Abstract
Ectonucleotidases modulate purinergic signaling by hydrolyzing ATP to adenosine. Here we characterized the impact of the cellular distribution of hepatic ectonucleotidases, namely nucleoside triphosphate diphosphohydrolase (NTPDase)1/CD39, NTPDase2/CD39L1, NTPDase8, and ecto-5′-nucleotidase/CD73, and of their specific biochemical properties, on the levels of P1 and P2 receptor agonists, with an emphasis on adenosine-producing CD73. Immunostaining and enzyme histochemistry showed that the distribution of CD73 (protein and AMPase activity) overlaps partially with those of NTPDase1, -2, and -8 (protein levels and ATPase and ADPase activities) in normal rat liver. CD73 is expressed in fibroblastic cells located underneath vascular endothelial cells and smooth muscle cells, which both express NTPDase1, in portal spaces in a distinct fibroblast population next to NTPDase2-positive portal fibroblasts, and in bile canaliculi, together with NTPDase8. In fibrotic rat livers, CD73 protein expression and activity are redistributed but still overlap with the NTPDases mentioned. The ability of the observed combinations of ectonucleotidases to generate adenosine over time was evaluated by reverse-phase HPLC with the recombinant rat enzymes at high “inflammatory” (500 μM) and low “physiological” (1 μM) ATP concentrations. Overall, ATP was rapidly converted to adenosine by the NTPDase1+CD73 combination, but not by the NTPDase2+CD73 combination. In the presence of NTPDase8 and CD73, ATP was sequentially dephosphorylated to the CD73 inhibitor ADP, and then to AMP, thus resulting in a delayed formation of adenosine. In conclusion, the specific cellular cocompartmentalization of CD73 with hepatic NTPDases is not redundant and may lead to the differential activation of P1 and P2 receptors, under normal and fibrotic conditions.
Keywords: P1 receptors, P2 receptors, ATP, fibrosis, carbon tetrachloride intoxication, nucleoside triphosphate diphosphohydrolases
via the activation of specific plasma membrane P1 and P2 receptors, extracellular nucleosides and nucleotides modulate various physiological functions in virtually all tissues, including blood vessels, heart, lungs, brain, smooth and skeletal muscle, etc. (9). In the liver, these signaling agents regulate key cellular pathways such as glucose metabolism, cell cycle progression, hepatic regeneration, cell volume regulation, ionic secretion, and scar formation (5, 19, 43, 44). To regulate extracellular nucleoside and nucleotide signaling and maintain tissue homeostasis, liver cells express various ectonucleotidases at their surface, including members of the ecto-nucleoside triphosphate diphosphohydrolase (NTPDase), ecto-nucleotide pyrophosphatase-phosphodiesterase, and alkaline phosphatase families, as well as ecto-5′-nucleotidase/CD73 (5, 19, 44). These ectoenzymes are thought to act in concert to sequentially dephosphorylate extracellular nucleotides and locally regulate their concentration in hepatic extracellular fluids (55).
At physiological pH, plasma membrane-bound NTPDases, namely NTPDase1/CD39, NTPDase2/CD39L1, and NTPDase8, represent the major liver ectonucleotidase activities (18, 45). NTPDases hydrolyze various nucleoside tri- and diphosphates (e.g., ATP and ADP) with distinct affinities and abilities. NTPDase1 hydrolyzes ATP and ADP equally well whereas NTPDase2 preferentially breaks down ATP over ADP, and NTPDase8 acts as a functional intermediate between NTPDase1 and NTPDase2 (18, 26, 31). In the liver, NTPDases are expressed in a cell-specific manner: NTPDase1 is expressed by vascular endothelial cells and smooth muscle cells (18, 45), NTPDase2 by cells in the vascular external layer and by portal fibroblasts (14, 46), whereas NTPDase8 is restricted to the canalicular membrane domain of hepatocytes (18).
The monophosphonucleosides (e.g., AMP) generated by NTPDase activity are ultimately converted into the corresponding nucleosides (e.g., adenosine) by ecto-5′-nucleotidase (12, 27, 49). Interestingly, the NTPDase substrates ATP and ADP are natural inhibitors of the monophosphatase activity of ecto-5′-nucleotidase (39). In the liver, ecto-5′-nucleotidase protein expression has been associated with the canalicular domain of hepatocyte plasma membrane and undefined cellular elements of the portal space (2, 47).
Because of their complementary biochemical properties, NTPDases and ecto-5′-nucleotidase represent central elements of liver purinergic signaling. However, the potential influence of the coordinated action of NTPDases and ecto-5′-nucleotidase on local hepatic purinergic signaling has yet to be established. In the present study, we have characterized the impact of the cellular distribution of these hepatic ectonucleotidases on the local levels of P1 and P2 receptor agonists. Here we show that ecto-5′-nucleotidase is coexpressed with NTPDase1, -2, and -8 in distinct liver compartments in normal and fibrotic rat liver. These specific associations of ecto-5′-nucleotidase with each hepatic NTPDase have the ability to differentially regulate the signaling pathways modulated by extracellular nucleotides and their metabolites in the liver.
EXPERIMENTAL PROCEDURES
Materials
Culture media were obtained from Invitrogen (Burlington, ON, Canada). Adenosine 5′-(α,β-methylene)diphosphate (α,β-methylene-ADP), 2-chloroacetaldehyde, 3,3′-diaminobenzidine (DAB), 1,4-diaza-bicyclo[2.2.2]octane (DABCO), nucleotides, levamisole, tetrabutylammonium hydrogen sulfate were provided by Sigma-Aldrich (Oakville, ON, Canada). The 4′,6-diamidino-2-phenylindole (DAPI) dye was purchased from Molecular Probes (Eugene, OR) and aqueous hematoxylin from Biomeda (Foster City, CA). Mouse monoclonal antibody to rat CD31/PECAM-1 (clone TLD-3A12) was purchased from BD Pharmingen (Mississauga, ON, Canada), to rat CD68 (clone ED-1) from AbD Serotec (Raleigh, NC), and those to mouse/rat α-smooth muscle actin (SMA; clone 1A4) and rat vimentin (clone V9) from Sigma-Aldrich. The antibodies to NTPDases and ecto-5′-nucleotidase can be obtained from ectonucleotidases-ab.com (Québec, QC, Canada).
Plasmids
The plasmids encoding rat NTPDase1 (NM_022587) (26), NTPDase2 (NM_172030) (30), NTPDase8 (AY536920) (18), and ecto-5′-nucleotidase/CD73 (NM_021576) (8) were used for antiserum generation and for cell transfection, as described below.
Animals and Antibody Production
Sprague-Dawley rats, Hartley guinea pigs and New Zealand rabbits were obtained from Charles River Laboratories (St-Constant, QC, Canada) or Harlan Sprague Dawley (Indianapolis, IN). All procedures were approved by the Canadian Council on Animal Care and by the Laval University Animal Ethics Advisory Committee and the Yale Institutional Animal Care and Use Committee. Genetic immunization was carried out by direct injection of a plasmid encoding rat ecto-5′-nucleotidase (8), as previously described with other antigens (18), in Hartley guinea pigs and New Zealand rabbits.
Chronic liver fibrosis was induced in rats by carbon tetrachloride (CCl4) intoxication. For CCl4 treatment, male rats were provided with drinking water containing 1% phenobarbital starting 2 wk before and until the end of CCl4 treatment, to upregulate cytochrome P-450 enzyme levels (24). Rats were injected with CCl4 (0.1 ml sc, CCl4 diluted 1:6 in olive oil) three times per week for 6–8 wk. For control experiments, rats were injected in identical fashion with olive oil alone. Livers were harvested at 6–8 wk and processed for enzyme histochemistry as described below.
Cell Transfection and Ectonucleotidase Activity Assays
Transfection of COS-7 cells using plasmids encoding either recombinant NTPDase1, -2, -8, or ecto-5′-nucleotidase and preparation of crude protein extracts were carried out as previously described (31). The COS-7 cell line was used because it exhibits very low basal levels of nucleotide- and nucleoside-metabolizing enzyme activities (Refs. 29, 51 and data not shown).
For assays at high ATP concentration (500 μM), ectonucleotidase-overexpressing crude protein extracts were added to obtain an enzymatic activity of 24 nmol Pi/min with ATP and AMP as substrate for NTPDases and ecto-5′-nucleotidase, respectively, as determined by the malachite green procedure according to Baykov et al. (3, 32). The enzymatic reactions were performed at 37°C for 150 min in Ringer buffer (120 mM NaCl, 5 mM KCl, 2.5 mM CaCl2, 2.5 mM MgCl2, 1.2 mM MgSO4, 25 mM NaHCO3, 10 mM dextrose, 80 mM Tris·HCl, pH 7.4) and the products of ATP hydrolysis were determined by reverse phase HPLC, as previously described (31).
For assays at low ATP concentration (1 μM), ectonucleotidase-overexpressing crude protein extracts were prepared with an ATPase (for NTPDase) and AMPase activity (for ecto-5′-nucleotidase) sufficient to convert ∼40% of the initial substrate within 5 min, as determined by HPLC. Enzymatic reactions were performed at 37°C for 30 min in a modified Krebs buffer (114 mM NaCl, 4.7 mM KCl, 1.2 mM KH2PO4, 1.16 mM MgSO4, 2.5 mM CaCl2, 2.5 MgCl2, 25.5 mM NaHCO3, 20 mM HEPES, pH 7.4). A modified Krebs buffer was used here instead of Ringer buffer because of interference with fluorescence detection observed with the latter. Briefly, 150-μl aliquots of reaction mixture were collected after various time intervals and transferred to an equal volume of ice-cold 5% (wt/vol) trichloroacetic acid (TCA) for deproteinization. The samples were centrifuged at 10,000 g for 3 min at 4°C and the supernatants were subjected to lipid extraction with diethyl ether (3:1, vol/vol; 5 cycles). The resulting samples were subsequently transformed to etheno(ε)-derivatives, according to a modified derivatization protocol by Levitt et al. (34, 36). This additional step allowed the determination of 50-fold lower concentrations of adenylated species by fluorescence detection.
In a representative derivatization assay, a 200-μl aliquot of the extracted sample was incubated for 60 min at 70°C in the presence of 1 M chloroacetaldehyde and 25 mM Na2HPO4, pH 4.0, in a final volume of 250 μl. The resulting fluorescent 1,N6-ethenoadenine derivatives (ε-ATP, ε-ADP, ε-AMP, and ε-adenosine) were separated by reverse-phase HPLC as previously described (31), where the mobile phase used contained 25 mM tetrabutylammonium hydrogen sulfate and 100 mM KH2PO4/K2HPO4, pH 7.0, in 10% (vol/vol) MeOH. All experiments were performed in triplicate. Samples to which crude protein extracts were added after reaction had been stopped (i.e., after TCA addition) were used as controls (baseline levels). Separation and detection of ethenylated derivatives were performed with an automated Waters and Beckman HPLC apparatus equipped with an RF-10AXL Shimadzu fluorescence detector (λexc = 307 nm; λem = 410 nm). Quantification and identification of the fluorescent adenylated derivatives were carried out by comparison with the corresponding ε-purine standards.
Immunoblotting
Protein preparation, SDS-PAGE fractionation, and electroblotting were performed as previously described (18). Following incubation with primary antibodies, proteins of interest were visualized with the appropriate horseradish peroxidase-conjugated secondary antibodies (anti-rabbit IgG, Amersham Biosciences, Boston, MA; anti-guinea pig IgG, Santa Cruz Biotechnology, Santa Cruz, CA) and the Chemiluminescent Reagent Plus (Perkin-Elmer, Boston, MA), according to the manufacturer's recommendations.
Immunofluorescence, Immunocytochemistry, and Enzyme Histochemistry
COS-7 cells (105/coverslip) or 6-μm sections of snap-frozen rat liver specimens were fixed with cold acetone:phosphate-buffered formalin (9:1). For immunofluorescence, fixed sections were incubated with the following polyclonal antibodies against rat enzymes: anti-NTPDase1 (rN1–6l) (18), anti-NTPDase2 (BZ3–4F) (14), anti-NTPDase8 (rN8–8c) (18), and anti-CD73 (either r5′NT-4c or r5′NT-9l), or corresponding preimmune serum samples. The appropriate Alexa Fluor-conjugated secondary antibodies (Molecular Probes) were then applied as previously described (18). In one set of double immunostaining experiments, rabbit rN1–6l and BZ3–4F antibodies were each used in combination with guinea pig r5′NT-4c serum, and the guinea pig rN8–8c antibody was used with rabbit r5′NT-9l serum. In the other set of experiments, mouse monoclonal antibodies against rat CD31/PECAM-1 (clone TLD-3A12), rat α-SMA (clone 1A4) and rat vimentin (clone V9) were each used in combination with the rabbit r5′NT-9l serum.
For immunocytochemistry, fixed cells were incubated with the polyclonal CD73 antiserum (r5′NT-4c and r5′NT-9l) and the appropriate biotinylated secondary antibodies (Jackson ImmunoResearch Laboratories, Westgrove, PA) before addition of Vectastain Elite ABC reagent (Vector Laboratories, Burlington, ON, Canada). All antibodies were used at a 1:1,000 working dilution, except for r5′NT-9L, which was used at a final concentration of 1:2,000. Endogenous peroxidase was quenched by incubating the fixed cells with 0.15% (vol/vol) H2O2 (in PBS). Endogenous avidin and biotin were blocked using the Avidin/Biotin Blocking Kit (Vector Laboratories). The peroxidase reaction was performed in a solution containing the DAB substrate (0.4 mg/ml) and 0.03% (vol/vol) H2O2 in PBS, and stopped with extensive water rinsing.
For enzyme histochemistry, ectonucleotidase activities were localized by using the Wachstein/Meisel lead phosphate method as modified by Braun et al. (see reference therein) (7). The enzymatic reaction was carried out by incubating liver sections with various nucleotides (200 μM) as substrates for 1 h. All experiments were performed in the presence of levamisole (5 mM), an inhibitor of tissue-nonspecific alkaline phosphatase (TNAP). In a separate set of experiments, α,β-methylene-ADP (1 mM) was used as an inhibitor of ecto-5′-nucleotidase. Control assays (not shown) were performed in the absence of nucleotide.
Nuclei were counterstained with either DAPI (immunofluorescence) or aqueous hematoxylin (immunocytochemistry and enzyme histochemistry). Mowiol 4-88 medium (supplemented with DABCO as antifade reagent for immunofluorescence only) was used as mounting medium.
Flow Cytometry and Isolation of Rat Liver Cell Populations
Hepatocytes, hepatic stellate cells, portal fibroblasts and nonparenchymal cell preparations were isolated from fresh rat liver tissue as previously described (14, 16). Fixed primary isolated cells were labeled by indirect immunofluorescence after incubation with a rabbit polyclonal serum to rat CD73 (r5′NT-9l) followed by Alexa Fluor 488-conjugated polyclonal anti-rabbit IgG antibody (Molecular Probes). Cell-surface expression of CD73 protein was assessed by fluorescence-activated cell sorting, using a FACSCalibur (BD Biosciences, Rockville, MD) sorter and Flowjo software Version 8.7 (TreeStar, Ashland, OR).
RESULTS
Although the hepatic expression of NTPDase1, -2, and -8 has previously been reported (14, 18, 45), the cellular localization of ecto-5′-nucleotidase/CD73 had not been clearly established. Therefore, we first looked at the distribution of ecto-5′-nucleotidase in relation to NTPDases, using enzyme histochemical and immunohistochemical methods.
Histochemical Detection of Ectonucleotidase Activities in Normal and Fibrotic Rat Liver
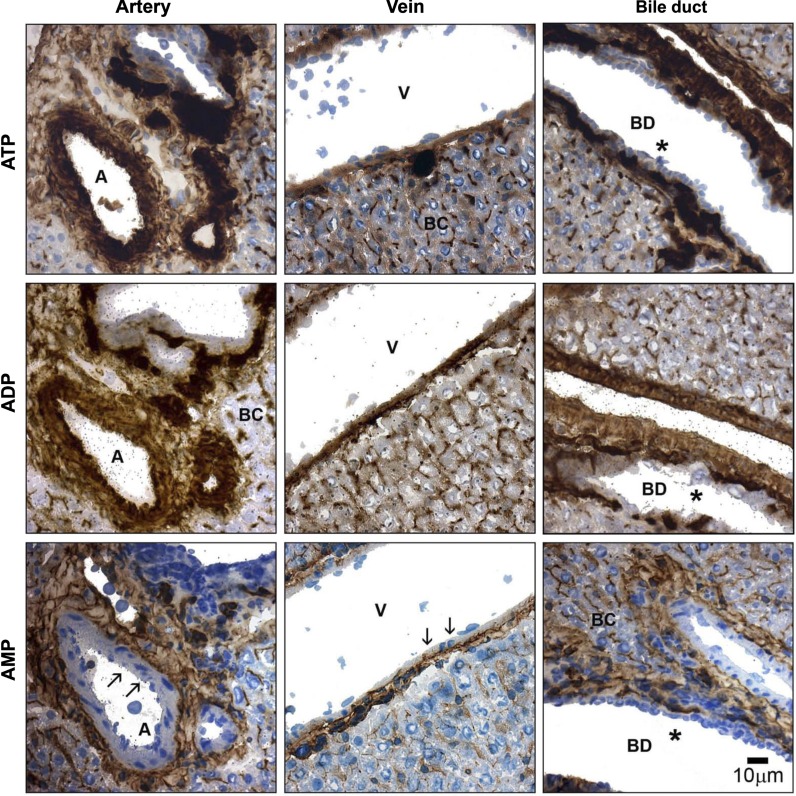
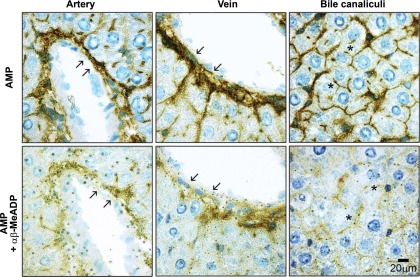
All enzyme histochemistry assays were performed in the presence of the TNAP inhibitor levamisole (5 mM) to exclude any contribution of this enzyme to the apparent catalytic activities measured with the nucleotide substrates. Nevertheless, no difference in ectonucleotidase activity could be detected when omitting levamisole from the reaction solution (data not shown), suggesting that TNAP activity was negligible under the experimental conditions used. In the rat liver, enzyme histochemical assays using ATP as substrate revealed a prominent staining activity in vascular endothelium and subendothelium, in the connective tissue adjacent to the portal/periportal space and surrounding central veins, in sinusoids, and in bile canaliculi (Fig. 1; ATP). No nucleotide hydrolysis was detected in ductular epithelium. Hepatic ADPase activity displayed a distribution pattern similar to ATPase activity, albeit with a slightly weaker staining intensity (Fig. 1; ADP). When AMP was applied as substrate, catalytic products were mainly detected in the subendothelium, in the connective tissue associated with the perivascular and periductular areas, and in the bile canaliculi. Note that ductular epithelium, vascular endothelium, as well as smooth muscle were devoid of AMPase activity (Fig. 1; AMP). Lack of endothelial AMPase activity was observed in blood vessels of all types and calibers. When α,β-methylene-ADP (1 mM) was added to specifically inhibit ecto-5′-nucleotidase activity, overall AMP hydrolysis was reduced in all regions, suggesting that this enzyme contributes to most, if not all, AMPase activity present in the rat liver at physiological pH (Fig. 2). In support of these findings, enzyme histochemical assays performed with other ecto-5′-nucleotidase substrates, namely CMP, GMP, and IMP, showed similar patterns of activity (not shown).
Fig. 1.
Histochemical detection of liver ectonucleotidase activities. Ectonucleotidase activities were located in serial sections of a normal rat liver, using various nucleotides (200 μM) as substrates. All 3 structural components of a classical hepatic triad are depicted, including branches of hepatic artery (A), portal vein (V), and intrahepatic bile duct (BD). ATPase and ADPase catalytic products are mainly detected in all vasculature (arterial > venous), in the connective tissue associated with the portal space, and in the bile canaliculi (BC) (top and middle). The AMP hydrolysis pattern is located in structures similar to those where ATPase and ADPase activities are found, although no signal is observed either in endothelium or in vascular smooth muscle (arrows, bottom). No ectonucleotidase activity is detected in cholangiocytes for any of the substrates tested (asterisks, right). All enzyme histochemical assays were performed in the presence of levamisole (5 mM) to inhibit tissue-nonspecific alkaline phosphatase (TNAP) activity. Scale bar, 10 μm.
Fig. 2.
Histochemical detection of liver ecto-5′-nucleotidase activity. Ecto-5′-nucleotidase activity was located in normal rat liver sections with AMP (200 μM) as substrate. Top: prominent signals for hepatic AMPase activity in the connective tissue associated with perivascular areas (arrows), and in bile canaliculi (asterisks). Bottom: ecto-5′-nucleotidase inhibitor α,β-methylene-ADP (αβ-MeADP; 1 mM) reduces AMPase activity in all structures (see arrows and asterisks). All enzyme histochemical assays were performed in the presence of the TNAP inhibitor levamisole (5 mM). Scale bar, 20 μm.
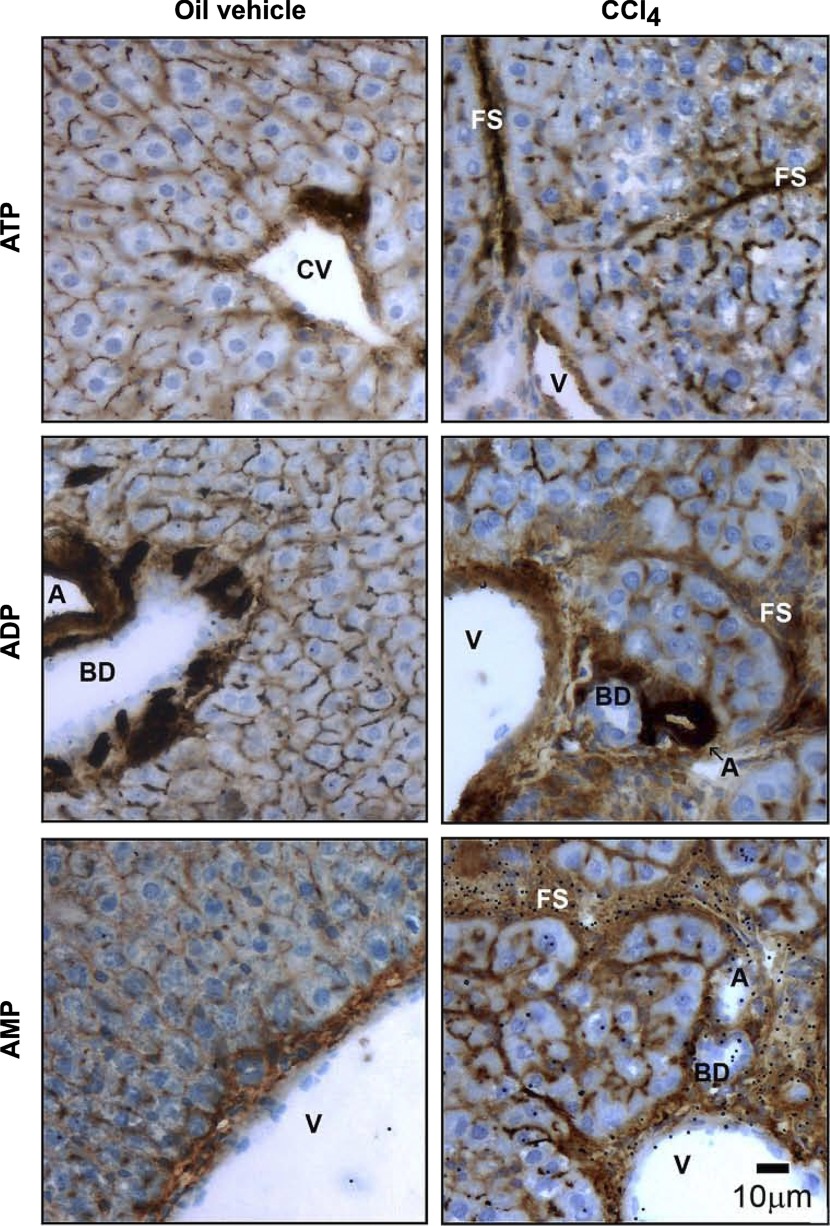
When enzyme histochemistry assays were performed in livers from rats subjected to CCl4 intoxication (Fig. 3), hepatic fibrosis induction was associated with obvious alterations of ectonucleotidase activity distribution. Strong signals for ATPase, ADPase, and AMPase activities were observed in fibrous septa surrounding hepatocyte nodules, in perivascular connective tissue areas, as well as in cells localized within hepatic sinusoids. Both ATPase and ADPase hydrolytic activities were still observed along the hepatocyte plasma membrane, with a more prominent but disrupted canalicular distribution, whereas AMPase activity was increased at all levels of the hepatocyte membrane. These observations, although qualitative, indicate that levels and distribution of ATPase, ADPase, and AMPase activities are regulated during hepatic fibrosis and suggest their involvement in the inflammatory liver response. This is in agreement with previous studies showing that transcriptional regulation of ectonucleotidase gene expression occurs during liver inflammation and fibrosis (15, 23, 40, 53).
Fig. 3.
Histochemical detection of liver ectonucleotidase activities in CCl4-induced hepatic fibrosis model. Ectonucleotidase activities were located in liver sections from vehicle (control) and CCl4-treated rats, using ATP, ADP, or AMP (200 μM) as substrate. All 3 structural components of a classical hepatic triad are depicted: branches of hepatic artery, portal vein, and the intrahepatic bile duct. ATPase, ADPase, and AMPase activities in control animals followed a distribution pattern similar to that described in Fig. 1. In contrast, in fibrotic animals, overlapping signals for ATP, ADP, and AMP hydrolytic products were detected in fibrous bands surrounding hepatic nodules. Moreover, staining for ATPase and ADPase activities was observed in hepatic sinusoids and hepatocyte basolateral membrane, and more prominently, although in a diffuse manner, in bile canaliculi. All enzyme histochemical assays were performed in the presence of the TNAP activity inhibitor levamisole (5 mM). CV, central vein; FS, fibrous septum. Scale bar, 10 μm.
Generation of Polyclonal Antibodies to Rat Ecto-5′-Nucleotidase/CD73 and Specificity
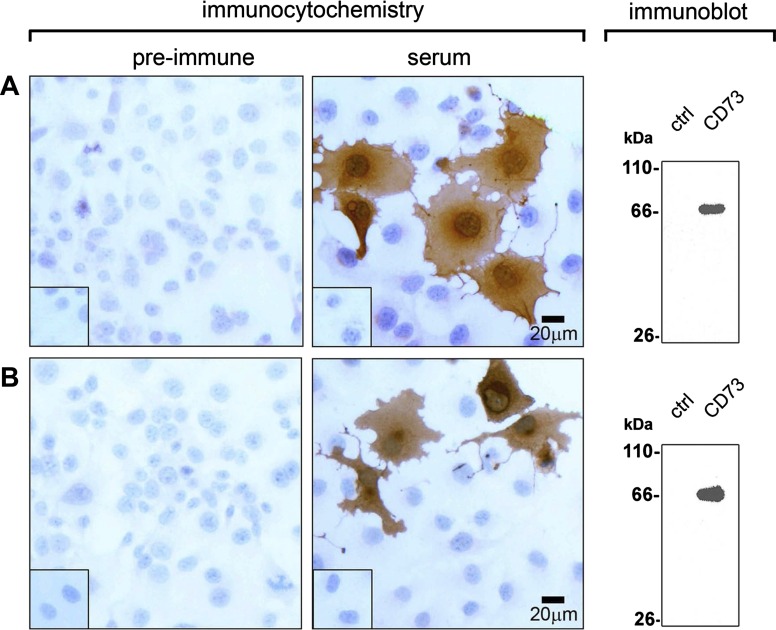
To determine the cellular distribution of ecto-5′-nucleotidase and the various NTPDases in the liver, we used a panel of polyclonal antibodies developed in our laboratory. The specificity of polyclonal sera to NTPDase1 (rN1–6l), NTPDase2 (BZ3–4F), and NTPDase8 (rN8–8c) has previously been validated (14, 18). Guinea pig and rabbit polyclonal antibodies directed against rat ecto-5′-nucleotidase have been generated and their specificity tested by the following immunological techniques. By immunocytochemistry, positive dark brown staining could be seen only with ecto-5′-nucleotidase-transfected COS-7 cells incubated in presence of either r5′NT-4c guinea pig serum or r5′NT-9l rabbit serum, respectively (Fig. 4, A and B; serum). The same antisera detected a strong protein band with a molecular weight of ∼66 kDa by immunoblotting in the lane corresponding to the crude protein extracts from ecto-5′-nucleotidase-transfected cells. No protein was detected in the lanes with crude protein extracts from nontransfected cells, confirming the specificity of both types of polyclonal antibodies (Fig. 4, A and B; immunoblot). Furthermore, both r5′NT-4c and r5′NT-9l antibodies displayed similar distribution patterns for hepatic ecto-5′-nucleotidase, as determined by immunohistochemistry and immunofluorescence (see next section; data not shown).
Fig. 4.
Specificity of the polyclonal antibodies to rat ecto-5′-nucleotidase. Immunocytochemistry was performed with intact nontransfected (insets) or transfected COS-7 cells with an expression vector encoding rat ecto-5′-nucleotidase. Cells were incubated with either preimmune serum or guinea pig (r5′NT-4c; top) or rabbit (r5′NT-9l; bottom) polyclonal antibody to ecto-5′-nucleotidase. Immunoreactivity is present only in transfected cells incubated with either antibody. Scale bar, 20 μm. Immunoblot analysis of lysates from nontransfected COS-7 cells (ctrl) next to lysates from COS-7 cells transfected with the ecto-5′-nucleotidase expression vector (CD73) shows a single band of ∼66 kDa found exclusively in lysates from transfected cells, again confirming the specificity of both guinea pig r5′NT-4c and rabbit r5′NT-9l sera. A: guinea pig antibody (r5′NT-4c; right) or its preimmune serum (left). B: rabbit antibody (r5′NT-9l; right) or its preimmune serum (left).
Immunolocalization of NTPDases and Ecto-5′-Nucleotidase in Normal and Fibrotic Rat Liver
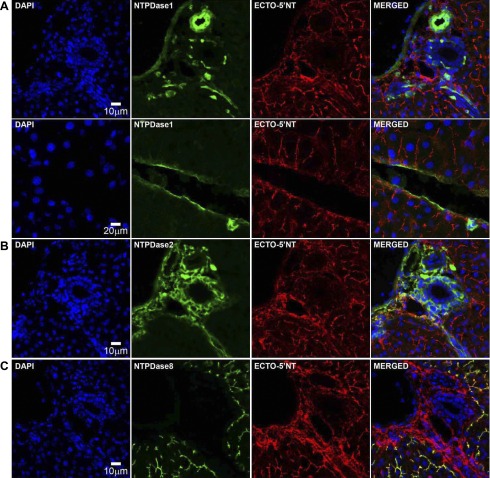
Immunofluorescence labeling in the rat liver with the polyclonal antibody rN1–6l showed prominent staining for NTPDase1 in vascular endothelium and smooth muscle cells as well as in liver resident macrophages, namely Kupffer cells (detected as CD68-positive; Fig. 5A and data not shown). As seen with BZ3–4F serum, NTPDase2 was detected in the subendothelial basement membrane cells, in portal fibroblasts, and in periductular “bundle-shaped” structures, in the connective tissue of portal space area, likely representing intrahepatic nerves (Fig. 5B). Staining with the rN8–8c polyclonal antibody showed that NTPDase8 expression was restricted to bile canaliculi (Fig. 4C). Both r5′NT-4c and r5′NT-9l polyclonal sera revealed strong ecto-5′-nucleotidase expression in the connective tissue associated with the perivascular and periductular areas, as well as in bile canaliculi, with a faint immunostaining in the basal membrane of hepatocytes (Fig. 4; ECTO-5′NT, all panels). Interestingly, ecto-5′-nucleotidase expression was absent from the vascular endothelium (Fig. 5; ECTO-5′NT, all panels). Double immunofluorescence staining showed that although ecto-5′-nucleotidase did not strictly colocalize with NTPDase1, both proteins were located in the same structures in adjacent cells, such as NTPDase1-positive Kupffer cells present in perivascular and/or periductular connective tissue, and on the basolateral surface of the endothelial cell layer in close contact with the perivascular extracellular matrix (Fig. 5A, merged). Likewise, although ecto-5′-nucleotidase did not colocalize with NTPDase2, both enzymes were expressed in the same vicinity in the perivascular and periductular connective tissue areas (Fig. 5B, merged). By contrast, NTPDase8 and ecto-5′-nucleotidase colocalized in bile canaliculi (Fig. 5C, merged).
Fig. 5.
Comparative distribution of liver nucleoside triphosphate diphosphohydrolases (NTPDases) and ecto-5′-nucleotidase (ECTO-5′NT). Double immunofluorescence labeling of NTPDases and ecto-5′-nucleotidase was performed in serial sections of rat liver. A: NTPDase1 staining is associated with the liver vascular network and Kupffer cells. B: NTPDase2 labeling is detected in the connective tissue of portal and septal spaces and perivascular areas. C: NTPDase8 signal is present exclusively in bile canaliculi. A–C: ecto-5′-nucleotidase immunostaining is observed in the connective tissue found in portal space or perivascular areas as well as in bile canaliculi. Double staining of NTPDase1 and ecto-5′-nucleotidase reveals that both ectoenzymes are expressed by different but adjacent cell populations in the vasculature, hepatic sinusoids, and hepatic extracellular matrix (A, MERGED). Similarly, NTPDase2 and ecto-5′-nucleotidase proteins are both present at the surface of distinct cell types of hepatic connective tissue (B, MERGED). Both NTPDase8 and ecto-5′-nucleotidase colocalize in bile canaliculi (C, MERGED). Scale bar, 10 μm except for A, bottom, where scale bar is 20 μm. DAPI, 4',6-diamidino-2-phenylindole.
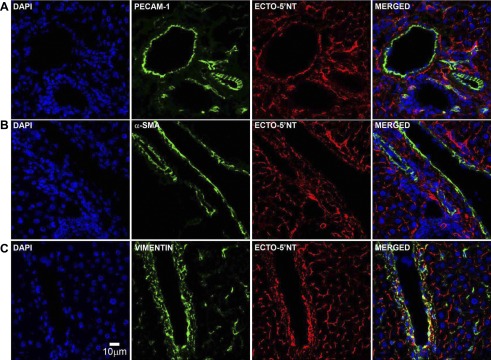
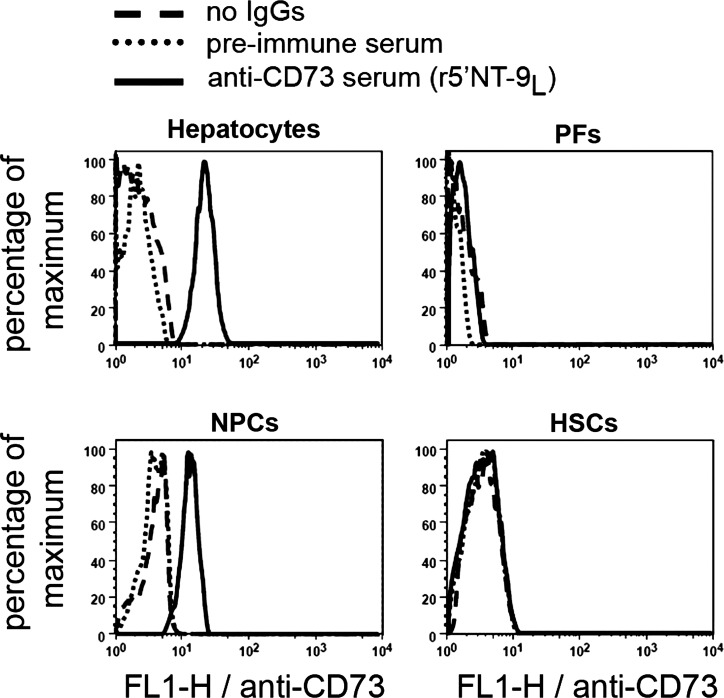
We further investigated ecto-5′-nucleotidase expression in the rat liver using the following cell markers: cell adhesion molecule PECAM-1 for vascular endothelium, α-SMA for smooth muscle cells and myofibroblasts, and vimentin for fibroblasts and other liver mesenchymal cells (42). Ecto-5′-nucleotidase labeling was clearly distinct from that observed for PECAM-1 and α-SMA, but partly overlapped with vimentin staining in the perivascular and periductular areas (Fig. 6; merged, all panels). We further confirmed the expression of ecto-5′-nucleotidase in primary liver cell preparations by fluorescence-activated cell sorting analysis. These data demonstrate that in normal rat liver, ecto-5′-nucleotidase is expressed by NTPDase8-positive hepatocytes and a nonparenchymal cell type that is distinct from hepatic stellate cells and NTPDase2-positive portal fibroblasts (Fig. 7).
Fig. 6.
Comparative distribution of liver ecto-5′-nucleotidase with cell markers. Double immunofluorescence labeling of ecto-5′-nucleotidase and cell markers for vascular endothelial cells [platelet/endothelial cell adhesion molecule-1 (PECAM-1)], myofibroblastic cells (α-smooth muscle actin), and nonmyofibroblastic cells (vimentin) was performed in rat liver sections. Although ecto-5′-nucleotidase immunostaining is clearly distinct from that of PECAM-1 and α-SMA, it partially overlaps with vimentin staining. Scale bar, 10 μm.
Fig. 7.
Flow cytometry analysis of ecto-5′-nucleotidase expression in liver primary cell populations. Cell populations were prepared as described in experimental procedures. Ecto-5′-nucleotidase (CD73) was labeled using rabbit polyclonal r5′NT-9l serum. Isolated hepatocytes and nonparenchymal cells (NPCs) are positive for ecto-5′-nucleotidase staining whereas portal fibroblasts (PFs) and hepatic stellate cells (HSCs) are mostly negative.
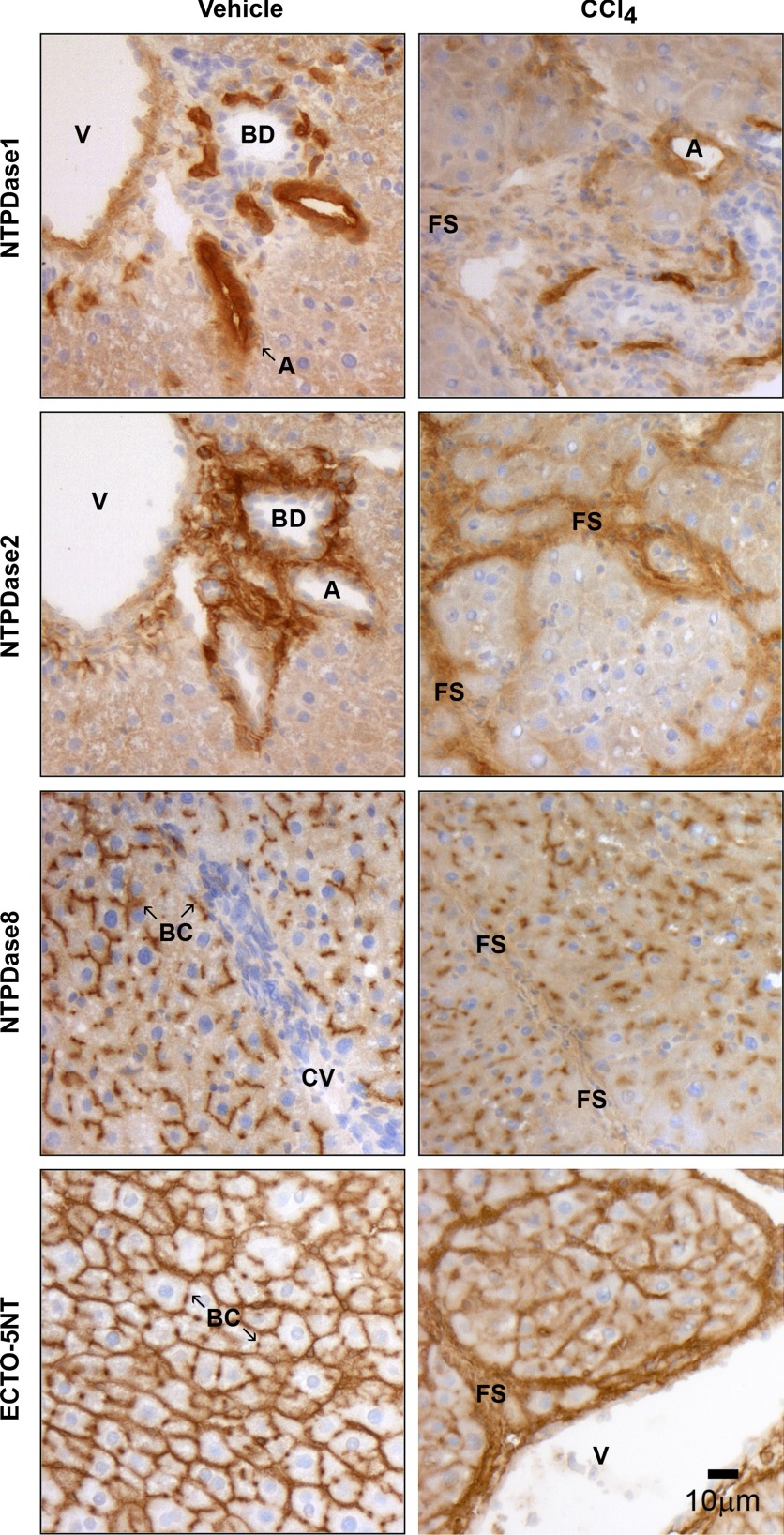
We also investigated the distribution of ectonucleotidases NTPDase1, -2, -8, and ecto-5′-nucleotidase in specimens from CCl4-induced fibrosis by immunohistochemistry (Fig. 8). Positive staining for NTPDase1 expression could be observed in several proliferating vascular structures and (likely) inflammatory cells in liver fibrotic areas and intersinusoidal spaces. NTPDase2 distribution was noticeably reorganized and present in fibrous septa surrounding hepatic nodules, as previously described (15). NTPDase8 expression, while still restricted to hepatocyte bile canaliculi, appeared rather reduced and diffuse in these structures compared with control liver. Finally, increased ecto-5′-nucleotidase expression was observed in fibrotic septa and sinusoidal spaces between hepatocytes. Also, its canalicular distribution at the hepatocyte membrane level was diffuse and less prominent relatively to control liver. Again, as noted in normal rat liver, the distribution profiles of NTPDase1, -2, -8, and ecto-5′-nucleotidase enzymes paralleled those described for ATPase, ADPase (NTPDases), and AMPase (ecto-5′-nucleotidase) activities in fibrotic livers.
Fig. 8.
Comparative distribution of liver NTPDases and ecto-5′-nucleotidase in CCl4-induced hepatic fibrosis model. Enzyme localization of the ectonucleotidases NTPDase1, -2, -8 and ecto-5′-nucleotidase was determined by immunohistochemistry in liver sections from vehicle (control) and CCl4-treated rats. In control animals, the expression patterns of NTPDase1, -2, -8 and ecto-5′-nucleotidase are similar to those described in Fig. 5. In contrast, in fibrotic animals, NTPDase1 expression is mainly associated with proliferating vascular structures. NTPDase2 expression is almost exclusively detected in perinodular fibrous areas. NTPDase8 protein is only found in bile canaliculi, although with a diffuse distribution. Immunoreactivity to ecto-5′-nucleotidase is localized at the level of both canalicular and basolateral membrane domains in hepatocytes, and prominently in fibrous septa surrounding hepatic fibrotic nodules. Scale bar, 10 μm.
Taken together, these data show that the histochemical localization of ATPase and ADPase activities correlates with the combined distribution of NTPDase1, -2, and -8, as detected by immunohistochemistry, whereas the AMPase activity corresponds to the immunohistological expression pattern of ecto-5′-nucleotidase in normal and fibrotic rat livers. In addition, our results suggest that ecto-5′-nucleotidase is expressed by a fibroblastic population that is devoid of NTPDase activity in normal rat liver.
HPLC Analysis of ATP Hydrolysis by Different Hepatic NTPDases in Combination with Ecto-5′-Nucleotidase
Based on the localization determined as above for NTPDase and ecto-5′-nucleotidase activities and protein expression, we evaluated how the net biochemical activity resulting from the pairing of ecto-5′-nucleotidase with either NTPDase1, NTPDase2, or NTPDase8 might affect the generation of nucleotide and nucleoside species in the liver. As described in experimental procedures, ATP hydrolysis assays were performed at a high, enzyme-saturating substrate concentration (500 μM) such as found under pathological conditions such as inflammation or hepatic failure (21, 25, 55), as well as at low more physiological substrate concentration (1 μM) (10, 33). Aliquots of the reaction mixtures were collected at different time points, and nucleotide and nucleoside contents were evaluated by HPLC. The time course of ATP hydrolysis and time-dependent generation of adenosine were analyzed for the different pairs of rat ectonucleotidases observed above for each liver structure.
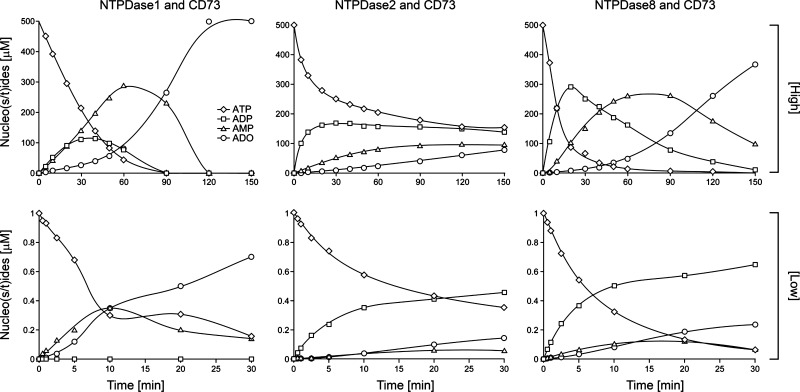
High ATP concentration (500 μM).
In the presence of NTPDase1 and ecto-5′-nucleotidase, ATP was hydrolyzed rapidly to adenosine, with transient increases of ADP and AMP concentrations (Fig. 9, NTPDase1 and CD73, high). Although AMP was rapidly produced, the formation of adenosine by ecto-5′-nucleotidase appeared to be slow during the first hour, likely because of the inhibitory effect of ATP and ADP transiently present in the medium at that early stage. When both ATP and ADP concentrations decreased afterward, adenosine formation increased until full conversion of AMP to adenosine. When ATP was incubated with NTPDase2 and ecto-5′-nucleotidase, it was readily converted into ADP, but very poorly into AMP, as predicted by the fact that ADP is a poor substrate of NTPDase2 (Fig. 9, NTPDase2 and CD73, high). Because of the protracted accumulation of ADP in the assay medium and the low abundance of AMP substrate for ecto-5′-nucleotidase, adenosine formation was accordingly limited. Of note is the observation that under the conditions described, rat NTPDase2 was active only for 50–60 min. The ATP hydrolysis profile obtained with the pairing of NTPDase8 with ecto-5′-nucleotidase showed a rapid ADP accumulation, peaking after 20 min, which was followed by the delayed generation of AMP, which reached a plateau between 50 and 120 min (Fig. 9, NTPDase8 and CD73, high). These observations concur with the fact that NTPDase8 activity is higher with ATP than with ADP. As a result, ecto-5′-nucleotidase activity was transiently inhibited by the presence of ADP, which delayed adenosine formation compared with the hydrolysis profile obtained with the NTPDase1 plus ecto-5′-nucleotidase pair.
Fig. 9.
ATP hydrolysis profile by the different combinations of ecto-5′-nucleotidase with hepatic NTPDases. Hydrolysis products of 500 μM (high) and 1 μM (low) ATP were analyzed over time, as indicated in experimental procedures. At high substrate concentration, ATP is rapidly and completely converted into adenosine in the presence of both NTPDase1 and ecto-5′-nucleotidase (CD73), whereas adenosine is hardly produced by the NTPDase2 + ecto-5′-nucleotidase combination. ATP hydrolysis in the presence of NTPDase8 + ecto-5′-nucleotidase combination generated a transient accumulation of ADP and AMP that resulted in a delayed (compared with the NTPDase1 + ecto-5′-nucleotidase combination) but substantial adenosine production (when compared with the NTPDase2 + ecto-5′-nucleotidase combination). At low substrate level, the incubation of ATP with NTPDase1 + ecto-5′-nucleotidase leads to its rapid dephosphorylation to adenosine without an increase in ADP level. In the presence of either NTPDase2 or NTPDase8 together with ecto-5′-nucleotidase, the accumulation of generated ADP was accompanied by poor AMP production because of limited ADP hydrolysis and thus resulted in a modest adenosine generation by ecto-5′-nucleotidase. ADO, adenosine.
Low ATP concentration (1 μM).
In the presence of the NTPDase1 and ecto-5′-nucleotidase pair, ATP was directly converted to AMP, ADP being rapidly hydrolyzed after it was formed (Fig. 9, NTPDase1 and CD73, low). Therefore, adenosine formation by ecto-5′-nucleotidase almost paralleled AMP appearance in the reaction mixture. After about 20 min, ∼80% of ATP added was hydrolyzed to adenosine. The ADP generated upon ATP hydrolysis by the NTPDase2 plus ecto-5′-nucleotidase pair accumulated in the reaction medium, with little AMP production, and as a result, adenosine was produced only in very small amounts (Fig. 9, NTPDase2 and CD73, low). The pattern of ATP hydrolysis by NTPDase8 plus ecto-5′-nucleotidase was very similar to the latter combination and resulted in the continuous accumulation of ADP (Fig. 9, NTPDase8 and CD73, low) with limited production of AMP and adenosine. In the presence of 1 μM ATP, the level of ATP and/or ADP produced was apparently too low to inhibit ecto-5′-nucleotidase. In this condition, the rate of adenosine formation was solely dependent on the substrate (AMP) generated. This contrasts with the data obtained at a high ATP concentration (Fig. 9 NTPDase8 and CD73, high) where the “higher” concentration of ADP generated was hampering the ability of ecto-5′-nucleotidase to hydrolyze AMP to adenosine, in agreement with our previous observations (35).
Note that in these assays we did not detect any degradation products of adenosine, indicating the lack of expression of adenosine-metabolizing enzymes in COS-7 cells, in keeping with the report of Tkacz et al. (51).
DISCUSSION
In the present study, we demonstrate a correlation between ecto-5′-nucleotidase/CD73 protein expression and AMPase activity in both normal and fibrotic rat livers. In normal rat liver, both attributes of ecto-5′-nucleotidase were detected in the canalicular and sinusoidal membrane domains of hepatocytes as well as in the connective tissue surrounding central veins or associated with the portal and larger septal space areas. Quite notably, ecto-5′-nucleotidase protein expression and AMPase activity were absent from the cell surface of hepatic vascular endothelium and bile duct epithelium in rat, as shown here, and in mouse (data not shown). The absence of ecto-5′-nucleotidase from murine endothelial cells contrasts with the vascular localization reported for its human ortholog (Ref. 1 and data not shown). Our examination of ecto-5′-nucleotidase distribution by double fluorescence immunostaining with specific cell markers as well as flow cytometry analysis of primary liver cells further indicates that ecto-5′-nucleotidase is mainly expressed in hepatocytes and nonmyofibroblastic (nonparenchymal) liver cells that are likely distinct from portal fibroblasts and hepatic stellate cells. Importantly, the facts that 1) levamisole, an inhibitor of alkaline phosphatase activity, had no effect on the AMPase activity observed (data not shown), 2) hydrolysis of AMP, CMP, GMP, and IMP (Figs. 1 and 2 and data not shown) all matched ecto-5′-nucleotidase immunolocalization using two different antibodies (Figs. 4 and 5 and data not shown), and 3) this activity was decreased by α,β-methylene-ADP, an ecto-5′-nucleotidase inhibitor (Fig. 2) suggest that ecto-5′-nucleotidase is responsible for most if not all nucleoside monophosphatase activity present in the liver. Ecto-5′-nucleotidase therefore appears to be the enzyme responsible for the formation of extracellular adenosine at physiological pH in the rat liver. This enzyme would therefore be expected to play a key role in the regulation of P1 receptor activation.
When we compared the distribution of ecto-5′-nucleotidase to that of hepatic NTPDase1, -2, and -8 with respect to the same parameters, i.e., protein expression and ectonucleotidase activity (14, 18) in normal rat liver, different combinations of NTPDase and ecto-5′-nucleotidase expression patterns were found: 1) NTPDase1 (vascular endothelium, smooth muscle cells, and Kupffer cells) and ecto-5′-nucleotidase, although not actually expressed in the same cells, were collectively present at the junction between the vascular tunica media and the perivascular connective tissue or at the sinusoidal face of hepatocyte membranes; 2) NTPDase2 (portal fibroblasts) and ecto-5′-nucleotidase were also expressed by neighboring cells located within the portal/periportal connective tissue; and finally 3) NTPDase8 and ecto-5′-nucleotidase were coexpressed by hepatocytes in bile canaliculi. It is noteworthy to mention that ecto-nucleotide pyrophosphatases/phosphodiesterases, which have the ability to hydrolyze tri- and diphosphate nucleosides in principle, are also expressed in the liver (48). However, their biochemical properties, namely a lower substrate affinity than NTPDases and optimal hydrolytic activity at alkaline rather than physiological pH, did not allow us to evaluate their potential contribution to liver purinergic signaling in our condition settings.
Induction of experimental liver fibrosis in CCl4-treated rats was associated with changes in the distribution of all hepatic ectonucleotidase activities. Overall, ATPase, ADPase, and AMPase activities were increased in liver fibrotic areas, in sinusoidal spaces, and in fibrous septa in the periphery of hepatic nodules, a hallmark of CCl4-induced fibrosis. In the same fibrotic livers, NTPDase1 protein was associated with proliferating blood vessels, NTPDase2 protein with the fibrous septa surrounding hepatic nodules, and NTPDase8 protein with structurally altered bile canaliculi of hepatocytes. Ecto-5′-nucleotidase protein expression was detected in both apical and sinusoidal domains of hepatocyte plasma membrane as well as in fibrous septa. Again, as observed in normal rat liver, hepatic AMPase activity and expression of ecto-5′-nucleotidase overlapped with the localization of hepatic ATPase and ADPase activities as well as the expression of NTPDase1, -2, and -8.
We addressed the potential impact of this specific distribution of NTPDases and ecto-5′-nucleotidase on extracellular nucleotide levels by performing a biochemical ATP hydrolysis assay in the presence of each pair formed by ecto-5′-nucleotidase with either NTPDase1, -2, or -8. It is noteworthy that the hydrolytic patterns observed in Fig. 9 are in agreement with the reported kinetic properties of each NTPDase (31) and with the fact that ATP and ADP are physiological inhibitors of ecto-5′-nucleotidase (35). Indeed, at 500 μM ATP, a significant decrease in ATP and ADP concentration was required to observe the effective production of adenosine from ecto-5′-nucleotidase. As expected, adenosine generation was fastest in the presence of NTPDase1, which rapidly hydrolyzed both ATP and ADP to the ecto-5′-nucleotidase substrate, AMP. In contrast, in the presence of NTPDase2, adenosine production was very low because of the very limited hydrolysis of ADP by this enzyme. The result obtained with NTPDase8 was somewhat intermediate between those observed for NTPDase1 and -2 at 500 μM ATP, which correlates with a transient production of ADP by NTPDase8 (18, 31), with adenosine production being inhibited when ATP and ADP levels exceed 50–100 μM, even at high AMP levels (200 μM). Interestingly, the situation was different for hydrolysis of low ATP concentration (1 μM), where ATP and ADP levels were insufficient to inhibit ecto-5′-nucleotidase, and as adenosine generation was limited only by the rate of AMP production by NTPDases. Of note, under the latter conditions, NTPDase8 behaved somewhat like NTPDase2 since it hydrolyzed ATP to ADP, with only minimal production of AMP, which greatly limited adenosine generation by ecto-5′-nucleotidase. This observation is in agreement with the Km value of NTPDase8 for ADP, which is much higher than for NTPDase1 (18, 31).
What functional importance could such a distribution of NTPDases and ecto-5′-nucleotidase have in the liver? At physiological pH, NTPDase1, -2, and -8 enzymatic activities account for the major bulk of hepatic ectonucleotidase activity, degrading nucleoside tri- and diphosphates into their monophosphate counterparts, which are essential for the activity of ecto-5′-nucleotidase, the rate-limiting ectoenzyme for adenosine formation (5). Thus the biochemical data presented here demonstrate that the precise expression pattern of the different NTPDases in the liver is of physiological relevance because they can distinctly modulate P1 and P2 receptor activation. Indeed, in a given cellular microenvironment, the coupling of each hepatic NTPDase species with its ecto-5′-nucleotidase complementary activity would feed extracellular purinergic mediators for different signal transduction pathways, according to the identity of the purinoceptors expressed at the cell membrane. In our assay, we used two different ATP concentrations, namely 1 μM, which is within the range of nucleotide levels measured in the conditioned media of liver cell lines (33), and reported for circulating nucleo(s/t)ides (50 nM to 1 μM) in both sinusoidal blood and bile flows (10), and 500 μM ATP, a value close to those potentially reached in the extracellular medium during inflammatory liver diseases (21, 25, 55). Our results show that the substrate concentration used can influence the net biochemical activity of the different NTPDase/ecto-5′-nucleotidase pairs, suggesting that the cellular context is an important parameter for the role of these ectonucleotidases in extracellular nucleotide signaling. Consequently, purinergic receptors expressed in the vicinity of a particular NTPDase/ecto-5′-nucleotidase combination might be differentially regulated depending on the prevailing biological conditions. Moreover, this hypothesis supports the notion that NTPDase1, -2, and -8 likely play nonredundant roles in the modulation of purinergic signaling, since ATP hydrolysis and adenosine generation profiles noticeably change as a function of the NTPDase species (see Fig. 9). Several liver functions could be affected by this specific distribution of NTPDases and ecto-5′-nucleotidase: 1) ion transport by cholangiocytes, an extracellular nucleotide-dependent mechanism that is essential to bile formation (20, 37–38, 57); 2) purine salvaging pathways in hepatocytes, as the liver represents the main source of nucleo(s/t)ides for extrahepatic tissues deficient in de novo purine biosynthesis (11); 3) cell proliferation, an important feature of liver tissue, which involves P2 receptor signaling in hepatocytes, sinusoidal endothelial cells, and portal fibroblasts (6, 22, 28, 50); 4) cell death, since stimulation of either P1 or P2 receptors in hepatocytes has been shown to induce apoptosis (52, 56); and finally 5) inflammation, since hepatocytes, liver immune cells such as Kupffer cells and T lymphocytes, portal fibroblasts, as well as hepatic stellate cells express different P1 and P2 receptors that might modulate their activation and/or deactivation states in liver diseases (4, 13, 17, 41, 54).
In summary, the specific distribution of NTPDase1, -2, and -8 in conjunction with ecto-5′-nucleotidase in the liver differentially regulates extracellular nucleo(s/t)ide levels and, therefore, may also differentially regulate the associated signaling pathways.
GRANTS
This work was supported by the Canadian Institutes of Health Research (CIHR; MOP-102472 to J. Sévigny) and by the National Institutes of Health (NIH/NIDDK R01 DK076735, R01 DK070849 and the Yale Liver Center DK 45710 awards to J. A. Dranoff). M. Fausther was the recipient of a Doctoral Scholarship from the Government of Gabon and is currently receiving the 2011 Roger L. Jenkins Post doctoral Fellowship from the American Liver Foundation, E. M. Soliman of a Student Research Grant from the Egyptian Government Ministry of Higher Education, G. Kauffenstein of fellowships from the Institut National de la Santé et de la Recherche Médicale in partnership with the Fonds de Recherche en Santé du Québec (FRSQ) and the Heart and Stroke Foundation of Canada in partnership with the CIHR and the Canadian Stroke Network, and J. Sévigny of a New Investigator award from the CIHR and a Junior 2 Scholarship from the FRSQ.
DISCLOSURES
No conflicts of interest, financial or otherwise, are declared by the author(s).
ACKNOWLEDGMENTS
The authors thank Dr. H. Zimmermann (J. W. Goethe University, Frankfurt, Germany) for providing the plasmid encoding for rat ecto-5′-nucleotidase/CD73 and Dr. M. J. Fernandes and V. Gagné (Rheumatology and Immunology Research Centre, CHUL Research Center) for technical assistance with confocal microscopy. We also thank Dr. R. Poulin (Scientific Proofreading and Writing Service, CHUQ Research Center) for editing this paper.
REFERENCES
- 1. Airas L, Salmi M, Jalkanen S. Lymphocyte-vascular adhesion protein-2 is a novel 70-kDa molecule involved in lymphocyte adhesion to vascular endothelium. J Immunol 151: 4228–4238, 1993. [PubMed] [Google Scholar]
- 2. Bachmann S, Ramasubbu K. Immunohistochemical colocalization of the alpha-subunit of neutrophil NADPH oxidase and ecto-5′-nucleotidase in kidney and liver. Kidney Int 51: 479–482, 1997. [DOI] [PubMed] [Google Scholar]
- 3. Baykov AA, Evtushenko OA, Avaeva SM. A malachite green procedure for orthophosphate determination and its use in alkaline phosphatase-based enzyme immunoassay. Anal Biochem 171: 266–270, 1988. [DOI] [PubMed] [Google Scholar]
- 4. Beldi G, Banz Y, Kroemer A, Sun X, Wu Y, Graubardt N, Rellstab A, Nowak M, Enjyoji K, Li X, Junger WG, Candinas D, Robson SC. Deletion of CD39 on natural killer cells attenuates hepatic ischemia/reperfusion injury in mice. Hepatology 51: 1702–1711, 2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Beldi G, Enjyoji K, Wu Y, Miller L, Banz Y, Sun X, Robson SC. The role of purinergic signaling in the liver and in transplantation: effects of extracellular nucleotides on hepatic graft vascular injury, rejection and metabolism. Front Biosci 13: 2588–2603, 2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Beldi G, Wu Y, Sun X, Imai M, Enjyoji K, Csizmadia E, Candinas D, Erb L, Robson SC. Regulated catalysis of extracellular nucleotides by vascular CD39/ENTPD1 is required for liver regeneration. Gastroenterology 135: 1751–1760, 2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Braun N, Lenz C, Gillardon F, Zimmermann M, Zimmermann H. Focal cerebral ischemia enhances glial expression of ecto-5′-nucleotidase. Brain Res 766: 213–226, 1997. [DOI] [PubMed] [Google Scholar]
- 8. Braun N, Zhu Y, Krieglstein J, Culmsee C, Zimmermann H. Upregulation of the enzyme chain hydrolyzing extracellular ATP after transient forebrain ischemia in the rat. J Neurosci 18: 4891–4900, 1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Burnstock G. Purine and pyrimidine receptors. Cell Mol Life Sci 64: 1471–1483, 2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Chari RS, Schutz SM, Haebig JE, Shimokura GH, Cotton PB, Fitz JG, Meyers WC. Adenosine nucleotides in bile. Am J Physiol Gastrointest Liver Physiol 270: G246–G252, 1996. [DOI] [PubMed] [Google Scholar]
- 11. Che MX, Gatmaitan Z, Arias IM. Ectonucleotidases, purine nucleoside transporter, and function of the bile canalicular plasma membrane of the hepatocyte. FASEB J 11: 101–108, 1997. [DOI] [PubMed] [Google Scholar]
- 12. Colgan SP, Eltzschig HK, Eckle T, Thompson LF. Physiological roles for ecto-5′-nucleotidase (CD73). Purinergic Signal 2: 351–360, 2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Di Virgilio F, Boeynaems JM, Robson SC. Extracellular nucleotides as negative modulators of immunity. Curr Opin Pharmacol 9: 507–513, 2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Dranoff JA, Kruglov EA, Robson SC, Braun N, Zimmermann H, Sévigny J. The ecto-nucleoside triphosphate diphosphohydrolase NTPDase2/CD39L1 is expressed in a novel functional compartment within the liver. Hepatology 36: 1135–1144, 2002. [DOI] [PubMed] [Google Scholar]
- 15. Dranoff JA, Kruglov EA, Toure J, Braun N, Zimmermann H, Jain D, Knowles AF, Sévigny J. Ectonucleotidase NTPDase2 is selectively down-regulated in biliary cirrhosis. J Investig Med 52: 475–482, 2004. [DOI] [PubMed] [Google Scholar]
- 16. Dranoff JA, Ogawa M, Kruglov EA, Gaca MD, Sévigny J, Robson SC, Wells RG. Expression of P2Y nucleotide receptors and ectonucleotidases in quiescent and activated rat hepatic stellate cells. Am J Physiol Gastrointest Liver Physiol 287: G417–G424, 2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Dranoff JA, Wells RG. Portal fibroblasts: underappreciated mediators of biliary fibrosis. Hepatology 51: 1438–1444, 2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Fausther M, Lecka J, Kukulski F, Lévesque SA, Pelletier J, Zimmermann H, Dranoff JA, Sévigny J. Cloning, purification, and identification of the liver canalicular ecto-ATPase as NTPDase8. Am J Physiol Gastrointest Liver Physiol 292: G785–G795, 2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Fausther M, Sévigny J. Extracellular nucleosides and nucleotides regulate liver functions via a complex system of membrane proteins. C R Biol 334: 100–117, 2011. [DOI] [PubMed] [Google Scholar]
- 20. Fiorotto R, Spirli C, Fabris L, Cadamuro M, Okolicsanyi L, Strazzabosco M. Ursodeoxycholic acid stimulates cholangiocyte fluid secretion in mice via CFTR-dependent ATP secretion. Gastroenterology 133: 1603–1613, 2007. [DOI] [PubMed] [Google Scholar]
- 21. Franke H, Illes P. Involvement of P2 receptors in the growth and survival of neurons in the CNS. Pharmacol Ther 109: 297–324, 2006. [DOI] [PubMed] [Google Scholar]
- 22. Gonzales E, Julien B, Serriere-Lanneau V, Nicou A, Doignon I, Lagoudakis L, Garcin I, Azoulay D, Duclos-Vallee JC, Castaing D, Samuel D, Hernandez-Garcia A, Awad SS, Combettes L, Thevananther S, Tordjmann T. ATP release after partial hepatectomy regulates liver regeneration in the rat. J Hepatol 52: 54–62, 2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Hart ML, Gorzolla IC, Schittenhelm J, Robson SC, Eltzschig HK. SP1-dependent induction of CD39 facilitates hepatic ischemic preconditioning. J Immunol 184: 4017–4024, 2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Hashimoto M, Kothary PC, Raper SE. Phenobarbital in comparison with carbon tetrachloride and phenobarbital-induced cirrhosis in rat liver regeneration. J Surg Res 81: 164–169, 1999. [DOI] [PubMed] [Google Scholar]
- 25. Hasko G, Linden J, Cronstein B, Pacher P. Adenosine receptors: therapeutic aspects for inflammatory and immune diseases. Nat Rev Drug Discov 7: 759–770, 2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Heine P, Braun N, Heilbronn A, Zimmermann H. Functional characterization of rat ecto-ATPase and ecto-ATP diphosphohydrolase after heterologous expression in CHO cells. Eur J Biochem 262: 102–107, 1999. [DOI] [PubMed] [Google Scholar]
- 27. Hunsucker SA, Mitchell BS, Spychala J. The 5′-nucleotidases as regulators of nucleotide and drug metabolism. Pharmacol Ther 107: 1–30, 2005. [DOI] [PubMed] [Google Scholar]
- 28. Jhandier MN, Kruglov EA, Lavoie EG, Sévigny J, Dranoff JA. Portal fibroblasts regulate the proliferation of bile duct epithelia via expression of NTPDase2. J Biol Chem 280: 22986–22992, 2005. [DOI] [PubMed] [Google Scholar]
- 29. Kaczmarek E, Koziak K, Sévigny J, Siegel JB, Anrather J, Beaudoin AR, Bach FH, Robson SC. Identification and characterization of CD39 vascular ATP diphosphohydrolase. J Biol Chem 271: 33116–33122, 1996. [DOI] [PubMed] [Google Scholar]
- 30. Kegel B, Braun N, Heine P, Maliszewski CR, Zimmermann H. An ecto-ATPase and an ecto-ATP diphosphohydrolase are expressed in rat brain. Neuropharmacology 36: 1189–1200, 1997. [DOI] [PubMed] [Google Scholar]
- 31. Kukulski F, Lévesque SA, Lavoie EG, Lecka J, Bigonnesse F, Knowles AF, Robson SC, Kirley TL, Sévigny J. Comparative hydrolysis of P2 receptor agonists by NTPDases 1, 2, 3 and 8. Purinergic Signal 1: 193–204, 2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Lavoie ÉG, Kukulski F, Lévesque SA, Lecka J, Sévigny J. Cloning and characterization of mouse nucleoside triphosphate diphosphohydrolase-3. Biochem Pharmacol 67: 1917–1926, 2004. [DOI] [PubMed] [Google Scholar]
- 33. Lazarowski ER, Boucher RC, Harden TK. Mechanisms of release of nucleotides and integration of their action as P2X- and P2Y-receptor activating molecules. Mol Pharmacol 64: 785–795, 2003. [DOI] [PubMed] [Google Scholar]
- 34. Lazarowski ER, Tarran R, Grubb BR, van Heusden CA, Okada S, Boucher RC. Nucleotide release provides a mechanism for airway surface liquid homeostasis. J Biol Chem 279: 36855–36864, 2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Lecka J, Rana MS, Sévigny J. Inhibition of vascular ectonucleotidase activities by the pro-drugs ticlopidine and clopidogrel favours platelet aggregation. Br J Pharmacol 161: 1150–1160, 2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Levitt B, Head RJ, Westfall DP. High-pressure liquid chromatographic-fluorometric detection of adenosine and adenine nucleotides: application to endogenous content and electrically induced release of adenyl purines in guinea pig vas deferens. Anal Biochem 137: 93–100, 1984. [DOI] [PubMed] [Google Scholar]
- 37. Masyuk AI, Gradilone SA, Banales JM, Huang BQ, Masyuk TV, Lee SO, Splinter PL, Stroope AJ, Larusso NF. Cholangiocyte primary cilia are chemosensory organelles that detect biliary nucleotides via P2Y12 purinergic receptors. Am J Physiol Gastrointest Liver Physiol 295: G725–G734, 2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Minagawa N, Nagata J, Shibao K, Masyuk AI, Gomes DA, Rodrigues MA, Lesage G, Akiba Y, Kaunitz JD, Ehrlich BE, Larusso NF, Nathanson MH. Cyclic AMP regulates bicarbonate secretion in cholangiocytes through release of ATP into bile. Gastroenterology 133: 1592–1602, 2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Naito Y, Lowenstein JM. 5′-Nucleotidase from rat heart membranes. Inhibition by adenine nucleotides and related compounds. Biochem J 226: 645–651, 1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Peng Z, Borea PA, Varani K, Wilder T, Yee H, Chiriboga L, Blackburn MR, Azzena G, Resta G, Cronstein BN. Adenosine signaling contributes to ethanol-induced fatty liver in mice. J Clin Invest 119: 582–594, 2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Peng Z, Fernandez P, Wilder T, Yee H, Chiriboga L, Chan ES, Cronstein BN. Ecto-5′-nucleotidase (CD73) -mediated extracellular adenosine production plays a critical role in hepatic fibrosis. FASEB J 22: 2263–2272, 2008. [DOI] [PubMed] [Google Scholar]
- 42. Popov Y, Schuppan D. Targeting liver fibrosis: strategies for development and validation of antifibrotic therapies. Hepatology 50: 1294–1306, 2009. [DOI] [PubMed] [Google Scholar]
- 43. Robson SC, Schuppan D. Adenosine: tipping the balance towards hepatic steatosis and fibrosis. J Hepatol 52: 941–943, 2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Robson SC, Sévigny J, Zimmermann H. The E-NTPDase family of ectonucleotidases: Structure, function, relationships and pathophysiological significance. Purinergic Signal 2: 409–430, 2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Sévigny J, Robson SC, Waelkens E, Csizmadia E, Smith RN, Lemmens R. Identification and characterization of a novel hepatic canalicular ATP diphosphohydrolase. J Biol Chem 275: 5640–5647, 2000. [DOI] [PubMed] [Google Scholar]
- 46. Sévigny J, Sundberg C, Braun N, Guckelberger O, Csizmadia E, Qawi I, Imai M, Zimmermann H, Robson SC. Differential catalytic properties and vascular topography of murine nucleoside triphosphate diphosphohydrolase 1 (NTPDase1) and NTPDase2 have implications for thromboregulation. Blood 99: 2801–2809, 2002. [DOI] [PubMed] [Google Scholar]
- 47. Song J, Bosch KS, Tigchelaar W, Van Den Munckhof RJ, Schellens JP, Van Noorden CJ, Frederiks WM. Demonstration of 5′-nucleotidase activity in unfixed cryostat sections of rat liver using a combined light- and electron-microscope procedure. Histochem J 27: 914–922, 1995. [PubMed] [Google Scholar]
- 48. Stefan C, Jansen S, Bollen M. Modulation of purinergic signaling by NPP-type ectophosphodiesterases. Purinergic Signal 2: 361–370, 2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Strater N. Ecto-5′-nucleotidase: Structure function relationships. Purinergic Signal 2: 343–350, 2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Thevananther S, Sun H, Li D, Arjunan V, Awad SS, Wyllie S, Zimmerman TL, Goss JA, Karpen SJ. Extracellular ATP activates c-jun N-terminal kinase signaling and cell cycle progression in hepatocytes. Hepatology 39: 393–402, 2004. [DOI] [PubMed] [Google Scholar]
- 51. Tkacz K, Cioroch M, Skladanowski AC, Makarewicz W. The cytotoxic effect of purine riboside on COS-7 cells. Adv Exp Med Biol 486: 355–359, 2000. [DOI] [PubMed] [Google Scholar]
- 52. Wen LT, Knowles AF. Extracellular ATP and adenosine induce cell apoptosis of human hepatoma Li-7A cells via the A3 adenosine receptor. Br J Pharmacol 140: 1009–1018, 2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Wood E, Broekman MJ, Kirley TL, Diani-Moore S, Tickner M, Drosopoulos JH, Islam N, Park JI, Marcus AJ, Rifkind AB. Cell-type specificity of ectonucleotidase expression and upregulation by 2,3,7,8-tetrachlorodibenzo-p-dioxin. Arch Biochem Biophys 407: 49–62, 2002. [DOI] [PubMed] [Google Scholar]
- 54. Yang P, Han Z, Chen P, Zhu L, Wang S, Hua Z, Zhang J. A contradictory role of A1 adenosine receptor in carbon tetrachloride- and bile duct ligation-induced liver fibrosis in mice. J Pharmacol Exp Ther 332: 747–754, 2010. [DOI] [PubMed] [Google Scholar]
- 55. Yegutkin GG. Nucleotide- and nucleoside-converting ectoenzymes: Important modulators of purinergic signalling cascade. Biochim Biophys Acta 1783: 673–694, 2008. [DOI] [PubMed] [Google Scholar]
- 56. Zoetewij JP, van de Water B, de Bont HJ, Nagelkerke JF. The role of a purinergic P2z receptor in calcium-dependent cell killing of isolated rat hepatocytes by extracellular adenosine triphosphate. Hepatology 23: 858–865, 1996. [DOI] [PubMed] [Google Scholar]
- 57. Zsembery A, Spirli C, Granato A, LaRusso NF, Okolicsanyi L, Crepaldi G, Strazzabosco M. Purinergic regulation of acid/base transport in human and rat biliary epithelial cell lines. Hepatology 28: 914–920, 1998. [DOI] [PubMed] [Google Scholar]