Abstract
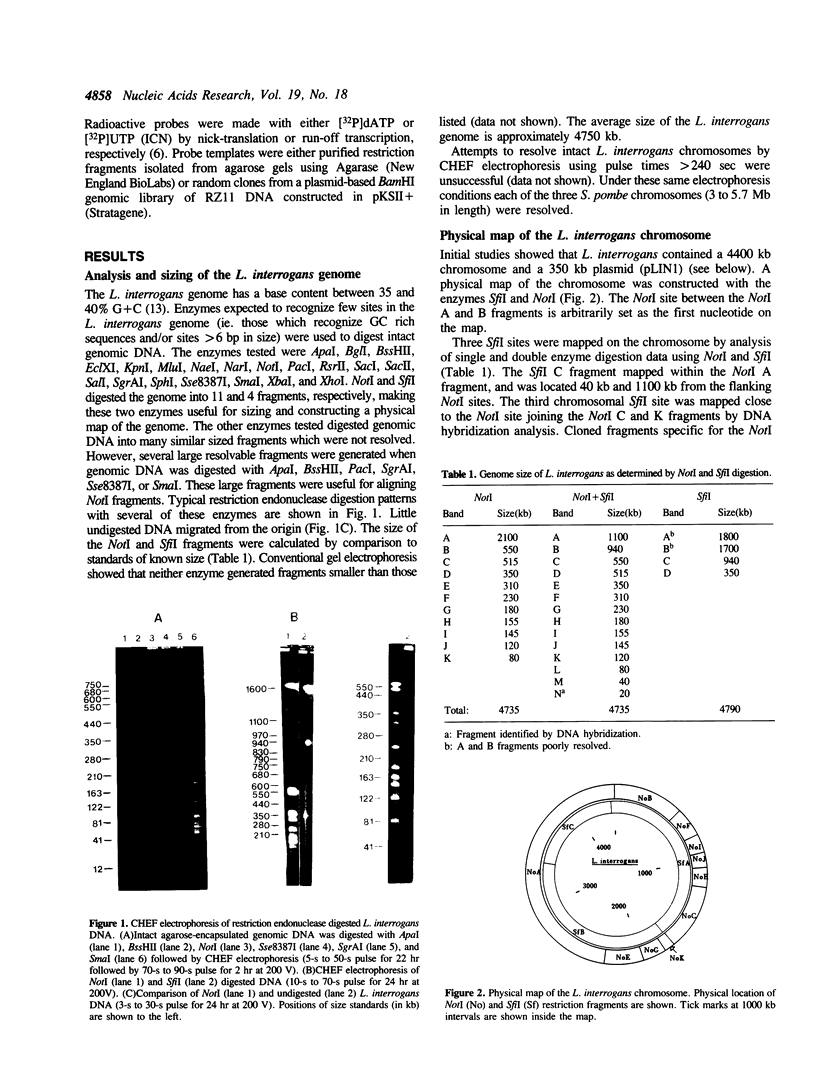
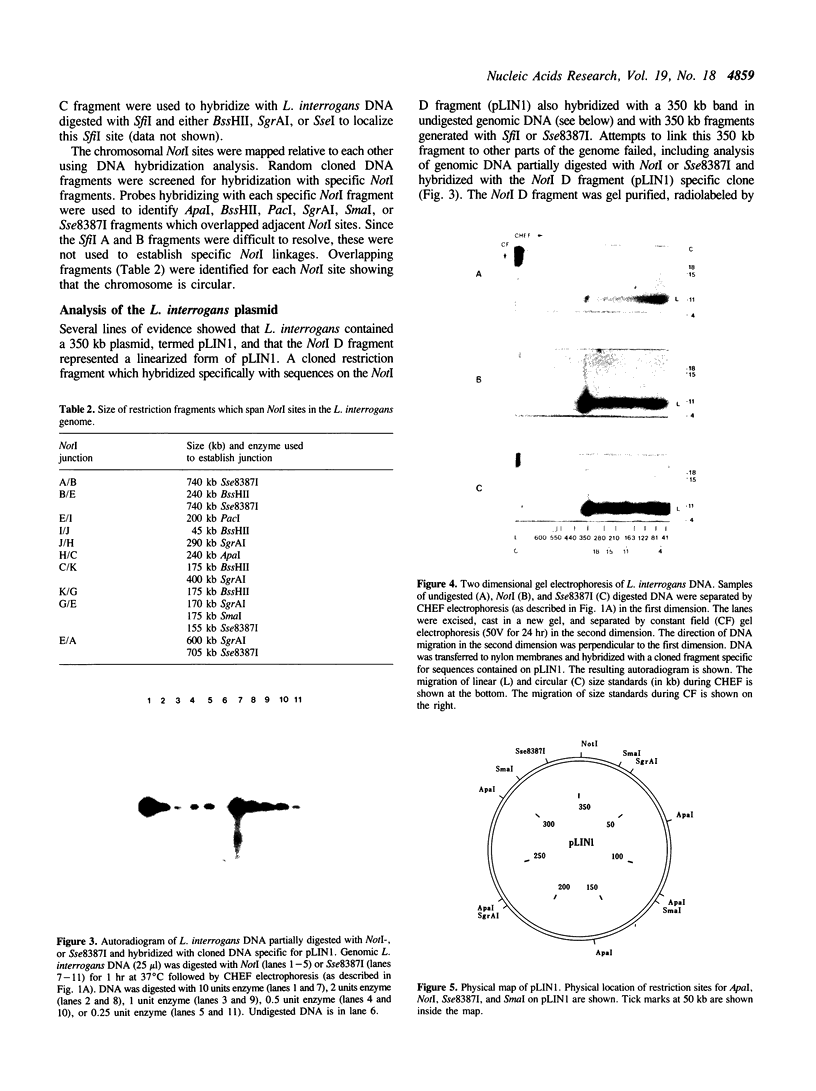
The size and physical structure of the Leptospira interrogans genome was characterized using contour-clamped homogenous electric field (CHEF) gel electrophoresis. The L. interrogans genome is approximately 4750 kb in size and is composed of two molecular species of DNA: a 4400 kb chromosome; and a 350 kb plasmid, pLIN1. A physical map of the chromosome was constructed with the restriction enzymes NotI and SfiI. A physical map of pLIN1 was constructed with ApaI, NotI, Sse83871, SgrAI, and SmaI. Both the L. interrogans chromosome and pLIN1 are circular.
Full text
PDF

Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Baril C., Richaud C., Baranton G., Saint Girons I. S. Linear chromosome of Borrelia burgdorferi. Res Microbiol. 1989 Oct;140(8):507–516. doi: 10.1016/0923-2508(89)90083-1. [DOI] [PubMed] [Google Scholar]
- Baril C., Saint Girons I. Sizing of the Leptospira genome by pulsed-field agarose gel electrophoresis. FEMS Microbiol Lett. 1990 Sep 1;59(1-2):95–99. doi: 10.1016/0378-1097(90)90038-r. [DOI] [PubMed] [Google Scholar]
- Chu G., Vollrath D., Davis R. W. Separation of large DNA molecules by contour-clamped homogeneous electric fields. Science. 1986 Dec 19;234(4783):1582–1585. doi: 10.1126/science.3538420. [DOI] [PubMed] [Google Scholar]
- Ferdows M. S., Barbour A. G. Megabase-sized linear DNA in the bacterium Borrelia burgdorferi, the Lyme disease agent. Proc Natl Acad Sci U S A. 1989 Aug;86(15):5969–5973. doi: 10.1073/pnas.86.15.5969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fox G. E., Stackebrandt E., Hespell R. B., Gibson J., Maniloff J., Dyer T. A., Wolfe R. S., Balch W. E., Tanner R. S., Magrum L. J. The phylogeny of prokaryotes. Science. 1980 Jul 25;209(4455):457–463. doi: 10.1126/science.6771870. [DOI] [PubMed] [Google Scholar]
- Haapala D. K., Rogul M., Evans L. B., Alexander A. D. Deoxyribonucleic acid base composition and homology studies of Leptospira. J Bacteriol. 1969 May;98(2):421–428. doi: 10.1128/jb.98.2.421-428.1969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kauc L., Mitchell M., Goodgal S. H. Size and physical map of the chromosome of Haemophilus influenzae. J Bacteriol. 1989 May;171(5):2474–2479. doi: 10.1128/jb.171.5.2474-2479.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levene S. D., Zimm B. H. Separations of open-circular DNA using pulsed-field electrophoresis. Proc Natl Acad Sci U S A. 1987 Jun;84(12):4054–4057. doi: 10.1073/pnas.84.12.4054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plasterk R. H., Simon M. I., Barbour A. G. Transposition of structural genes to an expression sequence on a linear plasmid causes antigenic variation in the bacterium Borrelia hermsii. Nature. 1985 Nov 21;318(6043):257–263. doi: 10.1038/318257a0. [DOI] [PubMed] [Google Scholar]
- Schwartz D. C., Cantor C. R. Separation of yeast chromosome-sized DNAs by pulsed field gradient gel electrophoresis. Cell. 1984 May;37(1):67–75. doi: 10.1016/0092-8674(84)90301-5. [DOI] [PubMed] [Google Scholar]
- Serwer P., Hayes S. J. Atypical sieving of open circular DNA during pulsed field agarose gel electrophoresis. Biochemistry. 1989 Jul 11;28(14):5827–5832. doi: 10.1021/bi00440a019. [DOI] [PubMed] [Google Scholar]
- Taylor K. A., Barbour A. G., Thomas D. D. Pulsed-field gel electrophoretic analysis of leptospiral DNA. Infect Immun. 1991 Jan;59(1):323–329. doi: 10.1128/iai.59.1.323-329.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker E. M., Arnett J. K., Heath J. D., Norris S. J. Treponema pallidum subsp. pallidum has a single, circular chromosome with a size of approximately 900 kilobase pairs. Infect Immun. 1991 Jul;59(7):2476–2479. doi: 10.1128/iai.59.7.2476-2479.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuerner R. L., Bolin C. A. Nucleic acid probe characterizes Leptospira interrogans serovars by restriction fragment length polymorphisms. Vet Microbiol. 1990 Sep;24(3-4):355–366. doi: 10.1016/0378-1135(90)90183-v. [DOI] [PubMed] [Google Scholar]
- Zuerner R. L., Bolin C. A. Repetitive sequence element cloned from Leptospira interrogans serovar hardjo type hardjo-bovis provides a sensitive diagnostic probe for bovine leptospirosis. J Clin Microbiol. 1988 Dec;26(12):2495–2500. doi: 10.1128/jcm.26.12.2495-2500.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]