or context-dependent cooperation in mice.
Through a retroviral integration mutagenesis strategy, Klusmann et al. showed that aberrant expansion of fetal, but not adult, megakaryocytic progenitors is dependent on IGF (53). This study emphasized the fetal origin of the target cell for transformation in DS-AMKL but did not address the requirement of trisomy 21, the initiating event of DS-leukemogenesis. We believe that our study is the first to address the functional requirement of trisomy 21 in the transformation process of Gata1s- and/or MPL W515L–expressing progenitors. Additional experiments will be required to address the fetal origin of the human disorder and to determine the IGF and calcineurin/NFAT signaling effects in trisomic Gata1s-expressing fetal liver progenitors. The absence of a fetal disease in our DS-AMKL model is not unprecedented, as mouse models of MLL rearrangements, which have their origins in the fetus, consistently develop adult not fetal leukemia.
Our study also specifically links trisomy for the 33 genes in the Ts1Rhr mice to the previously reported red blood cell phenotype observed in Ts65Dn, Ts1Cje, and Tc1 partially trisomic murine models (23–25). Interestingly, we observed a functional cooperation of Ts1Rhr with MPL W515L, leading to a leukocytosis and an increased hematocrit and increased Ter119+ cells in recipient mice (S. Malinge and J.D. Crispino, unpublished observations). These observations are in agreement with the enhanced expression of erythroid markers observed in DS-AMKL compared with that in non–DS-AMKL (26, 37).
Harmine, an inhibitor of DYRK1A kinase activity, altered the polyploidization of megakaryocytes established from murine and human primary cells in vitro (L. Diebold and S. Malinge, unpublished observations). Interestingly, the human AMKL cell line CHRF, established from a patient with an acquired trisomy 21, was sensitive to harmine in the same range as that of both of our DS-AMKL cell lines (Figure 7A). We also observed that pediatric AMKL samples with acquired trisomy 21 and GATA1 mutations were associated with DYRK1A overexpression (data not shown). These observations suggest that our study has broader significance, since trisomy 21 is one of the most common cytogenetic abnormalities seen in hematological malignancies. Moreover, the in vivo model of multistep DS leukemogenesis that we established will be a powerful tool to develop new small-molecule inhibitors of DYRK1A activity and assess new potential therapy for DS-AMKL.
Methods
Mouse experiments.
The partially trisomic Ts1Rhr murine model [also known as the Dp(16Cbr1-ORF9)1Rhr model] and Gata1s knockin mice (Gata1s mice; also known as Gata1∆ex2 mice), provided by R.H. Reeves (Johns Hopkins University School of Medicine, Baltimore, Maryland, USA) and S.H. Orkin (Dana-Farber Cancer Institute, Harvard, Massachusetts, USA), respectively, have been previously described (15, 31) and genotyped using Ts1Rhr-F 5′-CCGTCAGGACATTGTTGGA-3′ with Ts1Rhr-R 5′-CCGTAACCTCTGCCGTTCA-3′ and 5G1-7762 5′-GGGACAAAGGATGATGGAAGAAGAC-3′ with 3G1-8739 5′-GGTCTTCCTACAATAGTCTCTTGC-3′ for knockin Gata1s and 5G1-7762 with Gata1-ex3R 5′-TGCATCCAAACCCTCTGGC-3′ for the Gata1 wild-type locus. Peripheral blood (CBC) and flow cytometry analyses were performed at the specific time points indicated in the manuscript using Hemavet HV950FS (Drew Scientific) and BD LSRII, respectively. Murine cell populations were analyzed using Mac1-PE, Gr1-APC, Kit-PerCP-Cy5.5, CD41-PE, CD45.1-PerCP-Cy5.5, and CD45.2-APC antibodies (all from BD Biosciences); CD61/41-APC and CD42-PE (both form Emfret); and CD34-FITC and Sca1-PeCy7 antibodies (both from eBioscience) and analyzed using FlowJo 8.8.6 software. In vitro CFU assays were performed using M3234 and MegaCult (Stem Cell Technology), according to the manufacturer’s instructions. All experiments using the Gata1s/Ts1Rhr double-transgenic mice were done using males from at least the third generation from different founders and by comparing littermates to minimize background variations. For competitive transplants, 8-week-old lethally irradiated mice were transplanted in replicate experiments via retro-orbital injection with 15 × 103, 50 × 103, or 200 × 103 wild-type or Ts1Rhr Ly5.2 bone marrow donor cells in combination with 200 × 103 wild-type competitor cells (Ly5.1) (n = 8 mice per group). Engraftment was assessed at 8 weeks posttransplantation by flow cytometry for Ly5.1 and Ly5.2 markers on peripheral blood leukocytes. Stem cell frequencies were determined using L-Calc software (Stem Cell Technologies Inc.). All animals were maintained in microisolator housing within a barrier facility.
Plasmids.
Murine Dyrk1a and human CHAF1B and HLCS cDNAs were amplified from the Y10 murine megakaryocytic cell line and human DS-AMKL cells, respectively, and then subcloned into the MSCV-IRES-EGFP (Migr) plasmid for overexpression studies. The K179R mutation was done with the QuikChange Site-Directed Mutagenesis Kit (Stratagene, no. 200518) according to the manufacturer instructions and subcloned to the Migr plasmid. The murine Dyrk1a shRNA was subcloned from the pSM2c vector (Open Biosystem) into the GFP-containing Banshee for retroviral production (61).
Murine megakaryocyte cultures, overexpression, and knockdown studies.
Whole bone marrow cells were infected twice by retroviruses (Migr-Dyrk1a, Migr-Dyrk1a K179R, Migr-CHAF1B, and Migr-HLCS) for overexpression studies or by Banshee constructs containing shRNA Dyrk1a (Open Biosystem, from pSM2c no. V2MM_25405) for knockdown experiments, and cells were then resuspended in RPMI containing 10% FBS, penicillin-streptomycin, l-glutamine, 1 μM cortisol (Sigma-Aldrich, no. H6909), 10 ng/ml mouse IL-3 (mIL-3), 5 ng/ml mIL-6, and mouse SCF (mSCF) supernatant. On day 3, infected cells were cultured in RPMI1640, penicillin-streptomycin, l-glutamine, 10% FBS, 10 ng/ml mouse thrombopoietin (mTPO), and mSCF supernatants for an additional 3 days to promote megakaryocytic differentiation. After culture, cells were stained for flow cytometry experiments or separated by a BSA gradient (3%, 1.5%) for RNA extraction. Splenic megakaryocytes were enriched using CD41-PE antibody (BD Biosciences, no. 558040) and PE Selection Kit (StemCell Technology, no. 18551) for RNA or protein extraction.
Bone marrow transplantation.
Bone marrow transplant experiments of JAK3 wild-type, JAK3 A572V, MPL wild-type, and MPL W515L overexpression were performed as described previously (18, 19). JAK3 and MPL constructs were provided by D.G. Gilliland (Brigham and Women’s Hospital, Boston, Massachusetts, USA) and R. Levine (Memorial Sloan-Kettering Cancer Center, New York, New York, USA), respectively. Briefly, whole bone marrow cells were harvested from 6- to 8-month-old wild-type, Ts1Rhr, Gata1s, or Gata1s/Ts1Rhr (Ly5.2) donor mice at day 0, lysed for red blood cells for 5 minutes with ammonium chloride solution (StemCell Technology, no. 07850), washed with PBS, and then resuspended overnight in RPMI1640 containing 10% FBS, cortisol (Sigma-Aldrich, no. H6909), 10 ng/ml mIL-3, 5 ng/ml mIL-6, and mSCF supernatant. On day 1, cells were spin infected at 1,400 g, 33°C, for 99 minutes and resuspended overnight in fresh media. On day 2, 0.5 to 1 × 106 cells were spinoculated, resuspended for 5 hours in fresh media, and then injected through the retro-orbital vein into lethally irradiated Ly5.1 recipient mice (2 × 5.5 Gy). Proviral integration and clonality were assessed by Southern blot using 10 μg genomic DNA extracted from splenocytes (Puregene) from moribund mice, digested by BamH1, subjected to electrophoresis, and hybridized with an EGFP probe according to the standard protocols.
shRNA based screening.
Trisomic genes of interest were first screened for their expression in the human CMK and CMY DS-AMKL cells lines. Primer sequences used for the screening are shown in Supplemental Table 1. The expressed genes were then knocked down using pGIPZ shRNA purchased from Open Biosystem (Thermo Scientific) (Supplemental Table 2). Lentiviral particles were produced according the manufacturer’s instructions using psPAX2 (gag-pol) and pMD2.G (encoding VSV-G) helper vectors, and CMY cells were submitted to 2 rounds of spinoculation. On day 2, GFP-positive cells were incubated with 0.5 μg/ml Hoechst33342 (Invitrogen, no. H3570) for 90 minutes and/or stained with AnnexinV-PE (BD Biosciences, no. 556421) and CD41-APC (BD Biosciences, no. 559777). Infected cells were selected with 1 μg/ml puromycin for 4 days and expanded for additional 6 days prior to RNA extraction or were cultured in the same media, RPMI1640, 10% FBS, penicillin-streptomycin, and l-glutamine with 100 nM TPA, to chemically induce megakaryocytic differentiation. Cells were then stained for differentiation markers CD41 and CD42 (BD Biosciences, no. 551061) and DAPI to measure DNA content.
Immunohistochemistry.
Tissues were fixed for 24 hours in 10% formalin (Sigma-Aldrich, no. HT501320) and then placed in 70% ethanol prior paraffin embedding. Tissue sections were stained with anti-human von Willebrand factor, H&E, or reticulin. Briefly, tissue sections were deparaffinized and incubated for 20 minutes in target retrieval buffer (10 mM Tris, 1 mM EDTA, pH 9.0). Endogenous peroxidase activity was blocked with 3% hydrogen peroxide for 10 minutes. Tissue sections were blocked with normal serum from the VECTASTAIN ABC Elite Kit (Vector Laboratories, no. PK-6101) for 20 minutes before incubation overnight at 4°C with anti-human Von Willebrand Factor (1:1,200; Dako, no. A0082). Secondary biotinylated antibody and VECTASTAIN Elite ABC Reagent (Vector Laboratories) were applied as per manufacture directions. Positive cells were detected using diaminobenzidine (Sigma-Aldrich, no. D5905). All tissue sections were counterstained with Harris hematoxylin (Sigma-Aldrich, no. HHS16).
Cell lines, reagents, and Western blotting.
Human cell lines (CMK, CMY, CMS, CHRF, and K562) were cultured in RPMI1640, 10% FBS, penicillin-streptomycin, and l-glutamine. The murine cell lines Gata1s/MPLW515L and Gata1s/Ts1Rhr/MPLW515L were established by MPL W515L infection of Lin-negative bone marrow cells, selected for 4 days with 1 μg/ml puromycin, and cultured in complete RPMI, 1 μM cortisol, 10 ng/ml mIL-3, 5 ng/ml mIL-6, 5 ng/ml mTPO, and mSCF supernatant. Harmine hydrochloride inhibitor (MP Biomedicals, no. 210554) and cyclosporine A (Sigma-Aldrich, no. C3662) were incubated with human or primary murine cells for 3 days prior further analysis. Protein pellets were solubilized in 10 mM TRIS/HCl (pH 7.5), 1% Triton X-100, 20% glycerol, 5 mM EDTA, 50 mM NaCl, 50 mM NAF, 1 mM Na3OV4, and Complete lysis buffer; separated on a Bis-Tris polyacrylamide gel (Invitrogen); and transferred to a PVDF membrane (Millipore). Antibodies were anti-PhosphoSer326-NFATc2 (no. sc-32994), anti-NFATc2 (no. sc-7296), anti-NFATc4 (no. sc-13036), and anti-Hsc70 (no. sc-7298) (all from Santa Cruz Biotechnology) and anti-DYRK1A (Abnova, no. H00001859-M01).
Statistics.
Data are shown as mean ± SEM or SD as denoted. P values were calculated using the Student’s t test (2 tailed). P ≤ 0.05 was considered to be significant.
Study approval.
All animal experiments were approved by the Northwestern University Animal Care and Use Committee.
Supplementary Material
Acknowledgments
The authors thank Roger Reeves for the Ts1Rhr mice, Stuart Orkin and Zhe Li for the Gata1s knockin mice, and Mariona Arbones (Barcelona, Spain) for the Dyrk1a+/– mice. We thank Jeffrey Taub and Yubin Ge for the human AMKL gene expression profile data set. We thank Roger Reeves, Monika Stankiewicz, Lou Doré, Kim Rice, Stella Chou, and Sandra Ryeom for their advice and helpful discussions. We also thank the Robert H. Lurie Flow Cytometry Core Facility for assistance with flow cytometry. This work was supported by an R01 from the National Cancer Institute (CA101774). S. Malinge is a recipient of a postdoctoral fellowship from the Leukemia and Lymphoma Society.
Footnotes
Conflict of interest: The authors have declared that no conflict of interest exists.
Citation for this article: J Clin Invest. 2012;122(3):948–962. doi:10.1172/JCI60455.
See the related Commentary beginning on page 807.
References
- 1.Mitelman F, Heim S, Mandahl N. Trisomy 21 in neoplastic cells. Am J Med Genet Suppl. 1990;7:262–266. doi: 10.1002/ajmg.1320370752. [DOI] [PubMed] [Google Scholar]
- 2.Hasle H. Pattern of malignant disorders in individuals with Down’s syndrome. Lancet Oncol. 2001;2(7):429–436. doi: 10.1016/S1470-2045(00)00435-6. [DOI] [PubMed] [Google Scholar]
- 3.Baek KH, et al. Down’s syndrome suppression of tumour growth and the role of the calcineurin inhibitor DSCR1. Nature. 2009;459(7250):1126–1130. doi: 10.1038/nature08062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Reynolds LE, et al. Tumour angiogenesis is reduced in the Tc1 mouse model of Down’s syndrome. Nature. 2010;465(7299):813–817. doi: 10.1038/nature09106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sussan TE, Yang A, Li F, Ostrowski MC, Reeves RH. Trisomy represses Apc(Min)-mediated tumours in mouse models of Down’s syndrome. Nature. 2008;451(7174):73–75. doi: 10.1038/nature06446. [DOI] [PubMed] [Google Scholar]
- 6.Ryeom S, et al. Targeted deletion of the calcineurin inhibitor DSCR1 suppresses tumor growth. Cancer Cell. 2008;13(5):420–431. doi: 10.1016/j.ccr.2008.02.018. [DOI] [PubMed] [Google Scholar]
- 7.Malinge S, Izraeli S, Crispino JD. Insights into the manifestations, outcomes, and mechanisms of leukemogenesis in Down syndrome. Blood. 2009;113(12):2619–2628. doi: 10.1182/blood-2008-11-163501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lange B. The management of neoplastic disorders of haematopoiesis in children with Down’s syndrome. Br J Haematol. 2000;110(3):512–524. doi: 10.1046/j.1365-2141.2000.02027.x. [DOI] [PubMed] [Google Scholar]
- 9.Pine SR, Guo Q, Yin C, Jayabose S, Druschel CM, Sandoval C. Incidence and clinical implications of GATA1 mutations in newborns with Down syndrome. Blood. 2007;110(6):2128–2131. doi: 10.1182/blood-2007-01-069542. [DOI] [PubMed] [Google Scholar]
- 10.Roy A, Roberts I, Norton A, Vyas P. Acute megakaryoblastic leukaemia (AMKL) and transient myeloproliferative disorder (TMD) in Down syndrome: a multi-step model of myeloid leukaemogenesis. Br J Haematol. 2009;147(1):3–12. doi: 10.1111/j.1365-2141.2009.07789.x. [DOI] [PubMed] [Google Scholar]
- 11.Wechsler J, et al. Acquired mutations in GATA1 in the megakaryoblastic leukemia of Down syndrome. Nat Genet. 2002;32(1):148–152. doi: 10.1038/ng955. [DOI] [PubMed] [Google Scholar]
- 12.Hitzler JK, Cheung J, Li Y, Scherer SW, Zipursky A. GATA1 mutations in transient leukemia and acute megakaryoblastic leukemia of Down syndrome. Blood. 2003;101(11):4301–4304. doi: 10.1182/blood-2003-01-0013. [DOI] [PubMed] [Google Scholar]
- 13.Taub JW, et al. Prenatal origin of GATA1 mutations may be an initiating step in the development of megakaryocytic leukemia in Down syndrome. Blood. 2004;104(5):1588–1589. doi: 10.1182/blood-2004-04-1563. [DOI] [PubMed] [Google Scholar]
- 14.Hollanda LM, et al. An inherited mutation leading to production of only the short isoform of GATA-1 is associated with impaired erythropoiesis. Nat Genet. 2006;38(7):807–812. doi: 10.1038/ng1825. [DOI] [PubMed] [Google Scholar]
- 15.Li Z, Godinho FJ, Klusmann JH, Garriga-Canut M, Yu C, Orkin SH. Developmental stage-selective effect of somatically mutated leukemogenic transcription factor GATA1. Nat Genet. 2005;37(6):613–619. doi: 10.1038/ng1566. [DOI] [PubMed] [Google Scholar]
- 16.Lacout C, Pisani DF, Tulliez M, Gachelin FM, Vainchenker W, Villeval JL. JAK2V617F expression in murine hematopoietic cells leads to MPD mimicking human PV with secondary myelofibrosis. Blood. 2006;108(5):1652–1660. doi: 10.1182/blood-2006-02-002030. [DOI] [PubMed] [Google Scholar]
- 17.Wernig G, Mercher T, Okabe R, Levine RL, Lee BH, Gilliland DG. Expression of Jak2V617F causes a polycythemia vera-like disease with associated myelofibrosis in a murine bone marrow transplant model. Blood. 2006;107(11):4274–4281. doi: 10.1182/blood-2005-12-4824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Walters DK, et al. Activating alleles of JAK3 in acute megakaryoblastic leukemia. Cancer Cell. 2006;10(1):65–75. doi: 10.1016/j.ccr.2006.06.002. [DOI] [PubMed] [Google Scholar]
- 19.Pikman Y, et al. MPLW515L is a novel somatic activating mutation in myelofibrosis with myeloid metaplasia. PLoS Med. 2006;3(7):e270. doi: 10.1371/journal.pmed.0030270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dash A, Gilliland DG. Molecular genetics of acute myeloid leukaemia. Best Pract Res Clin Haematol. 2001;14(1):49–64. doi: 10.1053/beha.2000.0115. [DOI] [PubMed] [Google Scholar]
- 21.Chou ST, et al. Trisomy 21 enhances human fetal erythro-megakaryocytic development. Blood. 2008;112(12):4503–4506. doi: 10.1182/blood-2008-05-157859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tunstall-Pedoe O, et al. Abnormalities in the myeloid progenitor compartment in Down syndrome fetal liver precede acquisition of GATA1 mutations. Blood. 2008;112(12):4507–4511. doi: 10.1182/blood-2008-04-152967. [DOI] [PubMed] [Google Scholar]
- 23.Kirsammer G, et al. Highly penetrant myeloproliferative disease in the Ts65Dn mouse model of Down syndrome. Blood. 2008;111(2):767–775. doi: 10.1182/blood-2007-04-085670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Carmichael CL, et al. Hematopoietic defects in the Ts1Cje mouse model of Down syndrome. Blood. 2009;113(9):1929–1937. doi: 10.1182/blood-2008-06-161422. [DOI] [PubMed] [Google Scholar]
- 25.Alford KA, et al. Perturbed hematopoiesis in the Tc1 mouse model of Down syndrome. Blood. 2010;115(14):2928–2937. doi: 10.1182/blood-2009-06-227629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bourquin JP, et al. Identification of distinct molecular phenotypes in acute megakaryoblastic leukemia by gene expression profiling. Proc Natl Acad Sci U S A. 2006;103(9):3339–3344. doi: 10.1073/pnas.0511150103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ng AP, et al. Trisomy of Erg is required for myeloproliferation in a mouse model of Down syndrome. Blood. 2010;115(19):3966–3969. doi: 10.1182/blood-2009-09-242107. [DOI] [PubMed] [Google Scholar]
- 28.Ge Y, et al. The role of cytidine deaminase and GATA1 mutations in the increased cytosine arabinoside sensitivity of Down syndrome myeloblasts and leukemia cell lines. Cancer Res. 2004;64(2):728–735. doi: 10.1158/0008-5472.can-03-2456. [DOI] [PubMed] [Google Scholar]
- 29.Stankiewicz MJ, Crispino JD. ETS2 and ERG promote megakaryopoiesis and synergize with alterations in GATA-1 to immortalize hematopoietic progenitor cells. Blood. 2009;113(14):3337–3347. doi: 10.1182/blood-2008-08-174813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Park J, Song WJ, Chung KC. Function and regulation of Dyrk1A: towards understanding Down syndrome. Cell Mol Life Sci. 2009;66(20):3235–3240. doi: 10.1007/s00018-009-0123-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Olson LE, Richtsmeier JT, Leszl J, Reeves RH. A chromosome 21 critical region does not cause specific Down syndrome phenotypes. Science. 2004;306(5696):687–690. doi: 10.1126/science.1098992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Korbel JO, et al. The genetic architecture of Down syndrome phenotypes revealed by high-resolution analysis of human segmental trisomies. Proc Natl Acad Sci U S A. 2009;106(29):12031–12036. doi: 10.1073/pnas.0813248106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Morrison SJ, Hemmati HD, Wandycz AM, Weissman IL. The purification and characterization of fetal liver hematopoietic stem cells. Proc Natl Acad Sci U S A. 1995;92(22):10302–10306. doi: 10.1073/pnas.92.22.10302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Malinge S, et al. Activating mutations in human acute megakaryoblastic leukemia. Blood. 2008;112(10):4220–4226. doi: 10.1182/blood-2008-01-136366. [DOI] [PubMed] [Google Scholar]
- 35.Hussein K, et al. MPLW515L mutation in acute megakaryoblastic leukaemia. Leukemia. 2009;23(5):852–855. doi: 10.1038/leu.2008.371. [DOI] [PubMed] [Google Scholar]
- 36.Cornejo MG, et al. Constitutive JAK3 activation induces lymphoproliferative syndromes in murine bone marrow transplantation models. Blood. 2009;113(12):2746–2754. doi: 10.1182/blood-2008-06-164368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ge Y, et al. Differential gene expression, GATA1 target genes, and the chemotherapy sensitivity of Down syndrome megakaryocytic leukemia. Blood. 2006;107(4):1570–1581. doi: 10.1182/blood-2005-06-2219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Salek-Ardakani S, et al. ERG is a megakaryocytic oncogene. Cancer Res. 2009;69(11):4665–4673. doi: 10.1158/0008-5472.CAN-09-0075. [DOI] [PubMed] [Google Scholar]
- 39.Alvarez M, Estivill X, de la Luna S. DYRK1A accumulates in splicing speckles through a novel targeting signal and induces speckle disassembly. J Cell Sci. 2003;116(pt 15):3099–3107. doi: 10.1242/jcs.00618. [DOI] [PubMed] [Google Scholar]
- 40.Fotaki V, et al. Dyrk1A haploinsufficiency affects viability and causes developmental delay and abnormal brain morphology in mice. Mol Cell Biol. 2002;22(18):6636–6647. doi: 10.1128/MCB.22.18.6636-6647.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bain J, et al. The selectivity of protein kinase inhibitors: a further update. Biochem J. 2007;408(3):297–315. doi: 10.1042/BJ20070797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Aranda S, Laguna A, de la Luna S. DYRK family of protein kinases: evolutionary relationships, biochemical properties, and functional roles. FASEB J. 2011;25(2):449–462. doi: 10.1096/fj.10-165837. [DOI] [PubMed] [Google Scholar]
- 43.Kiani A, et al. Expression analysis of nuclear factor of activated T cells (NFAT) during myeloid differentiation of CD34+ cells: regulation of Fas ligand gene expression in megakaryocytes. Exp Hematol. 2007;35(5):757–770. doi: 10.1016/j.exphem.2007.02.001. [DOI] [PubMed] [Google Scholar]
- 44.Kyttala S, Habermann I, Minami T, Ehninger G, Kiani A. Regulation of Down syndrome critical region 1 expression by nuclear factor of activated T cells in megakaryocytes. Br J Haematol. 2009;144(3):395–408. doi: 10.1111/j.1365-2141.2008.07490.x. [DOI] [PubMed] [Google Scholar]
- 45.Tejedor F, et al. minibrain: a new protein kinase family involved in postembryonic neurogenesis in Drosophila. Neuron. 1995;14(2):287–301. doi: 10.1016/0896-6273(95)90286-4. [DOI] [PubMed] [Google Scholar]
- 46.Arron JR, et al. NFAT dysregulation by increased dosage of DSCR1 and DYRK1A on chromosome 21. Nature. 2006;441(7093):595–600. doi: 10.1038/nature04678. [DOI] [PubMed] [Google Scholar]
- 47.Canzonetta C, et al. DYRK1A-dosage imbalance perturbs NRSF/REST levels, deregulating pluripotency and embryonic stem cell fate in Down syndrome. Am J Hum Genet. 2008;83(3):388–400. doi: 10.1016/j.ajhg.2008.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Rainis L, et al. The proto-oncogene ERG in megakaryoblastic leukemias. Cancer Res. 2005;65(17):7596–7602. doi: 10.1158/0008-5472.CAN-05-0147. [DOI] [PubMed] [Google Scholar]
- 49.Sashida G, Bazzoli E, Menendez S, Liu Y, Nimer SD. The oncogenic role of the ETS transcription factors MEF and ERG. Cell Cycle. 2010;9(17):3457–3459. doi: 10.4161/cc.9.17.13000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Tsuzuki S, Taguchi O, Seto M. Promotion and maintenance of leukemia by ERG. Blood. 2011;117(14):3858–3868. doi: 10.1182/blood-2010-11-320515. [DOI] [PubMed] [Google Scholar]
- 51.Thoms JA, et al. ERG promotes T-acute lymphoblastic leukemia and is transcriptionally regulated in leukemic cells by a stem cell enhancer. Blood. 2011;117(26):7079–7089. doi: 10.1182/blood-2010-12-317990. [DOI] [PubMed] [Google Scholar]
- 52.Maenz B, Hekerman P, Vela EM, Galceran J, Becker W. Characterization of the human DYRK1A promoter and its regulation by the transcription factor E2F1. BMC Mol Biol. 2008;9:30. doi: 10.1186/1471-2199-9-30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Klusmann JH, et al. Developmental stage-specific interplay of GATA1 and IGF signaling in fetal megakaryopoiesis and leukemogenesis. Genes Dev. 2010;24(15):1659–1672. doi: 10.1101/gad.1903410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Lochhead PA, Sibbet G, Morrice N, Cleghon V. Activation-loop autophosphorylation is mediated by a novel transitional intermediate form of DYRKs. Cell. 2005;121(6):925–936. doi: 10.1016/j.cell.2005.03.034. [DOI] [PubMed] [Google Scholar]
- 55.Medyouf H, et al. Targeting calcineurin activation as a therapeutic strategy for T-cell acute lymphoblastic leukemia. Nat Med. 2007;13(6):736–741. doi: 10.1038/nm1588. [DOI] [PubMed] [Google Scholar]
- 56.Giraudier S, et al. Overexpression of FKBP51 in idiopathic myelofibrosis regulates the growth factor independence of megakaryocyte progenitors. Blood. 2002;100(8):2932–2940. doi: 10.1182/blood-2002-02-0485. [DOI] [PubMed] [Google Scholar]
- 57.Kiani A, et al. Expression and regulation of NFAT (nuclear factors of activated T cells) in human CD34+ cells: down-regulation upon myeloid differentiation. J Leukoc Biol. 2004;76(5):1057–1065. doi: 10.1189/jlb.0404259. [DOI] [PubMed] [Google Scholar]
- 58.Klusmann JH, et al. miR-125b-2 is a potential oncomiR on human chromosome 21 in megakaryoblastic leukemia. Genes Dev. 2010;24(5):478–490. doi: 10.1101/gad.1856210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Cai Z, et al. Haploinsufficiency of AML1 affects the temporal and spatial generation of hematopoietic stem cells in the mouse embryo. Immunity. 2000;13(4):423–431. doi: 10.1016/S1074-7613(00)00042-X. [DOI] [PubMed] [Google Scholar]
- 60.De Vita S, et al. Trisomic dose of several chromosome 21 genes perturbs haematopoietic stem and progenitor cell differentiation in Down’s syndrome. Oncogene. 2010;29(46):6102–6114. doi: 10.1038/onc.2010.351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Huang Z, et al. GATA-2 reinforces megakaryocyte development in the absence of GATA-1. Mol Cell Biol. 2009;29(18):5168–5180. doi: 10.1128/MCB.00482-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Ge Y, et al. GATA1, cytidine deaminase, and the high cure rate of Down syndrome children with acute megakaryocytic leukemia. J Natl Cancer Inst. 2005;97(3):226–231. doi: 10.1093/jnci/dji026. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.