Abstract
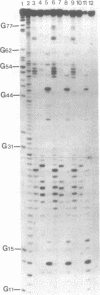
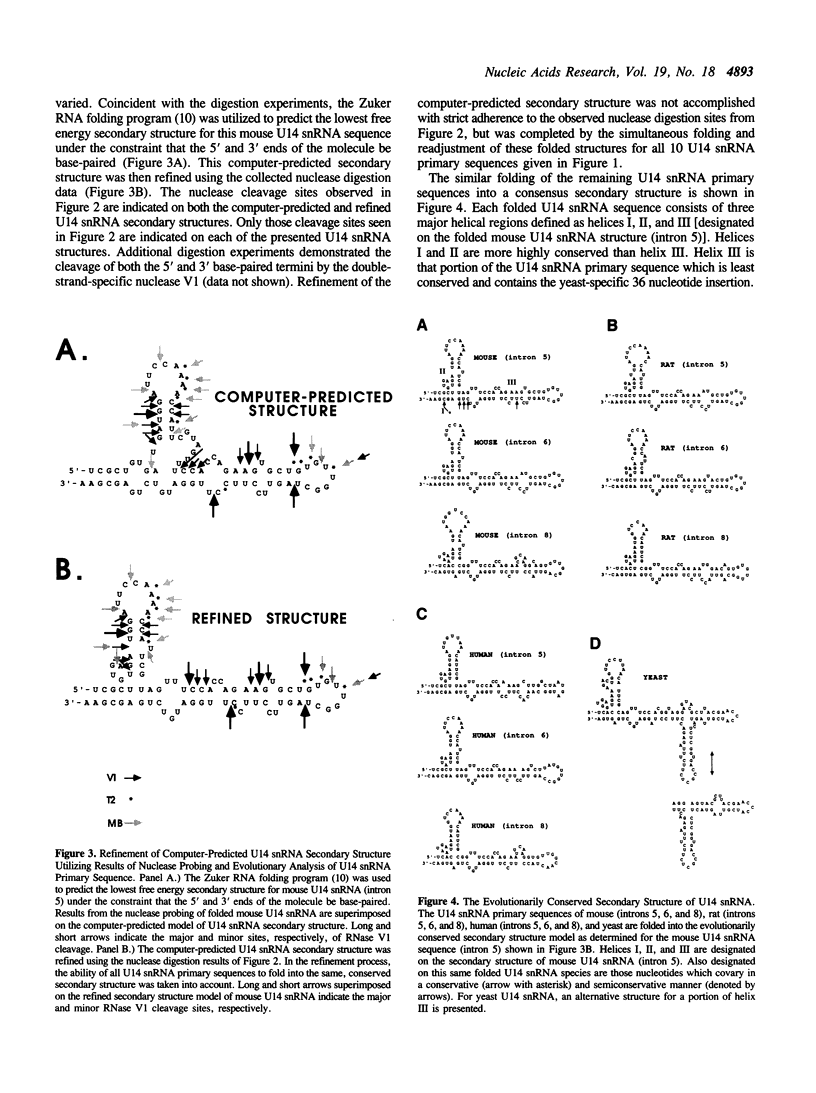
U14 snRNA is a small nuclear RNA that plays a role in the processing of eukaryotic ribosomal RNA. We have investigated the folded structure of this snRNA species using comparative analysis of evolutionarily diverse U14 snRNA primary sequences coupled with nuclease digestion analysis of mouse U14 snRNA. Covariant nucleotide analysis of aligned mouse, rat, human, and yeast U14 snRNA primary sequences suggested a basic folding pattern in which the 5' and 3' termini of all U14 snRNAs were base-paired. Subsequent digestion of mouse U14 snRNA with mung bean (single-strand-specific), T2 (single-strand-preferential), and V1 (double-strand-specific) nucleases defined the major and minor cleavage sites for each nuclease. This digestion data was then utilized in concert with the comparative sequence analysis of aligned U14 snRNA primary sequences to refine the secondary structure model suggested by computer-predicted folding. The proposed secondary structure of U14 snRNA is comprised of three major hairpin/helical regions which includes the helix of base-paired 5' and 3' termini. Strict and semiconservative covariation of specific base-pairs within two of the three major helices, as well as nucleotide changes that strengthen or extend base-paired regions, support this folded conformation as the evolutionary conserved secondary structure for U14 snRNA.
Full text
PDF


Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Jaeger J. A., Turner D. H., Zuker M. Predicting optimal and suboptimal secondary structure for RNA. Methods Enzymol. 1990;183:281–306. doi: 10.1016/0076-6879(90)83019-6. [DOI] [PubMed] [Google Scholar]
- Kass S., Tyc K., Steitz J. A., Sollner-Webb B. The U3 small nucleolar ribonucleoprotein functions in the first step of preribosomal RNA processing. Cell. 1990 Mar 23;60(6):897–908. doi: 10.1016/0092-8674(90)90338-f. [DOI] [PubMed] [Google Scholar]
- Li H. D., Zagorski J., Fournier M. J. Depletion of U14 small nuclear RNA (snR128) disrupts production of 18S rRNA in Saccharomyces cerevisiae. Mol Cell Biol. 1990 Mar;10(3):1145–1152. doi: 10.1128/mcb.10.3.1145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J., Maxwell E. S. Mouse U14 snRNA is encoded in an intron of the mouse cognate hsc70 heat shock gene. Nucleic Acids Res. 1990 Nov 25;18(22):6565–6571. doi: 10.1093/nar/18.22.6565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maxwell E. S., Martin T. E. A low-molecular-weight RNA from mouse ascites cells that hybridizes to both 18S rRNA and mRNA sequences. Proc Natl Acad Sci U S A. 1986 Oct;83(19):7261–7265. doi: 10.1073/pnas.83.19.7261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Padgett R. A., Grabowski P. J., Konarska M. M., Seiler S., Sharp P. A. Splicing of messenger RNA precursors. Annu Rev Biochem. 1986;55:1119–1150. doi: 10.1146/annurev.bi.55.070186.005351. [DOI] [PubMed] [Google Scholar]
- Sarge K. D., Maxwell E. S. Intermolecular hybridization of 5S rRNA with 18S rRNA: identification of a 5'-terminally-located nucleotide sequence in mouse 5S rRNA which base-pairs with two specific complementary sequences in 18S rRNA. Biochim Biophys Acta. 1991 Jan 17;1088(1):57–70. doi: 10.1016/0167-4781(91)90153-d. [DOI] [PubMed] [Google Scholar]
- Trinh-Rohlik Q., Maxwell E. S. Homologous genes for mouse 4.5S hybRNA are found in all eukaryotes and their low molecular weight RNA transcripts intermolecularly hybridize with eukaryotic 18S ribosomal RNAs. Nucleic Acids Res. 1988 Jul 11;16(13):6041–6056. doi: 10.1093/nar/16.13.6041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tyc K., Steitz J. A. U3, U8 and U13 comprise a new class of mammalian snRNPs localized in the cell nucleolus. EMBO J. 1989 Oct;8(10):3113–3119. doi: 10.1002/j.1460-2075.1989.tb08463.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zagorski J., Tollervey D., Fournier M. J. Characterization of an SNR gene locus in Saccharomyces cerevisiae that specifies both dispensible and essential small nuclear RNAs. Mol Cell Biol. 1988 Aug;8(8):3282–3290. doi: 10.1128/mcb.8.8.3282. [DOI] [PMC free article] [PubMed] [Google Scholar]