Abstract
Dysregulation of the G1/S transition in the cell cycle contributes to tumor development. The oncogenic transcription factors c-Jun and c-Myc are indispensable regulators at this transition, and their aberrant expression is associated with many malignancies. Degradation of c-Jun/c-Myc is a critical process for the G1/S transition, which is initiated upon phosphorylation by glycogen synthase kinase 3 β (GSK3β). However, a specific kinase or kinases responsible for priming phosphorylation events that precede this GSK3β modification has not been definitively identified. Here, we found that the dual-specificity tyrosine phosphorylation–regulated kinase DYRK2 functions as a priming kinase of c-Jun and c-Myc. Knockdown of DYRK2 in human cancer cells shortened the G1 phase and accelerated cell proliferation due to escape of c-Jun and c-Myc from ubiquitination-mediated degradation. In concert with these results, silencing DYRK2 increased cell proliferation in human cancer cells, and this promotion was completely impeded by codeprivation of c-Jun or c-Myc in vivo. We also found marked attenuation of DYRK2 expression in multiple human tumor samples. Downregulation of DYRK2 correlated with high levels of unphosphorylated c-Jun and c-Myc and, importantly, with invasiveness of human breast cancers. These results reveal that DYRK2 regulates tumor progression through modulation of c-Jun and c-Myc.
Introduction
The c-Jun and c-Myc transcription factors are critical promoters of cellular proliferation. Dysregulated expression and activation of these oncogenes are frequently observed in most human cancers. Control of their expression is thus vital to maintaining regulated cell proliferation in normal cells. Indeed, expressions of c-Jun and c-Myc are tightly governed at transcriptional, posttranscriptional, translational, and posttranslational levels (1, 2). These proteins rapidly accumulate when quiescent cells are stimulated into the cell cycle. This transient increase and subsequent decline in their protein levels potentiate the transition from the G1 phase into the S phase (G1/S transition) with progression through the cell cycle and continued proliferation (2, 3). The clearance of these oncoproteins in continuously cycling cells and the maintenance of low levels in quiescent cells are controlled by ubiquitin-mediated proteolysis (4, 5). In many cases, proteins that are targeted for degradation by the ubiquitin-proteasome pathway are first marked by phosphorylation of specific residues, allowing for their recognition by the ubiquitination machinery. Notably, previous studies have demonstrated that c-Jun and c-Myc are polyubiquitinated in a glycogen synthase kinase 3 β–dependent (GSK3β-dependent) fashion by an Skp–cullin–F-box protein ligase complex containing Fbw7 (6). GSK3β is a serine/threonine protein kinase that operates in the G1 phase to receive input from several signaling pathways (7). Growth factor–dependent inactivation of GSK3β promotes cell cycle entry by stabilizing proteins such as c-Jun and c-Myc (8–10). In this context, dysregulation of GSK3β has been implicated in tumorigenesis and cancer progression (11). GSK3 phosphorylates many of its substrates via a “priming phosphorylation” mechanism, recognizing the canonical phosphorylation motif S/TXXXpS/T (12). This motif contains the phospho-accepting Ser or Thr that is separated by 3 residues from phospho-Ser or phospho-Thr. A different kinase capable of the priming phosphorylation must first phosphorylate the substrate at the P+4 position, before GSK3 can phosphorylate the P0 residue. In this regard, priming kinases play critical roles in the regulation of molecular and biological activities for c-Jun and c-Myc. Degradation of c-Myc protein is controlled by ordered phosphorylation at 2 specific sites near the N terminus of the protein: Ser62 for a priming phosphorylation and Thr58 for the GSK3β phosphorylation. Surprisingly, Ser62 kinase or kinases remain inconsistent in spite of biological importance. Although both ERK and cdc2 kinase specifically phosphorylate Ser62 in vitro and cellular phosphorylation of Thr58/Ser62 is stimulated by mitogens, in vivo experiments do not support a role for these kinases in the phosphorylation of c-Myc (13, 14). Likewise, c-Jun is destroyed after sequential phosphorylation at Ser243 and Thr239 (10). Phosphorylation of Thr239 is catalyzed by GSK3β. However, as seen for c-Myc, a priming kinase or kinases phosphorylating Ser243 are as yet unidentified. Previously, GSK3 was reported to be capable of phosphorylation at both Thr239 and Ser243; a subsequent study concluded that GSK3 can phosphorylate at Thr239, and not Ser243 (15, 16). The results also demonstrated that Ser243 kinase or kinases are distinct from the MAPK superfamily, including ERK, JNK, and p38MAPK (16). While they implied the possibility that dual specificity tyrosine phosphorylation–regulated kinases (DYRKs) are capable of phosphorylating Ser243 in vitro, no data have been provided (16). These studies collectively suggest that priming kinases themselves for GSK3β in Fbw7-mediated degradation of c-Jun and c-Myc remains uncertain.
DYRK2 is a member of an evolutionarily conserved family of DYRKs that autophosphorylate a critical tyrosine in their activation loop, but function only as serine/threonine kinases toward their substrates (17). Until recently, the functional role for DYRK2 in cells has been largely obscure. A recent report indicated that DYRK2 functions as a scaffold for an E3 ligase complex and controls mitotic transition (18). Another study demonstrated that DYRK2 contributes to proteasomal degradation of the transcription factor GLI2 (19). These findings shed light on a role for DYRK2 in protein proteolysis. Our recent study revealed an unexpected role for DYRK2 in the regulation of the p53 tumor suppressor (20). We found that DYRK2 translocated from the cytoplasm to the nucleus in response to DNA damage. Subsequently, ataxia telangiectasia mutated (ATM) activated DYRK2, allowing it to phosphorylate p53 at Ser46, which is a crucial modification for induction of apoptosis. In this context, DYRK2 plays an indispensable role on the determination of cell fate in response to DNA damage. However, to our knowledge, there has been no physiological substrate identified for DYRK2 in cells besides p53.
Here, we demonstrate that DYRK2 is identified as a priming kinase responsible for phosphorylation of c-Jun/c-Myc. Importantly, priming phosphorylation by DYRK2 is a prerequisite for GSK3β phosphorylation to the G1/S transition. Moreover, silencing of DYRK2 contributes to accelerated proliferation by stabilizing c-Jun/c-Myc in vitro and in vivo. These findings thus support a coordinate regulation of c-Jun/c-Myc by DYRK2.
Results
DYRK2 affects cell proliferation by controlling the expression of c-Jun and c-Myc.
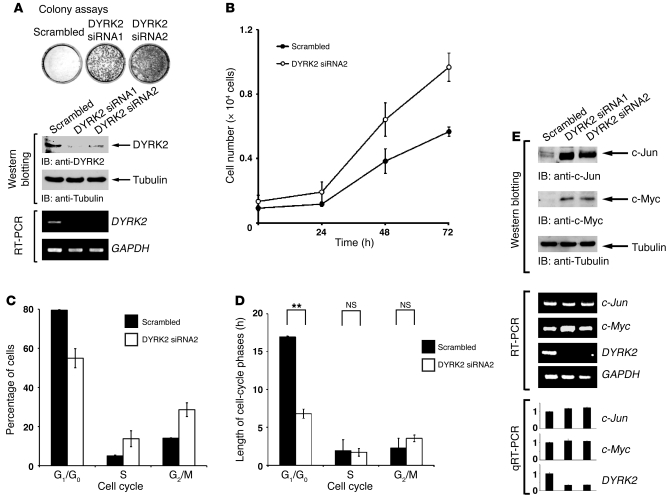
To examine the role of DYRK2 on cell proliferation, HeLa cells were transfected with 2 different siRNAs targeting DYRK2 (20). Colony formation assays revealed that depletion of DYRK2 elevated cell growth (Figure 1A). Similar results were obtained in U2OS cells (Supplemental Figure 1A; supplemental material available online with this article; doi: 10.1172/JCI60818DS1), indicating the independence of the p53 tumor suppressor. To confirm these results, we evaluated the growth rate of cells in the presence or absence of DYRK2 siRNAs by trypan blue dye exclusion assays. Analysis of the growth curves demonstrated that, in HeLa cells silenced for DYRK2, the exponential growth was markedly elevated compared with that in mocked cells. (Figure 1B). Comparable results were obtained in U2OS cells (Supplemental Figure 1B). To extend these findings, we explored cell cycle analysis by flow cytometry. Depletion of DYRK2 did not induce apoptosis, but led to a decrease in the percentage of cells in the G1 phase of the cell cycle and a concomitant increase in the number of cells in the S and G2/M phases (Figure 1C and Supplemental Figure 1C). We used the growth curves determined by trypan blue dye exclusion assays to estimate doubling times and combined both pieces of information to calculate the length of each phase of the cell cycle. The results demonstrated that knocking down DYRK2 leads to a significant reduction in length of the G1 phase of the cell cycle (Figure 1D and Supplemental Figure 1D). These findings indicated that DYRK2 regulates cell cycle progression at the G1 phase. In order to identify potential effectors that might cause this phenotype, we compared the expression status of cell cycle–associated genes between control cells and DYRK2-depleted cells. Toward this end, we found with surprise that silencing DYRK2 substantially augments the expression of c-Jun and c-Myc (Figure 1E and Supplemental Figure 1E), both of which affect cell proliferation by controlling the G1/S transition. To determine whether the increase in c-Jun and c-Myc depends on kinase activity of DYRK2, we prepared GFP-tagged plasmids expressing DYRK2 WT or the kinase-dead (KR) DYRK2 mutant, both of which are resistant to silencing by DYRK2 siRNA. Introduction of the resistant DYRK2 WT (designated GFP-rDYRK2 WT) in DYRK2-depleted cells abrogated the augmentation of c-Jun and c-Myc (Supplemental Figure 1F). In contrast, transduction of the resistant DYRK2-KR (GFP-rDYRK2-KR) had no remarkable effect on their expression levels (Supplemental Figure 1F), clearly demonstrating that the increase of these transcription factors requires kinase activity of DYRK2. To determine whether upregulation of c-Jun and c-Myc expression is regulated at transcriptional levels, we analyzed mRNA by RT-PCR. The demonstration that there is no significant increase of mRNA in cells silenced for DYRK2 (Figure 1E and Supplemental Figure 1E) indicates that the expression of these genes must be controlled at posttranslational levels.
Figure 1. Silencing of DYRK2 results in hyperproliferation by upregulation of c-Jun and c-Myc.
(A) HeLa cells were transfected with scrambled siRNA or DYRK2-specific siRNAs. After transfection, colony formation assays were performed. Cell lysates were immunoblotted with anti-DYRK2 or anti-tubulin. Total RNAs were analyzed by RT-PCR using DYRK2-specific or GAPDH-specific primers. (B) HeLa cells were transfected with scrambled siRNA or DYRK2 siRNA, and cell growth was analyzed by trypan blue exclusion assay. (C and D) HeLa cells were transfected with scrambled siRNA or DYRK2 siRNA. After transfection, cells were stained with propidium iodide and the cell cycle was analyzed using flow cytometry. **P < 0.01. (E) HeLa cells were transfected and analyzed by immunoblotting (IB) with anti–c-Jun (top panel), anti–c-Myc (2nd panel), or anti-tubulin (3rd panel). Total RNAs were analyzed with c-Jun–specific (4th and 8th panels), c-Myc–specific (5th and 9th panels), DYRK2-specific (6th and 10th panels), or GAPDH-specific (7th panel) primers. The result of quantitative RT-PCR was normalized for the level of GAPDH and represents the relative fold induction compared with control sample. The data were evaluated from 3 independent experiments, each performed in triplicate. Data represent mean ± SD. qRT-PCR, quantitative RT-PCR.
DYRK2 phosphorylates c-Jun and c-Myc, which is required for subsequent GSK3β phosphorylation.
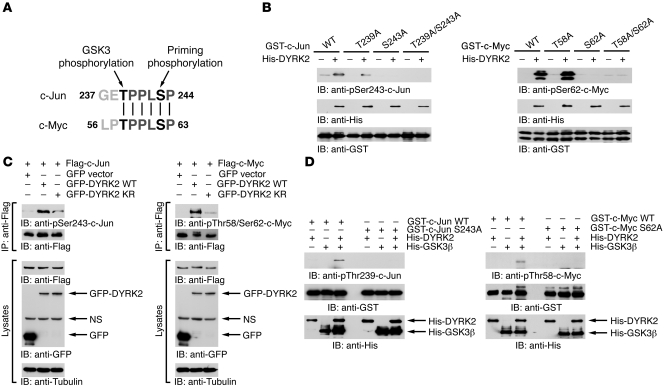
In order to define mechanisms in which DYRK2 contributes to the expression of c-Jun and c-Myc, we attempted to identify the potential phosphorylation sites in c-Jun and c-Myc. During this process, we eventually focused on Ser243 for c-Jun and Ser62 for c-Myc, both of which have been considered “priming phosphorylation” residues, since surrounding amino acid sequences of serine coincide with consensus phosphorylation sites of DYRK kinases (P-X-S/T-P) (Figure 2A and ref. 21). These colinear motifs, including serine, are highly conserved through evolution (10). More importantly, accumulating studies have revealed that phosphorylation at these residues is tightly integrated with appropriate control of their stability at the G1 phase (6). To determine whether DYRK2 can specifically phosphorylate these serine residues, recombinant DYRK2 was incubated with purified c-Jun210–310 or c-Myc1–100. The finding that kinase-active DYRK2 phosphorylated GST–c-Jun210–310-WT by detection with an anti–phospho–c-Jun(Ser243) antibody demonstrated that DYRK2 is a Ser243 kinase in vitro (Figure 2B). Similar phosphorylation was obtained with coincubation of DYRK2 and the GST–c-Jun210–310–T239A mutant in which Thr239 is substituted with Ala (Figure 2B). Notably, coincubation of DYRK2 with the GST–c-Jun210–310–S243A mutant or the GST–c-Jun210–310–T239A/S243A mutant completely abrogated reaction with the anti–phospho–c-Jun(Ser243) antibody (Figure 2B), indicating the specificity of this antibody against phospho-Ser243. We also performed in vitro kinase assays to explore DYRK2 phosphorylation of c-Myc at Ser62 by using anti–phospho–c-Myc(Ser62). As shown for c-Jun, DYRK2 specifically phosphorylated c-Myc at Ser62 in vitro (Figure 2B). To extend these findings to cellular DYRK2, 293T cells were transfected with Flag vector, Flag-DYRK2 WT, or the catalytically inactive Flag-DYRK2-KR mutant (20). Cell lysates were immunoprecipitated with anti-Flag followed by in vitro kinase assays using GST–c-Jun210–310 as the substrate. Expression of DYRK2 WT was associated with substantial phosphorylation of c-Jun at Ser243 (Supplemental Figure 2A). In contrast, Ser243 phosphorylation was completely abrogated by expression of the dominant negative DYRK2 mutant (Supplemental Figure 2A). Similar results were obtained in phosphorylation of c-Myc at Ser62 (Supplemental Figure 2A). These findings collectively support a role for DYRK2 as a kinase for the priming phosphorylation of c-Jun and c-Myc in vitro. To assess whether DYRK2 phosphorylates c-Jun and c-Myc in cells, 293T cells were cotransfected with Flag–c-Jun or Flag–c-Myc and GFP vector, GFP-DYRK2 WT, or the GFP-DYRK2-KR mutant. Immunoprecipitates with anti-Flag were analyzed by immunoblotting with the phospho-specific antibodies. Expression of DYRK2 WT enhanced phosphorylation of c-Jun at Ser243 (Figure 2C). Moreover, there was little if any induction of Ser243 phosphorylation in cells ectopically expressed with the dominant negative DYRK2, suggesting that kinase activity is required for DYRK2 phosphorylation of c-Jun at Ser243 (Figure 2C). Comparable findings were obtained in phosphorylation of c-Myc at Ser62 (Figure 2C). We previously demonstrated that DYRK2 is virtually localized in the cytoplasm and the nucleus; however, nuclear DYRK2 is constitutively degraded by the ubiquitination-proteasome machinery (22). Conversely, upon exposure to genotoxic stress, DYRK2 escaped from ubiquitination and was thereby stabilized in the nucleus. In this regard, to confirm that DYRK2 phosphorylates c-Jun and c-Myc in the cytoplasm, we examined phosphorylation of c-Jun and c-Myc by expressing the GFP-DYRK2-(KKR→NNN) mutant, in which key amino acids of nuclear localization signal were mutated with asparagine; thus, this mutant is localized exclusively in the cytoplasm (22). The GFP-DYRK2-(KKR→NNN) mutant phosphorylated c-Jun at Ser243 to levels equivalent to those of DYRK2 WT (Supplemental Figure 2B). Similar results were obtained in phosphorylation of c-Myc at Ser62 (Supplemental Figure 2B). These data suggest DYRK2 phosphorylation of c-Jun and c-Myc in the cytoplasm. We also found that DYRK2 interacts with c-Jun/c-Myc in cells (Supplemental Figure 2, B and C) and that the binding of DYRK2 to c-Myc is abrogated after exit from the G1 phase (Supplemental Figure 2D). To further define the prerequisite role for DYRK2 in subsequent GSK3β phosphorylation, GST–c-Jun210–310–WT or –S243A mutant was incubated with purified GSK3β in the presence or absence of recombinant DYRK2. Incubation of GST–c-Jun210–310–WT with GSK3β did not increase the reactivity of c-Jun with anti–phospho–c-Jun(Thr239) antibody (Figure 2D). However, prior phosphorylation of c-Jun by DYRK2 significantly enhanced the ability of GSK3β to phosphorylate c-Jun at Thr239 in vitro (Figure 2D). More importantly, incubation of the GST–c-Jun–S243A mutant with DYRK2 did not facilitate Thr239 phosphorylation by GSK3β (Figure 2D). Equivalent findings were obtained in phosphorylation of c-Myc at Thr58 (Figure 2D). These results demonstrated an indispensable role for DYRK2 on a priming function for GSK3β phosphorylation.
Figure 2. DYRK2 phosphorylates c-Jun and c-Myc at the priming phosphorylation residues.
(A) Sequence alignments around phosphorylation residues of c-Myc and c-Jun. (B) Recombinant DYRK2 was incubated with GST–c-Jun210–310 or GST–c-Myc1–100 in the presence of ATP. Reactants were analyzed by immunoblotting with anti–phospho–c-Jun(Ser243) (left upper panel), anti–phospho–c-Myc(Ser62) (right upper panel), anti-His (middle panels), or anti-GST (lower panels). (C) 293T cells were cotransfected with GFP vector, GFP-DYRK2 WT, or GFP-DYRK2-KR and Flag-tagged c-Jun (Flag–c-Jun) or c-Myc (Flag–c-Myc). Lysates were immunoprecipitated with anti-Flag agarose. Immunoprecipitates were then subjected to immunoblot analysis with anti–phospho–c-Jun(Ser243) (left top panel), anti–phospho–c-Myc(Thr58/Ser62) (right top panel), or anti-Flag (2nd panels). Lysates were also subjected to immunoblot analysis with anti-Flag (3rd panels), anti-GFP (4th panels), or anti-tubulin (bottom panels). (D) Recombinant DYRK2 and/or His-GSK3β were incubated with GST–c-Jun210–310 or GST–c-Myc1–100. Reaction products were analyzed with the indicated antibodies.
Abrogation of DYRK2 stabilizes c-Jun and c-Myc.
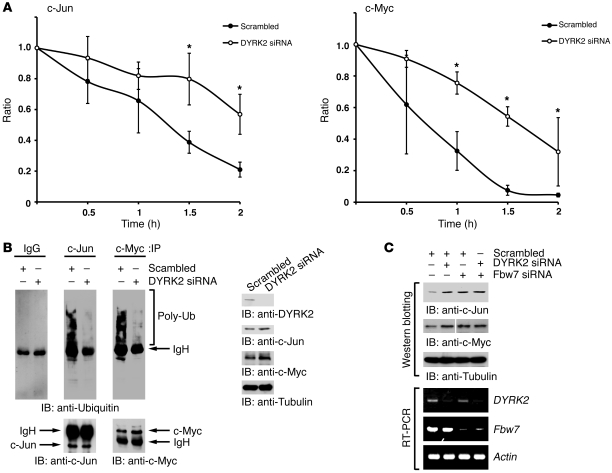
Accumulating studies have revealed that GSK3β phosphorylates and destabilizes c-Jun and c-Myc (12). In this context, we observed that phosphorylation by GSK3β requires prior phosphorylation by DYRK2, suggesting its indispensable involvement in the stability of c-Jun and c-Myc. To determine the direct role for DYRK2 on the half-life of c-Jun and c-Myc, U2OS cells were treated with an inhibitor of protein synthesis, cycloheximide (CHX). As expected, expression of c-Jun was unstable following treatment with CHX (Figure 3A). In sharp contrast, depletion of DYRK2 stabilized c-Jun even after inhibition of protein synthesis (Figure 3A). Comparable findings were obtained with c-Myc stability (Figure 3A). Previous studies have demonstrated that turnover of c-Jun and c-Myc is regulated by the ubiquitin-proteasome pathway. In this regard, to determine whether abrogation of DYRK2 impairs ubiquitination of c-Jun and c-Myc, U2OS cells were transfected with scrambled siRNA or DYRK2 siRNA in the presence of the proteasome inhibitor MG-132. Analysis with antiubiquitin demonstrated that constitutive ubiquitination of c-Jun and c-Myc was prevented in cells silenced for DYRK2 (Figure 3B). Moreover, ectopic expression of DYRK2 WT, but not the dominant negative DYRK2-KR mutant, increased ubiquitination of c-Myc (Supplemental Figure 3A). Intriguingly, however, ectopic expression of GSK3β or DYRK2 alone was insufficient to downregulate c-Jun/c-Myc (Supplemental Figure 3B). Cotransfection of GSK3β with DYRK2 robustly attenuated c-Jun/c-Myc expression (Supplemental Figure 3B). Destabilization of c-Jun/c-Myc was completely rescued by treatment of cells with MG-132 (Supplemental Figure 3B), suggesting that c-Jun and c-Myc were ubiquitinated and degraded in the proteasome following phosphorylation by DYRK2 and GSK3β. To further define an indispensable role for GSK3β on c-Jun/c-Myc ubiquitination, U2OS cells were transfected with HA vector, HA-GSK3β, or Flag-DYRK2 WT in the presence of MG-132. Coexpression of DYRK2 and GSK3β substantially enhanced ubiquitination of c-Jun/c-Myc compared with GSK3β or DYRK2 expression alone (Supplemental Figure 3, C and D), suggesting that dual phosphorylations by DYRK2 and GSK3β require full ubiquitination of c-Jun/c-Myc. Phosphorylated c-Jun/c-Myc are eventually ubiquitinated by Fbw7 for degradation (6). In this regard, we examined the effect of Fbw7 on DYRK2-mediated turnover of c-Jun/c-Myc. As previously shown, knockdown of DYRK2 increased expression of c-Jun/c-Myc (Figure 3C). Strikingly, depletion of Fbw7 elevated c-Jun/c-Myc expression regardless of DYRK2, to levels similar to those with DYRK2 depletion alone (Figure 3C). These findings clearly indicate that DYRK2 functions upstream of Fbw7. To determine the stability of the c-Jun/c-Myc mutant proteins in the priming sites, 293T cells were cotransfected with the Flag–c-Jun S243A mutant and scrambled siRNA or DYRK2 siRNA. Immunoblot analysis of anti-Flag revealed that the stability of the c-Jun S243A mutant remained constant regardless of DYRK2 status (Supplemental Figure 3E). Comparable results were obtained with the c-Myc S62A mutant (Supplemental Figure 3E). These findings suggest that mutations for the corresponding DYRK2 priming sites in c-Jun/c-Myc confer resistance to being phosphorylated by DYRK2/GSK3, followed by degradation. Taken together, these results collectively support a model in which sequential phosphorylation by DYRK2 followed by GSK3β destabilizes c-Jun and c-Myc to induce the Fbw7-mediated ubiquitination and subsequent proteasomal degradation.
Figure 3. DYRK2 regulates c-Jun and c-Myc stability.
(A) U2OS cells were transfected with scrambled siRNA or DYRK2 siRNA. After transfection, cells were incubated with 40 μg/ml CHX for the indicated times. Lysates were analyzed by immunoblotting with anti–c-Jun, anti–c-Myc, or anti-tubulin. The signals for c-Jun (left) or c-Myc (right) were scanned to compare the amount in control (time 0). *P < 0.05. Data represent mean ± SD. (B) U2OS cells were transfected with scrambled siRNA or DYRK2 siRNA and incubated with 10 μM MG-132 for 4 hours. Lysates were immunoprecipitated with rabbit IgG, anti–c-Jun, or anti–c-Myc followed by immunoblotting with anti-ubiquitin, anti–c-Jun, or anti–c-Myc. Lysates were also analyzed by immunoblotting with indicated antibodies (right panels). (C) U2OS cells were transfected with scrambled siRNA, DYRK2 siRNA, or Fbw7 siRNA. Lysates were immunoblotted with anti–c-Jun, anti–c-Myc, or anti-tubulin. The lanes separated by the white line were run on the same gel, but were noncontiguous. Total RNAs were analyzed by RT-PCR using DYRK2-specific, Fbw7-specific, or actin-specific primers.
Silencing DYRK2 accelerates the G1/S transition.
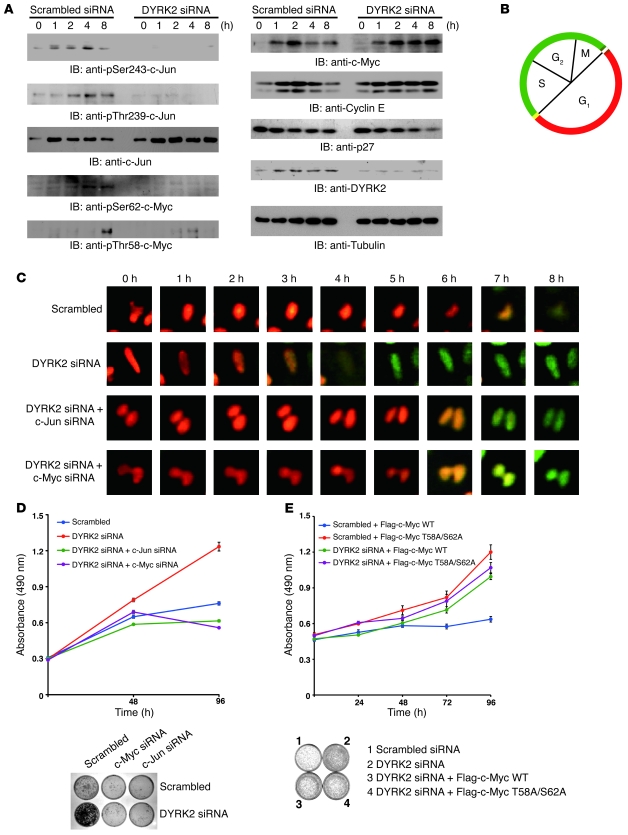
The findings that DYRK2 controls destruction of c-Jun and c-Myc provided us with the further notion that DYRK2 affects the G1/S transition coupled with c-Jun and c-Myc. c-Jun expression was increased at 1–2 hours and then markedly declined in control cells (Figure 4A). In contrast, in cells silenced for DYRK2, expression of c-Jun was constant from 1 to 8 hours (Figure 4A). Serum refeeding into serum-starved cells was also associated with a transient augmentation and subsequent decline of c-Myc expression (Figure 4A). In contrast, deprivation of DYRK2 abrogated downregulation of c-Myc expression (Figure 4A). We also assessed the phosphorylation status of endogenous c-Myc. The priming phosphorylation of endogenous c-Myc at Ser62 was detectable at 4 to 8 hours after serum stimulation (Figure 4A). In contrast, Ser62 phosphorylation was completely revoked in cells silenced for DYRK2, suggesting that endogenous DYRK2 is responsible for Ser62 phosphorylation. Phosphorylation of c-Myc at Thr58 by GSK3β occurred at 8 hours, thus indicating that this modification follows Ser62 phosphorylation by DYRK2 (Figure 4A). Importantly, the finding that depletion of DYRK2 impaired Thr58 phosphorylation demonstrated that DYRK2-mediated priming phosphorylation is a prerequisite for subsequent phosphorylation of c-Myc by GSK3β. Similar characteristics, albeit occurring prior to the phosphorylation of c-Myc, were observed in endogenous c-Jun phosphorylation (Figure 4A). Coincident with the expression levels of c-Jun or c-Myc, their target gene controlling the G1/S transition, cyclin E, exhibited enhanced and prolonged expression in cells silenced for DYRK2 (Figure 4A). As expected, degradation of G1/S cyclin/cdk inhibitor p27Kip1 by cyclin E/cdk2 phosphorylation was promoted in DYRK2-depleted cells (Figure 4A). To visualize the cell cycle progression, we utilized fluorescent, ubiquitination-based cell cycle indicator, (Fucci) technology, which can distinguish cells in the G1 phase from those in the S/G2/M phases (Figure 4B and ref. 23). Fucci-expressing HeLa cells were serum starved for synchronization prior to transfection with siRNAs. Cells were then stimulated with serum under time-lapse imaging using computer-assisted fluorescence microscopy. As expected, cell nuclei were visualized in red, indicating that cells were in the G1 phase. In control cells, the G1/S transition was around 8 hours after serum refeeding (Figure 4C). In contrast, onset of the S phase was at 4 hours after serum stimulation in DYRK2-depleted cells (Figure 4C). Intriguingly, depletion of c-Jun/c-Myc rescued aberrant S phase entry in cells silenced for DYRK2 (Figure 4C). These findings indicated that impairment of DYRK2 is associated with accelerated progression of the G1 phase. Serum-starved cells entered the S phase around 8 hours following serum stimulation. In contrast, depletion of DYRK2 accelerated the onset of S phase at 4 hours. To extend these findings, we analyzed the growth rate of U2OS cells by MTS assays. As shown previously (Figure 1B and Supplemental Figure 1B), knocking down DYRK2 alone accelerated cell growth. In contrast, deprivation of c-Jun/c-Myc normalized dysregulated cell growth elicited by DYRK2 depletion (Figure 4D). Similar results were obtained in colony formation assays with U2OS cells (Figure 4D) and HeLa cells (Supplemental Figure 4G). To determine whether priming phosphorylation of c-Myc affects proliferation of cells, we carried out MTS assays. Ectopic expression of the c-Myc T58A/S62A mutant, but not c-Myc–WT, accelerated proliferation of U2OS cells (Figure 4E). Moreover, DYRK2 silencing elevated cell growth regardless of c-Myc phosphorylation status, suggesting that abrogation of c-Myc priming phosphorylation by DYRK2 is associated with induction of aberrant cell growth (Figure 4E). Similar findings were obtained in colony formation assays (Figure 4E). Taken together, these results indicate that DYRK2 governs proper progression from G1 to S phase by controlling c-Jun and c-Myc via priming phosphorylation.
Figure 4. The G1/S transition is accelerated in DYRK2-depleted cells.
(A) U2OS cells were transfected with scrambled siRNAs or DYRK2 siRNAs, followed by serum starvation and stimulation for the indicated amount of time. Lysates were subjected to immunoblot analysis with the indicated antibodies. (B) Schematic depiction of Fucci system. (C) Fucci-expressing HeLa cells were transfected with scrambled siRNA or DYRK2 siRNA. Culture medium was changed to DMEM containing 0.25% serum. After refeeding with serum, cells were subjected to time-lapse fluorescence microscopy. (D) U2OS cells were transfected with scrambled siRNA, DYRK2 siRNA, c-Jun–specific siRNA, or c-Myc–specific siRNA. After transfection, cells were subjected to the MTS assay (upper panel) or the colony formation assay (lower panel). (E) U2OS cells were cotransfected with scrambled siRNA or DYRK2 siRNA and Flag–c-Myc WT or Flag–c-Myc T58A/S62A. After transfection, cells were subjected to the MTS assay (upper panel) or the colony formation assay (lower panel). Data represent mean ± SD.
DYRK2 depletion enhances tumor growth.
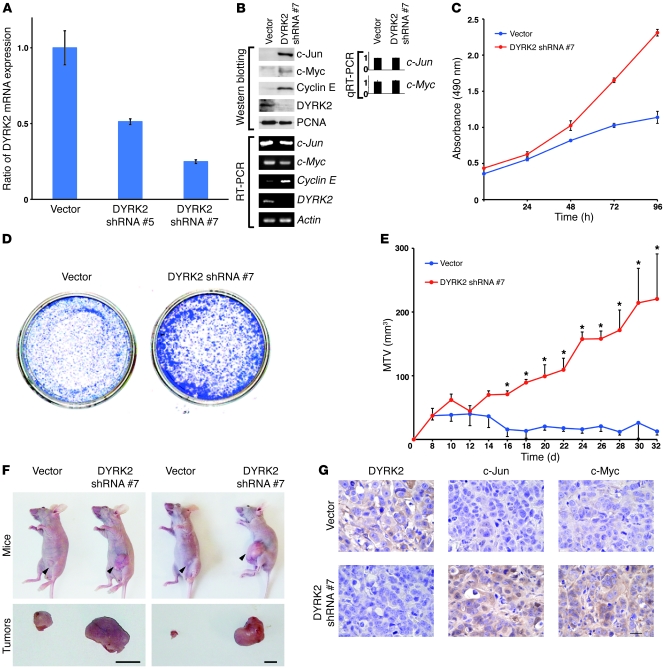
To determine whether abrogation of DYRK2 expression affects tumor cell growth, scrambled siRNA or DYRK2 siRNA was cotransfected together with GFP vector or DYRK2 variants into HeLa cells (Supplemental Figure 4A). Analysis of colony formation assays revealed that knockdown of DYRK2 was associated with aberrant proliferation (Supplemental Figure 4B). Significantly, this dysregulation of cell growth was overcome by ectopic expression of rDYRK2 WT, but not rDYRK2-KR (Supplemental Figure 4B). Comparable results were obtained in U2OS cells (Supplemental Figure 4, A and B). To extend these findings in vivo, transduced HeLa cells were injected subcutaneously into the flanks of nude mice. In concert with in vitro data from colony formation assays, impairment of DYRK2 in HeLa cells was associated with a significant acceleration of tumor growth (Supplemental Figure 4, C and D). Moreover, elevated tumor growth by endogenous DYRK2 silencing was markedly attenuated in exogenous expression of rDYRK2 WT, but not rDYRK2-KR (Supplemental Figure 4, C and D). These results indicate that DYRK2 controls tumor progression in a kinase activity–dependent manner. We assessed the expression levels of DYRK2 in enucleated tumors from HeLa xenografts. Both analyses of RT-PCR and immunostaining indicated that DYRK2 expression was attenuated in tumors (Supplemental Figure 4, E and F). These results provide evidence that transient transfection of DYRK2 siRNA into HeLa cells sustained knockdown efficiency in vivo by an unknown mechanism. In this context, a previous study reported that depletion of ILK expression prior to introduction of cells by siRNAs into the mouse xenograft model resulted in striking and sustained in vivo effects on tumor growth and apoptotic and angiogenic characteristics (24), implying a more permanent alteration by siRNA transfection of cells in vivo. To extend and confirm findings with transient siRNA transfection, we established stably DYRK2-depleted cells. MCF-7 cells were transfected with pSuper vector or pSuper DYRK2 shRNAs. Knockdown of DYRK2 was confirmed by RT-PCR (Figure 5, A and B) and immunoblotting (Figure 5B). Interestingly, the protein expression of c-Jun, c-Myc, and cyclin E were all upregulated in MCF-7 cells stably silenced for DYRK2 (Figure 5B). Importantly, transcripts of c-Jun and c-Myc remained unchanged, whereas transcript of cyclin E was markedly elevated in DYRK2-depleted cells (Figure 5B). These findings suggest that stable deprivation of DYRK2 hinders consistent degradation of c-Jun/c-Myc to augment their transcriptional target in the G1/S cyclin, cyclin E. In concert with the data obtained using siRNA transfection (Figure 1B and Supplemental Figure 1B), stable knockdown of DYRK2 strikingly elevated cell growth (Figure 5C). In addition, as previously shown in the transient transfection studies (Figure 1A, Supplemental Figure 1A, and Supplemental Figure 4B), colony formation assays revealed that stable knockdown of DYRK2 was associated with aberrant proliferation (Figure 5D). We further conducted in vivo tumorigenicity experiments with MCF-7 cells stably silenced for DYRK2. No marked tumor growth has been observed in mice injected with mocked MCF-7 cells (Figure 5, E and F). In stark contrast, similarly shown in xenograft studies with HeLa cells transiently transfected with DYRK2 siRNA (Supplemental Figure 4C), stable depletion of DYRK2 significantly accelerated tumor growth (Figure 5, E and F). As shown in HeLa xenografts (Supplemental Figure 4F), immunohistochemical analysis at sacrifice demonstrated that depletion of DYRK2 xenografts displayed dramatically elevated expression of c-Jun/c-Myc compared with controls, supporting the conclusion that DYRK2 is a negative regulator of c-Jun/c-Myc stability (Figure 5G). These findings demonstrate that knockdown of DYRK2 confers aberrant growth of various carcinoma cells in in vivo tumor models. To further assess whether DYRK2 abrogation–induced tumor cell proliferation requires c-Myc and c-Jun, scrambled siRNA or DYRK2 siRNA was cotransfected together with c-Myc siRNA or c-Jun siRNA into HeLa cells. Analysis of colony formation assays revealed that, as shown previously, depletion of DYRK2 conferred accelerated proliferation (Supplemental Figure 4G). Moreover, this dysregulation of cell growth was completely overcome by codeprivation of c-Myc or c-Jun. Consistent results were obtained in U2OS cells (Figure 4D). These data suggest that regulation of cell growth by DYRK2 depends upon c-Myc or c-Jun. To strengthen these findings in vivo, transfected HeLa cells were injected subcutaneously into the flanks of nude mice. Coincident with the data obtained from colony formation assays, codepletion of c-Myc or c-Jun completely cancelled facilitation of tumor growth driven by silencing of DYRK2 alone (Supplemental Figure 4, H and I; also see Supplemental Figure 4C). Taken together, these findings indicate that DYRK2 contributes to growth of carcinoma cells in in vivo tumor models.
Figure 5. Stable knockdown of DYRK2 elevates tumor growth in vitro and in vivo.
(A) MCF-7 cells were transfected with pSuper vector or pSuper-DYRK2 shRNA. To isolate stable shRNA-expressing cells, transfected cells were selected with puromycin. Knockdown efficiency of DYRK2 was monitored by quantitative RT-PCR. (B) Lysates form MCF-7 cells stably expressing vector (vector) or pSuper DYRK2 shRNA (DYRK2 shRNA #7) were analyzed by immunoblotting with indicated antibodies. Total RNAs were analyzed by RT-PCR. The result of quantitative RT-PCR was normalized for the level of Actin and represents the relative fold induction compared with control sample. The data were evaluated from 3 independent experiments, each performed in triplicate. (C and D) Cell growth was analyzed by the MTS assay (C) or the colony formation assay (D). (E and F) Transduced MCF-7 cells with 50% matrigel were inoculated into the opening of the lactiferous duct in the abdominal mammary gland in BALB/c nu/nu mice implanted with 17β-estradiol tablets. Tumor size was measured using calipers (n = 3). Data (maximum tumor volume [MTV]) indicate mean ± SD (E). *P < 0.05. Representative pictures of tumor-bearing nude mice (upper panels) and tumors (lower panels), which were taken 10 weeks after inoculation (F). Arrowheads indicate inoculated tumors. Scale bar: 10 mm. (G) Enucleated tumors were subjected to immunostaining with anti-DYRK2, anti–c-Jun, or anti–c-Myc. Scale bar: 50 μm.
DYRK2 expression is reduced or abolished in multiple human tumor tissues.
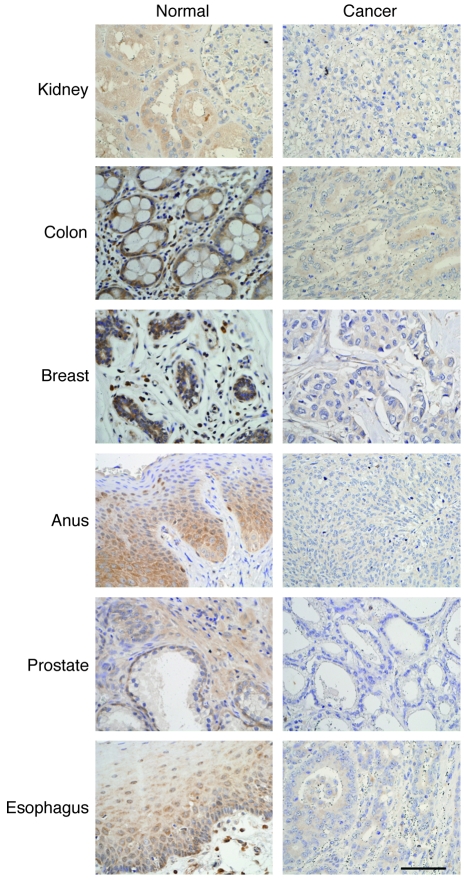
To assess the physiological relevance of our results to the biology of human cancers, we investigated DYRK2 expression using tissue microarrays derived from distinct human tumor tissues. Analysis of immunohistochemistry with anti-DYRK2 revealed that DYRK2 expression was positive and intact in all normal tissues (Figure 6). In contrast, DYRK2 was markedly attenuated or abolished in tumor samples originating from tissues such as breast, colon, esophagus, anus, prostate, kidney, and hypopharynx (Figure 6 and data not shown). These findings thus provide a potential correlation between downregulation of DYRK2 expression and carcinogenesis or tumor progression.
Figure 6. Downregulation of DYRK2 in various human tumor tissues.
Immunohistochemical staining of anti-DYRK2 was performed using multiple cancerous and corresponding normal tissue microarrays. Scale bar: 200 μm.
Downregulation of DYRK2 correlates with the invasiveness of human breast cancers.
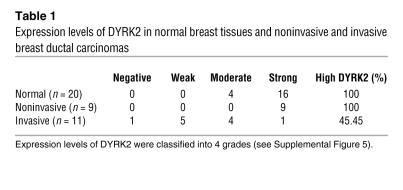
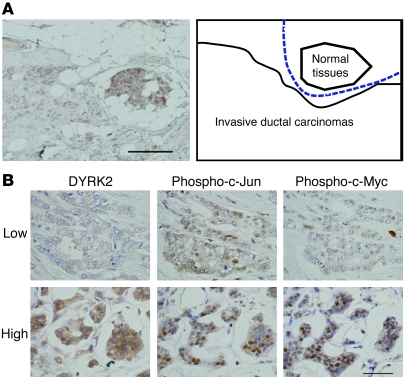
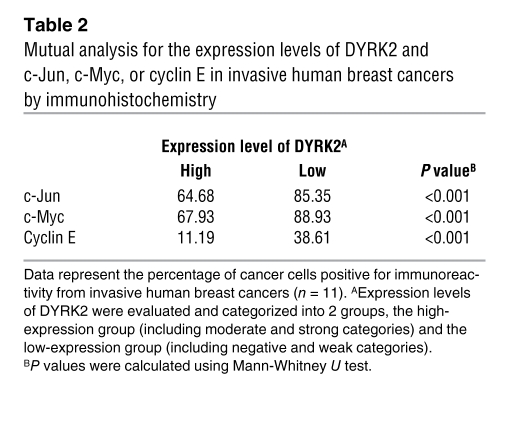
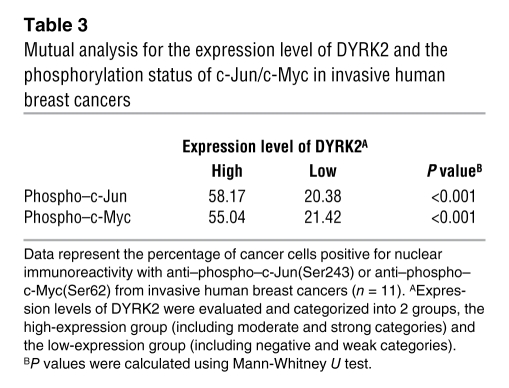
To further define a biological role for DYRK2 in carcinogenesis or tumor progression, we focused on human breast cancers, since aberrant activation of c-Jun and c-Myc affects progression of breast cancers (25, 26). To initially examine protein levels of DYRK2 in human breast cancers, we obtained samples of breast cancerous tissues and concomitant normal tissues from 20 patients with various stages of breast cancers. The ratios of DYRK2-positive cells were scored to categorize its expression levels into 4 grades, which were defined as follows: negative, 0; weak, greater than 0 to 1/3; moderate, greater than 1/3 to 2/3; and strong, greater than 2/3 to 1 (Supplemental Figure 5). In all normal tissues, DYRK2 was highly expressed to levels classified into moderate or strong, both of which were clustered as a high group (Table 1). Similar findings were obtained in noninvasive breast cancer tissues (Table 1). In contrast, in 6 out of 11 invasive breast cancer tissues, DYRK2 expression was significantly decreased to levels classified into weak or negative, both of which were categorized as a low group (Table 1 and Figure 7A). These results suggest the possibility that downregulation of DYRK2 contributes to the invasive potential of human breast cancers. We further examined the expression of c-Jun, c-Myc, and cyclin E in the invasive ductal breast cancer tissues. Positive expression was significantly higher in tissues in the low DYRK2 group compared with those in the high DYRK2 group (Table 2). This finding thus indicated that the accumulation of c-Jun, c-Myc, and cyclin E is inversely correlated with the expression level of DYRK2. Since DYRK2 phosphorylation of c-Jun and c-Myc targets them for degradation, we explored a potential correlation between expression levels of DYRK2 and phosphorylation levels of c-Jun and c-Myc in the invasive ductal breast cancer tissues. In samples from the high group of DYRK2, more than 50% of cells stained positive for phospho–c-Jun at Ser243 or phospho–c-Myc at Ser62 (Table 3 and Figure 7B). In stark contrast, the percentages of cells with phosphorylated c-Jun or c-Myc were significantly attenuated in samples from the low group of DYRK2 (Table 3 and Figure 7B). These findings support a model in which downregulation of DYRK2 stabilizes c-Jun and c-Myc to accelerate tumor progression and invasion (Figure 8).
Table 1 .
Expression levels of DYRK2 in normal breast tissues and noninvasive and invasive breast ductal carcinomas
Figure 7. DYRK2 inactivation is associated with the impairment of c-Jun/c-Myc phosphorylation and with aggressiveness of human breast cancers.
(A) Representative immunohistochemical staining of DYRK2 protein in the breast cancer specimen (left panel). Scale bar: 500 μm. Schematic depiction for the boundary of the normal tissues and invasive breast ductal carcinomas shown in the left panel (right panel). (B) Mutual analysis for the expression level of DYRK2 and the phosphorylation status of c-Jun/c-Myc in invasive human breast cancers. Representative immunohistochemical staining of anti-DYRK2, anti–phospho–c-Jun(Ser243), and anti–phospho–c-Myc(Ser62) are shown. Scale bar: 150 μm.
Table 2 .
Mutual analysis for the expression levels of DYRK2 and c-Jun, c-Myc, or cyclin E in invasive human breast cancers by immunohistochemistry
Table 3 .
Mutual analysis for the expression level of DYRK2 and the phosphorylation status of c-Jun/c-Myc in invasive human breast cancers
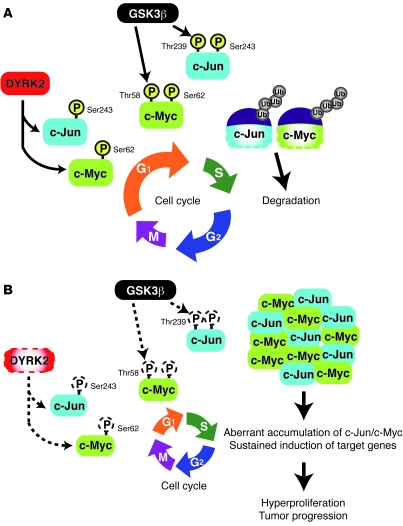
Figure 8. The model of DYRK2-mediated cell cycle regulation.
(A) At the late G1 phase, c-Myc and c-Jun are phosphorylated by DYRK2, which serves as a priming site for binding of GSK3β. GSK3β then obtains the license to phosphorylate (P) c-Jun and c-Myc via recognition of priming phosphorylation. Phosphorylated c-Jun and c-Myc bind to the ubiquitin ligase for their degradation. (B) In the absence of DYRK2, GSK3β is unable to phosphorylate c-Jun and c-Myc. Unphosphorylated c-Jun and c-Myc escape from the ubiquitin-mediated proteasomal degradation. Accumulation of c-Jun and c-Myc induces their target genes, such as cyclin E.
Discussion
We report here for what we believe is the first time that DYRK2 functions as a priming kinase of GSK3β for destroying c-Jun or c-Myc at the G1/S transition. Previous studies have demonstrated that c-Jun and c-Myc are coordinately regulated by GSK3β and the ubiquitin ligase Fbw7 (6). Growth stimulation in the early G1 phase induces Akt that phosphorylates and inactivates GSK3β (27). Conversely, Akt activity markedly diminishes in the late G1 phase, which triggers reactivation of GSK3β. In this model, GSK3β phosphorylation of c-Jun and c-Myc requires priming phosphorylation at Ser residues. Indeed, identification of priming kinases has been studied; however, little is known about physiological kinases. Moreover, to our knowledge, there is no report demonstrating that a single kinase contributes to priming phosphorylation of both c-Jun and c-Myc. Intriguingly, c-Jun and c-Myc were simultaneously destroyed at the late G1 phase. Given the fact that these oncogenes are regulated by the common kinase GSK3β and the common ubiquitin ligase Fbw7, it is conceivable that there should be a common priming kinase. In this context, the present study delineates DYRK2 kinase as a candidate. We further demonstrate indispensable prephosphorylation by DYRK2 in GSK3β-mediated degradation of c-Jun and c-Myc. In this regard, previous studies have shown that GSK3β activity inversely reflects expression levels of c-Jun and c-Myc in the G1 phase (10). Nonetheless, deprivation of DYRK2 rendered GSK3β unable to phosphorylate c-Jun and c-Myc, regardless of GSK3β activity. Collectively, these data suggest that sequential phosphorylation of DYRK2 followed by GSK3β constitutes a fail-safe mechanism that accurately regulates the degradation of these proteins.
While accumulating studies have ensured a requirement of priming phosphorylation for GSK3β-mediated destruction, biological roles for priming phosphorylation on c-Jun or c-Myc are controversial. A previous study showed that Ser62 phosphorylation stabilizes c-Myc (28). In contrast, other studies demonstrated that abrogation of Ser243 phosphorylation by mutation or dephosphorylation stabilizes c-Jun, resulting in an increase in c-Jun–induced gene expression (10, 29). These findings thus indicate opposite roles for priming phosphorylation: stabilization for c-Myc and destabilization for c-Jun. We are unable to distinctly explain this apparent inconsistency. In any case, however, priming phosphorylations of c-Jun and c-Myc are eventually triggered for Fbw7-mediated proteolysis. It should be noted that the study conducted by Sears et al. (28) was based on the overexpression of the c-Myc S62A mutant in quiescent cells. In this context, it could not exclude the possibility that conformational changes elicited by S62A mutation might render c-Myc more susceptible to being destroyed by Fbw7 or undefined ubiquitin ligases. Obviously, further studies are needed to clarify this issue. Instead, at least in the G1/S transition in proliferating cells, our data clearly demonstrate that loss of phosphorylation by DYRK2 allows c-Jun and c-Myc to escape from ubiquitin-mediated destruction. It is thus likely that DYRK2 phosphorylation of c-Jun and c-Myc is a licensing event that allows GSK3β to phosphorylate them, thus targeting them for degradation. Significantly, elevated and stable expression of c-Jun and c-Myc induced by stably knocking down DYRK2 exhibited dysregulation of transactivation targets, especially those controlling the G1/S transition, such as cyclin E. As a consequence, we observed accelerated proliferation, presumably due to premature progression through the G1 phase (Figure 1). In concert with these in vitro data, we revealed that downregulation of DYRK2 potentiates facilitation of tumor growth in xenograft models (Figure 5E and Supplemental Figure 4C). Strikingly, this growth advantage was completely abrogated by codeprivation with c-Jun or c-Myc in vivo (Supplemental Figure 4H). Taken together, our findings support a model in which DYRK2 controls cell proliferation via c-Jun and c-Myc.
As found for c-Jun and c-Myc, GSK3β phosphorylation recruits ubiquitin ligases for degradation of substrates, such as cdc25A, Mcl-1, PRLr, and cyclin D (30–34). However, in contrast to c-Jun and c-Myc, these substrates are modified by distinct ubiquitin ligases despite coordinate phosphorylation by GSK3β. While each of the priming kinases themselves remain unclear, we speculate that a priming phosphorylation, rather than GSK3 phosphorylation, determines the specificity for the ubiquitin ligase. In this regard, it is plausible that the recruitment of Fbw7 could predominately stipulate priming phosphorylation by DYRK2.
Dysregulated expression of c-Myc/c-Jun exerts significant roles on cell cycle progression. A reduction in c-Myc levels results in a lengthened G1 phase, with concomitant delayed expression of cyclin E, whereas ectopic expression of c-Myc leads to a shortened G1 phase mainly due to cyclin E overexpression (35–37). Furthermore, accumulating studies have demonstrated that overexpression of cyclin E shortens the G1 phase and accelerates the G1/S transition (38–41), indicating that cyclin E expression directly precedes the transition from the G1 to the S phase. Indeed, the cyclin E/cdk2 phosphorylation of p27Kip1 triggers its degradation to induce an exit from the G1 phase (42). This evidence supports our findings that sustained expression of c-Jun/c-Myc by DYRK2 knockdown accelerates G1/S transition attributable to elevated expression of cyclin E accompanied with expedited degradation of p27/Kip1 (Figure 4A). Importantly, a shortened G1 phase is one of the prominent characteristics in cancer progression. This may at least in part be explained by our results showing that impaired DYRK2 correlates with the invasiveness of breast cancers. A recent study demonstrated that DYRK2 phosphorylates katanin p60 for its degradation and controls mitotic transition in HeLa cells (18). Similarly, our findings revealed relative increments of the G2/M phase in HeLa cells depleted for DYRK2. However, calculated duration of the G2/M phase in DYRK2-deprived cells was comparable with that in mocked cells. Instead, duration of the G1 phase was markedly reduced in DYRK2-depleted HeLa cells (Figure 1D). Based on our findings, we thus concluded that DYRK2 controls the cell cycle at the G1 phase, whereas we cannot exclude the possibility that DYRK2 plays a role in mitosis. Indeed, the length of the G2/M phase was (not statistically significant but) still substantially augmented in cells silenced for DYRK2 (Figure 1D and Supplemental Figure 1D). These findings suggest the possibility that DYRK2 regulates the cell cycle at G1/S and/or G2/M transitions by distinct mechanisms.
Dysregulated expression of c-Myc/c-Jun also exerts critical influences on cancer progression. For example, high c-Myc expression by missense mutations in many human hematopoietic malignancies has been correlated with a poor prognosis (43). Intriguingly, the majority of the mutations reside within the aminoterminal transactivation domain of c-Myc including Thr58 and Ser62 in Burkitt’s lymphoma (44). Importantly, somatic mutations of c-Myc at Thr58 and Ser62 and their flanking amino acids confer increased oncogenic activity in Burkitt’s lymphoma (45), suggesting that phosphorylation of these sites leads to downregulation of c-Myc growth-promoting activity. Consistent with this is the finding that v-myc oncogenes frequently contain mutations in these phosphorylation sites (46, 47). Similar findings were observed with oncogenic v-Jun, in which the priming phosphorylation residue coinciding with c-Jun Ser243 is mutated to Phe (48). In contrast, another study demonstrated that normal mammary epithelial cells exhibited low c-Myc Ser62 phosphorylation, whereas invasive adenocarcinoma showed high Ser62 phosphorylation together with high c-Myc levels (49). It is conceivable that if GSK3β is inactivated, phospho-Ser62 is upregulated but phospho-Thr58 is downregulated; thus, c-Myc is accumulated, which is frequently observed in breast cancers (50). Notably, we found that impairment of priming phosphorylation in c-Myc and c-Jun by silencing DYRK2 increased cell proliferation as well as tumor growth. Collectively, lack of priming phosphorylation in c-Myc or c-Jun would be suspected for contribution to cancer development. In this context, our findings that robust correlation between DYRK2 inactivation and impaired phosphorylation of c-Jun and c-Myc in advanced human breast cancers imply that dysregulation of DYRK2 could trigger enhanced migration and invasion of tumors. In agreement with this model, accumulation of unphosphorylated c-Jun or c-Myc contributes to tumor progression. In this context, besides c-Jun and c-Myc, substrates for DYRK2 are largely unknown. It is thus possible that DYRK2 may phosphorylate unknown targets that are associated with cell proliferation or tumor progression. Considering this and given the previous findings that DYRK2 phosphorylates p53 at Ser46 to induce apoptosis in response to DNA damage (20), we are now investigating the hypothesis that DYRK2 exerts suppressive function for tumor development.
Until now, dysregulation of DYRK2 expression in human cancer has also been uncertain. DYRK2 is harbored at the locus on the chromosome 12q15 region, which is frequently amplified in various cancers. Importantly, MDM2 is mapped to the same locus, suggesting the possibility that DYRK2 and MDM2 exhibit genetic linkage on expression (51). Despite the amplification of copy number at 12q15, to our knowledge, no study has been carried out to assess DYRK2 expression at the protein levels. In contrast, accumulating reports employed upregulation of MDM2 expression at the protein levels in various tumor tissues. In this regard, our recent findings indicate that DYRK2 is constitutively degraded by an MDM2-mediated ubiquitination and proteasome machinery (22). It is thus conceivable that overexpressed MDM2 is associated with attenuation or abrogation of DYRK2 expression at the protein level even if the DYRK2 gene is amplified due to the 12q15 amplicon.
In conclusion, the present study demonstrates that DYRK2 controls cell cycle progression at the G1/S phase via phosphorylation of c-Jun and c-Myc. DYRK2 inactivation contributes to cell proliferation and tumor progression. We therefore propose that dissecting both DYRK2 expression and c-Jun/c-Myc phosphorylation could be valuable for the prognosis of invasiveness in tumors. Our results also emphasize the use of DYRK2 as a potential target of therapeutic intervention for advanced cancers, which may be achieved by restoring its expression and function.
Methods
Cell culture and treatment.
U2OS (human osteosarcoma) cells were cultured in RPMI 1640 medium supplemented with 10% heat-inactivated fetal bovine serum, 100 units/ml penicillin, 100 μg/ml streptomycin, and 2 mM l-glutamine. 293T (human embryonal kidney), HeLa (human cervical carcinoma), and MCF-7 (human mammary carcinoma) cells were grown in DMEM containing 10% heat-inactivated fetal bovine serum, 100 units/ml penicillin, 100 μg/ml streptomycin, and 2 mM l-glutamine. Fucci-expressing HeLa cells were provided by Riken Cell Bank. Cells were treated with 40 μg/ml CHX (Merck).
Plasmid construction.
Flag-tagged and GFP-tagged DYRK2 was constructed as described previously (20). c-Myc cDNA was cloned into the pcDNA3-Flag vector (52, 53). Flag-tagged c-Jun was a gift from S. Kitajima (Tokyo Medical and Dental University) (54). GSK3β cDNA was cloned into the pcDNA3-HA vector. Various mutations were introduced by site-directed mutagenesis. siRNA-resistant forms of DYRK2 were generated by introducing silent mutations in the targeting regions for DYRK2 siRNA.
Cell transfection.
Plasmids were transfected using FuGENE 6 (Roche). DYRK2-specific siRNAs (QIAGEN, Invitrogen), c-Jun siRNA (Invitrogen), c-Myc siRNA (Invitrogen), Fbw7 siRNA (Nippon EGT), and negative control siRNAs (QIAGEN) were used. Transfection of siRNAs was performed using Lipofectamine 2000 and Lipofectamine RNAi MAX (Invitrogen). To isolate stable transfectants, MCF-7 cells were transfected with pSuper vector (Oligoengine) or pSuper DYRK2 shRNA in the presence of puromycin.
Immunoblotting and immunoprecipitation.
Cells were washed twice in chilled PBS and resuspended in lysis buffer (50 mM Tris-HCl, pH 7.6, 150 mM NaCl, 10 mM NaF, 1 mM Na3VO4, 1 mM PMSF, 1 mM DTT, 10 μg/ml aprotinin, 1 μg/ml leupeptin, 1 μg/ml pepstatin A, and 1% NP-40). Cell extracts were centrifuged for 5 minutes at 4°C. Supernatants were separated by SDS-PAGE and transferred to nitrocellulose membranes. The membranes were incubated with anti–c-Jun (Santa Cruz Biotechnology Inc.), anti–phospho–c-Jun(Ser243) (Abcam), anti–phospho–c-Jun(Thr239) (Abcam), anti–c-Myc (Santa Cruz Biotechnology Inc.), anti–phospho–c-Myc(Thr58/Ser62) (Cell Signaling Technology), anti–phospho–c-Myc(Ser62) (MBL), anti–phospho–c-Myc(Thr58) (Abcam), anti-tubulin (Sigma-Aldrich), anti–cyclin E (Santa Cruz Biotechnology Inc.), anti-ubiquitin (Santa Cruz Biotechnology Inc.), anti-DYRK2 (Santa Cruz Biotechnology Inc.), anti-Flag (Sigma-Aldrich), anti-GFP (Nacalai Tesque), anti-PCNA (Santa Cruz Biotechnology Inc.), or anti-GST (Nacalai Tesque). Immune complexes were incubated with secondary antibodies and visualized by chemiluminescence (PerkinElmer). For endogenous c-Myc immunoprecipitation, cell lysates were incubated with anti–c-Myc (Santa Cruz Biotechnology Inc.) for 2 hours at 4°C and the beads were incubated with protein G–sepharose (Invitrogen). The beads were washed 3 times with the lysis buffer and boiled for 5 minutes. For immunoprecipitation of Flag-tagged proteins, lysates were incubated with anti-Flag agarose (Sigma-Aldrich). Immunoprecipitates were eluted with the Flag peptide (Sigma-Aldrich) as needed.
Cell growth assay.
The number of viable cells was counted by trypan blue dye exclusion using a hemocytometer. For MTS assay, cells were seeded in 96-well plates and transfected as described (55). MTS assays were performed using CellTiter 96 AQ One Solution Cell Proliferation Assay Kit (Promega). The absorbance was measured at 490 nm with the use of a multilabel counter (PerkinElmer). Doubling time for cells was calculated as described (56). The length of each phase of the cell cycle was calculated as a product of the doubling time and the percentage of cells in a given phase, as measured by FACS analysis.
In vitro kinase assay.
Recombinant His-DYRK2 and GST-GSK3β were obtained from Millipore and Invitrogen, respectively. Flag-DYRK2 was immunoprecipitated with anti-Flag agarose (Sigma-Aldrich) from 293T cells transfected with Flag-DYRK2. Immunoprecipitates were washed 3 times with lysis buffer and then with kinase buffer (20 mM HEPES, pH 7.0, 10 mM MgCl2, 0.1 mM Na3VO4, and 2 mM DTT). As substrates, GST–c-Jun (amino acid residues 210–310) and GST–c-Myc (amino acid residues 1–100) were purified from E. coli cultures with GST-sepharose beads (GE Healthcare). Immunoprecipitates, GST-GSK3β, or His-DYRK2 was incubated in kinase buffer with substrates and ATP for 20 minutes at 30°C (57). Samples were boiled for 5 minutes and analyzed by SDS-PAGE.
RT-PCR analysis.
Isolation of total RNA from cells or tumor tissues was performed using TRIsure (Nippon Genetics) according to the manufacturer’s instructions. 500 ng of total RNA was amplified using the SuperScript III One-Step RT-PCR System with Platinum Taq Kit (Invitrogen). The reaction for RT-PCR was as follows: cDNA synthesis at 55°C for 30 minutes, denaturation at 94°C for 2 minutes, followed by 30 cycles at 94°C for 15 seconds, 55°C for 30 seconds, and 68°C for 30 seconds, with a final extension at 68°C for 5 minutes. PCR products were separated by 2% agarose gels. For quantitative RT-PCR analysis, total RNA was reverse transcribed into cDNA using a PrimeScript 1st strand cDNA Synthesis Kit (Takara) following the manufacturer’s protocol. The PCR reaction was performed by using KAPA SYBR FAST ABI Prism 2X qPCR Master Mix (Nippon Genetics) according to the instruction manual. Primer sequences are described as follows: c-Jun, 5′-TGACTGCAAAGATGGAAACG-3′ and 5′-CAGGGTCATGCTCTGTTTCA-3′; c-Myc, 5′-TCAAGAGGCGAACACACAAC-3′ and 5′-GGCCTTTTCATTGTTTTCCA-3′; DYRK2, 5′-GGGGAGAAAACGTCAGTGAA-3′ and 5′-TCTGCGCCAAATTAGTCCTC-3′; cyclin E, 5′-ATCCTCCAAAGTTGCACCAG-3′ and 5′-AGGGGACTTAAACGCCACTT-3′; GAPDH, 5′-TCAAGGCTGAGAACGGGAAG-3′ and 5′-ATGGTGGTGAAGACGCCAGT-3′; and actin, 5′-GTGGCCGAGGACTTTGATTG-3′ and 5′-TGGAVTTGGGAGAGGCTGG-3′.
Colony formation assay.
Cells were washed with chilled PBS and fixed with 70% ethanol for 30 minutes at –20°C. After fixation, cells were stained using Giemsa Solution (Wako) for 10 minutes at room temperature. The cells were then washed with PBS twice.
Cell cycle analysis.
Cells were washed with chilled PBS and resuspended with PBS. Cells were incubated with RNase and propidium iodide for 30 minutes at 37°C. Data from the flow cytometry were acquired and analyzed using CytoSoft software (Guava Technologies).
Imaging of Fucci-expressing cells.
Fucci-expressing HeLa cells (23) were transfected with scrambled siRNA or DYRK2-specific siRNA. Serum-starved cells were released by stimulation with serum, and the time-lapse images were acquired at 20-minute intervals using a fluorescence microscope (Bio-Zero BZ-8000; Keyence) equipped with a phase difference lens (Nikon). For fluorescence imaging, the halogen lamp was used with excitation (BP520-540HQ) and emission (BP555-600HQ) filters. Image acquisition and analysis were performed using analyzer software (BZ-H1TL; Keyence).
In vivo tumorigenicity studies.
Seven-week-old nude mice (BALB/c, nu/nu, SLC) were housed under pathogen-free conditions. HeLa cells (1 × 107) were subcutaneously injected with a 26-gauge needle. For MCF-7 cell–derived xenografts, mice were implanted with the E2 pellet (Innovative Research of America) and inoculated with 5 × 106 cells suspended in matrigel (BD Biosciences) into the abdominal mammary gland. Tumor size was monitored every 2 days using calipers. Frozen or paraffin-embedded tumor samples were immunostained with anti-DYRK2, anti–c-Jun, or anti–c-Myc.
Immunohistochemistry.
Tissue microarrays were purchased from Provitro. Immunostaining was detected by the avidin–biotin–peroxidase method according to the manufacturer’s instructions (Vectastain ABC Kit; Vector Laboratories). For antigen retrieval, slides were microwaved for 20 minutes in sodium citrate buffer. After blocking endogenous peroxidase, the sections were incubated with normal serum and then incubated at 4°C overnight with anti-DYRK2 (Abgent), anti–c-Jun (Santa Cruz Biotechnology Inc.), anti–c-Myc (Santa Cruz Biotechnology Inc.), anti–cyclin E (Santa Cruz Biotechnology Inc.), anti–phospho–c-Jun(Ser243) (Abcam), and anti–phospho–c-Myc(Thr58/Ser62) (Cell Signaling Technology). Evaluation of the staining has been performed by pathologists using a light microscope. Scoring was based on examining all tumor cells on the slide. The evaluations were recorded as the proportion of positive cells in each of 4 proportion categories that were denoted as negative, weak, moderate, and strong. The categories were also divided into 2 groups, consisting of high-expression groups (including moderate and strong categories) and low-expression groups (including negative and weak categories). For phospho–c-Jun and phospho–c-Myc staining, only the nuclear staining was taken into consideration. For the evaluation, 3 fields at a magnification of ×400 were captured randomly from each slide, and the results are represented as the percentage of positive staining. In cases with carcinoma in situ and invasive carcinoma in the same specimen, only the invasive component was counted.
Statistics.
Statistical analysis was performed with 2-tailed t tests. Data represent the mean ± SD. The Mann-Whitney U test was also used to compare frequencies in the contingency tables. P < 0.05 was considered statistically significant.
Study approval.
Animal experiments were approved by the Animal Research Committee of Tokyo Medical and Dental University and were performed in accordance with established guidelines. The use of tissue specimens was reviewed and approved by the ethical committee of Tokyo Medical and Dental University. The samples were retrospectively acquired from the surgical pathology archives of Tokyo Medical and Dental University and did not directly involve human subjects; the study was considered no more than minimal risk. In accordance with Ethical Guidelines for Clinical Studies from the Ministry of Health, Labour and Welfare, Japan, informed consent has been obtained via disclosing information, along with specification that coded or anonymous leftover material is used for research and patients have been offered the option to opt out.
Supplementary Material
Acknowledgments
We thank Masabumi Shibuya, Tsuyoshi Osawa, Masashi Muramatsu, and Miori Inoue for technical assistance. We also thank Shigetaka Kitajima for c-Jun cDNA. This work was supported by grants from the Ministry of Education, Science and Culture of Japan (to N. Taira, Y. Miki, and K. Yoshida), the Yasuda Memorial Foundation (to N. Taira), the Uehara Memorial Foundation (to K. Yoshida), the Takeda Science Foundation (to K. Yoshida), the Sankyo Foundation of Life Science (to K. Yoshida), the Sato Memorial Foundation for Cancer Research (to K. Yoshida), the Ichiro Kanehara Foundation (to K. Yoshida), the Terumo Life Science Foundation (to K. Yoshida), the Foundation for Promotion of Cancer Research (to K. Yoshida), and the Kobayashi Foundation for Cancer Research (to K. Yoshida).
Footnotes
Conflict of interest: The authors have declared that no conflict of interest exists.
Citation for this article: J Clin Invest. 2012;122(3):859–872. doi:10.1172/JCI60818.
References
- 1.Hann SR. Role of post-translational modifications in regulating c-Myc proteolysis, transcriptional activity and biological function. Semin Cancer Biol. 2006;16(4):288–302. doi: 10.1016/j.semcancer.2006.08.004. [DOI] [PubMed] [Google Scholar]
- 2.Shaulian E, Karin M. AP-1 as a regulator of cell life and death. Nat Cell Biol. 2002;4(5):E131–E136. doi: 10.1038/ncb0502-e131. [DOI] [PubMed] [Google Scholar]
- 3.Pelengaris S, Khan M, Evan G. c-MYC: more than just a matter of life and death. Nat Rev Cancer. 2002;2(10):764–776. doi: 10.1038/nrc904. [DOI] [PubMed] [Google Scholar]
- 4.DeSalle LM, Pagano M. Regulation of the G1 to S transition by the ubiquitin pathway. . FEBS Lett. 2001;490(3):179–189. doi: 10.1016/S0014-5793(01)02121-4. [DOI] [PubMed] [Google Scholar]
- 5.Krek W. Proteolysis and the G1-S transition: the SCF connection. . Curr Opin Genet Dev. 1998;8(1):36–42. doi: 10.1016/S0959-437X(98)80059-2. [DOI] [PubMed] [Google Scholar]
- 6.Welcker M, Clurman BE. FBW7 ubiquitin ligase: a tumour suppressor at the crossroads of cell division, growth and differentiation. Nat Rev Cancer. 2008;8(2):83–93. doi: 10.1038/nrc2290. [DOI] [PubMed] [Google Scholar]
- 7.Cohen P, Frame S. The renaissance of GSK3. Nat Rev Mol Cell Biol. 2001;2(10):769–776. doi: 10.1038/35096075. [DOI] [PubMed] [Google Scholar]
- 8.Cross DA, Alessi DR, Cohen P, Andjelkovich M, Hemmings BA. Inhibition of glycogen synthase kinase-3 by insulin mediated by protein kinase B. Nature. 1995;378(6559):785–789. doi: 10.1038/378785a0. [DOI] [PubMed] [Google Scholar]
- 9.Doble BW, Woodgett JR. GSK-3: tricks of the trade for a multi-tasking kinase. J Cell Sci. 2003;116(pt 7):1175–1186. doi: 10.1242/jcs.00384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wei W, Jin J, Schlisio S, Harper JW, Kaelin WG., Jr The v-Jun point mutation allows c-Jun to escape GSK3-dependent recognition and destruction by the Fbw7 ubiquitin ligase. Cancer Cell. 2005;8(1):25–33. doi: 10.1016/j.ccr.2005.06.005. [DOI] [PubMed] [Google Scholar]
- 11.Luo J. Glycogen synthase kinase 3beta (GSK3beta) in tumorigenesis and cancer chemotherapy. Cancer Lett. 2009;273(2):194–200. doi: 10.1016/j.canlet.2008.05.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Frame S, Cohen P. GSK3 takes centre stage more than 20 years after its discovery. Biochem J. 2001;359(pt 1):1–16. doi: 10.1042/0264-6021:3590001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lutterbach B, Hann SR. Hierarchical phosphorylation at N-terminal transformation-sensitive sites in c-Myc protein is regulated by mitogens and in mitosis. Mol Cell Biol. 1994;14(8):5510–5522. doi: 10.1128/mcb.14.8.5510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lutterbach B, Hann SR. c-Myc transactivation domain-associated kinases: questionable role for map kinases in c-Myc phosphorylation. J Cell Biochem. 1999;72(4):483–491. doi: 10.1002/(SICI)1097-4644(19990315)72:4<483::AID-JCB4>3.0.CO;2-I. [DOI] [PubMed] [Google Scholar]
- 15.Boyle WJ, et al. Activation of protein kinase C decreases phosphorylation of c-Jun at sites that negatively regulate its DNA-binding activity. Cell. 1991;64(3):573–584. doi: 10.1016/0092-8674(91)90241-P. [DOI] [PubMed] [Google Scholar]
- 16.Morton S, Davis RJ, McLaren A, Cohen P. A reinvestigation of the multisite phosphorylation of the transcription factor c-Jun. EMBO J. 2003;22(15):3876–3886. doi: 10.1093/emboj/cdg388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yoshida K. Role for DYRK family kinases on regulation of apoptosis. Biochem Pharmacol. 2008;76(11):1389–1394. doi: 10.1016/j.bcp.2008.05.021. [DOI] [PubMed] [Google Scholar]
- 18.Maddika S, Chen J. Protein kinase DYRK2 is a scaffold that facilitates assembly of an E3 ligase. Nat Cell Biol. 2009;11(4):409–419. doi: 10.1038/ncb1848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Varjosalo M, et al. Application of active and kinase-deficient kinome collection for identification of kinases regulating hedgehog signaling. Cell. 2008;133(3):537–548. doi: 10.1016/j.cell.2008.02.047. [DOI] [PubMed] [Google Scholar]
- 20.Taira N, Nihira K, Yamaguchi T, Miki Y, Yoshida K. DYRK2 is targeted to the nucleus and controls p53 via Ser46 phosphorylation in the apoptotic response to DNA damage. Mol Cell. 2007;25(5):725–738. doi: 10.1016/j.molcel.2007.02.007. [DOI] [PubMed] [Google Scholar]
- 21.Himpel S, Tegge W, Frank R, Leder S, Joost HG, Becker W. Specificity determinants of substrate recognition by the protein kinase DYRK1A. J Biol Chem. 2000;275(4):2431–2438. doi: 10.1074/jbc.275.4.2431. [DOI] [PubMed] [Google Scholar]
- 22.Taira N, Yamamoto H, Yamaguchi T, Miki Y, Yoshida K. ATM augments nuclear stabilization of DYRK2 by inhibiting MDM2 in the apoptotic response to DNA damage. J Biol Chem. 2010;285(7):4909–4919. doi: 10.1074/jbc.M109.042341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sakaue-Sawano A, et al. Visualizing spatiotemporal dynamics of multicellular cell-cycle progression. Cell. 2008;132(3):487–498. doi: 10.1016/j.cell.2007.12.033. [DOI] [PubMed] [Google Scholar]
- 24.Durbin AD, Somers GR, Forrester M, Pienkowska M, Hannigan GE, Malkin D. JNK1 determines the oncogenic or tumor-suppressive activity of the integrin-linked kinase in human rhabdomyosarcoma. J Clin Invest. 2009;119(6):1558–1570. doi: 10.1172/JCI37958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chen Y, Olopade OI. MYC in breast tumor progression. Expert Rev Anticancer Ther. 2008;8(10):1689–1698. doi: 10.1586/14737140.8.10.1689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gee JM, Barroso AF, Ellis IO, Robertson JF, Nicholson RI. Biological and clinical associations of c-jun activation in human breast cancer. Int J Cancer. 2000;89(2):177–186. doi: 10.1002/(SICI)1097-0215(20000320)89:2<177::AID-IJC13>3.0.CO;2-0. [DOI] [PubMed] [Google Scholar]
- 27.Grimes CA, Jope RS. The multifaceted roles of glycogen synthase kinase 3beta in cellular signaling. Prog Neurobiol. 2001;65(4):391–426. doi: 10.1016/S0301-0082(01)00011-9. [DOI] [PubMed] [Google Scholar]
- 28.Sears R, Nuckolls F, Haura E, Taya Y, Tamai K, Nevins JR. Multiple Ras-dependent phosphorylation pathways regulate Myc protein stability. Genes Dev. 2000;14(19):2501–2514. doi: 10.1101/gad.836800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Huang CC, et al. Calcineurin-mediated dephosphorylation of c-Jun Ser-243 is required for c-Jun protein stability and cell transformation. Oncogene. 2008;27(17):2422–2429. doi: 10.1038/sj.onc.1210888. [DOI] [PubMed] [Google Scholar]
- 30.Kang T, et al. GSK-3 beta targets Cdc25A for ubiquitin-mediated proteolysis, and GSK-3 beta inactivation correlates with Cdc25A overproduction in human cancers. Cancer Cell. 2008;13(1):36–47. doi: 10.1016/j.ccr.2007.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ding Q, et al. Degradation of Mcl-1 by beta-TrCP mediates glycogen synthase kinase 3-induced tumor suppression and chemosensitization. Mol Cell Biol. 2007;27(11):4006–4017. doi: 10.1128/MCB.00620-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Maurer U, Charvet C, Wagman AS, Dejardin E, Green DR. Glycogen synthase kinase-3 regulates mitochondrial outer membrane permeabilization and apoptosis by destabilization of MCL-1. Mol Cell. 2006;21(6):749–760. doi: 10.1016/j.molcel.2006.02.009. [DOI] [PubMed] [Google Scholar]
- 33.Plotnikov A, et al. Oncogene-mediated inhibition of glycogen synthase kinase 3 beta impairs degradation of prolactin receptor. Cancer Res. 2008;68(5):1354–1361. doi: 10.1158/0008-5472.CAN-07-6094. [DOI] [PubMed] [Google Scholar]
- 34.Diehl JA, Cheng M, Roussel MF, Sherr CJ. Glycogen synthase kinase-3beta regulates cyclin D1 proteolysis and subcellular localization. Genes Dev. 1998;12(22):3499–3511. doi: 10.1101/gad.12.22.3499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hanson KD, Shichiri M, Follansbee MR, Sedivy JM. Effects of c-myc expression on cell cycle progression. Mol Cell Biol. 1994;14(9):5748–5755. doi: 10.1128/MCB.14.9.5748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Jansen-Durr P, et al. Differential modulation of cyclin gene expression by MYC. Proc Natl Acad Sci U S A. 1993;90(8):3685–3689. doi: 10.1073/pnas.90.8.3685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Karn J, Watson JV, Lowe AD, Green SM, Vedeckis W. Regulation of cell cycle duration by c-myc levels. Oncogene. 1989;4(6):773–787. [PubMed] [Google Scholar]
- 38.Ohtsubo M, Theodoras AM, Schumacher J, Roberts JM, Pagano M. Human cyclin E, a nuclear protein essential for the G1-to-S phase transition. . Mol Cell Biol. 1995;15(5):2612–2624. doi: 10.1128/mcb.15.5.2612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Perez-Roger I, Solomon DL, Sewing A, Land H. Myc activation of cyclin E/Cdk2 kinase involves induction of cyclin E gene transcription and inhibition of p27(Kip1) binding to newly formed complexes. Oncogene. 1997;14(20):2373–2381. doi: 10.1038/sj.onc.1201197. [DOI] [PubMed] [Google Scholar]
- 40.Resnitzky D, Gossen M, Bujard H, Reed SI. Acceleration of the G1/S phase transition by expression of cyclins D1 and E with an inducible system. . Mol Cell Biol. 1994;14(3):1669–1679. doi: 10.1128/mcb.14.3.1669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Richardson H, O’Keefe LV, Marty T, Saint R. Ectopic cyclin E expression induces premature entry into S phase and disrupts pattern formation in the Drosophila eye imaginal disc. Development. 1995;121(10):3371–3379. doi: 10.1242/dev.121.10.3371. [DOI] [PubMed] [Google Scholar]
- 42.Sheaff RJ, Groudine M, Gordon M, Roberts JM, Clurman BE. Cyclin E-CDK2 is a regulator of p27Kip1. Genes Dev. 1997;11(11):1464–1478. doi: 10.1101/gad.11.11.1464. [DOI] [PubMed] [Google Scholar]
- 43.Nesbit CE, Tersak JM, Prochownik EV. MYC oncogenes and human neoplastic disease. Oncogene. 1999;18(19):3004–3016. doi: 10.1038/sj.onc.1202746. [DOI] [PubMed] [Google Scholar]
- 44.Bhatia K, Huppi K, Spangler G, Siwarski D, Iyer R, Magrath I. Point mutations in the c-Myc transactivation domain are common in Burkitt’s lymphoma and mouse plasmacytomas. Nat Genet. 1993;5(1):56–61. doi: 10.1038/ng0993-56. [DOI] [PubMed] [Google Scholar]
- 45.Smith-Sorensen B, Hijmans EM, Beijersbergen RL, Bernards R. Functional analysis of Burkitt’s lymphoma mutant c-Myc proteins. J Biol Chem. 1996;271(10):5513–5518. doi: 10.1074/jbc.271.10.5513. [DOI] [PubMed] [Google Scholar]
- 46.Frykberg L, Graf T, Vennstrom B. The transforming activity of the chicken c-myc gene can be potentiated by mutations. Oncogene. 1987;1(4):415–422. [PubMed] [Google Scholar]
- 47.Symonds G, Hartshorn A, Kennewell A, O’Mara MA, Bruskin A, Bishop JM. Transformation of murine myelomonocytic cells by myc: point mutations in v-myc contribute synergistically to transforming potential. Oncogene. 1989;4(3):285–294. [PubMed] [Google Scholar]
- 48.Maki Y, Bos TJ, Davis C, Starbuck M, Vogt PK. Avian sarcoma virus 17 carries the jun oncogene. Proc Natl Acad Sci U S A. 1987;84(9):2848–2852. doi: 10.1073/pnas.84.9.2848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zhang X, et al. Mechanistic insight into Myc stabilization in breast cancer involving aberrant Axin1 expression. Proc Natl Acad Sci U S A. doi: 10.1073/pnas.1100764108. [published online ahead of print August 1, 2011]. doi:10.1073/pnas.1100764108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Armanious H, Deschenes J, Gelebart P, Ghosh S, Mackey J, Lai R. Clinical and biological significance of GSK-3beta inactivation in breast cancer-an immunohistochemical study. Hum Pathol. 2010;41(12):1657–1663. doi: 10.1016/j.humpath.2010.04.015. [DOI] [PubMed] [Google Scholar]
- 51.Italiano A, et al. HMGA2 is the partner of MDM2 in well-differentiated and dedifferentiated liposarcomas whereas CDK4 belongs to a distinct inconsistent amplicon. Int J Cancer. 2008;122(10):2233–2241. doi: 10.1002/ijc.23380. [DOI] [PubMed] [Google Scholar]
- 52.Liu H, Lu ZG, Miki Y, Yoshida K. Protein kinase C delta induces transcription of the TP53 tumor suppressor gene by controlling death-promoting factor Btf in the apoptotic response to DNA damage. Mol Cell Biol. 2007;27(24):8480–8491. doi: 10.1128/MCB.01126-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Yoshida K, Yamaguchi T, Shinagawa H, Taira N, Nakayama KI, Miki Y. Protein kinase C delta activates topoisomerase IIalpha to induce apoptotic cell death in response to DNA damage. Mol Cell Biol. 2006;26(9):3414–3431. doi: 10.1128/MCB.26.9.3414-3431.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Cai Y, et al. Homocysteine-responsive ATF3 gene expression in human vascular endothelial cells: activation of c-Jun NH(2)-terminal kinase and promoter response element. Blood. 2000;96(6):2140–2148. [PubMed] [Google Scholar]
- 55.Kimura J, Nguyen ST, Liu H, Taira N, Miki Y, Yoshida K. A functional genome-wide RNAi screen identifies TAF1 as a regulator for apoptosis in response to genotoxic stress. Nucleic Acids Res. 2008;36(16):5250–5259. doi: 10.1093/nar/gkn506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Lujambio A, et al. CpG island hypermethylation-associated silencing of non-coding RNAs transcribed from ultraconserved regions in human cancer. Oncogene. 2010;29(48):6390–6401. doi: 10.1038/onc.2010.361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Yoshida K, Yamaguchi T, Natsume T, Kufe D, Miki Y. JNK phosphorylation of 14-3-3 proteins regulates nuclear targeting of c-Abl in the apoptotic response to DNA damage. Nat Cell Biol. 2005;7(3):278–285. doi: 10.1038/ncb1228. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.