Abstract
Background
Strains of the endosymbiotic bacterium Wolbachia pipientis are extremely diverse both genotypically and in terms of their induced phenotypes in invertebrate hosts. Despite extensive molecular characterisation of Wolbachia diversity, little is known about the actual genomic diversity within or between closely related strains that group tightly on the basis of existing gene marker systems, including Multiple Locus Sequence Typing (MLST). There is an urgent need for higher resolution fingerprinting markers of Wolbachia for studies of population genetics, horizontal transmission and experimental evolution.
Results
The genome of the wMel Wolbachia strain that infects Drosophila melanogaster contains inter- and intragenic tandem repeats that may evolve through expansion or contraction. We identified hypervariable regions in wMel, including intergenic Variable Number Tandem Repeats (VNTRs), and genes encoding ankyrin (ANK) repeat domains. We amplified these markers from 14 related Wolbachia strains belonging to supergroup A and were successful in differentiating size polymorphic alleles. Because of their tandemly repeated structure and length polymorphism, the markers can be used in a PCR-diagnostic multilocus typing approach, analogous to the Multiple Locus VNTR Analysis (MLVA) established for many other bacteria and organisms. The isolated markers are highly specific for supergroup A and not informative for other supergroups. However, in silico analysis of completed genomes from other supergroups revealed the presence of tandem repeats that are variable and could therefore be useful for typing target strains.
Conclusions
Wolbachia genomes contain inter- and intragenic tandem repeats that evolve through expansion or contraction. A selection of polymorphic tandem repeats is a novel and useful PCR diagnostic extension to the existing MLST typing system of Wolbachia, as it allows rapid and inexpensive high-throughput fingerprinting of closely related strains for which polymorphic markers were previously lacking.
Background
Wolbachia pipientis (α-Proteobacteria) is an obligate endosymbionts of invertebrates, known to infect up to 70% of insect species, as well as spiders, terrestrial crustaceans and medically important filarial nematodes [1-5]. Many strains of Wolbachia found in insects manipulate their hosts by inducing feminisation, parthenogenesis, male killing or cytoplasmic incompatibility (CI) [6-9]; in contrast, the Wolbachia of nematodes are mutualists necessary for host reproduction [10]. Despite this great diversity of hosts and extended phenotypes, all strains of Wolbachia are currently recognised as the single species W. pipientis. Within this species, strains are clustered into at least eight divergent clades or 'supergroups', named A to K [11-15].
Several genes have been used for strain typing in Wolbachia. Initially, work focused on 16S rDNA[16], the genes encoding the cell division protein, ftsZ [11] and the Wolbachia surface protein, wsp [12]. Subsequent to the demonstration of widespread intra- and intergenic recombination betweens strains [17-19], two multi-locus sequence typing (MLST) systems were developed using different sets of a total of 14 Wolbachia genes [20,21]. The MLST approach uses partial nucleotide sequences of several ubiquitous loci with moderate rates of evolution to generate an allelic profile for tested strains. These profiles can be used to type novel isolates, while the relationships between strains may be inferred on the basis of either the allelic profiles themselves or the nucleotide sequences underlying them. MLST data have been used for both strain typing and evolutionary analyses of horizontal transfer events between host species of Wolbachia (e.g. [22,23]). Since most MLST primer sets cover housekeeping genes that are under purifying selection, these markers often cannot differentiate between closely related strains. Such difficulties have been revealed in the comparisons between wMel, wMelCS and wMelPop [20] or wMel and wAu within the ST-13 complex which appear indistinguishable in MLST loci [21,24]. These strains induce different phenotypes in their hosts, i.e. wMel induces CI in Drosophila, but wAu does not [25] and wMelPop induces lifespan reduction in its hosts but not wMel [26-28]. The divergence between MLST typing and actual genomic diversity within ST-13 was also raised when these closely related strains were compared for presence or absence of Wolbachia prophage WO-A and WO-B [24] and other genomic differences such as a large chromosomal inversion and differential IS5 insertion sites between wMel, wMelPop and wMelCS [29,30]. Furthermore, MLST can be time consuming and expensive for large population genetic studies as it requires sequencing of all MLST loci for many individuals. Recently other typing systems have been developed for bacteria that build on markers that contain Variable Number Tandem Repeats (VNTR). VNTRs consist of units of DNA (periods) that are tandemly repeated and vary in copy number between different isolates. These loci can be used for a PCR-based typing system and are increasingly being utilised in bacterial strain typing such as Multi Locus VNTR Analysis (MLVA) (e.g. [31-35]). MLVA offers a number of advantages, including highly polymorphic markers that allow fine-scale typing of very closely related isolates, rapid, high-throughput screening that is not dependent on sequencing, and potentially the fingerprinting of multiply infected hosts. The modular structure and evolution of these sites through tandem expansion and contraction also allows cladistic and phylogenetic inference.
Amplicon size polymorphic markers have previously been identified in Wolbachia genomes and include transposable element insertion sites [30,36,37], VNTRs [30,38-40] and genes encoding ankyrin repeat domains [36], but their efficiency for strain typing has not yet been compared. In this paper, we used some of these markers in order to estimate the feasibility of a MLVA system for Wolbachia. We isolated markers with tandem repeats from the wMel genome [41] and applied them to a number of Wolbachia strains from supergroups A, B and C to assess their applicability and resolution for Wolbachia strain typing. We chose two types of loci containing tandem repeats, two intergenic VNTR loci and two genes encoding proteins containing ankyrin repeats. The two VNTR loci, VNTR-105 and VNTR-141 were originally isolated from supergroup A strain wMel and were polymorphic between wMel, wMelCS and wMelPop isolates from different D. melanogaster lines [30]. VNTRs are also polymorphic between the closely related wAu from D. simulans and wWil from Drosophila willistoni [38], and serve as highly diagnostic marker sets for fingerprinting conspecific Wolbachia strains in the Drosophila paulistorum species cluster [39]. Recently, a polymorphic VNTR locus was isolated from supergroup B strain wPip [40]. Ankyrin repeat genes are abundant in the genomes of Wolbachia and a number of other intracellular bacteria [42,43]. The number and distribution of these repeats varies substantially between strains that induce different host phenotypes, suggesting that they may be involved in host manipulation [36]. We extended our analysis to include a wider range of Wolbachia strains from supergroup A, B and C in order to evaluate the usefulness of the four markers VNTR-105, VNTR-141, WD0550 and WD0766, originally isolated from wMel, in discriminating between Wolbachia strains.
Methods
Wolbachia strains and hosts
We used 14 supergroup A Wolbachia isolates from 8 different Drosophila species and 2 tephritid species, Rhagoletis cerasi, a host that is naturally infected, and Ceratitis capitata, microinjected with Wolbachia originating from R. cerasi (Table 1). Based on previous strain typing using 16S rRNA, ftsZ, wsp and some MLST loci, these 14 strains are moderately or closely related, yet they reveal different phenotypic characteristics, such as varying levels of CI induction (strong, weak, or non-CI inducers), and different CI rescue phenotypes (reviewed in [44]). Wolbachia DNA was isolated from Drosophila fly stocks reared on standard corn-flour-sugar-yeast medium at 25°C. Wolbachia-free controls D. melanogaster yw67c23T and D. simulans Riverside-DSRT were established by tetracycline treatment using standard techniques [45]. Wolbachia of R. cerasi was isolated from field collected samples from Austria and Hungary [46]. Wolbachia from C. capitata was isolated from the WolMed 88.6 lab line that was artificially infected with wCer2 from R. cerasi [47]. We also included strains from B (wNo, wBol1, wMau) and C (wDim) supergroups. wNo and wMau were isolated from D. simulans, wBol1 from Hypolimnas bolina [48] and wDim from dog heart worm Dirofilaria immitis [49].
Table 1.
List of Wolbachia strains.
| Strain | Supergroup | Host | Location | mod | res | Reference |
|---|---|---|---|---|---|---|
| wMel | A | D. melanogaster | USA | yes | yes | [75,76] |
| wMelCS | A | D. melanogaster | CantonS, USA | yes | yes | [30,70] |
| wMelPop | A | D. melanogaster | laboratory strain, USA | yes | yes | [26,27] |
| wAu | A | D. simulans | Coffs Harbour, Australia | no | no | [25] |
| wSan | A | D. santomea | Sao Tome, Africa | no* | yes | [77] |
| wYak | A | D. yakuba | Bom Successo, Africa | no* | yes | [77] |
| wTei | A | D. teissieri | Bom Successo, Africa | no* | yes | [77] |
| wWil | A | D. willistoni | Central and South America | no | n.d. | [38] |
| wSpt | A | D. septentriosaltans | Central and South America | n.d. | n.d. | [38] |
| wPro | A | D. prosaltans | Central and South America | n.d. | n.d. | [38] |
| wCer1 | A | R. cerasi | Hungary | n.d. | n.d. | [46,61] |
| wCer2 | A | R. cerasi | Austria | yes | yes | [46,61] |
| wCer2 | A | D. simulans | microinjected | yes | yes | [62] |
| wCer2 | A | C. capitata | microinjected | yes | yes | [47] |
| wRi | A | D. simulans | Riverside, USA | yes | yes | [16,45] |
| wHa | A | D. simulans | Hawaii, USA | yes | yes | [16,78] |
| wNo | B | D. simulans | Noumea | yes | yes | [79] |
| wMau | B | D. simulans | microinjected | no | yes | [80] |
| wBol1 | B | H. bolina | French Polynesia | yes¶ | yes¶ | [81] |
| wDim | C | Dirofilaria immitis | Queensland | no | no | [49] |
Modification/rescue phenotypes are included except for strains for which crossing phenotypes had not been determined (n.d.). Modification corresponds to the capacity of a strain to induce cytoplasmic incompatibility (CI) through sperm modification whereas rescue corresponds to the capacity to rescue CI in eggs fertilized by modified sperm [74]. The reference relates to the first description of the strain and/or the phenotype.
* wSan, wYak, wTei do not induce CI in their original hosts, yet can rescue CI induced by other strains [77], and induce CI in novel hosts upon artificial horizontal transfer through microinjection into D. simulans [23].
¶ CI only expressed in host genotypes that are resistant to the expression of male killing induced by wBol1 [48,81]
DNA extraction, PCR amplification and sequencing of molecular markers
Total genomic DNA was extracted from either freshly collected specimens or specimens stored in pure ethanol in a -20°C freezer. Extraction was carried out on pools of Drosophila flies and single individuals of Rhagoletis, Ceratitis, Hypolimnas and Dirofilaria. Flies were homogenized and extracted following either the Holmes-Bonner protocol [50] or the STE extraction method [16]. Wolbachia markers were amplified from total genomic DNA using specific primers (Table 2). The wsp gene was used as a quality control for DNA extraction and was amplified using the primers 81F and 691R, described in [12]. PCR cycling conditions were as follows: 94°C 3 min, (94°C 30 s, 50°C 30 s, 72°C 3 min) x 35 cycles, then 72°C 10 min. The reaction mixture contained 500 nM of each primer, 200 µM dNTPs, 1.5 mM MgCl2, 100 ng of DNA and 1 unit of Taq Polymerase (Promega) in a final volume of 20 µl. The reaction buffer contained 10 mM Tris pH 9.0, 50 mM KCl and 0.1% Triton X-100. PCR products were separated in 1% agarose gels, stained with ethidium bromide and gel-purified using gel extraction kits (QIAGEN). Purified DNA was cloned into the pGEM®-T-easy plasmid (Promega) and sequenced by Macrogen, in Korea, using T7, M13R, and internal primers, as required. Three independent PCRs were sequenced for each gene, checked and confirmed for consistency. Partial sequences of the VNTR-105, VNTR-141 and the ANK genes WD0550 and WD0766 from different Wolbachia strains have been deposited GenBank database (Table 3).
Table 2.
List of primers designed according to the wMel genome sequence to amplify VNTRs and ANK genes.
| Locus/primer | 5’ sequence | Reference |
|---|---|---|
| VNTR-141 for | ggagtattattgatatgcg | [30] |
| VNTR-141 rev | gactaaaggttagttgcat | [30] |
| VNTR-105 for | gcaattgaaaatgtggtgcc | [30] |
| VNTR-105 rev | atgacaccttacttaaccgtc | [30] |
| RO550F | ggccaccatgggatcagaatttgaag | [82] |
| RO550R | gatgacttatacgcagccccatag | [82] |
| RO766F | gaccaccatgaaatatgacaaattt | [82] |
| RO766R | tcaagtaagtgctttttctgtc | [82] |
Table 3.
GenBank accession numbers for VNTR and ANK sequences.
| Strain | VNTR-105 | VNTR-141 | WD0766 |
|---|---|---|---|
| wMel | JF797619 | JF797613 | NC_002978* |
| wMelCS | JF797618 | JF797611 | JF683428 |
| wMelPop | as wMelCS | JF797612 | JF683429 |
| wRi | n.d. | n.d. | NC_012416** |
| wAu | JF797617 | JF797608 | AY649753 |
| wSan | JN191623 | JN191622 | JF683435 |
| wWil | JF797616 | JF797607 | JF683433 |
| wSpt | JF797620 | JF797609 | JF683431 |
| wPro | n.d. | JF797610 | JF683430 |
| wCer1 | JF797615 | JF797606 | JF683434 |
| wCer2 | n.d. | JF797614 | JF683432 |
| wHa | n.d. | n.d. | JF683436 |
*wMel genome sequence
**wRi genome sequence
n.d. not determined
Selection of size variable markers
Polymorphic loci were previously identified from the sequenced genome of wMel of D. melanogaster ([41], GenBank reference sequence NC_002978) in silico by using Tandem Repeats Finder TRF (http://tandem.bu.edu/trf/trf.html) [51]. Two VNTR regions of interest, VNTR-105 and VNTR-141 were found to be polymorphic between different lines of D. melanogaster [30]. The TRF analysis also detected more candidate loci, including some genes encoding ANK domain repeats that can also contain tandemly repeated DNA, and are hence candidate markers for MLVA. Genes encoding ANK domain repeats were previously annotated [41] and variability was found in supergroup A and B Wolbachia strains [36]. All of the tandem repeats analysed here were amplified by using primers designed for the conserved flanking regions (single copy coding genes) of the repeats within wMel. We further extended the TRF analysis to other completed Wolbachia genomes, wRi ([52] NC_012416), wPip ([53] NC_010981) and wBm ([54] NC_006833) in order to highlight the potential of MLVA for more distantly related Wolbachia strains in silico. The TRF analysis also included the genomes of Anaplasma marginale strain St. Maries (CP_000030) and Ehrlichia ruminantium strain Welgevonden (NC_005295) and Neorickettsia risticii strain Illinois (NC_013009), the closest relatives of the genus Wolbachia [55], as well as a comparison with free living Escherichia coli K12 substrain MG1655 (NC_000913). The bacterial genomes were analysed in the basic mode of TRF (version 4.04), with alignment parameters for match, mismatch and indels set at 2, 7 and 7, respectively. The minimum alignment score to report repeats was set at 50, with a maximum period size of 500bp (Table 4).
Table 4.
Summary of Tandem Repeats Finder (TRF) analysis.
| Strain | genome size | TR | TR size in total (% genome) | mean TR period size (range) | mean number of repeats/TR (range) | mean TR internal match (%) |
|---|---|---|---|---|---|---|
| wMel | 1,267,812bp | 93 | 20,349bp (1.6%) | 80.9bp (10-291) | 2.7 (1.8-11.8) | 88.3 |
| wRi | 1,445,904bp | 94 | 16,667bp (1.1%) | 58.5bp (10-378) | 2.8 (1.8-8.8) | 87.5 |
| wPip | 1,482,530bp | 72 | 13,268bp (0.9%) | 68.5bp (12-399) | 2.8 (1.8-10.6) | 87.9 |
| wBm | 1,080,114bp | 11 | 1,032bp (0.1%) | 42.8bp (3-112) | 3.3 (1.9-15.7) | 89.0 |
| A. m. | 1,197,687bp | 54 | 8,541bp (0.7%) | 64.4bp (11-495) | 2.8 (1.9-11.2) | 91.1 |
| E. r. | 1,516,355bp | 201 | 95,290bp (6.3%) | 138.7bp (1-471) | 4.8 (1.8-65.1) | 91.6 |
| N. r. | 879,977bp | 27 | 5,569bp (0.6%) | 68.8bp (9-297) | 2.9 (1.9-4.9) | 88.4 |
| E. coli | 4,649,675bp | 89 | 17,807bp (0.38%) | 70.4bp (8-304) | 3.1 (1.9-12.5) | 90.1 |
Analysis in basic TRF basic mode included four completed Wolbachia genomes with strain names in bold, wMel (NCBI accession NC_002978), wRi (NC_012416), wPip (NC_010981) and wBm (NC_006833), and the genomes of Anaplasma marginale (A.m.) strain St. Maries (CP_000030), Ehrlichia ruminantium (E.r.) st. Welgevonden (NC_005295), Neorickettsia risticii (N.r.) st. Illinois (NC_013009) and Escherichia coli (E. coli) K12 substrain MG1655 (NC_000913). TRF detected several tandem repeats (TR) within the same genomic regions, as some tandem repeats contain internal repeats; the number of tandem repeats in column three does hence overrepresent the number of tandem repeat loci in the genome.
Sequence analysis
The analysis and assembly of the sequences was done using the EditSeq, SeqMan and MegAlign components of the Lasergene sequence analysis software package (DNAStar Inc., Madison, Wis.). The sequenced VNTR loci of the Wolbachia strains had to be manually aligned because of their long period length, internal repeats, SNPs and indels within individual VNTR periods. VNTR periods were searched for internal direct repeats, palindromic (dyad) repeats and secondary structures by using DNA Strider [56]. For ANK proteins, domain architecture was predicted using SMART v3.5 (Simple Modular Architecture Research Tool) (http://smart.embl-heidelberg.de/) [57,58] and TMHMM2 (http://www.cbs.dtu.dk/services/TMHMM/). We analysed the phylogenetic relationships between individual ANK repeats from WD0766 and their orthologs to investigate the mode of evolution of these repeats. All ANK repeats were extracted from the full length sequences of each gene and translated into amino acids. Gaps were inserted where necessary to correct for frameshifts. Sequences were aligned using T_coffee [59]. Maximum likelihood phylogenetic analysis of this alignment was performed using PhyML [60], with a JTT model of amino acid substitution, and a gamma model of rate heterogeneity with four rate classes and the gamma parameter estimated from the data. 1000 bootstrap replicates were performed.
Results and discussion
VNTR variability between strains of A-group Wolbachia
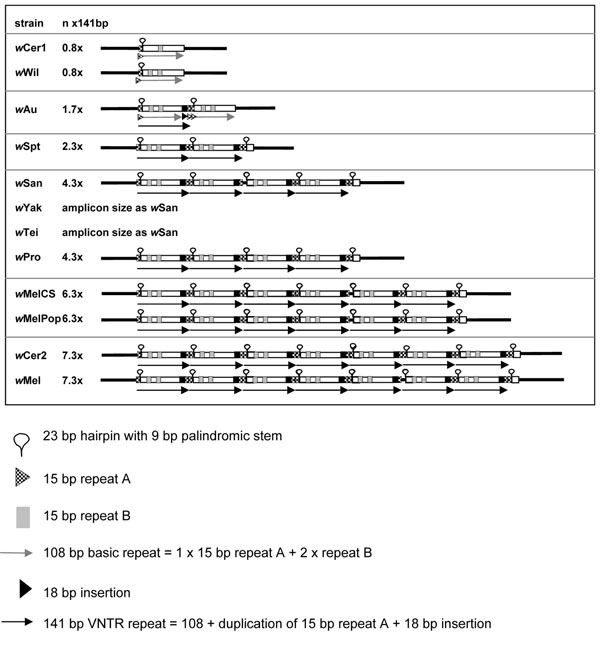
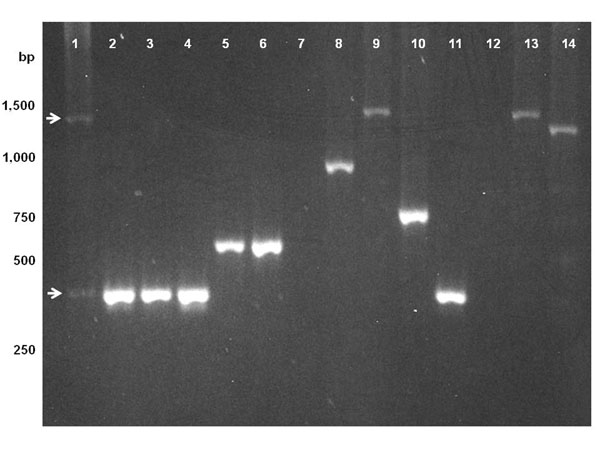
We isolated sequences for two Wolbachia VNTR loci, VNTR-141 and VNTR-105, with tandemly repeated periods of 141 and 105bp, respectively, for representative supergroup A Wolbachia strains. The loci had previously produced size polymorphic PCR fragments in isolates of wMel and wMelCS/wMelPop when amplified using primers that were designed to the flanking regions of the two VNTR loci of the sequenced wMel genome [30]. VNTR-141 is positioned between WD0096 and WD0098, and VNTR-105 is between WD1129 and WD1131 of the final wMel genome annotation (NCBI accession NC_002978, [41]). The basic 141bp period of VNTR-141 consists of the internal 15bp direct repeat A, a 23bp hairpin with a 9bp palindromic stem, an 18bp insertion and the internal 15bp direct repeat B (Figure 1 of this paper, and Figure 2E of [38]). Diagnostic VNTR-141 PCRs were run on DNA obtained from different Wolbachia hosts known to harbour very closely related strains of the symbiont that were not clearly distinguishable by using MLST [20,21,24]. The VNTR-141 fragments were sequenced and compared to the 141bp period of wMel. The shortest VNTR-141 alleles were amplified from wWil and wCer1: they contained only one single period consisting of a 108bp core period without the 18bp insertion, and missing the downstream 15bp A repeat. All other supergroup A strains produced VNTR-141 alleles containing different copy numbers of the 141bp period (Figure 1), i.e. 0.8 (wWil, amplicon size using the locus specific primers 387bp, wCer1 388bp), 1.7 (wAu 530bp), 2.3 (wSpt 643bp), 4.3 (wSan 889bp, wPro 925bp; wYak and wTei had similar amplicon sizes to wSan but were not sequenced), 6.3 (wMelCS 1189bp, wMelPop 1189bp) and 7.3 (wMel 1330bp, wCer2 1348bp for both original host R. cerasi and novel host C. capitata) (Figure 1). These polymorphic amplicons in VNTR-141 were visualised by standard PCR as different amplicon sizes on an agarose gel (Figure 2). Multiply infected R. cerasi [46,61] revealed two bands, with amplicons representing wCer1 and wCer2 (Figure 2). The VNTR alleles of wCer2 were assigned through comparisons with the isolates from the microinjected novel hosts D. simulans [62] and C. capitata [47]. Besides the internal deletions in the wWil and wCer1 periods, and variation in copy numbers, the sequence composition of the VNTR-141 periods are almost identical (i.e. 99%) within wMel and other strains, and hence highly conserved. For this reason a phylogenetic sequence analysis, other than the analysis of repeat numbers in cladistical approaches, is not informative.
Figure 1.
Schematic presentation of the VNTR-141 locus in ten wMel-like Wolbachia strains of Drosophila and R. cerasi. The complete 141bp period and the core 108bp period are shown as black and grey arrows, respectively; the 23bp hairpin as a lariat; the two 15bp inverted repeats A and B as dotted and grey boxes, respectively; and the 18bp insertion as a black arrow head.
Figure 2.
Diagnostic size difference for the VNTR-141 locus of Wolbachia. Lane 1: wCer1 and wCer2 doubly infected R. cerasi from Austria (the two arrows indicate the two faint bands for wCer1 and wCer2); 2-4: wWil infected D. willistoni from populations collected recently in Panama (Pan98), Mexico (Apa), and Equador (JS); lane 5-6: wAu infected D. simulans strain Coffs Harbor and Yaunde 6; lane 7: uninfected (tetracycline treated) controls = D. melanogaster yw67c23T; lane 8: wTei infected D. teissieri GN53; lane 9: wMel infected D. melanogaster yw67c23; lane 10: wSpt infected D. septentriosaltans; lane 11: wCer1 singly infected R. cerasi from Hungary; lane 12: uninfected (tetracycline treated) control = D. melanogaster line yw67c23T; lane 13: wMel infected D. melanogaster yw67c23; lane 14: wMelCS infected D. melanogaster Canton S.
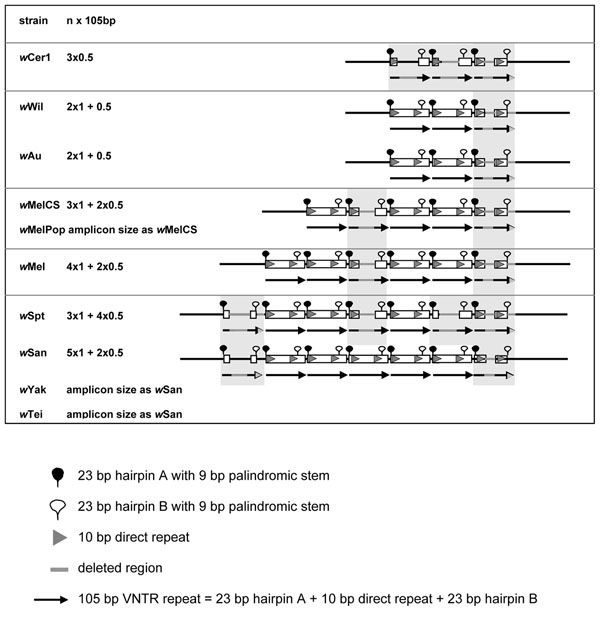
In contrast to VNTR-141, the basic period of VNTR-105 is 105bp long containing two 23bp hairpins with 9bp palindromic stem structures and one internal short repeat of 10bp (Figure 3). VNTR-105 of wMel contains four complete 105bp periods, and two with internal deletions of 25bp each. wMelCS and wMelPop lack one of the complete 105bp periods, i.e. contain three complete 105bp copies and two with internal deletions of 32bp (Figure 3). The tested supergroup A strains display different alleles in the VNTR-105 locus with amplicon sizes ranging from 3x0.5 copies (wCer1, amplicon size using the locus specific primers 998bp), 2.5 copies (wWil 1065bp, wAu 1065bp), 3+2x0.5 copies (wMelCS and wMelPop 1241bp), 4+2x0.5 copies (wMel 1347bp), 3+4x0.5 copies (wSpt 1408bp) and 5+2x0.5 copies (wSan, 1476bp; wYak and wTei had similar amplicon sizes to wSan but were not sequenced). wCer2 had a large amplicon for this VNTR locus and difficulties were experienced with accurately sequencing these large loci because of restrictions with read lengths, as well as problems in detecting an accurate overlap between forward and reverse sequences. VNTR-105 amplicon size differences can be easily resolved on agarose gels (data not shown). In comparison to VNTR-141, the structure of the VNTR-105 locus is less conserved within and between strains because of internal deletions, yet the period sequences are almost identical (i.e. 98%) within wMel and between other strains. For this reason a phylogenetic analysis of period sequence data is not appropriate, whereas the analysis of diagnostic characters such as copy numbers are more informative (Figure 3).
Figure 3.
Schematic presentation of the VNTR-105 locus in seven wMel-like Wolbachia strains of Drosophila. The complete 105bp period is shown as black arrows; the two 23bp hairpins A and B as full and empty lariats, respectively; the 15bp inverted repeat as grey boxes; and deleted sections in grey.
We extended our PCR analysis to a wider range of Wolbachia strains, including wRi and wHa, both supergroup A strains that are distantly related to wMel, as well as strains from supergroup B (wNo, wBol1, wMau) and C (wDim). None of these strains yielded PCR products for the tested VNTR primers, probably because of sequence divergence within the primer region or genome rearrangements [52-54]. Because of the latter it was not attempted to design primers of conserved coding regions in distantly related strains.
Evolution of repeats in VNTR loci
The individual periods of VNTR-141 and VNTR-105 respectively display high sequence conservation within and between strains, with variability in the copy numbers and internal deletions within some of the repeated periods. Two evolutionary processes may be shaping these loci with high variability in repeat copy numbers yet small sequence divergence. The accumulation of tandemly repeated periods may be facilitated through slippage and mispairing in the process of Wolbachia DNA replication and repair. Slipped-strand mispairing has previously been identified as a source for generation of repeat copies in general [63-65] and in E. ruminantium in particular, a genome with an elevated number of tandem repeats [66]. Palindromic sequences with the strong potential of forming secondary stem loops are well known to cause slipped-strand mispairing [67]. Hence we assume that the hairpins present in both Wolbachia VNTRs may trigger slippage in both these loci. The second evolutionary mechanism in action could be concerted evolution between different periods within the two loci, a phenomenon that has previously been observed in members of gene families that tend to be more similar within a species than between species because of the elimination or fixation of new point mutations [68]. The high structural turnover, triggering expansions and/or contractions of copy numbers in both VNTR loci of wMel-like Wolbachia, can thus be applied for simple and rapid but highly informative symbiont fingerprinting by standard PCR (Figure 2). We cannot infer directionality between expansion and contractions in the evolution of both loci. It is hence impossible to determine whether low copy numbers within the intergenic loci manifest an ancestral or derived state. It has been suggested though that tandem repeats go through cycles of gradual expansion followed by collapse of repeats [69]. It is hence adequate to state that closely related strains are more likely to have similar copy numbers, e.g. wMel and wMelCS. Interestingly, the CI inducing strains wCer2, wMel and wMelCS contain larger VNTR loci when compared to the non CI inducing wWil and wAu, with larger VNTR loci in wMel than wMelCS that coincide with stronger CI induction in wMel than wMelCS [70]. Furthermore increased copy numbers in one locus correspond with increased copy numbers in the second. Such a coincidence of intergenic tandem repeat variation with CI phenotype was also observed for supergroup B Wolbachia in C. pipiens[40]. Yet, these observations are not sufficiently supported by replication to conclude about any potential links between genotypes and phenotypes, but they warrant further structural and functional studies of the VNTR repeat expansions.
ANK gene variability between strains of A-group Wolbachia
Unlike most bacteria, genes that encode proteins with ANK repeats are extremely abundant in Wolbachia, representing up to 2-4% of the total number of genes in wMel [41], wRi [52] and wPip [53,71]. Some of the variability in these genes appears to correlate with crossing types in mosquitoes [72]. Several of the 23 ANK genes initially annotated in the wMel genome are highly variable between the CI-inducing strain wMel and the non-CI inducing related strain wAu [36]. These differences included point mutations, frameshifts and premature stop codons, presence/absence of transmembrane domains, disruption by insertion elements and variability in the number of predicted ANK repeats in the encoded proteins.
Based on earlier work [36], we performed an initial PCR screening (data not shown) using the most variable wMel ANK genes (WD0035, WD0294, WD0385, WD0498, WD0514, WD0550, WD0636, WD0766 and WD1213- also see results of TRF analysis below) in order to look for size differences across the Wolbachia strains used in this study. Some of the ANK genes could not be amplified in all strains, probably due to sequence divergence. For the ones that could be amplified, the non-phage related ANK genes WD0550 and in particular WD0766 were found to be the most variable in terms of size difference among the Wolbachia strains and they were selected for further analysis, with sequence data reported for WD0766 only.
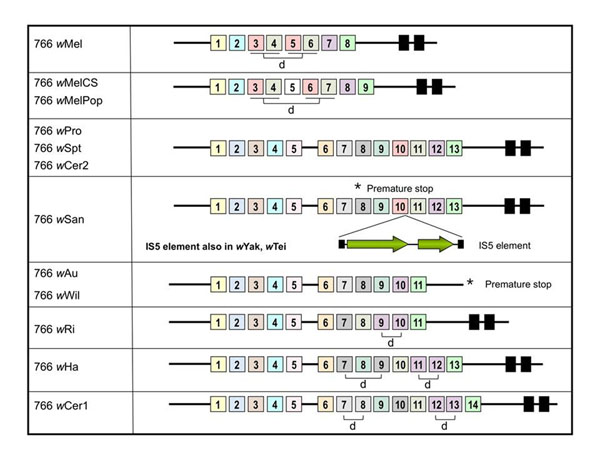
In wMel, WD0766 encodes a 51.8kDa protein containing eight ANK repeats and two transmembrane domains (TMDs) in the C-terminus. When this gene was sequenced in several Wolbachia strains, the number of predicted ANK repeats was found to be quite different among them, ranging from eight repeats in wMel to 14 in wCer1 (Figure 4). The wAu, wWil and wRi strains contained 11 ANK repeats, but the proteins were truncated by a premature stop codon that resulted in the elimination of the predicted TMDs in wAu and wWil. WD0766 in wSan is disrupted by a premature stop after the seventh ANK domain and contains a 918bp IS5 insertion element in the middle of its 10th ANK repeat (Figure 4). PCR results (data not shown) suggest that this IS5 insertion is also present in the orthologous gene in wYak and wTei, but these amplicons were not sequenced. The sequence of the wSan IS5 element is identical to that of the 13 IS5 elements present in the wMel genome [41]. Disruption of a Wolbachia ANK gene by an IS5 insertion element has previously been observed in the WD0385 gene from wAu (GenBank AY664873) [36], although in this case the insertion sequence differs by 5 nucleotides from the wMel and wSan IS5 elements. wSpt, wCer2 and wHa strains had the same structure for the WD0766 proteins (13 ANK domains + 2 TMDs), whereas the wCer1 protein contained 14 ANK domains and 2 TMDs.
Figure 4.
Domain architecture of the WD0766 ANK domain protein in Wolbachia strains. The location of ANK motifs (coloured boxes with numbers) was determined using SMART v3.5 (http://smart.embl-heidelberg.de/). Transmembrane domains (black boxes) were predicted using the TMHMM2 server. The presence of a frameshift in the wAu and wWil WD0766 gene creates a premature stop (*) that prevents the translation of the transmembrane domains. The wSan, wYak and wTei genes also contain a premature stop (*) that prevents the translation of 6 ANK domains and two transmembrane domains. These genes also contain an IS5 element insertion inside the 10th ANK domain. Some of the ANK repeat motifs are duplicated (d). The colour scheme corresponds to the DNA sequence similarity of the ANK repeat motifs (Figure 5).
WD0550 was also found to be variable among the strains analysed, although it was not as informative as WD0766. For this reason only a subset of strains was analysed for this locus in more detail. WD0550 codes for a 36.4kDa protein containing six predicted ANK repeats and has no TMDs. The protein contains six ANK repeats in wMel and wSpt, and eight repeats in wMelCS, wSan, wCer2, wAu and wWil (data not shown).
Evolution of repeats in WD0766
Orthologs of WD0766 encode for proteins containing different numbers of ANK repeats in different Wolbachia strains. Additional repeat copies may be gained by the duplication or loss of single or multiple repeats, and genes containing these repeats may also diverge due to loss or shuffling of repeat periods. To investigate the patterns of change in the number and order of ANK repeats in these proteins, we aligned the amino acid sequences of all individual repeats and performed a maximum likelihood analysis of the phylogenetic relationships between them (Figure 5). The tree shows clusters of typically six to ten repeats, separated by relatively long internal branches. Despite the large ratio of internal to tip branch lengths, bootstrap values on this tree are almost all extremely small, probably due to the short length of the alignment (34 residues). However, a clear pattern is observed wherein repeats in similar positions within multiple orthologs cluster together. For example, the first ANK repeat present in every ortholog clusters in a single clade, marked in yellow in Figures 4 and 5. A similar clustering is seen for the last repeat of every ortholog (marked in green), and for the second repeat in wMel and wMelPop/wMelCS with the fourth repeat of all other orthologs (marked in blue). Figure 4 shows the structure of each ortholog, with repeats that cluster together in the tree coloured in the same shade. Similar to VNTRs, ANK loci of Wolbachia provide highly informative and strain-specific marker sets that allow easy separation via PCR and high-resolution diagnosis of host infections (Figure 6).
Figure 5.
Maximum likelihood phylogeny of individual ANK repeats from WD0766 and its orthologs. Names indicate the strain of Wolbachia and the repeat number, as labelled in Figure 4. The scale bar corresponds to nucleotide substitutions per site.
Figure 6.
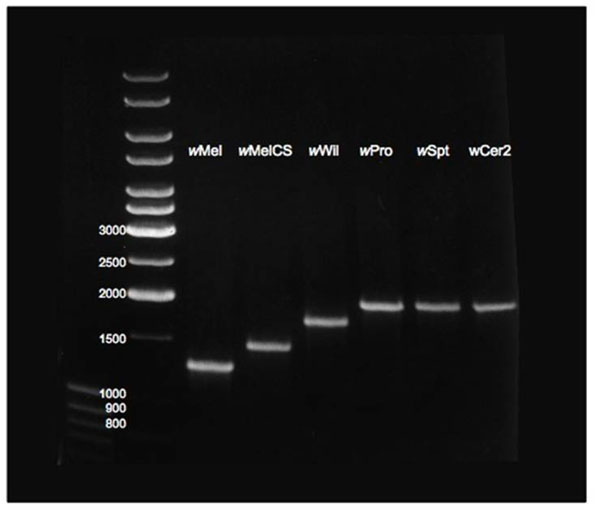
Diagnostic size polymorphism of the WD0766 gene. Isolates include Wolbachia of D. melanogaster (wMel, wMelCS), D. willistoni (wWil), D. prosaltans (wPro), D. septentriosaltans (wSpt) and D. simulans transinfected with Wolbachia from R. cerasi (wCer2).
A number of inferences about the evolution of the ANK repeats in these genes can be drawn from the tree in Figure 5 and the mapping of the phylogenetic data onto the modular structure of the genes. First, it is likely that the ancestral copy of this gene at the base of supergroup A already contained most of the repeats seen today, probably in a very similar linear order. Most of the clusters in the tree contain repeats from 7 or more of the orthologs, and the order of these orthologous repeats along the genes is highly similar. There is only one clear example of repeat shuffling: the eighth and ninth repeats in the wPro/wSan/wAu groups occur in the reverse order in wCer1 (as repeat periods 10 and 9), while wHa may represent an intermediate stage, with the repeats orthologous to wPro 8 and 9 followed by a second copy of a repeat orthologous to wPro 8. Secondly, at least some variation in repeat number is due to lineage-specific tandem duplication of a single repeat (e.g. repeats 7 and 8 in wCer1) or of multiple repeats (repeats 3-4 and 5-6 in wMel).
Extension of MLVA markers to other Wolbachia supergroups
In comparison to the MLST markers, the highly polymorphic markers used here have a major trade-off in the loss of universal applicability for all Wolbachia strains. Here we have focused on Wolbachia supergroup A and tested the primers of these markers in other supergroups but primers did not amplify the loci or the loci were not informative. The presence of VNTR loci was restricted to subsets of supergroup A while genes containing ANK domain repeats were found in all supergroup A strains. In silico analysis of three other completed genomes, wRi, wPip and wBm of supergroups A, B and D, respectively, revealed though that tandem repeated regions occur throughout these supergroups and may be of relevance for MLVA in other supergroups. As further genome data become available it will be possible to extend this to an even larger group of Wolbachia isolates. A TRF analysis of wMel revealed 93 sites with direct tandem repeats of periods ranging from 10bp to 291bp, with internal match percentages from 68% to 100% (Table 4). The larger wRi genome has a similar number of tandem repeats while wPip has a smaller set of tandem repeats. The tandem repeats of wMel, wRi and wPip have similar characteristics such as comparable period sizes, copy numbers as well as internal match ratios (Table 4). The number of tandem repeats in wBm is reduced by a factor of 10 when compared with the supergroup A and B Wolbachia, and the tandem periods appear to be shorter. This reduction in wBm is in accordance with the earlier described higher rate of secondary genome reduction in this strain [54]. Within the group of the closest relatives of the genus Wolbachia, the sequence of E. ruminantium revealed the highest content of tandem repeats for bacteria reported so far (Table 4), with size polymorphism in tandem repeats within the isolate that was used for genome sequencing the genome [66]. Our in silico analysis predicted the presence of variable tandem repeat markers in supergroup A strains and could hence readily be developed and tested on Wolbachia isolates from other supergroups. Highly polymorphic markers will be useful in population dynamic and population genetic studies similar to the ones undertaken in wMel-like strains [30,38,39]. We have not analysed the unfinished genome data sets of Wolbachia (e.g. [73]). A large proportion of tandem repeats are located in intergenic regions that tend to be assembled in genome sequencing projects last, yet their conserved flanking regions are required for the isolation of VNTR markers from total genomic extracts. A polymorphic VNTR locus has recently been reported for a supergroup B strain after applying a similar approach to wPip isolated from different C. pipiens populations [40].
Interestingly, our TRF analysis only detected five ANK repeat regions (WD0294, WD0385, WD0514, WD0550 and WD0766) of the 23 annotated genes encoding ANK repeat domains. Coincidentally, this group of genes includes the most variable genes encoding ANK repeat domains, suggesting that repeat extension/contraction is a strong diversifying mechanism in these genes.
Most of the primers designed for wMel ANK genes amplified expected PCR amplicons from supergroup A Wolbachia, but not from the majority of supergroup B, probably due to sequence divergence [36]. ANK domain genes are known to be present in other Wolbachia groups. In the B group mosquito strain wPip that infects mosquitoes there are 60 genes encoding ANK repeats, some of them also variable [53,71,72], whereas the fully sequenced D group wBm strain that infects the nematode Brugia malayi contains 5 ANK genes and 7 related pseudogenes [54]. Although wMel ANK genes were used as a reference in our study, another A group Wolbachia strain, wRi, contains 35 ANK genes, some of them very distinct from the wMel genes, probably as a result of duplications and recombination events [52]. Partial sequences of other A group strains have also revealed high numbers of ANK genes [73]. Thus, it seems clear that ANK genes are a signature feature in Wolbachia that can be potentially utilised to fingerprint closely related strains in A and other groups.
Conclusion
The identification of amplicon size polymorphic markers of Wolbachia provides a valuable addition to existing typing systems such as MLST, for the following three reasons: (1) The MLVA markers presented here display higher rates of evolution than the MLST loci, which are conserved protein encoding genes. Using MLVA, Wolbachia strains clustered in the same groups as in MLST typing, yet with a higher resolution that could be useful for different types of questions that MLST has not yet been able to target. These questions include the study of Wolbachia population genetics within infected species [30,38,39], and will further extend studies of horizontal transmission between host species for which MLST was originally developed [22]. Highly polymorphic markers will also be useful for experimental evolution of Wolbachia in order to track small genomic changes in short time frames. This higher resolution comes with the cost though, that markers are not universally applicable to the entire diversity of Wolbachia. (2) The majority of Wolbachia genomes are dotted with many different repeat regions which are highly appropriate to be targeted for the isolation of possible polymorphic markers. Tandem repeat markers such as the ones developed here can be tailored to individual studies. (3) MLVA markers are ideal for rapid and high-throughput DNA fingerprinting, as no sequencing is required. The markers are ideal to detect multiple infections in single PCR reactions if strains contain alleles with variable amplicon sizes. Our analysis of the evolution of the tandem repeat regions shows that they evolve by gain or loss of repeats. The variability in the number of ANK repeats, generally constituted by 33 amino acids each, creates size differences that are multiples of 99bp and, like VNTRs consisting of >100bp periods, can be clearly identified following simple PCR screenings without the need of initial sequencing or RFLP analyses as in the case of point mutations. The use of 2-3 highly variable markers per strain can generate easily readable fingerprints.
List of abbreviations used
CI: cytoplasmic incompatibility; MLVA: multiple locus variable number tandem repeat analysis; MLST: multiple locus sequence typing; VNTR: variable number tandem repeat; ANK: ankyrin domain; TRF: tandem repeats finder.
Competing interests
The authors declare that they have no competing interests.
Authors’ contribution
MR, IIO, WJM and SLO had the initial idea for this manuscript. MR, IIO, WJM and SLO designed the study. MR, IIO and WJM performed laboratory work. MR, IIO, WJM, MW performed data analysis. MR, IIO, WJM, MW and SLO wrote the manuscript. All authors approved the final manuscript.
Contributor Information
Markus Riegler, Email: m.riegler@uws.edu.au.
Iñaki Iturbe-Ormaetxe, Email: i.iturbeormaetxe@uq.edu.au.
Megan Woolfit, Email: m.woolfit@uq.edu.au.
Wolfgang J Miller, Email: wolfgang.miller@meduniwien.ac.at.
Scott L O’Neill, Email: scott.oneill@monash.edu.au.
Acknowledgements
We thank Sylvain Charlat, Kostas Bourtzis and the School of Veterinary Science, UQ, for supplying biological material, i.e. H. bolina, C. capitata and D. immitis, respectively. We thank the special edition editor Greg Hurst and two anonymous reviewers for their valuable comments. The research was supported by grants of the Australian Research Council ARC to MR, IIO, MW and SLO, and from COST Action FA-0701 and the research grant P22634-B17 of the Austrian Science Fund FWF to WJM.
This article has been published as part of BMC Microbiology Volume 11 Supplement 1, 2012: Arthropod symbioses: from fundamental studies to pest and disease mangement. The full contents of the supplement are available online at http://www.biomedcentral.com/1471-2180/12?issue=S1.
References
- Werren JH, Windsor D, Guo LR. Distribution of Wolbachia among neotropical arthropods. Proceedings of the Royal Society of London Series B-Biological Sciences. 1995;262(1364):197–204. doi: 10.1098/rspb.1995.0196. [DOI] [Google Scholar]
- West SA, Cook JM, Werren JH, Godfray HCJ. Wolbachia in two insect host-parasitoid communities. Molecular Ecology. 1998;7(11):1457–1465. doi: 10.1046/j.1365-294x.1998.00467.x. [DOI] [PubMed] [Google Scholar]
- Jeyaprakash A, Hoy MA. Long PCR improves Wolbachia DNA amplification: wsp sequences found in 76% of sixty-three arthropod species. Insect Molecular Biology. 2000;9(4):393–405. doi: 10.1046/j.1365-2583.2000.00203.x. [DOI] [PubMed] [Google Scholar]
- Hilgenboecker K, Hammerstein P, Schlattmann P, Telschow A, Werren JH. How many species are infected with Wolbachia? - A statistical analysis of current data. FEMS Microbiology Letters. 2008;281(2):215–220. doi: 10.1111/j.1574-6968.2008.01110.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arthofer W, Riegler M, Avtzis DN, Stauffer C. Evidence for low-titre infections in insect symbiosis: Wolbachia in the bark beetle Pityogenes chalcographus (Coleoptera, Scolytinae) Environmental Microbiology. 2009;11(8):1923–1933. doi: 10.1111/j.1462-2920.2009.01914.x. [DOI] [PubMed] [Google Scholar]
- O'Neill SL, Hoffmann AA, Werren JH. Influential passengers. Inherited microorganisms and arthropod reproduction. Oxford: Oxford University Press; 1997. [Google Scholar]
- Stouthamer R, Breeuwer JAJ, Hurst GDD. Wolbachia pipientis: microbial manipulator of arthropod reproduction. Annual Review of Microbiology. 1999;53:71–102. doi: 10.1146/annurev.micro.53.1.71. [DOI] [PubMed] [Google Scholar]
- Werren JH, Baldo L, Clark ME. Wolbachia: Master manipulators of invertebrate biology. Nature Reviews Microbiology. 2008;6(10):741–751. doi: 10.1038/nrmicro1969. [DOI] [PubMed] [Google Scholar]
- Schneider D, Miller WJ, Riegler M. In: Manipulative Tenants Bacteria associated with arthropods. Zchori-Fein E, Bourtzis K, editor. Boca Raton: CRC Press; 2011. Arthropods shopping for Wolbachia; pp. 149–173. [Google Scholar]
- Taylor MJ, Bandi C, Hoerauf A. Wolbachia bacterial endosymbionts of filarial nematodes. Adv Parasitol. 2005;60:245–284. doi: 10.1016/S0065-308X(05)60004-8. [DOI] [PubMed] [Google Scholar]
- Werren JH, Zhang W, Guo LR. Evolution and phylogeny of Wolbachia - reproductive parasites of arthropods. Proceedings of the Royal Society of London Series B-Biological Sciences. 1995;261(1360):55–63. doi: 10.1098/rspb.1995.0117. [DOI] [PubMed] [Google Scholar]
- Zhou WG, Rousset F, O'Neill S. Phylogeny and PCR-based classification of Wolbachia strains using wsp gene sequences. Proceedings of the Royal Society of London Series B-Biological Sciences. 1998;265(1395):509–515. doi: 10.1098/rspb.1998.0324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lo N, Paraskevopoulos C, Bourtzis K, O'Neill SL, Werren JH, Bordenstein SR, Bandi C. Taxonomic status of the intracellular bacterium Wolbachia pipientis. International Journal of Systematic and Evolutionary Microbiology. 2007;57(3):654–657. doi: 10.1099/ijs.0.64515-0. [DOI] [PubMed] [Google Scholar]
- Baldo L, Werren JH. Revisiting Wolbachia supergroup typing based on wsp: spurious lineages and discordance in MLST. Current Microbiology. 2007;55(1):81–87. doi: 10.1007/s00284-007-0055-8. [DOI] [PubMed] [Google Scholar]
- Ros VID, Fleming VM, Feil EJ, Breeuwer JAJ. How diverse is the genus Wolbachia? Multiple-gene sequencing reveals a putatively new Wolbachia supergroup recovered from spider mites (Acari: Tetranychidae) Applied and Environmental Microbiology. 2009;75(4):1036–1043. doi: 10.1128/AEM.01109-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Neill SL, Giordano R, Colbert AME, Karr TL, Robertson HM. 16S ribosomal RNA phylogenetic analysis of the bacterial endosymbionts associated with cytoplasmic incompatibility in insects. Proceedings of the National Academy of Sciences of the United States of America. 1992;89(7):2699–2702. doi: 10.1073/pnas.89.7.2699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiggins FM, von der Schulenburg JHG, Hurst GDD, Majerus MEN. Recombination confounds interpretations of Wolbachia evolution. Proceedings of the Royal Society of London Series B-Biological Sciences. 2001;268(1474):1423–1427. doi: 10.1098/rspb.2001.1656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiggins FM. The rate of recombination in Wolbachia bacteria. Molecular Biology and Evolution. 2002;19(9):1640–1643. doi: 10.1093/oxfordjournals.molbev.a004228. [DOI] [PubMed] [Google Scholar]
- Baldo L, Bordenstein S, Wernegreen JJ, Werren JH. Widespread recombination throughout Wolbachia genomes. Molecular Biology and Evolution. 2006;23(2):437–449. doi: 10.1093/molbev/msj049. [DOI] [PubMed] [Google Scholar]
- Paraskevopoulos C, Bordenstein SR, Wernegreen JJ, Werren JH, Bourtzis K. Toward a Wolbachia multilocus sequence typing system: Discrimination of Wolbachia strains present in Drosophila species. Current Microbiology. 2006;53(5):388–395. doi: 10.1007/s00284-006-0054-1. [DOI] [PubMed] [Google Scholar]
- Baldo L, Hotopp JCD, Jolley KA, Bordenstein SR, Biber SA, Choudhury RR, Hayashi C, Maiden MCJ, Tettelin H, Werren JH. Multilocus sequence typing system for the endosymbiont Wolbachia pipientis. Applied and Environmental Microbiology. 2006;72(11):7098–7110. doi: 10.1128/AEM.00731-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baldo L, Ayoub NA, Hayashi CY, Russell JA, Stahlhut JK, Werren JH. Insight into the routes of Wolbachia invasion: High levels of horizontal transfer in the spider genus Agelenopsis revealed by Wolbachia strain and mitochondrial DNA diversity. Molecular Ecology. 2008;17(2):557–569. doi: 10.1111/j.1365-294X.2007.03608.x. [DOI] [PubMed] [Google Scholar]
- Zabalou S, Apostolaki A, Pattas S, Veneti Z, Paraskevopoulos C, Livadaras I, Markakis G, Brissac T, Merçot H, Bourtzis K. Multiple rescue factors within a Wolbachia strain. Genetics. 2008;178(4):2145–2160. doi: 10.1534/genetics.107.086488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishmael N, Hotopp JCD, Loanidis P, Biber S, Sakamoto J, Siozios S, Nene V, Werren J, Boutriz K, Bordenstein SR. et al. Extensive genomic diversity of closely related Wolbachia strains. Microbiology. 2009;155(7):2211–2222. doi: 10.1099/mic.0.027581-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffmann AA, Clancy D, Duncan J. Naturally-occurring Wolbachia infection in Drosophila simulans that does not cause cytoplasmic incompatibility. Heredity. 1996;76:1–8. doi: 10.1038/hdy.1996.1. [DOI] [PubMed] [Google Scholar]
- Min KT, Benzer S. Wolbachia, normally a symbiont of Drosophila, can be virulent, causing degeneration and early death. Proceedings of the National Academy of Sciences of the United States of America. 1997;94(20):10792–10796. doi: 10.1073/pnas.94.20.10792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGraw EA, Merritt DJ, Droller JN, O'Neill SL. Wolbachia density and virulence attenuation after transfer into a novel host. Proceedings of the National Academy of Sciences of the United States of America. 2002;99(5):2918–2923. doi: 10.1073/pnas.052466499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McMeniman CJ, Lane RV, Cass BN, Fong AWC, Sidhu M, Wang YF, O'Neill SL. Stable introduction of a life-shortening Wolbachia infection into the mosquito Aedes aegypti. Science. 2009;323(5910):141–144. doi: 10.1126/science.1165326. [DOI] [PubMed] [Google Scholar]
- Sun LV, Riegler M, O'Neill SL. Development of a physical and genetic map of the virulent Wolbachia strain wMelPop. Journal of Bacteriology. 2003;185(24):7077–7084. doi: 10.1128/JB.185.24.7077-7084.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riegler M, Sidhu M, Miller WJ, O'Neill SL. Evidence for a global Wolbachia replacement in Drosophila melanogaster. Current Biology. 2005;15:1428–1433. doi: 10.1016/j.cub.2005.06.069. [DOI] [PubMed] [Google Scholar]
- Achtman M, Morelli G, Zhu P, Wirth T, Diehl I, Kusecek B, Vogler AJ, Wagner DM, Allender CJ, Easterday WR. et al. Microevolution and history of the plague bacillus, Yersinia pestis. Proceedings of the National Academy of Sciences of the United States of America. 2004;101(51):17837–17842. doi: 10.1073/pnas.0408026101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pourcel C, André-Mazeaud F, Neubauer H, Ramisse F, Vergnaud G. Tandem repeats analysis for the high resolution phylogenetic analysis of Yersinia pestis. BMC Microbiology. 2004;4 doi: 10.1186/1471-2180-4-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johansson A, Farlow J, Larsson P, Dukerich M, Chambers E, Byström M, Fox J, Chu M, Forsman M, Sjöstedt A. et al. Worldwide genetic relationships among Francisella tularensis isolates determined by multiple-locus variable-number tandem repeat analysis. Journal of Bacteriology. 2004;186(17):5808–5818. doi: 10.1128/JB.186.17.5808-5818.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yazdankhah SP, Lindstedt BA. In: Comparative Genomics Methods in Molecular Biolgy. Bergman NH, editor. Vol. 396. Totowa, NJ: Humana Press; 2007. Variable number tandem repeat typing of bacteria; pp. 395–405. [DOI] [PubMed] [Google Scholar]
- Vergnaud G, Pourcel C. Multiple locus variable number of tandem repeats analysis. Methods in molecular biology (Clifton, NJ) 2009;551:141–158. doi: 10.1007/978-1-60327-999-4_12. [DOI] [PubMed] [Google Scholar]
- Iturbe-Ormaetxe I, Burke GR, Riegler M, O'Neill SL. Distribution, expression, and motif variability of ankyrin domain genes in Wolbachia pipientis. Journal of Bacteriology. 2005;187:5136–5145. doi: 10.1128/JB.187.15.5136-5145.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duron O, Lagnel J, Raymond M, Bourtzis K, Fort P, Weill M. Transposable element polymorphism of Wolbachia in the mosquito Culex pipiens: evidence of genetic diversity, superinfection and recombination. Molecular Ecology. 2005;14(5):1561–1573. doi: 10.1111/j.1365-294X.2005.02495.x. [DOI] [PubMed] [Google Scholar]
- Miller WJ, Riegler M. Evolutionary dynamics of wAu-like Wolbachia variants in neotropical Drosophila spp. Applied and Environmental Microbiology. 2006;72(1):826–835. doi: 10.1128/AEM.72.1.826-835.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller WJ, Ehrman L, Schneider D. Infectious speciation revisited: Impact of symbiont-depletion on female fitness and mating behavior of drosophila paulistorum. PLoS Pathogens. 2010;6(12) doi: 10.1371/journal.ppat.1001214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petridis M, Chatzidimitriou D. Characterization of an intergenic polymorphic site (pp-hC1A_5) in Wolbachia pipientis (wPip) Molecular Ecology Resources. 2011;11(4):753–756. doi: 10.1111/j.1755-0998.2011.02991.x. [DOI] [PubMed] [Google Scholar]
- Wu M, Sun LV, Vamathevan J, Riegler M, Deboy R, Brownlie JC, McGraw EA, Martin W, Esser C, Ahmadinejad N. et al. Phylogenomics of the reproductive parasite Wolbachia pipientis wMel: a streamlined genome overrun by mobile genetic elements. Public Library of Science Biology. 2004;2(3):327–341. doi: 10.1371/journal.pbio.0020069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mosavi LK, Cammett TJ, Desrosiers DC, Peng ZY. The ankyrin repeat as molecular architecture for protein recognition. Protein Science. 2004;13(6):1435–1448. doi: 10.1110/ps.03554604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan X, Lührmann A, Satoh A, Laskowski-Arce MA, Roy CR. Ankyrin repeat proteins comprise a diverse family of bacterial type IV effectors. Science. 2008;320(5883):1651–1654. doi: 10.1126/science.1158160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merçot H, Poinsot D. Infection by Wolbachia: from passengers to residents. Comptes Rendus - Biologies. 2009;332(2-3):284–297. doi: 10.1016/j.crvi.2008.09.010. [DOI] [PubMed] [Google Scholar]
- Hoffmann AA, Turelli M, Simmons GM. Unidirectional incompatibility between populations of Drosophila simulans. Evolution. 1986;40:692–701. doi: 10.2307/2408456. [DOI] [PubMed] [Google Scholar]
- Riegler M, Stauffer C. Wolbachia infections and superinfections in cytoplasmically incompatible populations of the European cherry fruit fly Rhagoletis cerasi (Diptera, Tephritidae) Molecular Ecology. 2002;11(11):2425–2434. doi: 10.1046/j.1365-294x.2002.01614.x. [DOI] [PubMed] [Google Scholar]
- Zabalou S, Riegler M, Theodorakopoulou M, Stauffer C, Savakis C, Bourtzis K. Wolbachia-induced cytoplasmic incompatibility as a means for insect pest population control. Proceedings of the National Academy of Sciences of the United States of America. 2004;101:15042–15045. doi: 10.1073/pnas.0403853101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dyson EA, Hurst GD. Persistence of an extreme sex-ratio bias in natural population. Proceedings of the National Academy of Sciences USA. 2004;101:6520–6523. doi: 10.1073/pnas.0304068101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sironi M, Bandi C, Sacchi L, DiSacco B, Damiani G, Genchi C. Molecular evidence for a close relative of the arthropod endosymbiont Wolbachia in a filarial worm. Molecular and Biochemical Parasitology. 1995;74(2):223–227. doi: 10.1016/0166-6851(95)02494-8. [DOI] [PubMed] [Google Scholar]
- Holmes DS, Bonner J. Preparation, molecular weight, base composition and secondary structure of giant nuclear ribonucleic acid. Biochemistry. 1973;12(12):2330–2338. doi: 10.1021/bi00736a023. [DOI] [PubMed] [Google Scholar]
- Benson G. Tandem repeats finder: a program to analyse DNA sequences. Nucleic Acid Research. 1999;27(2):573–580. doi: 10.1093/nar/27.2.573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klasson L, Westberg J, Sapountzis P, Näslund K, Lutnaes Y, Darby AC, Veneti Z, Chen L, Braig HR, Garrett R. et al. The mosaic genome structure of the Wolbachia wRi strain infecting Drosophila simulans. Proceedings of the National Academy of Sciences of the United States of America. 2009;106(14):5725–5730. doi: 10.1073/pnas.0810753106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klasson L, Walker T, Sebaihia M, Sanders MJ, Quail MA, Lord A, Sanders S, Earl J, O'Neill SL, Thomson N. et al. Genome evolution of Wolbachia strain wPip from the Culex pipiens group. Molecular Biology and Evolution. 2008;25(9):1877–1887. doi: 10.1093/molbev/msn133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foster J, Ganatra M, Kamal I, Ware J, Makarova K, Ivanova N, Bhattacharyya A, Kapatral V, Kumar S, Posfai J. et al. The Wolbachia genome of Brugia malayi: endosymbiont evolution within a human pathogenic nematode. Public Library of Science Biology. 2005;3(4):e121. doi: 10.1371/journal.pbio.0030121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riegler M, O'Neill SL. In: The Prokaryotes: A handbook on the biology of bacteria. Dworkin M, Falkow S, Rosenberg E, Schleifer KH, Stackebrandt E, editor. Vol. 5. New York: Springer; 2006. The genus Wolbachia; pp. 547–561. Proteobacteria: Alpha and Beta Subclass. [Google Scholar]
- Douglas SE. DNA strider - an inexpensive sequence analysis package for the Macintosh. Molecular Biotechnology. 1995;3(1):37–45. doi: 10.1007/BF02821333. [DOI] [PubMed] [Google Scholar]
- Letunic I, Copley RR, Schmidt S, Ciccarelli FD, Doerks T, Schultz J, Ponting CP, Bork P. SMART 4.0: towards genomic data integration. Nucleic Acids Research. 2004;32(DATABASE ISS.):D142–D144. doi: 10.1093/nar/gkh088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schultz J, Milpetz F, Bork P, Ponting CP. SMART, a simple modular architecture research tool: identification of signaling domains. Proceedings of the National Academy of Sciences of the United States of America. 1998;95(11):5857–5864. doi: 10.1073/pnas.95.11.5857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Notredame C, Higgins DG, Heringa J. T-coffee: a novel method for fast and accurate multiple sequence alignment. Journal of Molecular Biology. 2000;302(1):205–217. doi: 10.1006/jmbi.2000.4042. [DOI] [PubMed] [Google Scholar]
- Guindon S, Gascuel O. A simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood. Systematic Biology. 2003;52(5):696–704. doi: 10.1080/10635150390235520. [DOI] [PubMed] [Google Scholar]
- Arthofer W, Riegler M, Schneider D, Krammer M, Miller WJ, Stauffer C. Hidden Wolbachia diversity in field populations of the European cherry fruit fly, Rhagoletis cerasi (Diptera, Tephritidae) Molecular Ecology. 2009;18(18):3816–3830. doi: 10.1111/j.1365-294X.2009.04321.x. [DOI] [PubMed] [Google Scholar]
- Riegler M, Charlat S, Stauffer C, Merçot H. Wolbachia transfer from Rhagoletis cerasi to Drosophila simulans: investigating the outcomes of host-symbiont coevolution. Applied and Environmental Microbiology. 2004;70(1):273–279. doi: 10.1128/AEM.70.1.273-279.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levinson G, Gutman GA. High frequencies of short frameshifts in poly-CA/TG tandem repeats borne by bacteriophage M13 in Escherichia coli K-12. Nucleic Acids Research. 1987;15(13):5323–5338. doi: 10.1093/nar/15.13.5323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pâques F, Leung WY, Haber JE. Expansions and contractions in a tandem repeat induced by double-strand break repair. Molecular and Cellular Biology. 1998;18(4):2045–2054. doi: 10.1128/mcb.18.4.2045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rocha EPC. DNA repeats lead to the accelerated loss of gene order in bacteria. Trends in Genetics. 2003;19(11):600–603. doi: 10.1016/j.tig.2003.09.011. [DOI] [PubMed] [Google Scholar]
- Collins NE, Liebenberg J, De Villiers EP, Brayton KA, Louw E, Pretorius A, Faber FE, Van Heerden H, Josemans A, Van Kleef M. et al. The genome of the heartwater agent Ehrlichia ruminantium contains multiple tandem repeats of actively variable copy number. Proceedings of the National Academy of Sciences of the United States of America. 2005;102(3):838–843. doi: 10.1073/pnas.0406633102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weitzmann MN, Woodford KJ, Usdin K. DNA secondary structures and the evolution of hypervariable tandem arrays. Journal of Biological Chemistry. 1997;272(14):9517–9523. doi: 10.1074/jbc.272.14.9517. [DOI] [PubMed] [Google Scholar]
- Dover G. Molecular drive: A cohesive mode of species evolution. Nature. 1982;299(5879):111–117. doi: 10.1038/299111a0. [DOI] [PubMed] [Google Scholar]
- Amos W. In: Microsatellites Evolution and Applications. Goldstein DB, Schlötterer C, editor. Oxford: Oxford University Press; 1999. A comparative approach to the study of microsatellite evolution; pp. 66–79. [Google Scholar]
- Yamada R, Floate KD, Riegler M, O'Neill SL. Male development time influences the strength of Wolbachia-induced cytoplasmic incompatibility expression in Drosophila melanogaster. Genetics. 2007;177(2):801–808. doi: 10.1534/genetics.106.068486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker T, Klasson L, Sebaihia M, Sanders MJ, Thomson NR, Parkhill J, Sinkins SP. Ankyrin repeat domain-encoding genes in the wPip strain of Wolbachia from the Culex pipiens group. BMC Biology. 2007;5 doi: 10.1186/1741-7007-5-39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sinkins SP, Walker T, Lynd AR, Steven AR, Makepeace BL, Godfray HC, Parkhill J. Wolbachia variability and host effects on crossing type in Culex mosquitoes. Nature. 2005;436:257–260. doi: 10.1038/nature03629. [DOI] [PubMed] [Google Scholar]
- Salzberg SL, Hotopp JC, Delcher AL, Pop M, Smith DR, Eisen MB, Nelson WC. Serendipitous discovery of Wolbachia genomes in multiple Drosophila species. Genome Biology. 2005;6(3):R23. doi: 10.1186/gb-2005-6-3-r23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Werren JH. Biology of Wolbachia. Annual Review of Entomology. 1997;42:587–609. doi: 10.1146/annurev.ento.42.1.587. [DOI] [PubMed] [Google Scholar]
- Hoffmann AA. Partial cytoplasmic incompatibility between two Australian populations of Drosophila melanogaster. Entomologia Experimentalis Et Applicata. 1988;48:61–67. doi: 10.1111/j.1570-7458.1988.tb02299.x. [DOI] [Google Scholar]
- Reynolds KT, Hoffmann AA. Male age, host effects and the weak expression or nonexpression of cytoplasmic incompatibility in Drosophila strains infected by maternally transmitted Wolbachia. Genetical Research. 2002;80(2):79–87. doi: 10.1017/s0016672302005827. [DOI] [PubMed] [Google Scholar]
- Zabalou S, Charlat S, Nirgianaki A, Lachaise D, Merçot H, Bourtzis K. Natural Wolbachia infections in the Drosophila yakuba species complex do not induce cytoplasmic incompatibility but fully rescue the wRi modification. Genetics. 2004;167(2):827–834. doi: 10.1534/genetics.103.015990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Neill SL, Karr TL. Bidirectional incompatibility between conspecific populations of Drosophila simulans. Nature. 1990;348:178–180. doi: 10.1038/348178a0. [DOI] [PubMed] [Google Scholar]
- Merçot H, Llorente B, Jacques M, Atlan A, Montchampmoreau C. Variability within the Seychelles cytoplasmic incompatibility system in Drosophila simulans. Genetics. 1995;141(3):1015–1023. doi: 10.1093/genetics/141.3.1015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giordano R, O'Neill SL, Robertson HM. Wolbachia infections and the expression of cytoplasmic incompatibility in Drosophila sechellia and D. mauritiana. Genetics. 1995;140(4):1307–1317. doi: 10.1093/genetics/140.4.1307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hornett EA, Duplouy AMR, Davies N, Roderick GK, Wedell N, Hurst GDD, Charlat S. You can't keep a good parasite down: evolution of a male-killer suppressor uncovers cytoplasmic incompatibility. Evolution. 2008;62(5):1258–1263. doi: 10.1111/j.1558-5646.2008.00353.x. [DOI] [PubMed] [Google Scholar]
- Yamada R, Iturbe-Ormaetxe I, Brownlie JC, O'Neill SL. Functional test of the influence of Wolbachia genes on cytoplasmic incompatibility expression in Drosophila melanogaster. Insect Molecular Biology. 2011;20(1):75–85. doi: 10.1111/j.1365-2583.2010.01042.x. [DOI] [PubMed] [Google Scholar]