Abstract
Protein kinases are essential signaling molecules with a characteristic bilobal shape that has been studied for over fifteen years. Despite the number of crystal structures available, little study has been directed away from the prototypical functional elements of the kinase domain. We have performed a structural alignment of thirteen active-conformation kinases and discovered the presence of six water molecules that occur in conserved locations across this group of diverse kinases. Molecular dynamics simulations demonstrated that these waters confer a great deal of stability to their local environment and to a key catalytic residue. Our results highlight the importance of novel elements within the greater kinase family and suggest that conserved water molecules are necessary for efficient kinase function.
Keywords: free energy, hydrogen bonding, molecular dynamics, protein stability, structural alignment
Introduction
In protein kinases, phosphotransfer capability requires at least two well defined conserved elements in addition to the polypeptide: an active site metal cofactor, usually magnesium or manganese, and ATP. The only element found in crystallography data with potential conservation between kinases that has not been examined in detail is water. It was first noted by Shaltiel, Cox and Taylor1 that different crystal structures of protein kinase A (PKA) had water molecules in virtually identical locations. They noted several waters within the active site, interacting with both conserved and non-conserved residues, as well as ATP and the active site manganese, and the authors suggested a critical role for these waters in the efficient function of PKA.
That water molecules play critical roles in enzymes/proteins has been known for some time. Condensation and hydrolysis reactions are two clear examples of water functioning as a crucial molecular element – a product or reactant. In chemical reactions water molecules can also function as transition state intermediates. Another role for water at the molecular level is as a structural element, interconnecting the protein through hydrogen bonds and maintaining/stabilizing the positions of residues and/or the fold.2 These structural waters are often conserved within a protein family, i.e. they perform the same functions and occur in nearly identical three-dimensional locations with reference to their associated structures. Structurally conserved waters have been found in several classes of proteins, including ribonuclease T1,3,4 serine proteases,5 Rossmann fold dinucleotide binding proteins,6 MHC class 1 proteins7 and aspartic proteinases.8 In these studies, conserved waters are often found in or near an enzyme's active site, suggesting they play an important function in active site stability, flexibility, ligand coordination and residue positioning, hence their evolutionary conservation. Rodríguez-Almazán and colleagues9 have shown recently how even a conservative amino acid change can disrupt conserved water molecules and the connective networks they form, drastically altering enzyme function.
Protein kinases can be divided into two interdependent functional regions: the ATP binding/phosphotransfer region, and the second where substrate binding occurs. These regions are obviously interconnected but they can be discussed separately. The area where ATP is bound, and the tertiary phosphate positioned for transfer, is bordered principally by the 5-stranded β sheet, helix C, the magnesium-positioning loop and the catalytic loop, with the area between them forming the active site (Figure 1). It is in the immediate area of the transferable phosphate where the greatest conservation is found.10,11 Six fully conserved residues play very important roles in positioning this key element, all of which are absolutely critical for efficient function.12 The ATP-positioning lysine is located on β–strand 3 and interacts with the α- and β- phosphates of ATP, and this lysine is critically stabilized by a glutamic acid originating from helix C. The catalytic aspartic acid that initiates phosphotransfer is located on the catalytic loop directly beneath the γ-phosphate. The γ-phosphate itself is directly and indirectly coordinated by three residues and one or two divalent cations. The magnesium-positioning aspartic acid arises from the magnesium-positioning loop and indirectly positions the γ-phosphate through a magnesium or manganese ion. Similarly, an asparagine residue located on the catalytic loop also indirectly positions this phosphate through a secondary metal ion. And the last fully conserved residue is a lysine located just downstream of the catalytic residue that directly aids in positioning the γ-phosphate, although this residue is exchanged for an arginine in tyrosine kinases. While the area of phosphotransfer is relatively well-studied and understood, in contrast the substrate binding region is not. Substrates are bound within a cleft formed from the catalytic loop, the P+1 loop, helix D, and parts of helices F and H. There is little residue conservation within this area,11 which is entirely consistent with its function in substrate binding. However, understanding this area is critical as it mediates substrate binding and contains the information that confers substrate specificity. In neither the substrate binding groove, nor the well-studied active site, has the role of water molecules and the conserved functional roles they might play been examined.
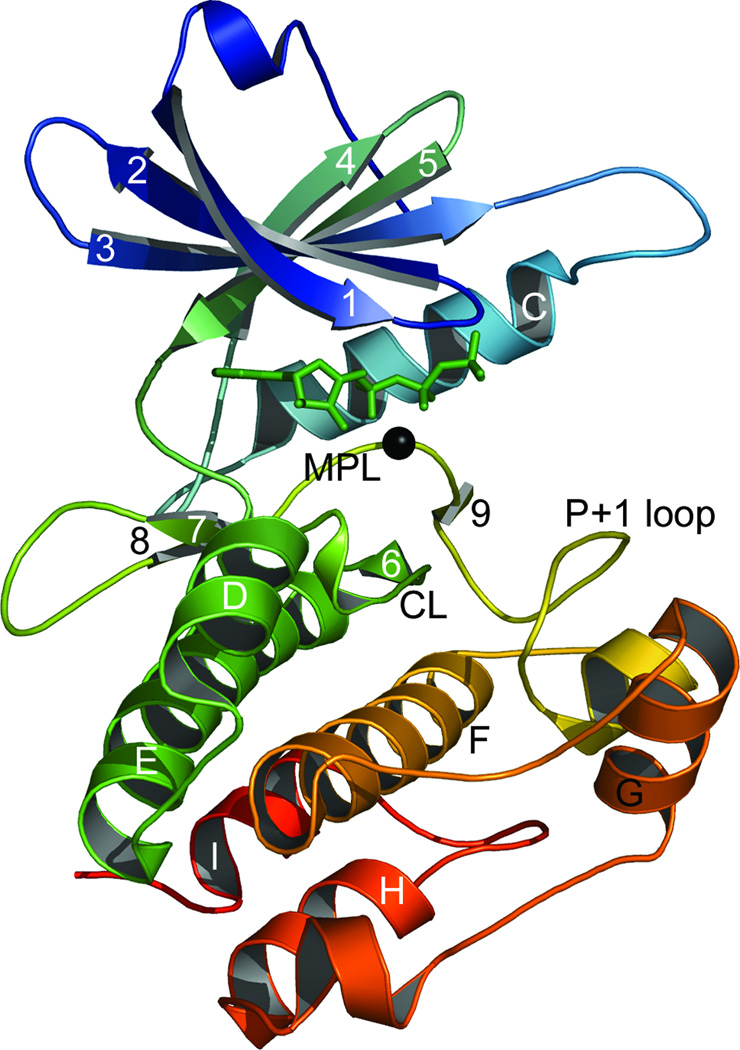
Figure 1.
The crystal structure of the active conformation death-associated protein kinase.24 β-strands, helices and prominent loops are labelled in accordance with Knighton, et al.25 The non-hydrolyzable ATP analog ANP is shown in green sticks and manganese ion as a black sphere. The small lobe comprises the β-sheet, helix C and the interveening loops. The remainder of the protein, below ANP, constitutes the large lobe. CL: catalytic loop; MPL: magnesium positioning loop.
Here we report on an examination of water conservation across a diverse array of kinases from a variety of species to better establish and elucidate the importance of individual water molecules for kinase function. In addition to an analysis of crystallography data, we have performed molecular dynamics simulations to examine the potential roles of conserved waters within the protein kinase domain. Our results reveal the presence of six conserved water molecules in protein kinases. Two waters are located within the heart of the active site, interacting with some of the aforementioned key functional residues. Both of these waters have large free energy benefits and simulations demonstrate how one aids in coordinating the absolutely critical magnesium-positioning aspartic acid. We also discovered a collection of dynamic yet conserved waters trapped within a closed pocket that lend dramatic stability to the substrate binding cleft. And there are a further two water molecules for which we cannot postulate clear reasons for their conservation. In all, our results suggest that conserved waters are crucial to the efficient function of the kinase family and reveal in general the different circumstances and behaviors associated with water molecules in proteins.
Methods
Data Set
Kinase structures representing active conformations were retrieved from the Protein Data Bank. A kinase was considered “active” if it had any necessary phosphorylations, ATP or a non-hydrolyzable ATP analog, and had at least one divalent cation in the active site. Crystal structures with resolutions poorer than the diameter of water (2.8 Å) were rejected. Thirteen structures met these criteria (Table I).
Table I.
Protein kinase structures in active conformations. The number of water molecules in each structure within 3.2 Å of a nitrogen, oxygen or sulpfur atom is listed in the final column.
| Kinase | Full Name | PDB Code | Species | Resolution (Å) | No. Waters |
|---|---|---|---|---|---|
| Akt2 | RAC-β serine/threonine-protein kinase | 1O6K | H. sapiens | 1.7 | 224 |
| CDK2 | cell division protein kinase 2 | 1JST | H. sapiens | 2.6 | 31 |
| CK1 | casein kinase I | 1CSN | S. pombe | 2.0 | 83 |
| CK2 | casein kinase II subunit α | 1LP4 | Z. mays | 1.86 | 174 |
| DAPK | death-associated protein kinase | 1IG1 | H. sapiens | 1.8 | 190 |
| IRK | insulin receptor tyrosine kinase | 1IR3 | H. sapiens | 1.9 | 160 |
| MAPK p38-γ | mitogen-activated protein kinase p38-γgamma | 1CM8 | H. sapiens | 2.4 | 62 |
| PhK | phosphorylase kinase | 1PHK | O. cuniculus | 2.2 | 86 |
| Pim-1 | proto-oncogene serine/threonine-protein kinase Pim-1 | 1XR1 | H. sapiens | 2.1 | 21 |
| PKA | protein kinase A | 1ATP | M. musculus | 2.2 | 68 |
| Rio2 | Rio2 serine kinase | 1ZAO | A. fulgidus | 1.84 | 223 |
| Sky1P | SR protein kinase | 1Q97 | S. cerevisiae | 2.3 | 57 |
| TAO2 | thousand and one amino acid protein 2 | 1U5R | R. norvegicus | 2.1 | 152 |
Structural Alignment and Conserved Water Identification
Structures were aligned using a sequence order independent structural alignment algorithm we have developed.11 After alignment, water molecules were sought for in identical locations between structures. For each structure all water molecules within 3.2 Å of a nitrogen, oxygen or sulfur atom were included in the search. Waters from different structures that fell within a sphere of 2 Å radius were deemed to occupy the same location. Only waters found in at least ten of the thirteen structures were considered conserved. Normalized B-factors were calculated as (Bi-<B>)/σ(B), where Bi is the B-factor of water i, <B> the mean of all waters, and σ(B) the standard deviation. ASA was calculated with the program GETAREA 1.113 using a probe radius of 1.4. Internal water surfaces were defined using the CASTp server with default parameters.14 A residue was considered fully conserved if an identical or similar residue could be found in all structures within a sphere of 2 Å radius. Partially conserved residues need only be found in ten of thirteen structures. Conserved categories must be present in all structures (see Supplementary Table II).
Molecular Dynamics
The coordinates of the initial structure used in this study was the crystal structure of protein kinase A (1ATP)15 obtained from the protein data bank (PDB). The hydrogens were added to the heavy atoms using the Leap module in the AMBER 816 suite of programs. Furthermore, the atomic charges and force field parameters of the bound ATP were obtained from the work of Meagher et al.17 The complex was solvated with TIP3P18 water molecules such that the solvent was placed up to about 10 Å away from the protein complex to fill a truncated octahedron box. Three chloride ions were placed around the complex using the Leap module in AMBER 8 to obtain electrostatic neutrality. All molecular dynamics simulations were performed with the sander module in AMBER 8.
The system was equilibrated by using a multistage equilibration protocol. At the start of the equilibration, 100 kcal/mol/Å2 harmonic constraints were placed only on the complex. The water and ions were minimized for 1000 steps. This was followed by another 1000 step minimization on the entire system with a 50 kcal/mol/Å2 constraint placed on the complex. With 50 kcal/mol/Å2 constraint on the complex, the entire system was heated from 100 to 300 K over 50 picoseconds. Finally, the entire system was simulated for another 50 picoseconds without any constraints applied at 300 K. After equilibration, the final equilibrated structure was used to carry out 10 nanoseconds MD simulations in the NPT ensemble with periodic boundary conditions at a constant temperature of 300 K and pressure of 1 bar. The SHAKE algorithm19 was applied to all bonds involving hydrogen atoms and an integration time step of 2.0 femtoseconds (fs) was used in solving Newton’s equation of motion. Long-range electrostatic interactions were evaluated using the particle mesh Ewald (PME) summation method.20 The non-bonded interactions were subjected to a 9 Å cutoff that distinguished between the direct space and the reciprocal space in PME. The MD trajectories were collected at every one picosecond (ps) time interval. The resulting trajectories were used to calculate the atomic positional fluctuation of the oxygen atom of the bound water molecules in order to estimate the harmonic force constants used in the free energy simulations, as previously described.21
Free Energy Calculations
The free energy calculations were performed using the double-decoupling method, as previously described.21 The double-decoupling free energy simulations involve gradually turning off the electrostatic and van der Waals interactions of the bound water molecule from the rest of the system. This involves two sets of simulations: the transfer of the water molecule from bulk water to the gas phase, and transferring the water molecule from the binding pocket of the protein complex to the gas phase during which the water molecule is always constrained to occupy the binding site as defined by the coordinate system of the complex. To perform the change from H2Osol to H2Ogas, the energy term is progressively mapped from U(sol) to U(gas) along a chosen path, where U is the potential energy function. This chosen path is mapped as a function of a coupling parameter λ that varies from 0 to 1, where U can be written in terms of λ as U(λ) = (1- λ)k U(0) + [1-(1- λ)k]U(1), and U(0) = U(sol) and U(1) = U(gas). k = 1 when decoupling the electrostatic interactions and k = 6 when decoupling the van der Waals interactions. The free energy difference between the two states is calculated by the thermodynamic integration approach using 12-points Gaussian quadrature integration at discrete points of λi of 0.00922, 0.04794, 0.11505, 0.20634, 0.31608, 0.43738, 0.56262, 0.68392, 0.79366, 0.88495, 0.95206, and 0.99078 along the path. The simulation for each value of λ was initially equilibrated for 200.0 picoseconds, and the data sampling was also performed for 200.0 picoseconds after the equilibration. All of the free energy simulations were performed under the same conditions as described above for the molecular dynamics simulation, except for the fact that a time step of 1.0 fs, instead of 2.0 fs, was used when evaluating the free energy component of the van der Waals interactions. Three independent runs were performed for each free energy calculation. The final free energy was then estimated by averaging over the calculated free energies from the three independent simulations. Also the error was estimated by calculating the standard error using the standard deviation of the calculated free energies from the three independent simulations.
Results and Discussion
Conserved Waters
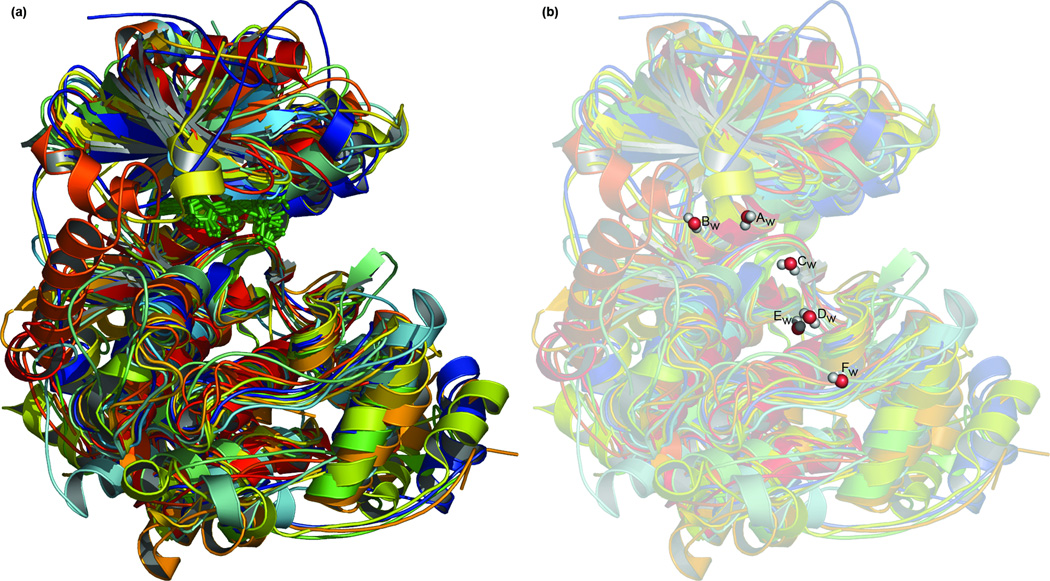
We present our results with reference to the death associated protein kinase (DAPK) and PKA. The structure of DAPK is shown in Figure 1 for purposes of orientation. Thirteen kinases in active conformations (Table I) were aligned (Figure 2(a)) and six conserved water molecules identified (Figure 2(b) and Table II). These waters have low average B-factors, reflecting a general lack of mobility in these molecules and confidence in their placement. These waters bind both to side-chain and main-chain atoms (Supplementary Table I). All but ten of the fifty-seven side-chain interactions are with conserved residues. Water interactions with main-chain atoms occur evenly between conserved and non-conserved residues. Between structures, water interactions with main-chain residues are entirely with equivalent parts of the polypeptide. Waters Aw, Cw, Dw, and Fw have low accessible surface areas.
Figure 2.
An active conformation protein kinase structural alignment and consensus. (a) The structural alignment of the thirteen kinases listed in Table I. The ATP or non-hydrolyzable ATP analog of each structure is shown in green sticks. Each kinase is colored uniquely. (b) The conserved water molecules are shown superimposed above the alignment, coloured as red spheres with attached hydrogens in white. Waters are labelled as in Table II.
Table II.
Conserved waters. Identifiers from each structure are listed, along with the normalized B-factors and the acessible surface area (ASA) in Å2.
| Kinase | ||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Water | Akt2 | CDK2 | CK1 | CK2 | DAPK | IRK | p38-γ | PhK | Pim-1 | PKA | Rio2 | Sky1P | TAO2 | |
| Aw | 31 | 5 | 411 | 20 | 3405 | 44 | 2006 | 401 | 400 | 11 | 44 | |||
| B | −1.207 | 0.191 | −1.483 | −1.230 | −1.402 | −0.834 | −0.786 | −2.053 | - | −0.081 | −1.981 | - | −1.074 | |
| ASA | 0.05 | 3.04 | 3.21 | 0.58 | 0.30 | 1.25 | 1.00 | 0.93 | 0.65 | 1.04 | 1.30 | |||
| Bw | 34 | 3 | 409 | 428 | 45 | 2106 | 426 | 404 | 91 | 53 | ||||
| B | −1.394 | −0.279 | −1.031 | - | −0.475 | −0.557 | −0.304 | −1.338 | - | 0.070 | - | −1.058 | −0.538 | |
| ASA | 0.55 | 0.88 | 0.82 | 8.09 | 1.61 | 2.39 | 0.00 | 9.18 | 0.00 | 10.52 | ||||
| Cw | 114 | 8 | 441 | 10 | 413 | 29 | 2042 | 405 | 25 | 403 | 21 | |||
| B | −1.433 | −0.025 | −1.394 | −1.548 | −1.351 | −0.844 | −0.603 | −1.360 | 0.302 | −1.324 | - | - | −1.510 | |
| ASA | 0.15 | 0.04 | 0.25 | 0.00 | 0.00 | 0.07 | 0.06 | 0.14 | 0.24 | 0.09 | 0.00 | |||
| Dw | 97 | 427 | 23 | 435 | 7 | 2124 | 14 | 410 | 110 | 29 | ||||
| B | −1.325 | - | −0.951 | −1.464 | −1.017 | −1.453 | 1.510 | - | 0.206 | −0.857 | −0.573 | - | −1.567 | |
| ASA | 1.53 | 1.25 | 0.00 | 0.79 | 0.28 | 0.00 | 0.96 | 1.31 | 3.90 | 0.96 | ||||
| Ew | 98 | 6 | 444 | 11 | 404 | 1 | 2136 | 415 | 8 | 431 | 8 | 1 | ||
| B | −1.141 | −0.788 | −0.197 | −1.672 | −1.604 | −1.940 | 0.440 | −1.687 | 0.738 | −1.406 | - | −1.830 | −2.098 | |
| ASA | 4.81 | 0.00 | 2.43 | 0.00 | 1.21 | 1.72 | 0.00 | 1.32 | 5.27 | 3.08 | 0.00 | 1.18 | ||
| Fw | 153 | 13 | 65 | 403 | 46 | 2015 | 413 | 24 | 568 | 132 | 110 | 2 | ||
| B | −1.819 | −2.526 | - | −0.978 | −1.684 | −0.827 | 0.633 | −1.669 | −0.131 | −0.503 | 0.201 | −1.384 | −2.025 | |
| ASA | 0.00 | 0.00 | 0.00 | 0.00 | 0.08 | 0.00 | 0.00 | 0.00 | 0.00 | 24.43 | 0.00 | 1.07 | ||
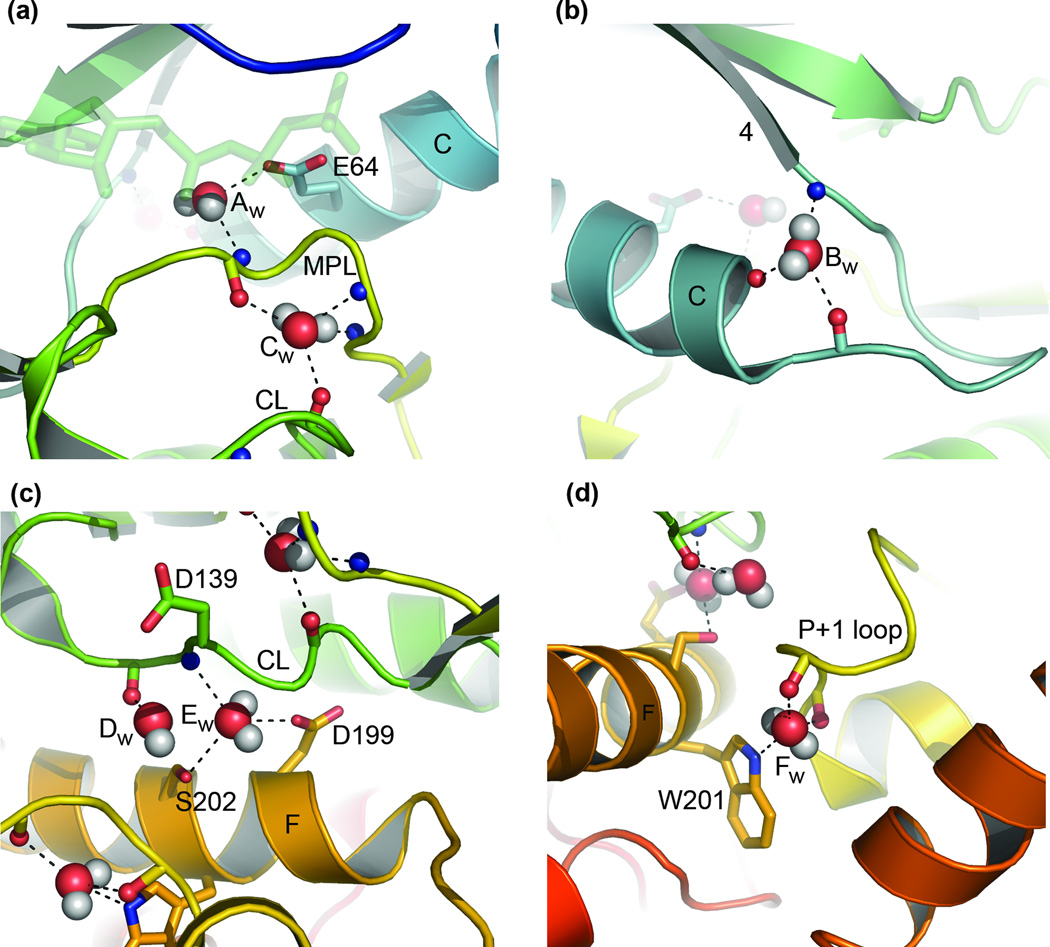
Of the waters consistently found by Shaltiel, Cox and Taylor1 across several different crystal structures of PKA, we also found ASCW B (their nomenclature) as conserved across the protein kinase family (our Aw), as is ASCW C (our Cw). Although they only reported these as potentially interacting with single atoms, both can interact with two (using our distance cut-off, which was also slightly stricter). Interestingly, unlike these authors, we find no conserved waters interacting with either ATP or magnesium/manganese. The functions of these waters in PKA are not likely to be substituted for by amino-acid residues in other kinases, as the region around the γ-phosphate is a very highly conserved area. However, while the active-site divalent cations necessary for catalytic activity are consistently placed throughout the structures we have used, there is a moderate degree of variability in the positioning of the γ-phosphate. We do not know what to attribute this to but variability in the position of this phosphate would be accompanied by variability in the local water environment, and this would manifest as a lack of water molecule conservation in this area. Whether this lack of conservation is real and in contradiction to the findings of Shaltiel, Cox and Taylor, or instead is an artifact of the data set we are examining, we cannot say. When more active-conformation kinase structures become available this issue can be better answered.
The conserved water molecules from DAPK are shown in Figure 3 as an example of the typical interactions that are occurring. Water molecule Aw (Figure 3(a)) interacts with the fully conserved glutamic acid E64 of helix C and connects it to the main chain of the magnesium positioning loop (Supplementary Table II lists conserved residues). Molecule Bw (Figure 3(b)) can potentially interact with three atoms, connecting the terminus of helix C with the end of the loop preceding β-strand 4. Molecule Cw (Figure 3(a)) can interconnect the magnesium positioning loop through three main-chain atoms and join these to the main chain of the catalytic loop. Molecule Dw (Figure 3(c)) is only bound to the structure in one place, through the carbonyl of the residue following the catalytic base D139. Molecule Ew (Figure 3(c)) can interact with the side chains of two partially conserved residues from helix F and connect these to the main chain of the catalytic loop. Lastly, water molecule Fw (Figure 3(d)) can connect the side chain of the partially conserved tryptophan W201 from helix F with two main-chain atoms of the P+1 loop.
Figure 3.
Conserved waters in DAPK. Each of the conserved waters is labelled as in Table II. Dashes indicate potential hydrogen bonds to oxygen and nitrogen atoms within 3.2 Å. Oxygens are colored red, nitrogens blue and hydrogens white. Side chains are shown as sticks, as is the bond between the main-chain carbonyl atoms. CL: catalytic loop; MPL: magnesium positioning loop.
From these results we hypothesized that these water molecules may be conserved because of the connectivity they create within the protein kinase domain, and hence the stability they lend to the fold, and/or because of the roles they play in positioning/stabilizing key residues crucial to the efficient function of these enzymes.
Molecular Dynamics and Free Energy
To further elucidate the functions of these water molecules and probe their apparent stabilizing role, molecular dynamics simulations were performed and free energies calculated for the structure of protein kinase A (PKA).
Water Aw
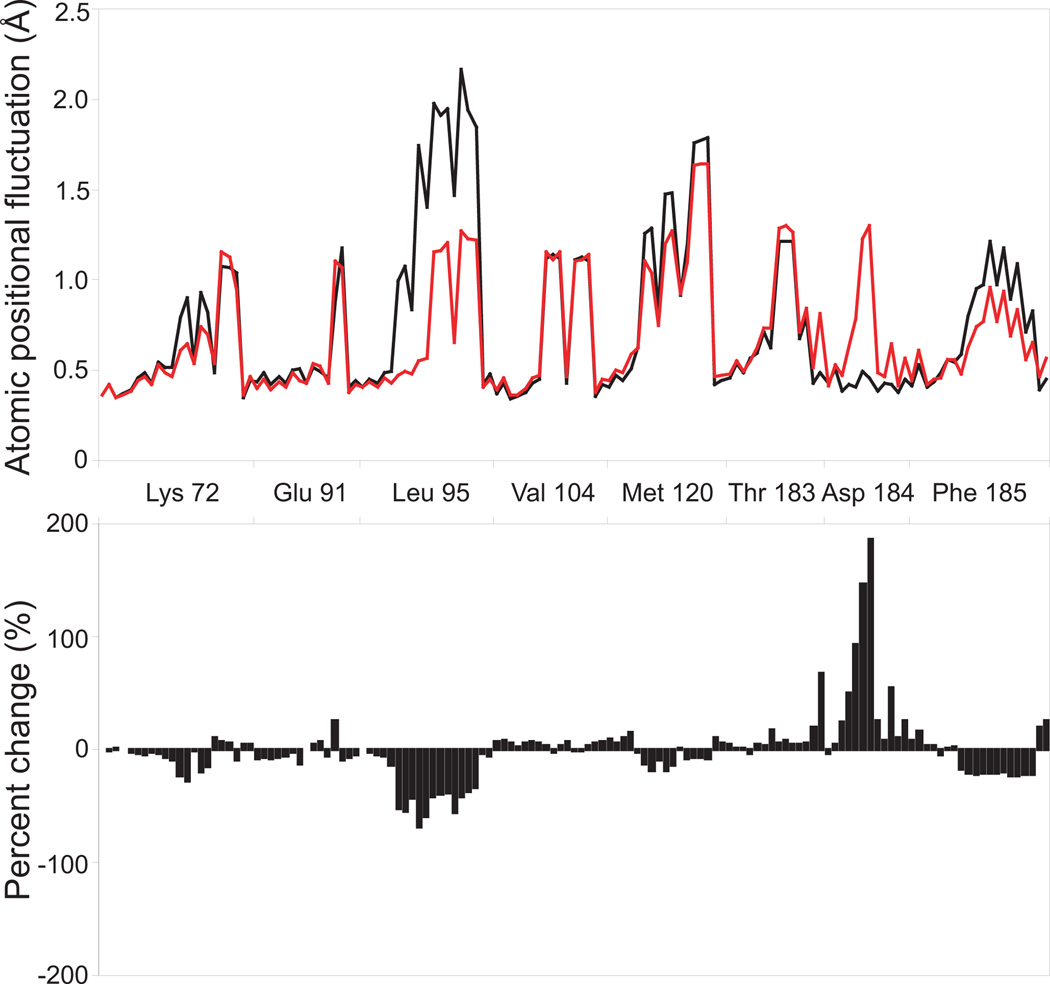
This water is internal and does not exchange with bulk solvent during the time course of simulation. It is fixed within its pocket, with an average fluctuation in position of 1.03 Å. Its free energy contribution is highly favorable at −2.1±0.4 kcal/mol. We also performed simulations to observe the effects of this water molecule on nearby atoms. Simulations were performed with and without this water, and the average positional fluctuation of all atoms from adjacent residues were measured (Figure 4). The conserved leucine of helix C (PKA: 95, DAPK: 68) and the conserved phenylalanine of the magnesium positioning loop (PKA: 185, DAPK:162) show increased stability in the absence of this water. This is not unexpected as these are hydrophobic residues. In the absence of this water the fully conserved and functionally critical aspartic acid found on the magnesium positioning loop (PKA: 184, DAPK: 161) shows a great increase in fluctuation. This aspartic acid chelates the active site magnesium/manganese, thereby aiding in its positioning and the orientation of the γ-phosphate. A lack of stability in this residue is likely to detrimentally affect catalysis.
Figure 4.
Water Aw mediated stabilization/destabilization. Molecular dynamics simulations were performed on protein kinase A in the presence (black) and absence (red) of this water. The root-mean-square fluctuation of all atoms in the vicinty of this water is shown in the top panel and the percent difference beneath.
Water Bw
Water Bw is found on the surface of the protein. It is accessible to and exchanges with bulk solvent. The occupancy for the site this water is found in is ~10%. The residence time for waters in this site is ~2.5 picoseconds. These results suggest this water is not playing an obvious beneficial role in so far as regional stability is concerned, however we cannot rule out that it may be positively involved in the “fluidity”/movement of its local environment, hence its loosely bound nature and simultaneous conservation.
Water Cw
This water is internal and does not exchange with bulk solvent. Its atomic positional fluctuation is only ~0.78 Å. In other proteins in our dataset (apart from PKA) this water is not adjacent to any other waters but only contacts atoms of the polypeptide. This is not the case in PKA, where it is found in contact with a second water. This creates unfavorable conditions, resulting in a free energy contribution of 0.4±1.6 kcal/mol. However, we believe the second water (424) to be erroneously assigned, as it displays a large overlap with the carbonyl oxygen of tyrosine 164 and has a high average B-factor. When the second water is removed, the free energy contribution of water Cw is very favorable: −5.1±0.7 kcal/mol. This significant lowering of free energy is of obvious benefit in a region so crucial to function as the catalytic loop.
Waters Dw and Ew
These two waters are found in a pocket that contains four additional waters and will be discussed together. This pocket is found in all structures and contains between two and six water molecules. Only those found in the positions of Dw and Ew are consistently placed. In the first 1–2 nanoseconds of the simulations two water molecules of PKA exit through a mouth that opens between three hydrophobic residues (PKA: leucine 198, leucine 205 and isoleucine 209). The mouth then closes trapping the remaining four waters. The waters in this pocket do not exchange with bulk solvent but do exchange with one another. Their average positional fluctuation is ~2.30 Å. The site occupied by water Dw is occupied ~19% of time, while site Ew is occupied ~54% of the time. The respective residence times of waters in these sites are 27.8 picoseconds and 46.6 picoseconds. The free energy contribution of adding one water into this pocket is −4.1±1.2 kcal/mol, followed by a second −3.8±1.4 kcal/mol, third −2.6±1.8 kcal/mol and fourth −3.8±1.9 kcal/mol. Their presence obviously lends a great deal of stability.
In several of the structures, water Dw interacts with a carboxyl oxygen from the catalytic aspartic acid and a main-chain carbonyl oxygen from the adjacent downstream residue. From the orientations of these atoms in crystals it appears that this water may aid in positioning the catalytic aspartic acid. However, the crystal structure orientation of the carbonyl is strained and high energy, and during the equilibration stage of the simulations this functional group flips inwards towards the active site and does not change position.
An important point emerges from consideration of waters Dw and Ew: none of the water molecules we have studied is permanently trapped, nor are hydrogen bonds made permanent. Water molecules, even those found in internal pockets, will exchange with bulk solvent. This occurs over nanosecond to microsecond timescales,22 outside of the range of our simulations, and results from large-scale fluctuations in structure.23 Exchange between water molecules in deep pockets can be rapid and have a small enthalpic cost when the water is weakly hydrogen bonded.22 Hydrogen bonds must obviously be broken prior to exchange. But even when a water molecule is present in a closed pocket and in contact with the polypeptide, they often have great rotational freedom.22 Interactions between such waters and the polypeptide should be viewed as transient, not permanent. These points do not contradict the notion of water-molecule mediated stability. Although on/off, hydrogen bonds between water and other functional groups may only need to exist for a continuous series of brief instants to stabilize and orient atoms of the polypeptide. What we see in crystal structures, and even to a degree in molecular dynamics simulations, are the average or end effects of a water molecule's presence. Differences between waters, such as rate of exchange and occupancy, should be considered against one another rather than on their own when assessing any particular water's functional role.
Water Fw
This water is internal and does not exchange with bulk solvent. It has the most restricted motion at ~0.53 Å. Its free energy contribution is −0.4±0.7 kcal/mol. As with water Aw the dynamics of adjacent residues was also examined in the presence and absence of this water. However, all atoms with the exception of one showed an increase or decrease in atomic positional fluctuation of less than 10%. The exception showed an ~25% decrease in fluctuation. The location of this water in the substrate binding groove may suggest that its interaction with adjacent residues/regions participates beneficially in this process, facilitating residue movement or stability on substrate binding, or assisting in large-scale domain motions. We examined the apo(free) enzyme state and found a negative free energy for this water (−2.9±0.5), alternatively suggesting it may play a beneficial role in stabilizing this region prior to ATP or substrate binding. As we are only examining short timescales of a single context in our simulations, an understandable solution may exist in another unexplored context.
Conclusion
Our examination of active-conformation protein kinases has revealed the presence of six conserved water molecules that create a high degree of inter-protein connectivity. Two of these waters are located adjacent to the magnesium-positioning and catalytic loops, and three more are located within the relatively unstudied substrate binding groove. Molecular dynamics simulations on these waters and their local environments suggest that one water molecule (Aw) plays a key role in stabilizing an important catalytic residue and that (very) favorable free energy benefits (and hence stability) are conferred by five of the six waters we uncovered. In our study, if we consider conserved waters against conserved amino acids in protein kinases, then we find water is more abundant than all of the twenty amino acids except leucine. The lack of reported proteins with conserved water molecules is striking in this regard. Although the waters we have discovered are not directly involved in the phosphotransfer mechanism, and hence cannot be considered cofactors or ligands, our results suggest they may be essential structural and functional elements necessary for the efficient function of protein kinases.
Supplementary Material
Acknowledgements
We wish to thank Dr. Lynn Megeney for critical reading of the manuscript. J.D.R.K. was supported by a Canadian Institutes of Health Research (CIHR) Canadian Graduate Scholarship and a Multiple Sclerosis Society of Canada (MSSC) Studentship. D.H. was supported in part by the Georgia Cancer Coalition (GCC). J.A.M. was supported in part by the NSF, NIH, HHMI, NBCR, CTBP, SDSC and Accelrys. This work was supported by grants from CIHR and the MSSC to R.K.
References
- 1.Shaltiel S, Cox S, Taylor SS. Conserved water molecules contribute to the extensive network of interactions at the active site of protein kinase A. Proc Natl Acad Sci U S A. 1998;95(2):484–491. doi: 10.1073/pnas.95.2.484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Levy Y, Onuchic JN. Water mediation in protein folding and molecular recognition. Annu Rev Biophys Biomol Struct. 2006;35:389–415. doi: 10.1146/annurev.biophys.35.040405.102134. [DOI] [PubMed] [Google Scholar]
- 3.Loris R, Langhorst U, De Vos S, Decanniere K, Bouckaert J, Maes D, Transue TR, Steyaert J. Conserved water molecules in a large family of microbial ribonucleases. Proteins. 1999;36(1):117–134. doi: 10.1002/(sici)1097-0134(19990701)36:1<117::aid-prot10>3.0.co;2-h. [DOI] [PubMed] [Google Scholar]
- 4.Malin R, Zielenkiewicz P, Saenger W. Structurally conserved water molecules in ribonuclease T1. J Biol Chem. 1991;266(8):4848–4852. [PubMed] [Google Scholar]
- 5.Sreenivasan U, Axelsen PH. Buried water in homologous serine proteases. Biochemistry. 1992;31(51):12785–12791. doi: 10.1021/bi00166a011. [DOI] [PubMed] [Google Scholar]
- 6.Bottoms CA, Smith PE, Tanner JJ. A structurally conserved water molecule in Rossmann dinucleotide-binding domains. Protein Sci. 2002;11(9):2125–2137. doi: 10.1110/ps.0213502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ogata K, Wodak SJ. Conserved water molecules in MHC class-I molecules and their putative structural and functional roles. Protein Eng. 2002;15(8):697–705. doi: 10.1093/protein/15.8.697. [DOI] [PubMed] [Google Scholar]
- 8.Prasad BV, Suguna K. Role of water molecules in the structure and function of aspartic proteinases. Acta Crystallogr D Biol Crystallogr. 2002;58(Pt 2):250–259. doi: 10.1107/s0907444901018327. [DOI] [PubMed] [Google Scholar]
- 9.Rodriguez-Almazan C, Arreola R, Rodriguez-Larrea D, Aguirre-Lopez B, Tuena de Gomez-Puyou M, Perez-Montfort R, Costas M, Gomez-Puyou A, Torres-Larios A. Structural basis of triosephosphate isomerase deficiency: Mutation E104D is related to alterations of a conserved water network at the dimer interface. J Biol Chem. 2008 doi: 10.1074/jbc.M802145200. [DOI] [PubMed] [Google Scholar]
- 10.Hanks SK, Hunter T. Protein kinases 6. The eukaryotic protein kinase superfamily: kinase (catalytic) domain structure and classification. Faseb J. 1995;9(8):576–596. [PubMed] [Google Scholar]
- 11.Knight JD, Qian B, Baker D, Kothary R. Conservation, variability and the modeling of active protein kinases. PLoS ONE. 2007;2(10):e982. doi: 10.1371/journal.pone.0000982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gibbs CS, Zoller MJ. Rational scanning mutagenesis of a protein kinase identifies functional regions involved in catalysis and substrate interactions. J Biol Chem. 1991;266(14):8923–8931. [PubMed] [Google Scholar]
- 13.Fraczkiewicz R, Braun W. Exact and efficient analytical calculation of the accessible surface areas and their gradients for macromolecules. J Comput Chem. 1998;19:319–333. [Google Scholar]
- 14.Dundas J, Ouyang Z, Tseng J, Binkowski A, Turpaz Y, Liang J. CASTp: computed atlas of surface topography of proteins with structural and topographical mapping of functionally annotated residues. Nucleic Acids Res. 2006;34:W116–W118. doi: 10.1093/nar/gkl282. (Web Server issue) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zheng J, Trafny EA, Knighton DR, Xuong NH, Taylor SS, Ten Eyck LF, Sowadski JM. 2.2 A refined crystal structure of the catalytic subunit of cAMP-dependent protein kinase complexed with MnATP and a peptide inhibitor. Acta Crystallogr D Biol Crystallogr. 1993;49(Pt 3):362–365. doi: 10.1107/S0907444993000423. [DOI] [PubMed] [Google Scholar]
- 16.Case DA, Cheatham TE, Darden T, 3rd, Gohlke H, Luo R, Merz KM, Jr, Onufriev A, Simmerling C, Wang B, Woods RJ. The Amber biomolecular simulation programs. J Comput Chem. 2005;26(16):1668–1688. doi: 10.1002/jcc.20290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Meagher KL, Redman LT, Carlson HA. Development of polyphosphate parameters for use with the AMBER force field. J Comput Chem. 2003;24(9):1016–1025. doi: 10.1002/jcc.10262. [DOI] [PubMed] [Google Scholar]
- 18.Jorgensen WL, Chandrasekhar J, Madura JD. Comparison of Simple Potential Functions for Simulating Liquid Water. J Chem Phys. 1983;79:926. [Google Scholar]
- 19.Ryckaert JP, Ciccotti G, Berendsen HJC. Numerical integration of the cartesian equations of motion of a system with constraints: molecular dynamics of n-alkanes. J Comp Phys. 1977;23:327–341. [Google Scholar]
- 20.Darden T, York D, Pedersen L. Particle mesh Ewald. An Nlog(N) method for Ewald sums in large systems. J Chem Phys. 1993;98:10089–10092. [Google Scholar]
- 21.Hamelberg D, McCammon JA. Standard free energy of releasing a localized water molecule from the binding pockets of proteins: double-decoupling method. J Am Chem Soc. 2004;126(24):7683–7689. doi: 10.1021/ja0377908. [DOI] [PubMed] [Google Scholar]
- 22.Denisov VP, Venu K, Peters J, Hörlein HD, Halle B. Orientational disorder and entropy of water in protein cavities. J Phys Chem B. 1997;101:9380–9389. [Google Scholar]
- 23.Denisov VP, Peters J, Horlein HD, Halle B. Using buried water molecules to explore the energy landscape of proteins. Nat Struct Biol. 1996;3(6):505–509. doi: 10.1038/nsb0696-505. [DOI] [PubMed] [Google Scholar]
- 24.Tereshko V, Teplova M, Brunzelle J, Watterson DM, Egli M. Crystal structures of the catalytic domain of human protein kinase associated with apoptosis and tumor suppression. Nat Struct Biol. 2001;8(10):899–907. doi: 10.1038/nsb1001-899. [DOI] [PubMed] [Google Scholar]
- 25.Knighton DR, Zheng JH, Ten Eyck LF, Ashford VA, Xuong NH, Taylor SS, Sowadski JM. Crystal structure of the catalytic subunit of cyclic adenosine monophosphate-dependent protein kinase. Science. 1991;253(5018):407–414. doi: 10.1126/science.1862342. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.