Abstract
Background
Wolbachia and Cardinium are endosymbiotic bacteria infecting many arthropods and manipulating host reproduction. Although these bacteria are maternally transmitted, incongruencies between phylogenies of host and parasite suggest an additional role for occasional horizontal transmission. Consistent with this view is the strong evidence for recombination in Wolbachia, although it is less clear to what extent recombination drives diversification within single host species and genera. Furthermore, little is known concerning the population structures of other insect endosymbionts which co-infect with Wolbachia, such as Cardinium. Here, we explore Wolbachia and Cardinium strain diversity within nine spider mite species (Tetranychidae) from 38 populations, and quantify the contribution of recombination compared to point mutation in generating Wolbachia diversity.
Results
We found a high level of genetic diversity for Wolbachia, with 36 unique strains detected (64 investigated mite individuals). Sequence data from four Wolbachia genes suggest that new alleles are 7.5 to 11 times more likely to be generated by recombination than point mutation. Consistent with previous reports on more diverse host samples, our data did not reveal evidence for co-evolution of Wolbachia with its host. Cardinium was less frequently found in the mites, but also showed a high level of diversity, with eight unique strains detected in 15 individuals on the basis of only two genes. A lack of congruence among host and Cardinium phylogenies was observed.
Conclusions
We found a high rate of recombination for Wolbachia strains obtained from host species of the spider mite family Tetranychidae, comparable to rates found for horizontally transmitted bacteria. This suggests frequent horizontal transmission of Wolbachia and/or frequent horizontal transfer of single genes. Our findings strengthens earlier reports of recombination for Wolbachia, and shows that high recombination rates are also present on strains from a restrictive host range. Cardinium was found co-infecting several spider mite species, and phylogenetic comparisons suggest also horizontal transmission of Cardinium among hosts.
Background
Wolbachia and Cardinium are intracellular bacteria infecting a wide range of arthropod species. They have been classified as reproductive parasites, being able to manipulate their host's reproductive system in order to promote their own transmission [1-3]. Recently, beneficial effects of Wolbachia have been identified as well, as Wolbachia can protect hosts against virus infection [4,5]. Cardinium may also exert beneficial effects [6] and in many other cases the effect of Wolbachia or Cardinium is unknown. Wolbachia is well studied and is widespread among arthropods and nematodes. It is estimated that around 66% of all insects are infected with Wolbachia [7]. This diverse genus has been subdivided into 11 “supergroups” (A-K) on the basis of molecular phylogenetic analysis [8-13]. Cardinium was more recently discovered and has so far been found in 6-7% of all arthropods, though seems to be more common in Chelicerates than in insects [2,14-18]. Wolbachia and Cardinium have been found co-infecting the same host species [2,15,17-21].
Although Wolbachia and Cardinium are generally considered to be clonally inherited via vertical transmission, there is now a large body of molecular evidence for discordant phylogenies of host and endosymbiont [22-29]. Distantly related Wolbachia or Cardinium strains can infect closely related host species, and closely related strains may infect distantly related host species. Such patterns suggest horizontal transmission of bacteria (or at least of some bacterial genes) between hosts, although direct evidence for horizontal transmission is rare [30-32].
Horizontal transfer has been further supported by evidence for recombination [33]. For Wolbachia, recombination has been found between genes (intergenic) as well as within genes (intragenic). Intergenic recombination is evident from inconsistencies between gene trees [34-36]. Intragenic recombination has been observed within the genes wsp, ftsZ, and gltA and within and between supergroups A and B [34,37-41]. Recently, a genomic comparison of A-group Wolbachia strains by Klasson et al. [42] showed highly recombining genomes, implying frequent horizontal gene transfer.
Cardinium has been less well investigated: although there is some evidence for inconsistent phylogenies between Cardinium and their hosts, which is compatible with horizontal transmission, only a few studies have been performed, mainly focusing on a single gene (16S rDNA) [2,15,18,21,23], and there is currently no evidence regarding recombination in Cardinium.
Phylogenetic and evolutionary studies on Wolbachia have mainly focused on samples representing a wide range of host species [26,34,37,38,43,44]. Based on two genes, Jiggins [38] showed that among strains from a wide range of host species, the rate of recombination is similar to that of a horizontally transmitted bacterium (Cowdria ruminantium). It remains however unclear to what extent these conclusions will be supported by the analyses of much more tightly defined samples such as those recovered from closely related host genera, or even from a single host species from a single geographical and temporal source. Most current studies which address this have used only one or two genes or a restricted number of species or populations [31,36,41,45]. A study by Baldo et al. [22] included a more detailed study of the extent of recombination and horizontal transfer in a single spider genus and revealed that horizontal transfer explains a large part of the Wolbachia distribution patterns within the genus. Exact rates of recombination among Wolbachia strains have however not been inferred so far, which makes it difficult to draw direct comparisons with rates found for other bacteria. Recombination rates can be obtained from multilocus sequence data. Strains that differ at only a single locus are grouped into clonal complexes. Subsequently, the allele sequences are examined to determine whether single allelic variants within a clonal complex result from point mutation or homologous recombination [46].
We present here a detailed study of the diversity of Wolbachia and Cardinium in the phytophagous spider mite family Tetranychidae, by analyzing strains recovered from seven Bryobia species, Tetranychus urticae, and Petrobia harti. We consider strain diversity between tetranychid host species, within single host species (investigating multiple populations; up to 20 populations for B. kissophila) and within single populations and individuals. Both Wolbachia and Cardinium have been reported from this family. Wolbachia has been detected in at least six asexual and one sexual Bryobia species and strains from both supergroup B and K have been found [12,47,48]. Supergroup K is a new supergroup that has only been detected in Bryobia so far [12]. We investigate intra- and intergenic recombination in Wolbachia (four genes) and Cardinium (two genes), and quantify the rate of recombination relative to mutation for Wolbachia, by analyzing the variation between pairs of very closely related strains. We compare this endosymbiont diversity to the degree of host congruence (co-speciation), host mitochondrial DNA diversity, and geographical distribution.
Results
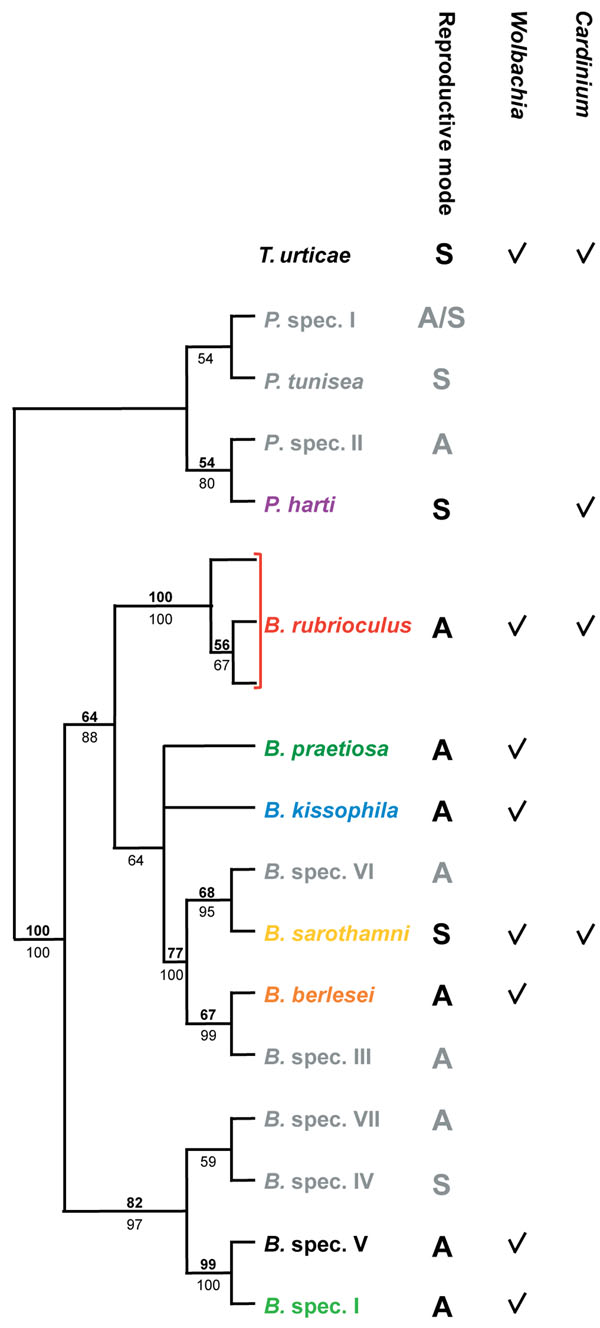
We included Wolbachia strains from seven Bryobia species (B. berlesei, B. kissophila, B. praetiosa, B. rubrioculus, B. sarothamni, B. spec. I, and B. spec. V) and T. urticae, and Cardinium strains from B. rubrioculus, B. sarothamni, T. urticae, and P. harti (Figure 1 and Additional file 1) [49]. We were unable to reliably determine the infection status of the other Bryobia host species (Figure 1) due to the lack of adequate material and/or inconsistent amplification of the bacterial genes, therefore these species were excluded from further analyses. The dataset includes strains from sexually (B. sarothamni, T. urticae, P. harti) and asexually (the remaining species) reproducing species.
Figure 1.
Phylogenetic relationship between the tetranychid host species from which Wolbachia and Cardinium strains were obtained. Maximum likelihood cladogram (28S rDNA) of the genus Bryobia and four outgroup species of the genus Petrobia is shown [49]. Tetranychus urticae was depicted separately as the exact position of T. urticae relative to the other host species was not studied so far. The genus Tetranychus belongs to another subfamily (Tetranychinae) than Bryobia and Petrobia (both Bryobiinae) of the family Tetranychidae. The mode of reproduction is given for each host species (A=asexual, S=sexual) in a separate column, and the subsequent columns indicate from which host species Wolbachia and or Cardinium strains were included in this study. Species names are colored as in Figure 2, 4, 5, and Additional file 3. Host species in grey were not included in this study. Numbers above branches (bold) indicate ML bootstrap values based on 1,000 replicates, numbers below branches (plain) depict Bayesian posterior probabilities (only values larger than 50 are indicted).
Multiple infections
Wolbachia and Cardinium were found co-infecting B. rubrioculus, B. sarothamni, and T. urticae. In B. rubrioculus and B. sarothamni, Wolbachia and Cardinium strains were obtained from doubly infected individuals, whereas in T. urticae they were obtained from singly infected individuals (Additional file 1). Multiple Wolbachia strains infecting a single host individual were not detected, and neither were multiple Cardinium strains. Sequence chromatograms revealed no double peaks and cloning and sequencing of eleven PCR products did not reveal multiple infections.
Wolbachia diversity
Sequences from the four Wolbachia genes (wsp, ftsZ, groEL, and trmD) were recovered for 65 Wolbachia infected individuals, except for wsp from B. spec. V (ITA11). The Wolbachia strain infecting B. spec. V belongs to the newly described supergroup K [12], which is highly divergent from supergroup B strains infecting other tetranychid mites. We excluded the supergroup K strain from phylogenetic and recombination analyses. No insertions or deletions were found within ftsZ, groEL, and trmD. Within wsp small indels (3-9bp) were found in a few strains but all sequences could be unambiguously aligned.
The sequenced Wolbachia strains reveal a high diversity. From the 64 Wolbachia strains (excluding the supergroup K Wolbachia strain in B. spec. V), 36 strains (sequence types; STs) were found unique (Additional file 2). Between 11 (groEL) and 18 (trmD) alleles were found per locus (Table 1). Nucleotide diversity was 5-11 times higher for wsp than for the other loci (Table 1). The dN/dS ratio was < 1 for all loci, indicating that the genes where not subjected to positive selection. The wsp gene also revealed a high rate of intragenic recombination (see below), with two sites identified within hyper variable region 1 (HVR1) under positive selection (HyPhy: codons 20 and 30; unpublished data). Despite this high nucleotide diversity and recombination rate for wsp, we found the most alleles for trmD.
Table 1.
Diversity observed for four Wolbachia genes and two Cardinium genes.
| Locus | Size (bp) | Alleles | Variable sites | π | p-distance max. (%) | dN/dS ratio | ||
|---|---|---|---|---|---|---|---|---|
| n | % | |||||||
| Wolbachia(n=64) | wsp | 525 | 13 | 155 | 29.52 | 0.1030 | 20.08 | 0.60 |
| ftsZ | 507 | 14 | 20 | 3.94 | 0.0126 | 2.37 | 0.07 | |
| groEL | 491 | 11 | 18 | 3.67 | 0.0087 | 1.83 | 0.29 | |
| trmD | 453 | 18 | 34 | 7.51 | 0.0176 | 4.42 | 0.23 | |
| Cardinium(n=15) | CLO | 407 | 6 | 15 | 3.69 | 0.0151 | 2.22 | - |
| gyrB | 631 | 8 | 127 | 20.13 | 0.0839 | 14.9 | 0.06 | |
π = nucleotide diversity
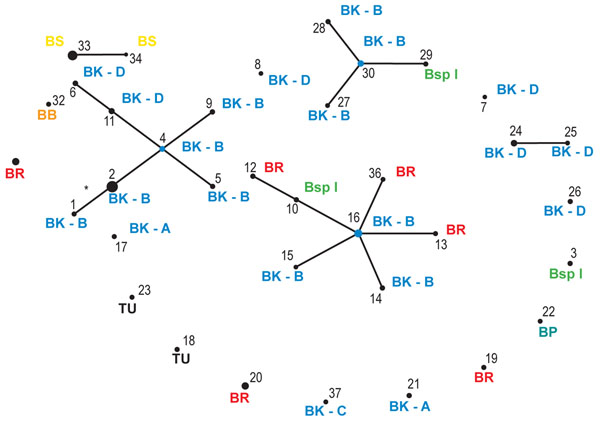
Forty-four out of the 64 strains were grouped into five clonal complexes (I-V; Figure 2 and Table 2). All other strains differed at more than one locus from the strains in these complexes. A total of 17 alleles deviated from the alleles from the founding genotypes within the clonal complexes (Table 2). A significant higher number of these variant alleles were found for trmD compared to the other loci (Table 2; Chi-square test; p=0.003), which is consistent with the observation that this locus contains the most alleles.
Figure 2.
Schematic overview of the clonal relatedness of the Wolbachia STs as predicted by eBURST. Each ST is represented by a black dot, the size of which is proportional to the number of strains of that ST. STs that differ at a single locus are linked by lines. Only one variant is likely due to a mutational event (indicated by *), the other variants are most likely due to recombination events. STs that are not linked to other STs do not share at least four identical alleles with any other ST. Host species name in which each ST was detected is indicated: BB=B. berlesei; BK=B. kissophila (A-D indicate different COI clades, see text); BP=B. praetiosa; BR=B. rubrioculus; BS=B. sarothamni; BspI= B. spec. I; TU=T. urticae.
Table 2.
Clonal complexes found forWolbachia
| Complex | I | II | III | IV | V | |||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| STa | 4 | 9 | 5 | 2 | 1 | 11 | 6 | 30 | 29 | 28 | 27 | 16 | 15 | 14 | 13 | 36 | 10 | 12 | 24 | 25 | 33 | 34 |
| wsp | 1 | - | - | - | - | - | 5 18 | 12 | - | - | - | 5 | 3 8 | - | - | - | - | 4 16 | 12 | - | 6 | - |
| ftsZ | 2 | - | - | - | 1 1 | - | - | 10 | - | - | - | 3 | - | - | - | - | - | - | 10 | - | 14 | - |
| groEl | 8 | - | - | 4 4 | 4 4 | - | - | 8 | - | - | - | 8 | - | 4 4 | - | - | - | - | 12 | 11 2 | 3 | - |
| trmD | 1 | 3 1* | 10 15 | - | - | 6 7 | 6 7 | 1 | 14 9 | 6 7 | 2 1* | 1 | - | - | 5 2* | 17 9 | 15 8 | 15 8 | 8 | - | 9 | 11 1* |
| Speciesb | BK-B | BK-B | BK-B | BK-B | BK-B | BK-D | BK-D | BK-B | BspI | BK-D | BK-B | BK-D | BK-D | BK-D | BR | BR | BspI | BR | BK-D | BK-D | BS | BS |
| Freq.c | 2 | 1 | 1 | 12 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 2 | 1 | 1 | 1 | 1 | 1 | 1 | 3 | 1 | 8 | 1 |
a Allelic variants within each clonal complex are depicted, listed per sequence type (ST). Likely recombinational changes are depicted in plain text, and putative mutations are shown in bold. Disputable cases are highlighted in italics (possible recombinational changes). For each allelic variant the allele number is given, with in superscript the number of polymorphic sites between the allelic variant and the typical allele of the clonal group. * indicates non-unique mutations (in cases where one or two mutations were found).
b Host species name in which each ST was detected is indicated (for abbreviations see legend Figure 2).
c The frequency of each sequence type is listed.
Recombination between Wolbachia strains
We investigated intergenic recombination by analysis of allelic variation within the clonal complexes. This approach reveals whether variant alleles arose by point mutation or by recombination. Of the 17 variant alleles, four differed from the typical allele in the clonal complex by a single nucleotide change (Table 2). Three of these single nucleotide changes, however, were non-unique. Two other alleles differed by two nucleotide changes, and could be either derived by point mutations or recombination (the chance of two independent mutations both occurring in one out of four genes is 0.25). One of these variant alleles was found elsewhere in the dataset, implicating recombination. All other allelic variants differed from the founder alleles at four or more sites and were considered as putative recombinational imports. Ignoring alleles with one non-unique and two nucleotide changes, the estimated ratio of recombinational events to mutational events per gene fragment is 11:1. If we include non-unique changes as recombinational imports, and unique changes as point mutations, the ratio is 15:2. We therefore conclude that new alleles were 7.5 to 11 times more likely to be generated by recombination than by point mutation. This is a conservative estimate because single nucleotide changes were attributed to point mutation and not to recombination, although recombination between similar alleles could result in a single nucleotide change. Further, a high rate of recombination is consistent with the observed incongruence between the four gene tree topologies (Additional file 3).
Intragenic recombination is another process that may contribute to the origin of new Wolbachia genotypes. We detected intragenic recombination within the trmD and wsp genes (Figure 3). The alignment of wsp genes shows that the polymorphic sites are not randomly distributed, but clearly shows a mosaic pattern consistent with recombination. Intragenic recombination is not restricted to Wolbachia strains from the same host species, but also involves strains infecting different host species. For example, the wsp sequence obtained from Wolbachia in B. sarothamni (all populations) is a recombinant between the wsp sequences obtained from Wolbachia in B. kissophila (FR13) and T. urticae (T3) (Figure 3).
Figure 3.
Examples of recombination within trmD and wsp. Only polymorphic sites are shown (position in alignment is given on top). Sequences are named by their sample code (Additional file 1) and abbreviated host species name (see legend Figure 2). Each sequence may have been found in different populations or host species, see phylogenies of trmD and wsp in Additional file 3. Different shadings indicate possible recombinant regions (see results). Differences and identities (dots) compared to the middle sequence are shown. * = also detected in BspI, BK-A, BK-C, and BP. ^ = also detected in BR.
Cospeciation of Wolbachia and host species
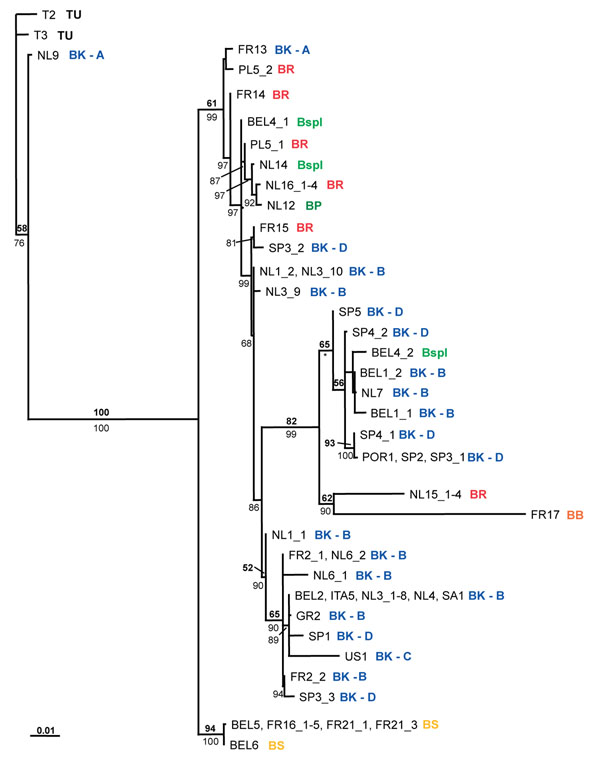
Examination of the concatenated Wolbachia phylogeny reveals that there is generally a lack of cospeciation between host and parasite (Figure 4). Wolbachia strains obtained from a single host species do not clearly cluster. For example, strains from B. rubrioculus are found at different places in the phylogeny. The same is true for strains from B. spec. I. On the other hand, the Wolbachia phylogeny is not completely random with respect to host species. Some Wolbachia strains from B. kissophila cluster together, whilst others cluster with strains from B. spec I (BEL4_2) or B. rubrioculus (FR15). Two B. kissophila-derived strains (NL9 and FR13) are very divergent from all other B. kissophila strains. In the exceptional case of B. sarothamni, the same Wolbachia genotype was found in all five populations (from Belgium and France; except for a minor difference in trmD for BEL6; Figure 2, 4, and Table 2). This strain was not found in any of the other species, although it closely resembles the Wolbachia strain infecting B. berlesei at three of the four genes (wsp is highly divergent between the two strains). Bryobia sarothamni and B. berlesei share the same host plant species, Cytisus scoparius, which potentially facilitates horizontal transmission of both Wolbachia strains and genes.
Figure 4.
Concatenated ML phylogeny for Wolbachia, based on four genes (wsp, ftsZ, groEL, and trmD). Thirty-six unique strains are shown. Sample code (Additional file 1) and host species name in which each strain was detected are indicated (for abbreviations see legend Figure 2). ML bootstrap values (top number, bold) and Bayesian posterior probabilities (bottom number, plain) are depicted (only values larger than 50 are indicated). * = the topology within this clade is slightly different for the MrBayes topology. The bar at the bottom indicates a branch length of 10% likelihood distance. Independent phylogenies for each gene are depicted in Additional file 3.
Finally, a single B. praetiosa individual was investigated. Although this species was found to harbor a unique Wolbachia strain, this strain shares each of its alleles with strains in (multiple) other host species. Although allelic identity by descent cannot be ruled out without more detailed analysis, this observation is also consistent with frequent inter-allele recombination. Within the other species, divergent Wolbachia strains were found between populations and also within populations (Figure 4). In five B. rubrioculus mite populations, six divergent Wolbachia strains were found: population PL5 contains two divergent Wolbachia strains. For B. spec. I three Wolbachia strains were detected in two populations: two individuals from BEL4 harbor highly divergent Wolbachia strains (mainly due to differences at wsp and ftsZ).
Correlation between Wolbachia and host mitochondrial diversity or geographical location
It has been suggested that infection by Wolbachia affects host mitochondrial diversity and that mitochondrial haplotypes and Wolbachia haplotypes may be linked [50-53]. As this has serious implications for population studies based on mtDNA [54], we were motivated to examine this possibility for B. kissophila. High levels of diversity at the mitochondrial COI locus were observed within B. kissophila, which resolved into four clades (A-D) [49]. However, there was little evidence for correlation between the COI haplotypes and the Wolbachia strains (Figure 2 and 4). A total of 20 populations were investigated for B. kissophila, and a highly divergent set of Wolbachia strains was found within this species. Twenty-one Wolbachia strains were found, four of which were shared between populations. Within several populations (BEL1, FR2, NL1, NL3, NL6, SP3, and SP4) more than one Wolbachia strain was detected.
Bryobia kissophila COI clade A was highly divergent from all other COI clades, and contains Wolbachia strains that are divergent from the ones found in the other clades. However, the two investigated populations belonging to clade A (NL9 and FR13) harbor divergent Wolbachia strains. Also, some alleles of these strains are shared with other B. kissophila clades (for groEL and trmD) or with other Bryobia species (for all four genes) (Additional file 3). Wolbachia strains from clade B, C, and D show a mixture of different Wolbachia strains. There is no correlation with COI haplotype, although there are no strains shared among populations belonging to different COI clades.
There is a similar lack of congruence between Wolbachia strain diversity and geographic location of the host populations. Very distant populations may harbor identical Wolbachia strains (e.g., BEL2 and SA1; B. kissophila), while nearby populations harbor very divergent Wolbachia strains (e.g., NL15 and NL16; B. rubrioculus). Also within populations divergent strains are found.
Cardinium diversity
Cardinium diversity was investigated by sequencing part of the 16S rDNA and gyrB gene. Sequences were successfully recovered from all Cardinium infected individuals and all sequences could be unambiguously aligned. No insertions or deletions were found within gyrB. Within 16S rDNA, one insertion and one deletion (both 1bp) were found. For 16S rDNA six alleles were found, with 3.7% variable sites, a maximum p-distance of 2.2%, and a nucleotide diversity of 0.015 (Table 1). Diversity for gyrB was much higher, with eight alleles, 20.1% variable sites, a maximum p-distance of 14.9%, and a nucleotide diversity of 0.084.
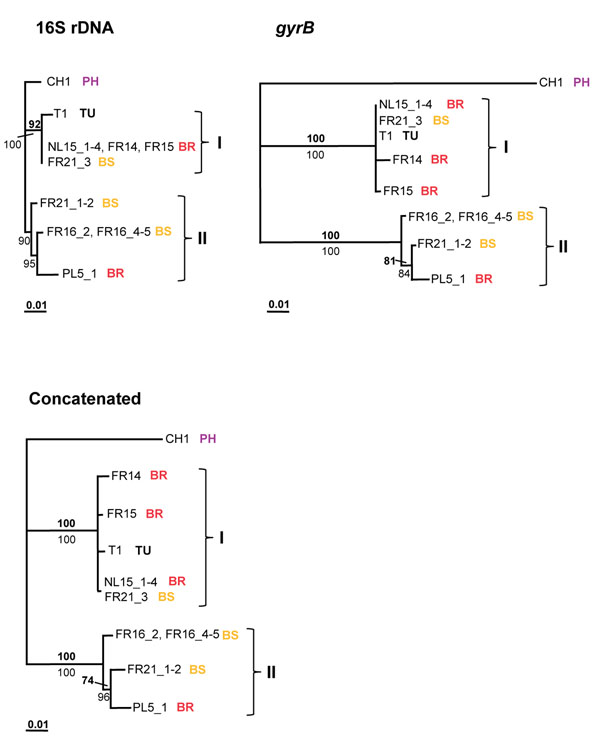
In total, eight strains were detected within eight populations, belonging to four mite species, and phylogenetic analysis resolved these eight stains into two major clades (Figure 5). The Cardinium strain found in P. harti (CH1) is divergent from two other clades (named I and II), which were detected in B. sarothamni and B. rubrioculus (both clade I and II), and in T. urticae (clade I). These two clades are highly supported. Clade I and II differed at 1.7% of nucleotide sites for 16S rDNA and at 10.6% for gyrB, while differences within clades are small (<1.2% for both genes). Generally, there is congruence between the phylogenies obtained for 16S rDNA and gyrB which suggests less recombination than in Wolbachia, although the evidence is equivocal. However, there is no obvious association between Cardinium genotype and host species. Clade I contains strains found in three B. rubrioculus populations and in one T. urticae and one B. sarothamni population, while clade II contains highly related strains found in two B. sarothamni populations and one B. rubrioculus population. One strain was found infecting two host species: B. rubrioculus (NL15_1-4) and B. sarothamni (FR21_3). Other strains belonging to B. sarothamni population FR21 group within clade II (FR21_1-2). These patterns imply horizontal transfer of strains (or genes) between and within host species.
Figure 5.
16S rDNA, gyrB, and concatenated ML phylogenies for Cardinium. Sample code (Additional file 1) and host species name in which each strain was detected are indicated: BR=B. rubrioculus; BS=B. sarothamni; PH= P. harti. Two clades are named I and II. ML bootstrap values (top number, bold) and Bayesian posterior probabilities (bottom number, plain) are depicted (only values larger than 50 are indicated). The bar at the bottom indicates a branch length of 10% likelihood distance.
Discussion
This detailed study of reproductive parasites in nine tetranychid mite species reveals a high genetic diversity. Wolbachia strains belonging to two highly divergent supergroups (B and K) were detected (see also [12]). The diversity within supergroup B was high, with 36 unique strains found in 64 investigated individuals. The level of recombination detected is extremely high, supporting the mosaic genome structure of Wolbachia [42]. Cardinium was less frequently found in the mites than Wolbachia, but also showed a high level of diversity, with eight unique strains detected in 15 individuals on the basis of only two genes.
Wolbachia diversity
We investigated Wolbachia diversity at a fine scale with respect to host diversity, by comparing strains from nine closely related host species, all belonging to the family Tetranychidae, and mainly from the genus Bryobia. Our study shows that even within a single host genus there exists a high level of Wolbachia diversity. Wolbachia strains belonging to two highly divergent supergroups (B and K) were detected. Even within Wolbachia supergroup B, 36 unique STs were obtained from 64 infected hosts. Although there was little correlation between host species and Wolbachia strains, strains were not distributed randomly among different species (Figure 2 and 4), so that a certain level of specificity was observed. Strains within clonal complex I were restricted to B. kissophila and within clonal complex V to B. sarothamni. Other complexes however contain strains from different host species. It is striking that many alleles are shared among the different STs, even from different host species, indicating that recombination contributes substantially to the genetic diversity of Wolbachia. Recombination is further evidenced by the many phylogenetic conflicts observed among the individual gene trees and a high recombination rate compared to mutation rate. Analysis of the variant alleles in the clonal complexes reveals that the rate of recombination compared to point mutation in the diversification of lineages ranges between 7.5:1 and 11:1.
The observed recombination rate and diversity is much higher than what would be expected for clonal organisms. Recombination is rare in other clonally inherited, obligate intracellular bacteria [55,56]. The high recombination rate we found is comparable to rates of horizontally transmitted human pathogens. For example, for Streptococcus pneumoniae a recombination to mutation ratio of 10:1 was found, for Neisseria meningitidis a ratio of 5:1 [57]. Horizontal transmission of Wolbachia has been observed, but examples are rare [30-32]. Although many studies based on molecular data have suggested extensive horizontal gene transfer of Wolbachia [22,25,35,36,42,43], it is unclear if bacteria are transmitted horizontally, or if the transfer concerns single genes, possibly via bacteriophages [58]. The high rate of recombination found in this study, the observation that individual alleles are shared among Wolbachia strains from different host species but complete STs are not, and the fact that Wolbachia is mainly clonally inherited, suggest that individual genes rather than complete bacteria are exchanged. Alternatively, transfer of bacteria leading to mixed infections and subsequent recombination may explain these observations. Although our cloning data suggest that mixed infections are rare, this possibility cannot be excluded (see also [59]). The observation that the trees are not completely random with respect to host species suggests that vertical transmission does occur [26,43].
Homologous recombination in bacteria can occur by transformation, conjugation, or transduction. Conjugation and transformation require physical contact, or close proximity, of donor DNA and recipient bacteria. Ecological circumstances may create opportunities for recombination, e.g., Wolbachia strains from B. sarothamni and B. berlesei share three of the four alleles, and their mite hosts feed on the same host plant species (we found both species co-occurring on the same individual plant). Other ecological interactions have been suggested as means for bacteria or gene exchange, e.g., host-parasite interactions or double Wolbachia infections [28,36,45]. However, in many other cases, opportunities for recombination are less obvious. Transduction involving vectors (e.g., plasmids, phages, or viruses) is a more likely manner of gene exchange. Good vector candidates are bacteriophages, as these have been isolated from Wolbachia infected populations [60-62] and seem to be common in Wolbachia genomes [42,63]. Phylogenetic analyses suggest that the bacteriophage WO is horizontally transferred between different Wolbachia strains, and is able to infect new Wolbachia hosts [60,61,64]. Other, free-living, bacteria might even be involved in phage-transfer. We also noted the presence of a bacteriophage in an individual of B. spec. I. The bacteriophage sequence, detected coincidentally with groEL primers, appeared similar to the sequence of the Wolbachia bacteriophage WOcauB1 from Cadra cautella (GenBank: AB161975; 12% p-distance) [65], and to part of the sequenced genome (located within the gene dnaA) of Wolbachia from Drosophila melanogaster (GenBank: AE017196; 11% p-distance).
With strict vertical transmission, strong linkage disequilibrium between host mtDNA and Wolbachia would be expected. However, recombination may uncouple such associations, and could be a reason for the observed lack of congruence between host mtDNA and Wolbachia STs. There are some signs of congruence, with related host strains (with identical COI sequences) sharing identical or closely related Wolbachia strains, but due to the high rate of recombination such associations are broken up rather quickly.
Cardinium diversity
For Cardinium, the two investigated genes showed highly similar phylogenies, giving no clear evidence for intergenic recombination. Also, no signs of intragenic recombination were found. There was however no congruence between Cardinium strains and associated host species: similar strains were found in B. rubrioculus, B. sarothamni, and T. urticae. Only the strain infecting P. harti was clearly distinct from all other strains. The sharing of strains among different host species, and the occurrence of divergent strains in one host population (FR21), suggest that horizontal transmission is also prevalent for Cardinium. Horizontal transmission seemed also to explain diversity patterns found for Cardinium infecting Cybaeus spiders [17]. Patterns of recombination and horizontal transfer should however be further studied including more genes. An MLST set for Cardinium is desirable, for reliable strain typing and for investigating patterns of recombination, horizontal transmission, or host manipulation. This requires the use of several independent markers, sufficiently distant from each other within the genome.
Phenotypic effects of Wolbachia and Cardinium in spider mites
We analyzed Wolbachia and Cardinium strains from both asexual and sexual host species. Weeks and Breeuwer [48] showed that Wolbachia is involved in causing asexuality in at least two species: B. praetiosa and an unidentified species. Wolbachia is possibly causing asexuality in the other infected asexual Bryobia species as well. The general observation is that all individuals within the asexual Bryobia species are infected with Wolbachia. No males have ever been observed, neither in cultures nor in the field, and additional lab experiments including at least 20 individuals per species (except for B. berlesei) show a fixed infection with Wolbachia (unpublished data). Moreover, Weeks and Breeuwer [48] analyzed 240 B. kissophila, 144 B. praetiosa, and 24 B. rubrioculus individuals and found all individuals infected with Wolbachia. We detected Cardinium in one asexual species, B. rubrioculus. This species is doubly infected with both Wolbachia and Cardinium, although Cardinium was not found in all individuals. It is unclear if Cardinium is having an effect on the host species, but it is unlikely that it induces the asexuality as not all individuals are infected. We detected both Wolbachia and Cardinium in the sexually reproducing species B. sarothamni and T. urticae. Both species appear polymorphic for infection with both bacteria. Cardinium induces strong CI in B. sarothamni, while no effect for Wolbachia has been found so far [47]. Previously, Wolbachia was found inducing CI in T. urticae [66-69], but no effect of Cardinium on T. urticae was found so far [68]. We detected only Cardinium in P. harti, but Weeks et al. [2] also report Wolbachia from P. harti. The effects of both Wolbachia and Cardinium in P. harti, and T. urticae require further investigation.
Conclusions
We found a relatively high rate of recombination for Wolbachia strains obtained from host species of the family Tetranychidae. Considering the fact that Wolbachia is widely distributed among arthropods, we investigated strains from a restrictive host range. It remains to be investigated if our findings present a general pattern and if similar recombination rates will be found among strains from other restricted host ranges. Our study of diversity within Cardinium revealed incongruencies among host and bacterial phylogenies, confirming earlier findings. Analysis of additional genes is needed to investigate recombination rates within this reproductive parasite.
Methods
DNA isolation, amplification, and sequencing
We analyzed Wolbachia and Cardinium strains from seven Bryobia species (34 populations), T. urticae (three populations), and P. harti (one population) (Figure 1 and Additional file 1). Samples were collected between May 2004 and November 2006 from eight European countries, and from South Africa, the United States, and China. For each host population, information on mitochondrial (part of the COI gene) and nuclear (part of the 28S rDNA gene) diversity was obtained as described in Ros et al. [49] and used for species identification. Mites were either set up as cultures in the lab or stored in 96% ethanol. DNA was extracted from single mites using the CTAB extraction method as previously described [54] or using the NucleoSpin Kit (Macherey-Nagel, Düren, Germany) following manufacturers’ instructions.
For Wolbachia, four genes were amplified and sequenced: wsp, flsZ, groEL, and trmD. Wsp was amplified and sequenced using the primers wsp-81F and wsp-691R [70]. FtsZ and groEl were amplified and sequenced as described in Ros et al. [49]. TrmD was amplified and sequenced using the primers trmD-F 5’-GAACTATTCTCTTTGCCGGAAAAGC-3’ and trmD-R 5’-CACTGCTCAGGTCTAGTATATTGAGG-3’.These primers were designed from available Wolbachia and Rickettsia genome sequences [71-73] and were shown to reliably amplify products from strains representative of supergroups A and B (data not shown; samples kindly donated by Dr. Robert Butcher).
For Cardinium, two genes were amplified: 16S rDNA and gyrB. 16S rDNA was amplified and sequenced directly using the primers CLOf and CLOr1 [2]. GyrB was amplified using primers from Groot and Breeuwer [74], cloned, and subsequently sequenced. Amplified fragments were separated from non-specific products by running the PCR products on a 1% agarose in 1x TAE gel and excising the fragments from the gel. Fragments were purified using the method of Boom et al. [75]. Products were first cloned and subsequently sequenced following the cloning protocol described below, with 1-2 clones sequenced per sample using M13 forward and reverse primers.
PCR amplifications were performed in 25 μl reactions containing 1X Super Taq buffer (HT BioTechnology, Cambridge, UK), 0.5 mg/ml bovine serum albumin (BSA), 1.25 mM MgCl2, 0.2 mM dNTP’s, 160 nM of each primer, 1 u of Super Taq (HT BioTechnology), and 2.5 μl of DNA extract. For ftsZ, groEL, and trmD, no MgCl2 was added and for 16S rDNA no MgCl2 and BSA was added. PCR cycling profile for wsp and ftsZ was 35 cycles of 30 sec. at 95 °C, 30 sec. at 51 °C, and 1 min. at 72 °C, for groEL and trmD 35 cycles of 1 min. at 95 °C, 1 min. at 49 °C, and 1.5 min. at 72 °C, for Cardinium 16S rDNA 35 cycles of 40 sec. at 95 °C, 40 sec. at 57 °C, and 45 sec. at 72 °C, and for gyrB 35 cycles of 1 min. at 95 °C, 1 min. at 50 °C, and 1 min. at 72 °C. Products (2 μl) were visualized on a 1% agarose gel stained with ethidium bromide in 0.5X TBE buffer (45mM Tris base, 45mM boric acid, and 1 mM EDTA, pH 8.0). PCR products were purified using a DNA extraction kit (Fermentas, St. Leon-Rot, Germany). The purified products were directly sequenced using the ABI PRISM BigDye Terminator Sequence Kit (Applied Biosystems, Nieuwerkerk a/d IJssel, The Netherlands). Both strands of the products were sequenced using the same primers as used in the PCR amplification. Sequences were run on an ABI 3700 automated DNA sequencer.
All new unique sequence data have been submitted to the GenBank under accession numbers: JN572802-JN572888 (see Additional file 4).
Testing for multiple infections
We tested for multiple infections by checking all sequences for the presence of double peaks in the chromatograms and for differences between forward and reverse sequences. Additionally, 11 PCR products were cloned and subsequently sequenced (two for wsp, groEL, trmD, and gyrB, and three for ftsZ) (Additional file 1). This approach would reveal multiple infections by Wolbachia or Cardinium. PCR products selected for cloning were cleaned using the method of Boom et al. [75]. The cleaned products were ligated into vectors and transformed into bacteria using the pGEM-T Easy Vector System and JM109 competent cells (Promega, Madison WI, US). Plasmids were recovered for 3-11 colonies per sample, using mini-preparation procedures [76]. Plasmids were sequenced using the M13 forward and reverse primers.
Data assembling and phylogenetic analyses
Sequences were aligned using ClustalX version 1.8.0 with default settings [77] and modified in BioEdit version 7.0.7 [78]. We excluded one Wolbachia strain (ITA11) from subsequent analyses, as this strain represents a separate supergroup and is highly divergent from all other strains (see results). We analyzed alignments of 525bp for wsp, 557bp for ftsZ, 491bp for groEL, 453bp for trmD, 407bp for Cardinium 16S rDNA, and 631bp for gyrB. Nucleotide diversity was calculated in MEGAv4.0 [79]. The program SNAP (http://www.hiv.lanl.gov) [80] was used to calculate the rate of nonsynonymous to synonymous substitutions (dN/dS). To determine the overall selection pressures faced by each gene, the SLAC method within the HyPhy package was used [81]. Phylogenetic analyses were performed using Neighbor-Joining (NJ), Maximum Likelihood (ML), and Bayesian methods, for each gene separately and for a concatenated dataset of four genes for Wolbachia and two genes for Cardinium. PAUP* version 4.0b10 [82] was used to select the optimal evolution model by critically evaluating the selected parameters [83] using the Akaike Information Criterion (AIC) [84]. For the protein coding genes, we tested if the likelihood of models could be further improved by incorporating specific rates for each codon position [85]. This approach suggested the following models: wsp (submodel of GTR + G with rate class 'a b c c a c'),ftsZ (K3P+I), groEL (submodel of GTR with rate class 'a a b b a b'), trmD (HKY with site-specific rates for each codon position), 16S rDNA (submodel of GTR with rate class 'a a b c a c'), gyrB (submodel of GTR with rate class 'a b c d b a' and site-specific rates), the concatenated Wolbachia dataset (submodel of GTR + I + G with rate class ‘a b c c b d’), and the concatenated Cardinium dataset (submodel of GTR + G with rate class ‘a b c a b c’). Under the selected models, parameters and tree topology were optimized using the successive approximations approach [86]. NJ analyses (p-distances) and ML analyses (heuristic search, random addition of sequences with 10 replicates, TBR branch swapping) were performed in PAUP. Robustness of nodes was assessed with 100 NJ- resp. ML-bootstrap replicates. However, as PAUP does not allow for site-specific rates in bootstrap analysis, ML bootstrapping for trmD and gyrB was performed with gamma distributed rates, with 100 bootstrap replicates. Bootstrap values were then plotted on the phylogeny obtained with the original model with site-specific rates. Bayesian analyses were performed as implemented in MrBayes 3.1.2 [87]. Models used were GTR + G (wsp), GTR + I (ftsZ), GTR (groEL, 16S rDNA), and GTR with separate rates for each codon position (trmD, gyrB). For the concatenated dataset, the same models were used for each gene partition. Analyses were initiated from random starting trees. Two separate Markov Chain Monte Carlo (MCMC) runs, each composed of four chains (one cold and three heated), were run for 6,000,000 generations (7,000,000 generations for the concatenated Wolbachia set). The cold chain was sampled every 100 generations, the first 15,000 generations were discarded afterwards (burn-in of 25%). Posterior probabilities were computed from the remaining trees. We checked whether the MCMC analyses ran long enough using the program AWTY [88]. Stationarity was assumed when there was convergence between the two MCMC runs and when the cumulative posterior probabilities of splits stabilized; in all analyses 6,000,000 generations proved sufficient. The concatenated Wolbachia dataset however, showed no convergence or stabilization of probabilities (not even after 15,000,000 generations). This is most likely due to the extensive recombination present within this dataset.
Analysis of recombination
Evidence for recombination within Wolbachia and Cardinium was obtained by comparing topologies of different genes. For Wolbachia, we also quantified the relative impact of recombination compared to point mutation over short-term clonal diversification. Following standard MLST protocol [89], we assigned allele identifiers for each unique sequence at a particular locus, and an “ST” (sequence type) for each unique allelic profile. We used eBURST version 3 [90] (Figure 2) to identify closely related pairs or clusters (clonal complexes). All members assigned to a clonal complex share identical alleles at three of the four loci with at least one other ST member of the complex. By comparing, for each ST within a clonal complex, the sequence of the deviating allele with the allele of the founding genotype, it is possible to estimate how many STs have arisen by de novo point mutation (i.e. a novel change at a single base) or homologous recombination (a single non-unique change or multiple nucleotide changes) [46].
Additionally, single gene alignments for Wolbachia and Cardinium were checked for signs of intragenic recombination using the software package RDP3 [91] and by visual inspection. Programs used in the RDP3 software package were RDP, Geneconv, Bootscan, MaxChi, Chimaera, and Sister Scanning. Analyses were run with default settings, except for window- and stepsizes: these were varied during independent analyses. Analyses were performed for total datasets and reduced datasets (removal of highly similar strains). This analysis was performed for each of the four Wolbachia genes and for the two Cardinium genes.
Authors’ contributions
VIDR collected samples, carried out the molecular studies and analyzed data. VMF carried out the molecular studies and analyzed data. VIDR, VMF, JAJB and EJF conceived the study and VIDR, JAJB and EJF drafted the manuscript. All authors read and approved the final manuscript.
Competing interests
The authors declare that they have no competing interests.
Supplementary Material
List of tetranychid samples in which Wolbachia and/or Cardinium strains were detected.
Allelic profiles for each of the 37 unique Wolbachia STs.
Wolbachia gene phylogenies (wsp, ftsZ, groEL, and trmD).
GenBank accession numbers.
Contributor Information
Vera I D Ros, Email: vera.ros@wur.nl.
Vicki M Fleming, Email: vicki.fleming@ndm.ox.ac.uk.
Edward J Feil, Email: e.feil@bath.ac.uk.
Johannes A J Breeuwer, Email: j.a.j.breeuwer@uva.nl.
Acknowledgments
We thank Tom Groot, Maria Nomikou, Cécile Fauvelot, Jeroen Swinkels, and Petra Wilbrink for assisting in sample collection, Betsie Voetdijk and Sangeeta Jessurun for assistance with cloning and sequencing, Louis Lie for help with maintaining cultures, and Jan van Arkel for help with the figures. We thank Robert Butcher and Tim Karr for useful discussions and Steph Menken for useful discussions and valuable comments on the manuscript. This research was funded by a grant from The Netherlands Organisation for Scientific Research (NWO; ALW4PJ/03-25).
This article has been published as part of BMC Microbiology Volume 11 Supplement 1, 2012: Arthropod symbioses: from fundamental studies to pest and disease mangement. The full contents of the supplement are available online at http://www.biomedcentral.com/1471-2180/12?issue=S1.
References
- O'Neill SL, Hoffmann AA, Werren JH. Influential passengers. Inherited microorganisms and arthropod reproduction. New York: Oxford University Press; 1997. [Google Scholar]
- Weeks AR, Velten R, Stouthamer R. Incidence of a new sex-ratio-distorting endosymbiotic bacterium among arthropods. Proc Roy Soc Lond B. 2003;270:1857–1865. doi: 10.1098/rspb.2003.2425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Werren JH, Baldo L, Clark ME. Wolbachia: master manipulators of invertebrate biology. Nat Rev Microbiol. 2008;6:741–751. doi: 10.1038/nrmicro1969. [DOI] [PubMed] [Google Scholar]
- Hedges LM, Brownlie JC, O'Neill SL, Johnson KN. Wolbachia and virus protection in Insects. Science. 2008;322:702–702. doi: 10.1126/science.1162418. [DOI] [PubMed] [Google Scholar]
- Teixeira L, Ferreira A, Ashburner M. The bacterial symbiont Wolbachia induces resistance to RNA viral infections in Drosophila melanogaster. PLoS Biol. 2008;6:2753–2763. doi: 10.1371/journal.pbio.1000002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weeks AR, Stouthamer R. Increased fecundity associated with infection by a Cytophaga-like intracellular bacterium in the predatory mite, Metaseiulus occidentalis. Proc Roy Soc Lond B. 2004;271:S193–S195. doi: 10.1098/rsbl.2003.0137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hilgenboecker K, Hammerstein P, Schlattmann P, Telschow A, Werren JH. How many species are infected with Wolbachia? - a statistical analysis of current data. FEMS Microbiol Lett. 2008;281:215–220. doi: 10.1111/j.1574-6968.2008.01110.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bandi C, Anderson TJC, Genchi C, Blaxter ML. Phylogeny of Wolbachia in filarial nematodes. Proc Roy Soc Lond B. 1998;265:2407–2413. doi: 10.1098/rspb.1998.0591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bordenstein S, Rosengaus RB. Discovery of a novel Wolbachia supergroup in isoptera. Curr Microbiol. 2005;51:393–398. doi: 10.1007/s00284-005-0084-0. [DOI] [PubMed] [Google Scholar]
- Casiraghi M, Bordenstein SR, Baldo L, Lo N, Beninati T, Wernegreen JJ, Werren JH, Bandi C. Phylogeny of Wolbachia pipientis based on gltA, groEL and ftsZ gene sequences: clustering of arthropod and nematode symbionts in the F supergroup, and evidence for further diversity in the Wolbachia tree. Microbiol. 2005;151:4015–4022. doi: 10.1099/mic.0.28313-0. [DOI] [PubMed] [Google Scholar]
- Lo N, Casiraghi M, Salati E, Bazzocchi C, Bandi C. How many Wolbachia supergroups exist? Mol Biol Evol. 2002;19:341–346. doi: 10.1093/oxfordjournals.molbev.a004087. [DOI] [PubMed] [Google Scholar]
- Ros VID, Fleming VM, Feil EJ, Breeuwer JAJ. How diverse is the genus Wolbachia? Multiple-gene sequencing reveals a putatively new Wolbachia supergroup recovered from spider mites (Acari: Tetranychidae) Appl Environ Microbiol. 2009;75:1036–1043. doi: 10.1128/AEM.01109-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Werren JH, Windsor D, Guo LR. Distribution of Wolbachia among neotropical arthropods. Proc Roy Soc Lond B. 1995;262:197–204. doi: 10.1098/rspb.1995.0196. [DOI] [Google Scholar]
- Chang J, Masters A, Avery A, Werren JH. A divergent Cardinium found in daddy long-legs (Arachnida: Opiliones) J invertebr Pathol. 2010;105:220–227. doi: 10.1016/j.jip.2010.05.017. [DOI] [PubMed] [Google Scholar]
- Duron O, Hurst GDD, Hornett EA, Josling JA, Engelstädter J. High incidence of the maternally inherited bacterium Cardinium in spiders. Mol Ecol. 2008;17:1427–1437. doi: 10.1111/j.1365-294X.2008.03689.x. [DOI] [PubMed] [Google Scholar]
- Martin OY, Goodacre SL. Widespread infection by the bacterial endosymbiont Cardinium in arachnids. J Arachnol. 2009;37:106–108. doi: 10.1636/SH08-05.1. [DOI] [Google Scholar]
- Perlman SJ, Magnus SA, Copley CR. Pervasive associations between Cybaeus spiders and the bacterial symbiont Cardinium. J Invert Pathol. 2010;103:150–155. doi: 10.1016/j.jip.2009.12.009. [DOI] [PubMed] [Google Scholar]
- Zchori-Fein E, Perlman SJ. Distribution of the bacterial symbiont Cardinium in arthropods. Mol Ecol. 2004;13:2009–2016. doi: 10.1111/j.1365-294X.2004.02203.x. [DOI] [PubMed] [Google Scholar]
- Enigl M, Schausberger P. Incidence of the endosymbionts Wolbachia, Cardinium and Spiroplasma in phytoseiid mites and associated prey. Exp Appl Acarol. 2007;42:75–85. doi: 10.1007/s10493-007-9080-3. [DOI] [PubMed] [Google Scholar]
- Gotoh T, Noda H, Ito S. Cardinium symbionts cause cytoplasmic incompatibility in spider mites. Heredity. 2006;98:13–20. doi: 10.1093/jhered/esl056. [DOI] [PubMed] [Google Scholar]
- Nakamura Y, Kawai S, Yukuhiro F, Ito S, Gotoh T, Kisimoto R, Yanase T, Matsumoto Y, Kageyama D, Noda H. Prevalence of Cardinium bacteria in planthoppers and spider mites and taxonomic revision of "Candidatus Cardinium hertigii" based on detection of a new Cardinium group from biting midges. App Environ Microbiol. 2009;75:6757–6763. doi: 10.1128/AEM.01583-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baldo L, Ayoub NA, Hayashi CY, Russell JA, Stahlhut JK, Werren JH. Insight into the routes of Wolbachia invasion: high levels of horizontal transfer in the spider genus Agelenopsis revealed by Wolbachia strain and mitochondrial DNA diversity. Mol Ecol. 2008;17:557–569. doi: 10.1111/j.1365-294X.2007.03608.x. [DOI] [PubMed] [Google Scholar]
- Cordaux R, Michel-Salzat A, Bouchon D. Wolbachia infection in crustaceans: novel hosts and potential routes for horizontal transmission. J Evol Biol. 2001;14:237–243. doi: 10.1046/j.1420-9101.2001.00279.x. [DOI] [Google Scholar]
- Jeong G, Lee K, Choi J, Hwang S, Park B, Kim W, Choi Y, Park I, Kim J. Incidence of Wolbachia and Cardinium endosymbionts in the Osmia community in Korea. J Microbiol. 2009;47:28–32. doi: 10.1007/s12275-009-0198-3. [DOI] [PubMed] [Google Scholar]
- Lachowska D, Kajtoch L, Knutelski S. Occurrence of Wolbachia in central European weevils: correlations with host systematics, ecology, and biology. Ent Exp Appl. 2010;135:105–118. doi: 10.1111/j.1570-7458.2010.00974.x. [DOI] [Google Scholar]
- Stahlhut JK, Desjardins CA, Clark ME, Baldo L, Russell JA, Werren JH, Jaenike J. The mushroom habitat as an ecological arena for global exchange of Wolbachia. Mol Ecol. 2010;19:1940–1952. doi: 10.1111/j.1365-294X.2010.04572.x. [DOI] [PubMed] [Google Scholar]
- van Meer MMM, Witteveldt J, Stouthamer R. Phylogeny of the arthropod endosymbiont Wolbachia based on the wsp gene. Insect Mol Biol. 1999;8:399–408. doi: 10.1046/j.1365-2583.1999.83129.x. [DOI] [PubMed] [Google Scholar]
- Vavre F, Fleury F, Lepetit D, Fouillet P, Bouletreau M. Phylogenetic evidence for horizontal transmission of Wolbachia in host-parasitoid associations. Mol Biol Evol. 1999;16:1711–1723. doi: 10.1093/oxfordjournals.molbev.a026084. [DOI] [PubMed] [Google Scholar]
- Werren JH, Zhang W, Guo LR. Evolution and phylogeny of Wolbachia -reproductive parasites of arthropods. Proc Roy Soc Lond B. 1995;261:55–63. doi: 10.1098/rspb.1995.0117. [DOI] [PubMed] [Google Scholar]
- Heath BD, Butcher RDJ, Whitfield WGF, Hubbard SF. Horizontal transfer of Wolbachia between phylogenetically distant insect species by a naturally occurring mechanism. Curr Biol. 1999;9:313–316. doi: 10.1016/S0960-9822(99)80139-0. [DOI] [PubMed] [Google Scholar]
- Huigens ME, Luck RF, Klaassen RHG, Maas MFPM, Timmermans MJTN, Stouthamer R. Infectious parthenogenesis. Nature. 2000;405:178–179. doi: 10.1038/35012066. [DOI] [PubMed] [Google Scholar]
- Huigens ME, de Almeida RP, Boons PAH, Luck RF, Stouthamer R. Natural interspecific and intraspecific horizontal transfer of parthenogenesis-inducing Wolbachia in Trichogramma wasps. Proc Roy Soc Lond B. 2004;271:509–515. doi: 10.1098/rspb.2003.2640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishmael N, Dunning Hotopp JC, Ioannidis P, Biber S, Sakamoto J, Siozios S, Nene V, Werren J, Bourtzis K, Bordenstein SR, Tettelin H. Extensive genomic diversity of closely related Wolbachia strains. Microbiology. 2009;155:2211–2222. doi: 10.1099/mic.0.027581-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baldo L, Bordenstein S, Wernegreen JJ, Werren JH. Widespread recombination throughout Wolbachia genomes. Mol Biol Evol. 2006;23:437–449. doi: 10.1093/molbev/msj049. [DOI] [PubMed] [Google Scholar]
- Jiggins FM, von der Schulenburg JHG, Hurst GDD, Majerus MEN. Recombination confounds interpretations of Wolbachia evolution. Proc Roy Soc Lond B. 2001;268:1423–1427. doi: 10.1098/rspb.2001.1656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Werren JH, Bartos JD. Recombination in Wolbachia. Curr Biol. 2001;11:431–435. doi: 10.1016/S0960-9822(01)00101-4. [DOI] [PubMed] [Google Scholar]
- Baldo L, Lo N, Werren JH. Mosaic nature of the Wolbachia surface protein. J Bacteriol. 2005;187:5406–5418. doi: 10.1128/JB.187.15.5406-5418.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiggins FM. The rate of recombination in Wolbachia bacteria. Mol Biol Evol. 2002;19:1640–1643. doi: 10.1093/oxfordjournals.molbev.a004228. [DOI] [PubMed] [Google Scholar]
- Keller GP, Windsor DM, Saucedo JM, Werren JH. Reproductive effects and geographical distributions of two Wolbachia strains infecting the Neotropical beetle, Chelymorpha alternans Boh. (Chrysomelidae, Cassidinae) Mol Ecol. 2004;13:2405–2420. doi: 10.1111/j.1365-294X.2004.02213.x. [DOI] [PubMed] [Google Scholar]
- Malloch G, Fenton B. Super-infections of Wolbachia in byturid beetles and evidence for genetic transfer between A and B super-groups of Wolbachia. Mol Ecol. 2005;14:627–637. doi: 10.1111/j.1365-294X.2005.02432.x. [DOI] [PubMed] [Google Scholar]
- Reuter M, Keller L. High levels of multiple Wolbachia infection and recombination in the ant Formica exsecta. Mol Biol Evol. 2003;20:748–753. doi: 10.1093/molbev/msg082. [DOI] [PubMed] [Google Scholar]
- Klasson L, Westberg J, Sapountzis P, Nasiund K, Lutnaes Y, Darby AC, Veneti Z, Chen LM, Braig HR, Garrett R, Bourtzis K, Andersson SGE. The mosaic genome structure of the Wolbachia wRi strain infecting Drosophila simulans. PNAS. 2009;106:5725–5730. doi: 10.1073/pnas.0810753106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frost CL, Fernández-Marín H, Smith JE, Hughes WO. Multiple gain and losses of Wolbachia symbionts across a tribe of fungus-growing ants. Mol Ecol. 2010;19:4077–4085. doi: 10.1111/j.1365-294X.2010.04764.x. [DOI] [PubMed] [Google Scholar]
- Jiggins FM, Bentley JK, Majerus MEN, Hurst GDD. How many species are infected with Wolbachia? Cryptic sex ratio distorters revealed to be common by intensive sampling. Proc Roy Soc Lond B. 2001;268:1123–1126. doi: 10.1098/rspb.2001.1632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verne S, Johnson M, Bouchon D, Grandjean F. Evidence for recombination between feminizing Wolbachia in the isopod genus Armadillidium. Gene. 2007;397:58–66. doi: 10.1016/j.gene.2007.04.006. [DOI] [PubMed] [Google Scholar]
- Feil EJ, Maiden MCJ, Achtman M, Spratt BG. The relative contributions of recombination and mutation to the divergence of clones of Neisseria meningitidis. Mol Biol Evol. 1999;16:1496–1502. doi: 10.1093/oxfordjournals.molbev.a026061. [DOI] [PubMed] [Google Scholar]
- Ros VID, Breeuwer JAJ. The effects of, and interactions between, Cardinium and Wolbachia in the doubly infected spider mite Bryobia sarothamni. Heredity. 2009;102:413–422. doi: 10.1038/hdy.2009.4. [DOI] [PubMed] [Google Scholar]
- Weeks AR, Breeuwer JAJ. Wolbachia-induced parthenogenesis in a genus of phytophagous mites. Proc Roy Soc Lond B. 2001;268:2245–2251. doi: 10.1098/rspb.2001.1797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ros VID, Breeuwer JAJ, Menken SBJ. Origins of asexuality in Bryobia mites (Acari: Tetranychidae) BMC Evol Biol. 2008;8:153. doi: 10.1186/1471-2148-8-153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahrens ME, Shoemaker D. Evolutionary history of Wolbachia infections in the fire ant Solenopsis invicta. BMC Evol Biol. 2005;5:35. doi: 10.1186/1471-2148-5-35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dean MD, Ballard JWO. High divergence among Drosophila simulans mitochondrial haplogroups arose in midst of long term purifying selection. Mol Phyl Evol. 2005;36:328–337. doi: 10.1016/j.ympev.2005.02.016. [DOI] [PubMed] [Google Scholar]
- Hurst GDD, Jiggins FM. Problems with mitochondrial DNA as a marker in population, phylogeographic and phylogenetic studies: the effects of inherited symbionts. Proc Roy Soc Lond B. 2005;272:1525–1534. doi: 10.1098/rspb.2005.3056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rasgon JL, Cornel AJ, Scott TW. Evolutionary history of a mosquito endosymbiont revealed through mitochondrial hitchhiking. Proc Roy Soc Lond B. 2006;273:1603–1611. doi: 10.1098/rspb.2006.3493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ros VID, Breeuwer JAJ. Spider mite (Acari: Tetranychidae) mitochondrial COI phylogeny reviewed: host plant relationships, phylogeography, reproductive parasites and barcoding. Exp Appl Acarol. 2007;42:239–262. doi: 10.1007/s10493-007-9092-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamas I, Klasson L, Canback B, Naslund AK, Eriksson AS, Wernegreen JJ, Sandstrom JP, Moran NA, Andersson SGE. 50 million years of genomic stasis in endosymbiotic bacteria. Science. 2002;296:2376–2379. doi: 10.1126/science.1071278. [DOI] [PubMed] [Google Scholar]
- Wernegreen JJ. Endosymbiosis: Lessons in conflict resolution. PLoS Biol. 2004;2:307–311. doi: 10.1371/journal.pbio.0020307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feil EJ, Enright MC, Spratt BG. Estimating the relative contributions of mutation and recombination to clonal diversification: a comparison between Neisseria meningitidis and Streptococcus pneumoniae. Res Microbiol. 2000;151:465–469. doi: 10.1016/S0923-2508(00)00168-6. [DOI] [PubMed] [Google Scholar]
- Charlat S, Mercot H. Did Wolbachia cross the border? Trends Ecol Evol. 2001;16:540–541. [Google Scholar]
- Arthofer W, Riegler M, Schneider D, Krammer M, Miller WJ, Stauffer C. Hidden Wolbachia diversity in field populations of the European cherry fruit fly, Rhagoletis cerasi (Diptera, Tephritidae) Mol Ecol. 2009;18:3816–3830. doi: 10.1111/j.1365-294X.2009.04321.x. [DOI] [PubMed] [Google Scholar]
- Bordenstein SR, Wernegreen JJ. Bacteriophage flux in endosymbionts (Wolbachia): Infection frequency, lateral transfer, and recombination rates. Mol Biol Evol. 2004;21:1981–1991. doi: 10.1093/molbev/msh211. [DOI] [PubMed] [Google Scholar]
- Gavotte L, Henri H, Stouthamer R, Charif D, Charlat S, Bouletreau M, Vavre F. A survey of the bacteriophage WO in the endosymbiotic bacteria Wolbachia. Mol Biol Evol. 2007;24:427–435. doi: 10.1093/molbev/msl171. [DOI] [PubMed] [Google Scholar]
- Masui S, Kamoda S, Sasaki T, Ishikawa H. Distribution and evolution of bacteriophage WO in Wolbachia, the endosymbiont causing sexual alterations in arthropods. J Mol Evol. 2000;51:491–497. doi: 10.1007/s002390010112. [DOI] [PubMed] [Google Scholar]
- Kent BN, Salichos L, Gibbons JG, Rokas A, Newton IL, Clark ME, Bordenstein SR. Complete bacteriophage transfer in a bacterial endosymbiont (Wolbachia) determined by targeted genome capture. Genome Biol Evol. 2011;3:209–218. doi: 10.1093/gbe/evr007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chafee ME, Funk DJ, Harrison RG, Bordenstein SR. Lateral phage transfer in obligate intracellular bacteria (Wolbachia): Verification from natural populations. Mol Biol Evol. 2010;27:501–505. doi: 10.1093/molbev/msp275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujii Y, Kubo T, Ishikawa H, Sasaki T. Isolation and characterization of the bacteriophage WO from Wolbachia, an arthropod endosymbiont. Biochem Biophys Res Commun. 2004;317:1183–1188. doi: 10.1016/j.bbrc.2004.03.164. [DOI] [PubMed] [Google Scholar]
- Breeuwer JAJ. Wolbachia and cytoplasmic incompatibility in the spider mites Tetranychus urticae and T. turkestani. Heredity. 1997;79:41–47. doi: 10.1038/hdy.1997.121. [DOI] [Google Scholar]
- Gotoh T, Noda H, Hong XY. Wolbachia distribution and cytoplasmic incompatibility based on a survey of 42 spider mite species (Acari: Tetranychidae) in Japan. Heredity. 2003;91:208–216. doi: 10.1038/sj.hdy.6800329. [DOI] [PubMed] [Google Scholar]
- Gotoh T, Sugasawa J, Noda H, Kitashima Y. Wolbachia-induced cytoplasmic incompatibility in Japanese populations of Tetranychus urticae (Acari: Tetranychidae) Exp Appl Acarol. 2007;42:1–16. doi: 10.1007/s10493-007-9072-3. [DOI] [PubMed] [Google Scholar]
- Vala F, Breeuwer JAJ, Sabelis MW. Wolbachia-induced 'hybrid breakdown' in the two-spotted spider mite Tetranychus urticae Koch. Proc Roy Soc Lond B. 2000;267:1931–1937. doi: 10.1098/rspb.2000.1232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braig HR, Zhou WG, Dobson SL, O'Neill SL. Cloning and characterization of a gene encoding the major surface protein of the bacterial endosymbiont Wolbachia pipientis. J Bacteriol. 1998;180:2373–2378. doi: 10.1128/jb.180.9.2373-2378.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andersson SGE, Zomorodipour A, Andersson JO, Sicheritz-Ponten T, Alsmark UCM, Podowski RM, Näslund AK, Eriksson AS, Winkler HH, Kurland CG. The genome sequence of Rickettsia prowazekii and the origin of mitochondria. Nature. 1998;396:133–140. doi: 10.1038/24094. [DOI] [PubMed] [Google Scholar]
- McLeod MP, Qin X, Karpathy SE, Gioia J, Highlander SK, Fox GE, McNeill TZ, Jiang HY, Muzny D, Jacob LS, Hawes AC, Sodergren E, Gill R, Hume J, Morgan M, Fan GW, Amin AG, Gibbs RA, Hong C, Yu XJ, Walker DH, Weinstock GM. Complete genome sequence of Rickettsia typhi and comparison with sequences of other rickettsiae. J Bacteriol. 2004;186:5842–5855. doi: 10.1128/JB.186.17.5842-5855.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu M, Sun LV, Vamathevan J, Riegler M, Deboy R, Brownlie JC, McGraw EA, Martin W, Esser C, Ahmadinejad N, Wiegand C, Madupu R, Beanan MJ, Brinkac LM, Daugherty SC, Durkin AS, Kolonay JF, Nelson WC, Mohamoud Y, Lee P, Berry K, Young MB, Utterback T, Weidman J, Merman WC, Paulsen IT, Nelson KE, Tettelin H, O'Neill SL, Eisen JA. Phylogenomics of the reproductive parasite Wolbachia pipientis wMel: A streamlined genome overrun by mobile genetic elements. PLoS Biol. 2004;2:327–341. doi: 10.1371/journal.pbio.0020327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Groot TVM, Breeuwer JAJ. Cardinium symbionts induce haploid thelytoky in most clones of three closely related Brevipalpus species. Exp Appl Acarol. 2006;39:257–271. doi: 10.1007/s10493-006-9019-0. [DOI] [PubMed] [Google Scholar]
- Boom R, Sol CJA, Salimans MMM, Jansen CL, Wertheim-van Dillen PME, Van der Noordaa J. Rapid and simple method for purification of nucleic-acids. J Clin Microbiol. 1990;28:495–503. doi: 10.1128/jcm.28.3.495-503.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sambrook J, Fritsch EF, Maniatis T. Molecular cloning. Cold Spring Harbor, New York: Cold Spring Harbor Press; 1989. [Google Scholar]
- Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG. The CLUSTAL_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 1997;25:4876–4882. doi: 10.1093/nar/25.24.4876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall TA. BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucl Acids Symp Ser. 1999;41:95–98. [Google Scholar]
- Tamura K, Dudley J, Nei M, Kumar S. MEGA4: Molecular evolutionary genetics analysis (MEGA) software version 4.0. Mol Biol Evol. 2007;24:1596–1599. doi: 10.1093/molbev/msm092. [DOI] [PubMed] [Google Scholar]
- Korber B. In: Computational and evolutionary analysis of HIV molecular sequences. Rodrigo AG, Learn GH, editor. Dordrecht, Netherlands: Kluwer Academic Publishers; 2000. HIV sequence signatures and similarities; pp. 55–72. [Google Scholar]
- Kosakovsky Pond SL, Frost SDW, Muse SV. HyPhy: hypothesis testing using phylogenies. Bioinformatics. 2005;21:676–679. doi: 10.1093/bioinformatics/bti079. [DOI] [PubMed] [Google Scholar]
- Swofford DL. PAUP*, Phylogenetic Analysis Using Parsimony (*and Other Methods) Sunderland, MA: Sinauer Associates; 2002. [Google Scholar]
- Swofford DL, Sullivan J. In: The phylogenetic handbook A practical approach to DNA and protein phylogeny. Salemi M, Vandamme A-M, editor. Cambridge: Cambridge University Press; 2003. Phylogeny inference based on parsimony and other methods using PAUP*; pp. 160–206. [Google Scholar]
- Akaike H. New look at statistical-model identification. IEEE Autom Contr. 1974;19:716–723. doi: 10.1109/TAC.1974.1100705. [DOI] [Google Scholar]
- Shapiro B, Rambaut A, Drummond AJ. Choosing appropriate substitution models for the phylogenetic analysis of protein-coding sequences. Mol Biol Evol. 2006;23:7–9. doi: 10.1093/molbev/msj021. [DOI] [PubMed] [Google Scholar]
- Sullivan J, Abdo Z, Joyce P, Swofford DL. Evaluating the performance of a successive-approximations approach to parameter optimization in maximum-likelihood phylogeny estimation. Mol Biol Evol. 2005;22:1386–1392. doi: 10.1093/molbev/msi129. [DOI] [PubMed] [Google Scholar]
- Ronquist F, Huelsenbeck JP. MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics. 2003;19:1572–1574. doi: 10.1093/bioinformatics/btg180. [DOI] [PubMed] [Google Scholar]
- Wilgenbusch JC, Warren DL, Swofford DL. AWTY: A system for graphical exploration of MCMC convergence in Bayesian phylogenetic inference. 2004. http://ceb.csit.fsu.edu/awty [DOI] [PubMed]
- Maiden MCJ, Bygraves JA, Feil E, Morelli G, Russell JE, Urwin R, Zhang Q, Zhou JJ, Zurth K, Caugant DA, Feavers IM, Acthman M, Spratt BG. Multilocus sequence typing: A portable approach to the identification of clones within populations of pathogenic microorganisms. PNAS. 1998;95:3140–3145. doi: 10.1073/pnas.95.6.3140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feil EJ, Li BC, Aanensen DM, Hanage WP, Spratt BG. eBURST: Inferring patterns of evolutionary descent among clusters of related bacterial genotypes from multilocus sequence typing data. J Bacteriol. 2004;186:1518–1530. doi: 10.1128/JB.186.5.1518-1530.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin DP, Williamson C, Posada D. RDP2: recombination detection and analysis from sequence alignments. Bioinformatics. 2005;21:260–262. doi: 10.1093/bioinformatics/bth490. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
List of tetranychid samples in which Wolbachia and/or Cardinium strains were detected.
Allelic profiles for each of the 37 unique Wolbachia STs.
Wolbachia gene phylogenies (wsp, ftsZ, groEL, and trmD).
GenBank accession numbers.