Abstract
The implications of autophagy-related genes in serious neural degenerative diseases have been well documented. However, the functions and regulation of the family genes in embryonic development remain to be rigorously studied. Here, we report on for the first time the important role of atg5 gene in zebrafish neurogenesis and organogenesis as evidenced by the spatiotemporal expression pattern and functional analysis. Using morpholino oligo knockdown and mRNA overexpression, we demonstrated that zebrafish atg5 is required for normal morphogenesis of brain regionalization and body plan as well as for expression regulation of neural gene markers: gli1, huC, nkx2.2, pink1, β-synuclein, xb51 and zic1. We further demonstrated that ATG5 protein is involved in autophagy by LC3-II/LC3I ratio and rapamycin-induction experiments, and that ATG5 is capable of regulating expression of itself gene in the manner of a feedback inhibition loop. In addition, we found that expression of another autophagy-related gene, atg12, is maintained at a higher constant level like a housekeeping gene. This indicates that the formation of the ATG12–ATG5 conjugate may be dependent on ATG5 protein generation and its splicing, rather than on ATG12 protein in zebrafish. Importantly, in the present study, we provide a mechanistic insight into the regulation and functional roles of atg5 in development of zebrafish nervous system.
Keywords: atg5, zebrafish, autophagy, neurogenesis, expression regulation, feedback inhibition, neural gene, rapamycin
Introduction
Autophagy is an evolutionarily conserved intracellular mechanism by which long-lived proteins and organelles that are no longer needed can be degraded for rapid cell turnover.1 This process is a kind of programmed cell death (PCD), called PCD type II (PCD I indicating apoptosis).1,2 As a physiological mechanism, autophagy is implicated in the whole living process, including embryonic development, individual growth, and death. Embryo morphogenesis is involved in structural remodeling and requires high dynamic changes in cell movement, cell category and cell amount. This means that rapid cell turnover is necessary for embryonic development. Since autophagy can mediate the bulk of unrequired protein and organelle renovation quickly, it plays a key role in cell fate decision and differentiation, including cell proliferation, sorting, migration, information transfer, differentiation and death;3-5 it is also necessary for maintaining neurons in a physiological state and ensuring cell survival by degradation of abnormal protein aggregates in nerve tissue.2 Excessively high or low autophagy levels can lead to pathological processes. Accumulation of neuroproteins can cause neurodegenerative diseases, such as Alzheimer disease (AD), Parkinson disease (PD), Huntington disease and stroke, which result from a reduced level of autophagy.6-11
In the autophagy gene superfamily, autophagy-related gene 5 (atg5) is a key gene. ATG5 protein can conjugate to ATG12 and then form a complex with the multimeric protein ATG16. The ATG12–ATG5-ATG16 multimeric complex plays an essential role in autophagy.12,13 The occurrence and alteration of autophagy pathways are dependent on enhanced or reduced atg5 levels. Hungry mice with atg5 knockout not only live half as long as normal mice, but they also develop functional dysregulation of the hepatic tissues and progressive neurodegenerative conditions;10,14,15 this implies that autophagy offers functional amino acid turnover in undernutrition. Atg5 knockdown in the nervous tissue of mice after 10.5 d led to growth retardation, weight loss, ubiquitin-proteasome accumulation, and motor nerve damage; after 16 d, inclusion bodies developed in hepatic cells.16-18 Atg5 also benefited T-cell survival and proliferation.19 Overexpression of atg5 in T cells may contribute to inflammatory demyelination in multiple sclerosis.20 Upregulation of atg5 expression has potential therapeutic effects on neurodegenerative conditions, such as Parkinson and Huntington diseases.21 However, thus far, no studies have been reported about the expression pattern and function of atg5 in neurogenesis and the embryonic developmental process. The relationship of the autophagy pathway to neural genes in embryonic development remains unclear.
Zebrafish offer superior characteristics for in vivo experiments since the developmental mechanism of tissues and organs is very similar to mammals in terms of structure, function and genetic sequences. The embryo body is transparent, and the fish has high fecundity, undergoes rapid embryonic development, and can be easily observed. As a result, zebrafish have been used widely in basic sciences and in medical sciences for disease models as well as in procedures such as screening of lead compounds.22,23 In this study, the expression pattern and functions of atg5 and its potential target genes were studied in zebrafish embryonic development.
Results
Sequence homologous analysis of zebrafish atg5
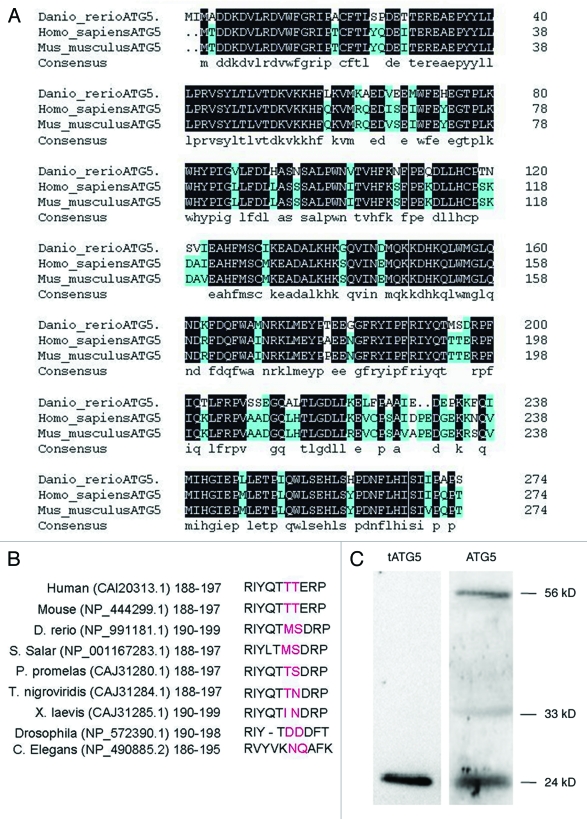
High conservation of atg5 coding sequences in a wide variety of species is probably due to its important role in autophagy. We cloned atg5 coding sequences from zebrafish lines Tuebingen and AB, and they showed 100% identity with each other (data not shown) (GenBank:HQ450378.1). Multiple sequences alignment online indicated that only five bases were different from zebrafish atg5 sequence registered in GenBank (NP_991181.1), and all the change occurred at the third base of the codons, which signifies synonymous variation and identical amino acids.
The whole amino acid sequence of zebrafish atg5 was compared with those of human and mouse, which indicates the sequence similarity of over 90% and identity of 82% (http://blast.ncbi.nlm.nih.gov/Blast.cgi#4757798) (Fig. 1A). Then, we analyzed sequences of the calpain cleavage sites in ATG5 proteins among nine species and found that the amino acid at the calpain cleavage site in zebrafish is the same as in Salmo salar; in other species, variation at the site is also obvious, such as in Pimephales promelas, Xenopus laevis and Drosophila (Fig. 1B). However, both ends of the flanking sequence are conserved (Fig. 1B).24 The ATG5 of a 33 kD protein can be cut into 24 kD at the site by calpain selective splicing. These two forms of ATG5 function as a molecular switch to guide the cell into autophagy or apoptosis, respectively.25 In order to confirm that zebrafish ATG5 protein also exists in the two forms, western blot was performed to detect the forms of ATG5 with two ATG5 antibodies which were differentially recognizing ATG5 24 kD, 33 kD and 56 kD of ATG12–ATG5 conjugate respectively. The result demonstrates that three forms of ATG5 protein, the truncated 24 kD, the full-length 33 kD alone and the 56 kD ATG12–ATG5 conjugate do exist in zebrafish (Fig. 1C). These in vivo studies suggest that calpain selective splicing for ATG5 protein also occurs in zebrafish (Fig. 1C).
Figure 1 (See previous page).
Sequence homologous analysis of zebrafish ATG5 protein. (A) Whole sequence alignment of ATG5 proteins between mouse, human and zebrafish orthologs. Zebrafish ATG5 amino acid sequence is putative based on a coding sequence cloned in this study (HQ450378.1). The other two atg5 sequences are from GeneBank as follows: human (CAI_20313.1) and mouse (NP_444299.1). Amino acids shaded in black are identities; in azure-like are positive ones. (B) Comparison of calpain cutting sites of ATG5 proteins. Red amino acids show the putative cut site of calpain. (C) Identification of the calpain cutting sites in zebrafish ATG5 protein by protein gel blot. The protein sample was from 48 hpf wild-type embryo. The results indicate that ATG5 protein exists in three forms in zebrafish: the truncated 24 kD that should be resulted from calpain selective splicing of 33 kD ATG5, the 33 kD whole length ATG5 alone and 56 kD conjugate of ATG5 with ATG12. Anti-tATG5 and anti-ATG5 antibodies were diluted at 1:200 (Abgent, AP1812a and AP1812b).
Spatiotemporal expression pattern of atg5 during embryonic development
Expression stage pattern of atg5 gene
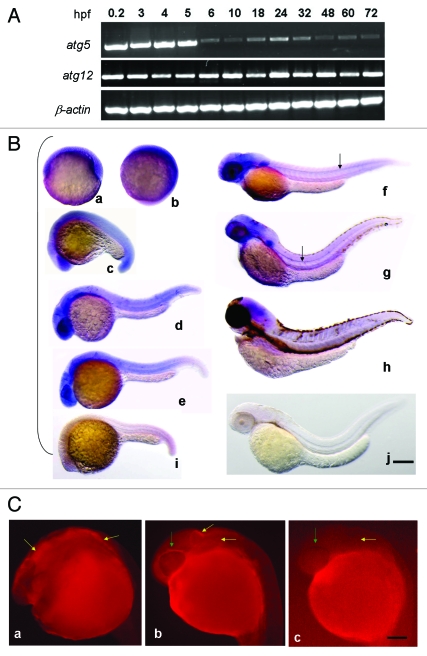
The expression pattern of atg5 in zebrafish embryonic development was tested by RT-PCR. The result shows that atg5 transcription was dynamically sustained throughout the whole process. The highest level obviously occurs in the one-cell zygote, which is a maternal product. In the subsequent process, the transcription level decreased to basal level at 6–10 h postfertilization (hpf), which indicates that the maternal product was almost exhausted. After 18 hpf, atg5 transcription increased again, reaching the highest level at 24 hpf, then decreased again to a basal level by 72 hpf (Fig. 2A). The concurrence of the atg5 high expression stage with the key period of embryo neurogenesis and organogenesis at 18–24 hpf suggests that atg5 may be implicated in neurula formation in embryonic development in zebrafish.26 In contrast to atg5, the expression level of another autophagy-related gene, atg12, shows higher and stable level during the whole development process (Fig. 2A).
Figure 2.
Spatiotemporal expression of autophagy related gene atg5 during zebrafish embryonic development. (A) Expression stage-pattern of atg5 during zebrafish embryonic development. Atg5 and atg12 mRNAs were detected as early as 0.2 hpf and throughout the whole development up to 72 hpf with RT-PCR in different developmental stages (hpf). β-actin was used as a loading control. (B) Atg5 transcription mainly in brain region and the central neural system. Antisense atg5 RNA probe was used for whole-mount in situ hybridization on zebrafish embryos at 6 hpf (a), 10 hpf (b), 18 hpf (c), 24 hpf (d), 30 (e), 48 hpf (f), 72 hpf (g), 96 hpf (h); and sense atg5 RNA probe was used for embryos at 22 hpf (i) and 48 hpf (j) as negative controls.Scale bar: 400 μm. (C) Protein of ATG5 presented in brain region in zebrafish embryo. Whole-mount immunofluorescent histochemistry was performed to reveal ATG5 protein localization on 24 hpf wild-type embryo with anti-ATG5 polyclonal antibody (Abcam, 1:500 dilution) (a,b), without anti-ATG5 polyclonal antibody as a control (c). Yellow arrows indicate ATG5 positive area in brain in (a and b), negative in (c). Green arrows show ATG5 positive signal in eye circumferential in (b), and negative in (c). Scale bar: 250 µm
High transcription of atg5 mainly in the encephalic region and the central neural system
To examine the distribution of atg5 expression, whole-mount in situ hybridization of wild-type zebrafish embryos was performed. Essentially, up to the tailbud stage (10 hpf), high signals of the atg5 transcript were displayed in a diffuse manner around the whole embryonic area. After 18 hpf, the atg5 signal began to be focused and enhanced on the forebrain (containing the eye region), midbrain, hindbrain and tail end, and a moderate level was scattered in the trunk. The highest level of atg5 transcription at 24–30 hpf was evident mainly in the telencephalon, eyes, midbrain, midbrain-hindbrain boundary and ventral area of the hindbrain. From 48 to 96 hpf, positive regions and levels were diminished, though the head still retained the highest intensity (Fig. 2B). In addition, the atg5 positive signal obviously appeared in the floor plate along the anterior-posterior axis of the embryonic midline at 48 hpf and 72 hpf (Fig. 2B, f and g) with slowly shortening toward the head, and disappeared by 96 hpf. So we infer that atg5 gene is likely to be implicated in the formation of the cranial nerve and the central nervous system in neurula development.
Protein of ATG5 localized in the brain region
To confirm the final localization of the ATG5 signal, whole-mount in situ immunofluorescence was used to detect ATG5 protein in zebrafish 24 hpf embryos. The results show that the ATG5 protein signal also appeared in the position corresponding to those of atg5 transcription, including the forebrain, midbrain and hindbrain (Fig. 2C); yet, the signal was hardly evident in the embryonic trunk. The presence of both mRNA and the protein in the brain tissues further confirms that ATG5 probably participates in early brain development in zebrafish.
Functional study of atg5 gene
Downregulation of atg5 gene resulted in head shrinkage and central axis shortening
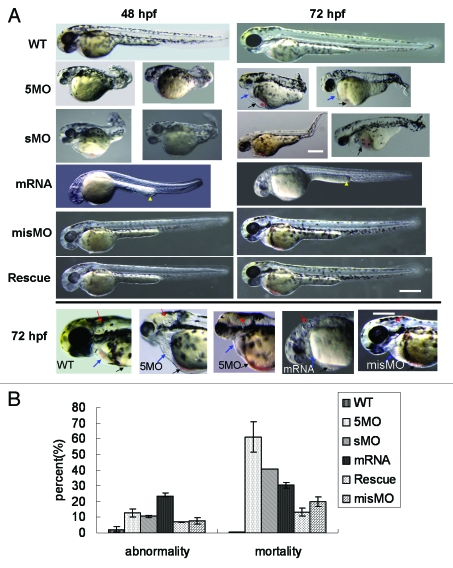
To investigate atg5 function on embryonic development, two kinds of atg5 morpholino oligos, 5MOatg5 (5MO) and sMOatg5 (sMO) were separately injected into embryos at the one- to four-cell stage, in which 5MO can complementarily bind to the initial sequence of atg5 mRNA and inhibit translation of ATG5 protein; sMO can particularly bind to the splicing site at intron 2/exon 3 which leads to deletion of 27 amino acids and frameshift mutation of the succeeded reading frame sequence. The results from both 5MO and sMO injection displayed the same embryonic over-dorsalized deformity, such as smaller and incomplete head and eyes, absent otoliths or three otoliths, abnormal heart structure with less blood and congestion in the ventral common cardinal vein (CCV) where the yolk slightly intra-sinked and vacuolated (Fig. 3A). The body trunk, tail and yolk extension were significantly shortened and somite was deficient. The evidence suggests that atg5 knockdown is implicated in the development of the body plan, particularly in the neural system, heart, vascular tissues, and anterior-posterior axis. We found 60% and 41% mortality in the 5MO and sMO groups respectively (4 dpf) (Fig. 3B), implying that ATG5 protein is probably an important regulator in the formation of essential embryonic body structures.
Figure 3.
Dysregulation of atg5 leading to embryonic abnormal phenomenon in zebrafish. (A) Abnormal phenotypes caused by atg5 dysregulation at 48 hpf and 72 hpf. 5MO and sMO row: the 5MOatg5 or sMOatg5 injected embryos display dorsalization-like abnormalities; mRNA row: the embryos were injected with atg5 capped-mRNA, and their development was retarded and slightly ventralized; misMO row: the embryos were injected with a control MO; Rescue row: the embryos were injected with a mixture of 5MOatg5 and atg5 mRNA; the bottom panel presents a comparison of the heads and hearts of 72 hpf embryos injected with 5MOatg5, capped mRNA or misMO. Blue arrows indicate the heart and pericardial sac; black arrows show congestion in the ventral CCV and the entrapped and vacuolated yolk; yellow arrowheads show thickened posterior yolk extension; red arrows indicate the structure of the otic capsule. Above scale bar: 400 μm (one picture of sMO group has its own bar); below scale bar: 300 μm. (B) Statistics of teratogenesis and mortality on 4-dpf embryos. Each treatment group compared with wild-type, p < 0.01 (ANOVA Tests).
Atg5 gene overexpression led to embryo development retardation and absence of the body pigment
Embryos injected by atg5 capped-mRNA developed malformations. The whole embryo growth presented infantile or retarded phenotypes, such as a nearly colorless body, small otolith, ventralized optic vesicles, short of brain partition and insufficient differentiation of the trunk and abdomen, thickened posterior yolk extension, and reduced clarity of the yolk and yolk extension (Fig. 3A). Deformity and mortality at 4 dpf were about 25% and 30%, respectively (Fig. 3B).
Rescue of atg5 gene function—5MO coinjection with capped mRNA
To confirm atg5 function, a rescue experiment was performed by coinjecting 5MO and atg5 capped-mRNA. The result indicated that more embryos presented the normal phenotype than the MO group or overexpression group separately; some of the embryo phenotypes were improved and similar to that of the wild type. Deformity and mortality decreased to about 8% and 15%, respectively (Fig. 3B), lower than either the both MOs or capped-mRNA alone injection. This suggests that improvement of the malformation benefits from the rescue action by coinjection of 5MO plus the capped mRNA, which indicates that the teratogenesis resulted from dysregulation of the atg5 level.
Atg5 activity regulating the transcription level of atg5 itself in a feedback inhibition loop and linking to autophagy
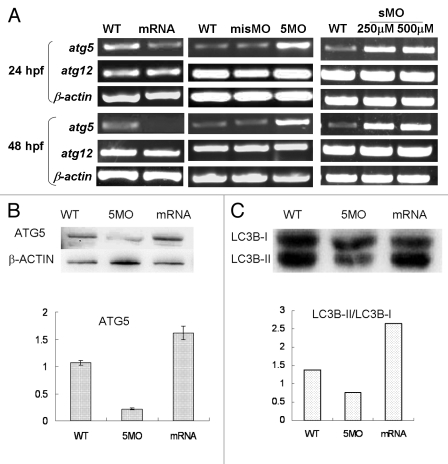
To determine how atg5 expression is regulated in the neurula development, transcription levels of atg5 and atg12 were tested with RT-PCR under conditions of atg5 gene down- or upregulated. Results show that the atg5 level not only did not decrease, but also clearly increased in 5MOatg5 or sMOatg5 injected embryos at 24 hpf and 48 hpf. However in embryos injected with atg5 capped-mRNA, atg5 mRNA level was markedly downregulated. Consistently, the atg12 level didn’t change under the both conditions (Fig. 4A). In contrast, western blotting showed that ATG5 protein (ATG12–ATG5 conjugate) was decreased in the 5MO injected group but obviously elevated in the mRNA-injected group (Fig. 4B). This contrary evidence suggests that the ATG5 protein level probably can regulate atg5 itself transcription in a feedback inhibition loop.
Figure 4.
Atg5 expression is regulated by itself protein in feedback inhibition. Embryos were injected 5MO, sMO or atg5 mRNA at 1–4 cell stage and collected for the detection. (A) RT-PCR results show that atg5 transcription level was down- or upregulated by atg5 mRNA or 5MO, sMO injection at 24 hpf and 48 hpf respectively, but atg12 was not affected. (B) Western blot results indicate that ATG5 protein (ATG5-ATG12 conjugate) down- or upregulated by 5MO or atg5 mRNA injection respectively at 48 hpf embryos, which is opposite to atg5 transcription. Anti-ATG5 antibody was diluted at 1:500 (Abcam, ab54033). misMO was as an injection control, β-ACTIN as a loading control, and WT as an untreated control. (C) Western blotting results show that the ratio of LC3B-II/LC3B-I is regulated by ATG5 protein at 48 hpf embryos, WT as a control. Anti-human LC3B antibody (Sigma, L7543) was diluted at 1/500.
Further, in order to explore whether the ATG5 activity really associates with autophagy, an autophagy marker LC3B was detected by western blot. The results showed the protein ratio of LC3B-II/LC3B-I was raised or reduced following ATG5 up- or downregulated respectively, which means ATG5 protein participating in embryonic neural development by implicated in autophagy pathway in zebrafish (Fig. 4C).
Atg5 activity can change the transcription pattern of some neural genes
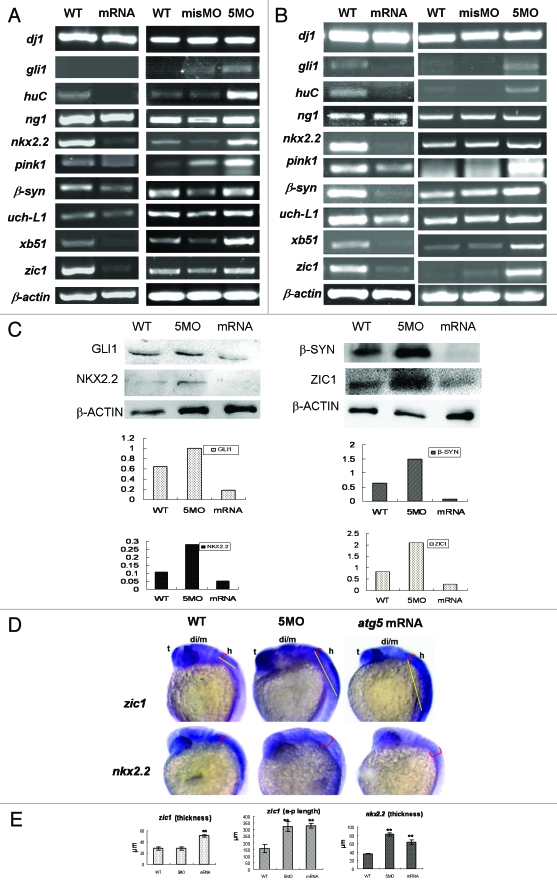
Respecting those analyses of ATG5 activity involved in embryonic neural development, we sought to determine whether atg5 could regulate expression of neural genes, and several important neural genes, such as dj1, gli1, neurogenin1 (ng1), huC, nkx2.2, pink1, β-synuclein, uch-L1, xb51, and zic1 were tested in transcriptional level under the condition of atg5 up- or downregulated respectively. The result showed that the expression level of these neural genes had significant increase except for dj1, ng1, and uch-L1 at a higher constant level in 5MO-injection group (Fig. 5A and B). In contrast, in embryos injected with atg5 capped-mRNA, mRNA levels of the gli1, huC, nkx2.2, pink1, β-synuclein, xb51 and zic1 were markedly downregulated, except for the three same invariable genes as in the 5MO-treated group (Fig. 5A and B). This evidence consistent with the atg5 change in transcription suggests that variation of ATG5 activity probably is able to affect those neural gene transcriptions. Then, some of them such as gli1, nkx2.2, zic1 and β-syn, were checked in protein variation, and the results showed the similar tendency to their mRNA change (Fig. 5C).
Figure 5 (See opposite page).
Atg5 activity can impact expression pattern of some neural genes in transcription and protein levels. RT-PCR shows that seven of the ten neural genes were down- or upregulated by atg5 mRNA or 5MO injection respectively at 24 hpf embryos (A) and at 48 hpf embryos (B). Protein gel blot results of 48 hpf embryo indicate that GLI1, NKX2.2, β-SYN and ZIC1 protein were up- or downregulated by 5MO or atg5 mRNA injection respectively (C), which is consistent with the transcription tendency. Hybridization in situ shows that gene zic1 and nkx2.2 signals were affected in intensity and distribution by atg5 level at 24 hpf (D). Histograms (E) show averages and standard deviations (SD) which are quantification of the variant phenotypes in D. For zic1 gene expression, zic1 signal in the dorsal-ventral axis of the hind brain (labeled as red semi-brackets, thickness in E) extended ventrally with about 1.7 times in atg5 mRNA injection (n = 12; 12/17#) compared with WT group (n = 12; 12/15#), and no obvious change in MO group (n = 9; 9/15#); in the anterior-posterior zone of the hind brain, zic1 signals (labeled as yellow lines in D, a-p length in E) in both mRNA group (n = 10; 10/17#) and MO group (n = 10; 10/15#) elongated backwardly at about two times compared with that in WT group (n = 12; 12/15#). For nkx2.2 gene expression, compared with WT group (n = 8; 8/12#), nkx2.2 signal in the dorsal-ventral axis of the hind brain (labeled as red semi-brackets, thickness in E) extended at about two times and 1.8 times in MO group (n = 8; 8/13#) and mRNA group (n = 9; 9/16#) respectively. **p < 0.01 (Oneway ANOVA Tests). #: in a fraction, a numerator represents number of the embryos selected randomly for the statistics analysis, a denominator indicates total embryos in a group. The error bars represent standard deviation (SD) in (E).
Further, hybridization in situ was used to reveal the spatial variations of nkx2.2 and zic1 activity in the embryonic brain when atg5 transcription was knockdown or overexpressed; nkx2.2 and zic1 are hallmarks for the ventral brain and dorsal brain, respectively. The results indicate that the activity of the two genes was for the most part limited in the embryonic brain and affected by increases or decreases of atg5 activity as with the RT-PCR result. In the atg5-mRNA injected group, the gene signals become attenuated and suffused evenly from the dorsal to the ventral region (Fig. 5D); nkx2.2 was spread around the whole brain area; its signal in dorsal-ventral axis of the hindbrain extended about 1.8 times as in WT control (Fig. 5E). zic1 signal became weaker in the diencephalon and midbrain than in wild-type embryos, but diffused on the whole hindbrain and enlarged backward closely to the trunk at least 1.7 times in the dorsal-ventral axis and more than two times in the anterior-posterior axis (Fig. 5D and E). In the 5MO groups, the gene transcriptions were enhanced, but their differential expression patterns were also disturbed: zic1 still resided in dorsal hindbrain but extended caudally about two times in the hindbrain and no obvious change in the dorsal-ventral axis; nkx2.2 was diffused around the whole brain over two times in the dorsal-ventral axis as in wild-type embryos (Fig. 5D and E). Statistical analysis showed their significant differences (Fig. 5E). This evidence suggests that the ATG5 signal may function as an important regulator to affect the expression level and pattern of the neural genes in a manner of feedback inhibition loop in zebrafish neural system development.
Rapamycin influence on embryonic development and on transcription of atg5 and some neural genes in zebrafish
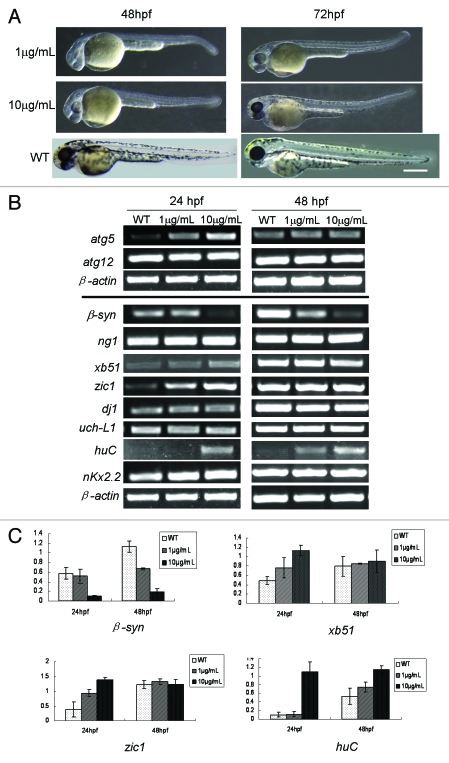
In view of rapamycin as a reagent for autophagy induction,27,28 we probed whether rapamycin can impact zebrafish embryonic development and atg5 expression. The results showed that the rapamycin-treated embryos presented the similar immature phenotype (Fig. 6A) as those injected with atg5 mRNA (Fig. 3A). Transcription levels of atg5, atg12 and some neural marker genes were examined in rapamycin-treated embryos. The results indicated that expression of atg5 and neural genes, xb51, zic1 and huC were upregulated mainly at 24 hpf embryo; however, β-synuclein was downregulated, and atg12, dj1, nkx2.2 and uch-L1 were nearly not disturbed by rapamycin at 24- and 48 hpf (Fig. 6B). It appears that these gene transcriptions at 24 hpf embryos are more sensitive than at 48 hpf in responding to rapamycin induction. The evidence suggests that the ATG5-related autophagy pathway may be affected by rapamycin too, but some neural genes’ responses are opposite to the regulation by overexpressing atg5 mRNA (Figs. 5A and 5B). The result means that ATG5 level is not a unique regulator or determining factor for the neural gene expression, particularly for those neural developmental genes. Signaling network of the neural system has much intercrossing linkage and is complicated. One gene's expression can be regulated by multiple regulators, and vice versa, and one regulator or inducer can play differential or opposite roles to different downstream genes, such as enhancement or inhibition. So, probably, the opposite responses of huC, xb51 and zic1 to atg5-upregulation may be a balanced result of ATG5 activity and rapamycin treatment in spite of RAPA raising atg5 level.
Figure 6.
RAPA leads to embryo development delayed and affects expression of atg5 and some neural genes. (A) Phenotype of rapamycin-treated embryos. Embryos treated by rapamycin from 6 hpf to 48 hpf or 72 hpf. The embryos show immature embryonic body, shorting of pigment, small eyes and brain, deficient heart, enlarged yolk and bended tail. WT indicates an un-treated embryo as a normal control. Scale bar: 400 μm. (B and C) Rapamycin can affect transcription of atg5 and some neural genes. Zebrafish embryos were exposed to rapamycin water from 6 hpf to 24 or 48 hpf respectively and collected. Then RT-PCR was performed and a representative RT-PCR showed in (B); and the bands were scanned according to their density; the relative ratio represents density mean values of three repeated RT-PCR results (C). Atg5 gene and neural genes, xb51, zic1 and huC were upregulated by rapamycin mainly at 24 hpf embryo. However β-syn was downregulated, and atg12, dj1 and uch-L1 were nearly not disturbed by rapamycin at 24- and 48 hpf.
Discussion
Atg5 expression pattern and function are correlated with neurula developmental process
We present here the first report of atg5 expression pattern and function in zebrafish embryonic development. We found that dynamic expression of the atg5 gene is concurrent with nerve system developmental process in zebrafish. The zygote atg5 expression was enhanced after 18 hpf and reached the greatest level at 24 hpf. Hybridization in situ and immunofluorescence further confirmed a high atg5 signal in the brain region at 24 hpf, then later focused on the midbrain-hindbrain and floor plate at 48–72 hpf. It is known that 24 hpf is a critical time for neurogenesis and organogenesis, which maintain an equal pace with high atg5 transcription. This indicates that atg5 transcription is probably correlated with embryonic nerve system development.
We investigated the embryo phenotype with MO-injected, capped mRNA-injected, and rescue by coinjection of the both molecules. Overdorsalized deformed embryos were produced by MOs knockdown, whereby the neural system and body structure were lethally impaired. Conversely, atg5 overexpression led to embryo hypodevelopment, with an infantile head and lack of body pigmentation. A lesser degree of malformation was obtained by coinjection of 5MO plus the capped mRNA, which implies that the teratogenesis is resulted from atg5 dysregulation. These results are in accordance with studies in mice. Deficiency of autophagy-related genes in mice can cause death early in embryogenesis29 as well as a lower birth weight and neonatal death within a day of birth.17,30 Our results together with these data provide a strong basis for the theory that autophagy genes, particularly atg5, play an important role in the regulation of neurula development.
Regulation of atg5 gene expression in a feedback inhibition loop and underlying autophagy
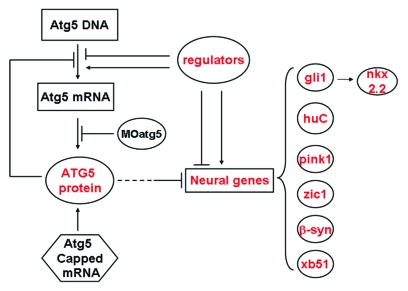
We investigated how the atg5 gene expression was regulated. From our results, we speculate that a regulation mechanism of atg5 expression is probably—at least partially—in the manner of a feedback inhibition loop (Fig. 7). We found that changes in the ATG5 protein level were the reverse of changes in atg5 transcription level, but this is reasonable. Atg5 transcription is likely to be negatively regulated by its own protein. The higher atg5 transcript signal probably resulted from the reduction in ATG5 protein; in other words, less ATG5 protein reactivated host cell atg5 transcription and produced more atg5 transcript copies, as evidenced by the 5MO and sMO injected treatments. However, in the experiment, more atg5 mRNA from host cells was not translated into protein, probably owing to excess 5MO still having a sustained suppression.31 Thus, in the embryos subjected to MO injection, atg5 mRNA was elevated and the protein downregulated. On the other hand, in the atg5 mRNA-injected group, the ATG5 protein quantity was apparently increased directly from the ectogenous capped mRNA, but the transcript level diminished, probably owing to excessive ATG5 protein feedback inhibition and the injected mRNA degrading quickly in the subsequent developmental process.
Figure 7.
A model for explaining expression regulation of atg5 gene and some neural genes in zebrafish embryos.
A couple of studies have shown that there are two forms of ATG5 protein: truncated ATG5 alone and the ATG12–ATG5 conjugate, which the former ATG5 exerts pro-apoptosis, and the latter heterodimer is involved in autophagy.24,25 Our western blot suggests that ATG5 appears in three forms, the 24 kD truncated, the whole length of 33 kD and the conjugate of 56 kD (Fig. 1C), and among them two forms of the 56 kD and 24 kD are major; the 33 kD is little only, which means that atg5-related autophagy and apoptosis may be coexisted in zebrafish embryonic development. In order to confirm autophagy is really involved in the embryonic neural development, the ratio of LC3-II/LC3-I, which is a specific marker for autophagic vacuole,32,33 was detected under conditions of atg5 up- and downregulated using western blot. The result demonstrates that the LC3-II/LC3-I ratio raised or reduced is consistent with ATG5 protein level. Other evidence from the rapamycin experiment also supports that atg5 is involved in the neural developing by autophagic process: rapamycin enhanced atg5 expression and caused embryonic development delayed or infantile, which is similar to atg5 overexpression. So, we think that ATG5-related autophagy is involved in the neural development even though apoptosis may be also implicated.
In addition, some studies have reported that both ATG5 and ATG12 were key factors in regulating the formation of autophagic vacuoles (AV) within eukaryotic cells.34 However, our study showed no change in atg12 expression in either atg5 gene knockdown or overexpression, even in rapamycin treating or during the whole embryonic developing process; instead, it maintained a constant higher expression level, which indicates that atg12 probably is a housekeeping gene and atg5 is a key regulator involved in the ATG12–ATG5 conjugate formation in the zebrafish neurula.
Atg5 activity regulating transcription of neural marker genes in embryonic neurula development
To understand how atg5 affects neural genes in neurogenesis, transcription of some neural marker genes was measured by RT-PCR. We found an expression association between atg5 and these neural genes—gli1, huC, nkx2.2, pink1, β-synuclein, xb51 and zic1—which are tightly regulated in spatiotemporal patterning in conjunction with elevated or decreased atg5 transcription. So we suppose that atg5 may act in the formation of the embryonic neural system, mediated by acting upstream of the neural genes to regulate their transcription in the same manner as atg5 itself (Fig. 5A and 5B). In other words, ATG5 protein, probably as a negative regulator of these genes, exerts transcription regulation in zebrafish neurula development. The gli1 gene functions by maintaining neural progenitors at the midbrain-hindbrain boundary35 and by inducing the motor neuron in zebrafish.36 GLI1 protein, as a zinc finger transcription factor, can activate nkx2.2 and other Hh signaling responding gene transcription.37-39 Nkx2.2 functions in the ventral brain and posterior neural tissue, which complements the zic1 pattern in the zebrafish brain.40 So, in our study, gli1 was downregulated by atg5 overexpression, and this probably in turn downregulated nkx2.2, which may be a reason for the poor brain partition and infantile brain development. As a brain dorsal marker gene, zic1 is expressed throughout the dorsal brain and in the optic stalk in wild-type embryo.41 In 5MO-injected embryos, zic1 expression is enhanced in the whole dorsal brain, particularly in the hindbrain extended posteriorly to the trunk; however, it was reduced in the diencephalon/midbrain and suffused ventrally and backwardly in the whole hindbrain, with poor brain division in the atg5 capped-mRNA-injected embryos. This result is supported by a study which indicated that zebrafish zic1 could control the midline formation and forebrain patterning; zic1 loss of function led to ventralization of the optic vesicle and defects of the forebrain midline in zebrafish embryos.42 The other neural marker genes—huC, β-synuclein, pink1, uch-L1 and xb51—are also responding for formation of the nerve system in zebrafish. Xb51 is mainly expressed in the dorsal telencephalon at 24 hpf and is a hallmark of Alzheimer disease;43 huC is a pan-neuronal marker in the early neural development.44 Pink1 gene is involved in the formation of the semicircular canal, and its dysregulation is correlated with Parkinson disease.45 Zebrafish uch-L1 is expressed in neuronal cells in the ventral diencephalon and other brain domains.46 β-synuclein is expressed strongly in the nervous system, such as the eyes, ventral brain, and dorsal somites at 48 hpf.47 The genes in this study were up- or downregulated depending on the atg5 level, so we think these genes exerting their action partly as ATG5 effectors. For example, otic capsule deformity, including otolith number, size and the semicircular canal, is perhaps mediated directly by pink1 up- or downregulation,48 and the immature brain probably resulted from the combined inhibition of these genes, which can be downregulated by the atg5 overexpression.
Together with these previous findings, the observations in our study provide strong support for the hypothesis that autophagy-related gene atg5 plays an important role by regulating expression of some key neural genes and itself by means of a feedback inhibition loop in the development of the embryonic nervous system, particularly in brain formation in zebrafish.
In summary, we first examined the zebrafish atg5 gene spatiotemporal expression pattern, which coincides with neurogenesis. By functional analysis, we understand that atg5 plays an important role in neurogenesis and organogenesis. By detection of the gene transcription and translation, we find that variations in ATG5 protein level are capable of regulating the expression of some key neural genes and of the atg5 gene by a feedback inhibition loop. Atg12 transcription generally maintains an excess level, and atg5 expression is controlled tightly under a spatiotemporal pattern (Fig. 2). Thus, we infer that formation of an ATG12–ATG5 conjugate may be dependent on ATG5 protein production and its splicing, and not on the ATG12 protein in zebrafish development. Besides, atg5 signaling and neural genes are also affected by rapamycin. Finally, based on the results of this study, we provide a mechanistic insight into the regulation of atg5 expression and action in development of zebrafish nervous system.
Materials and Methods
Zebrafish maintenance and embryo collection
Zebrafish (Danio rerio) wild-type Tuebingen (TU) strain were raised at 28.5 ± 1°C and staged according to the literature.26 The TU strain of fish was originally obtained from the College of Life Sciences, Beijing University. Embryos were obtained by natural mating; synchronous embryos at the appropriate stage were collected. They were fixed with 4% paraformaldehyde in phosphate-buffered saline for in situ hybridization or immersed in Trizol reagent for mRNA isolation. Embryos after 24 hpf were discolored by incubating in phenylthiourea (PTU) (Sigma, P3755) before being fixed.
RT-PCR assays
Total RNA was extracted from whole embryos at different stages of development using Trizol reagent (Invitrogen, 15596-026). First-strand cDNAs were synthesized by reverse transcription using the M-MLV RTase cDNA Synthesis Kit (TaKaRa, D6130). For the polymerase chain reaction (PCR) amplification, 1 ng cDNA was used for each reaction with different primer pairs. β-actin was amplified as a PCR template (loading) control. PCR conditions were 94°C (30 sec), 52°C (30 sec), and 72°C (60 or 120 sec based on the gene-fragment length) for 25 (for β-actin) or 30 cycles.
Drug treatment
A stock solution of rapamycin (RAPA, provided by National Institutes for Food and Drug Control) in DMSO was added to the embryo media at final concentrations of 1 μg/mL and 10 μg/mL at embryo 6 hpf and upto 24 hpf or 48 hpf respectively.49 And wild-type embryo was used as negative control.
Microinjection of morpholino oligo and capped mRNA
The atg5 morpholino oligo (5MOatg5, sMO) and misMO were designed and bought from Genetools LLC (http://www.gene-tools.com), in which 5MOatg5 was used for inhibition of atg5 translation by binding to atg5 initiation sites with the sequence 5′- CAT CCT TGT CAT CTG CCA TTA TCA T-3′; the control misMO’ sequence was 5′- CAT CgT TcT CAT CTc CCA TaA TgAT-3′. The sMOatg5′ sequence was 5′- GTG CCC TTA AAA CCA AAA ATA ACAC-3′. The 5MO, sMO and misMO of 0.5 nL with 250 μM or 500 μM was injected into embryos at the one- to four-cell stage.
Atg5 whole-length cDNA sequence was cloned using RT-PCR from 24 hpf wild-type embryos according to the atg5 gene sequence logged on GenBank (HQ450378.1) and inserted into pBluescript KS plasmid. Capped mRNA of atg5 was synthesized with the Capped mRNA kit (Ambion, AM1348) and 175 pg was injected into one- to four-cell-stage embryos.
Whole-mount in situ hybridization
Preparation of RNA probe. For in vitro synthesis of the RNA probe, target gene fragments, such as zebrafish atg5, nkx2.2 and zic1 sequences, using PCR (the parameters are presented in Table 1), were cloned into pBluescript KS plasmid as the templates. Then, antisense and sense RNA probes were synthesized as described in the kit instructions of the DIG RNA labeling kit (Roche, 1175041).
Table 1. Primer pairs in this study.
| Gene name | Primer sequence | Ta (°C) | PCR size (bp) |
|---|---|---|---|
|
atg5 |
F: 5′ ATGATAATGGCAGATGACAAGG 3′ R: 5′ TCAGTCACTCGGTGCAGG 3′ |
54 |
828 |
|
atg12 |
F: 5′ ATGTCTGACAACGCAGAATC 3′ R: 5′ TCATCCCCAGGCCTGAGACTT 3′ |
52 |
363 |
|
β-synuclein |
F: 5′ ATGGATGTTTTTATGAAGGGGC 3′ R: 5′ TTACGCCTCGGGCTCATAATCCTGG 3′ |
56 |
384 |
|
dj1 |
F: 5′ATGGCCGGTAAAAGAGCGTTAGTGA 3′ R: 5′TTAGTCTTTCAGGATGAGCGGG 3′ |
57 |
570 |
|
gli1 |
F: 5′ CCATCATAATCTCCCTCATA 3′ R: 5′ GTGGCAGTTCGTCTCATAAA 3′ |
55 |
598 |
|
huC |
F: 5′ CTTTGTACGTCAAGAATGG 3′ R: 5′ CATGTTAAAGAGCAATAGTGAC 3′ |
54 |
1086 |
|
nkx2.2 |
F: 5′ CCAGAACATGTCGTTGAC 3′ R: 5′ TTTGTTCACCAAGTCCAC 3′ |
53 |
822 |
|
neurogenin 1 |
F: 5′ AGGTTATCAACAATGGAG 3′ R: 5′ TCACATTAATAGATGCTAGG 3′ |
53 |
644 |
|
pink1 |
F: 5′ GCTCACAGAGACCTCAAATCAGA 3′ R: 5′CAGTGTGTAGAAGGGGTTTGG 3′ |
55 |
291 |
|
uch-L1 |
F: 5′ ATGGAGTGGAAACCGATGGAAATA 3′ R: 5′TCAGGCTTTGCAGAGAGCA 3′ |
54 |
657 |
|
xb51 |
F: 5′ ATGGATTGCTTTGAAGAG 3′ R: 5′ CACTAGTTATTGTTGAGGAC 3′ |
51 |
1049 |
|
zic1 |
F: 5′ CTGAAGATGCTCTTGGAC 3′ R: 5′ TTACACGTACCATTCATTA 3′ |
52 |
1335 |
| β-actin | F: 5′ AGGGAAATCGTGGGTGACATCAAA 3′ R: 5′ ACTCATCGTACTCCTGCTTGCTGA 3′ |
55 | 478 |
Embryo whole-mount in situ hybridization. Embryos developed over 24 hpf were pretreated with PTU to suppress pigmentation before being fixed. Then, embryos were fixed with 4% paraformaldehyde overnight at 4°C. Embryos from 6–72 hpf stages were pretreated by DNase prior to starting the hybridization to remove genomic DNA pseudo-positive interference. In situ hybridization was performed as described by Westerfield.50
Whole-mount immunohistochemistry
Embryos were collected at 24 hpf and pretreated with proteinase K (Tiangen, RT403). The procedure basically followed that of Higashijima et al.51 Briefly, embryos were incubated in the primary antibody of rabbit anti-human ATG5 polyclonal antibody (Abcam, ab54033) at a dilution of 1/500 overnight at 4°C, then incubated with a rhodamine-conjugated anti-rabbit secondary antibody at a dilution of 1/2,000 overnight. Control embryos were incubated only with the secondary antibody at the same dilution overnight. Images were taken using an inverted phase-contrast fluorescent microscope (Olympus, IX 51).
Western blot
The immunoblotting procedure has been previously described in the literature.25 Briefly, total protein of zebrafish embryos was extracted with the Tissue Protein Rapid Miniprep Kit (TIANDZ, 90707-50) using protease inhibitors and separated in 12% sodium dodecyl sulfate-PAGE (SDS-PAGE). The protein bands were then transferred to a nitrocellulose membrane that was blocked with tris buffered saline (TBS) containing 10% skim milk for 1 h at room temperature. The membranes were incubated with primary antibody anti-human ATG5 (Abcam, ab54033); anti-human tATG5 and anti-human ATG5 antibodies (abgent, AP1812a and AP1812b)52,53 diluted at 1:200. Anti-human LC3B antibody (sigma, L7543), anti-human ZIC1, anti-human β-SYNUCLEIN, anti-human GLI1 and anti-human NKX2.2 antibodies (Abcam, ab58080, ab76111, ab49314 and ab85675) and mouse anti-human β-ACTIN antibody (Zhongshan Goldbridge, TA09) at a dilution of 1:1,000 overnight at 4°C and then treated with horseradish peroxidase-conjugated secondary antibodies. The membranes were visualized by enhanced chemiluminescence using the Supersignal® West Pico chemiluminescent substrate (Thermo, 34080) with the AlphaEase® FC Imaging System. β-actin was used as a loading control.
Statistical analysis
Data in bars represent mean ± s.d in histograms. The means and standard deviations are derived from at least triplicates. Statistical analyses were performed using Oneway ANOVA Tests and p-values < 0.05 were considered as significant. The embryos used in the quantification were randomly selected.
Acknowledgment
This study was supported by The National Natural Science Foundation of China (No. 30772681), and National S&T Major Special Project on Major New Drug Innovation (Item Number: 2009ZX09301-003-6-2 and 2012ZX09301002-001-021 ) grant. And we also thank Weixian Wang and Jie Meng for their efforts in microinjection and fish administration, and thank professor Bo Zhang (Beijing University) provided zebrafish TU stain seedling.
Glossary
Abbreviations
- PCD
programmed cell death
- atg5
autophagy-related gene 5
- TU
Tuebingen
- hpf
hour postfertilization
- WT
wild type
- kD
Kilo Dalton
- RAPA
rapamycin
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Footnotes
Previously published online: www.landesbioscience.com/journals/autophagy/article/18040
References
- 1.Kametaka S, Matsuura A, Wada Y, Ohsumi Y. Structural and functional analyses of APG5, a gene involved in autophagy in yeast. Gene. 1996;178:139–43. doi: 10.1016/0378-1119(96)00354-X. [DOI] [PubMed] [Google Scholar]
- 2.Ravikumar B, Rubinsztein DC. Role of autophagy in the clearance of mutant huntingtin: A step towards therapy? Mol Aspects Med. 2006;27:520–7. doi: 10.1016/j.mam.2006.08.008. [DOI] [PubMed] [Google Scholar]
- 3.Cecconi F, Levine B. The role of autophagy in mammalian development: cell makeover rather than cell death. Dev Cell. 2008;15:344–57. doi: 10.1016/j.devcel.2008.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Penaloza C, Lin L, Lockshin RA, Zakeri Z. Cell death in development: shaping the embryo. Histochem Cell Biol. 2006;126:149–58. doi: 10.1007/s00418-006-0214-1. [DOI] [PubMed] [Google Scholar]
- 5.Baehrecke EH. Autophagy SEPArates germline and somatic cells. Cell. 2009;136:207–8. doi: 10.1016/j.cell.2009.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Boland B, Nixon RA. Neuronal macroautophagy: From development to degeneration. Mol Aspects Med. 2006;27:503–19. doi: 10.1016/j.mam.2006.08.009. [DOI] [PubMed] [Google Scholar]
- 7.Funderburk SF, Marcellino BK, Yue Z. Cell “self-eating” (autophagy) mechanism in Alzheimer's disease. Mt Sinai J Med. 2010;77:59–68. doi: 10.1002/msj.20161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cherra SJ, Chu CT. Autophagy in neuroprotection and neurodegeneration: A question of balance. Future Neurol. 2008;3:309–23. doi: 10.2217/14796708.3.3.309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cheung ZH, Ip NY. The emerging role of autophagy in Parkinson's disease. Mol Brain. 2009;2:29. doi: 10.1186/1756-6606-2-29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Komatsu M, Waguri S, Chiba T, Murata S, Iwata J, Tanida I, et al. Loss of autophagy in the central nervous system causes neurodegeneration in mice. Nature. 2006;441:880–4. doi: 10.1038/nature04723. [DOI] [PubMed] [Google Scholar]
- 11.Williams A, Sarkar S, Cuddon P, Ttofi EK, Saiki S, Siddiqi FH, et al. Novel targets for Huntington’s disease in an mTOR-independent autophagy pathway. Nat Chem Biol. 2008;4:295–305. doi: 10.1038/nchembio.79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Matsushita M, Suzuki NN, Obara K, Fujioka Y, Ohsumi Y, Inagaki F. Structure of Atg5.Atg16, a complex essential for autophagy. J Biol Chem. 2007;282:6763–72. doi: 10.1074/jbc.M609876200. [DOI] [PubMed] [Google Scholar]
- 13.Fujioka Y, Noda NN, Nakatogawa H, Ohsumi Y, Inagaki F. Dimeric coiled-coil structure of Saccharomyces cerevisiae Atg16 and its functional significance in autophagy. J Biol Chem. 2010;285:1508–15. doi: 10.1074/jbc.M109.053520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kuma A, Hatano M, Matsui M, Yamamoto A, Nakaya H, Yoshimori T, et al. The role of autophagy during the early neonatal starvation period. Nature. 2004;432:1032–6. doi: 10.1038/nature03029. [DOI] [PubMed] [Google Scholar]
- 15.Komatsu M, Waguri S, Ueno T, Iwata J, Murata S, Tanida I, et al. Impairment of starvation-induced and constitutive autophagy in Atg7-deficient mice. J Cell Biol. 2005;169:425–34. doi: 10.1083/jcb.200412022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hara T, Nakamura K, Matsui M, Yamamoto A, Nakahara Y, Suzuki-Migishima R, et al. Suppression of basal autophagy in neural cells causes neurodegenerative disease in mice. Nature. 2006;441:885–9. doi: 10.1038/nature04724. [DOI] [PubMed] [Google Scholar]
- 17.Winslow AR, Rubinsztein DC. Autophagy in neurodegeneration and development. Biochim Biophys Acta. 2008;1782:723–9. doi: 10.1016/j.bbadis.2008.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chae M, Rhee GS, Jang IS, Kim K, Lee JH, Lee SY, et al. ATG5 expression induced by MDMA (ecstasy), interferes with neuronal differentiation of neuroblastoma cells. Mol Cells. 2009;27:571–5. doi: 10.1007/s10059-009-0075-2. [DOI] [PubMed] [Google Scholar]
- 19.Pua HH, Dzhagalov I, Chuck M, Mizushima N, He YW. A critical role for the autophagy gene Atg5 in T cell survival and proliferation. J Exp Med. 2007;204:25–31. doi: 10.1084/jem.20061303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Alirezaei M, Fox HS, Flynn CT, Moore CS, Hebb AL, Frausto RF, et al. Elevated ATG5 expression in autoimmune demyelination and multiple sclerosis. Autophagy. 2009;5:152–8. doi: 10.4161/auto.5.2.7348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Crews L, Spencer B, Desplats P, Patrick C, Paulino A, Rockenstein E, et al. Selective molecular alterations in the autophagy pathway in patients with Lewy body disease and in models of alpha-synucleinopathy. PLoS ONE. 2010;5:e9313. doi: 10.1371/journal.pone.0009313. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 22.Kari G, Rodeck U, Dicker AP. Zebrafish: An Emerging Model System for Human Disease and Drug Discovery. Clin Pharmacol Ther. 2007;82:70–80. doi: 10.1038/sj.clpt.6100223. [DOI] [PubMed] [Google Scholar]
- 23.Best JD, Alderton WK. Zebrafish: An in vivo model for the study of neurological diseases. Neuropsychiatr Dis Treat. 2008;4:567–76. doi: 10.2147/ndt.s2056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Codogno P, Meijer AJ. Atg5: more than an autophagy factor. Nat Cell Biol. 2006;8:1045–7. doi: 10.1038/ncb1006-1045. [DOI] [PubMed] [Google Scholar]
- 25.Yousefi S, Perozzo R, Schmid I, Ziemiecki A, Schaffner T, Scapozza L, et al. Calpain-mediated cleavage of Atg5 switches autophagy to apoptosis. Nat Cell Biol. 2006;8:1124–32. doi: 10.1038/ncb1482. [DOI] [PubMed] [Google Scholar]
- 26.Kimmel CB, Ballard WW, Kimmel SR, Ullmann B, Schilling TF. Stages of embryonic development of the Zebrafish. Dev Dyn. 1995;203:253–310. doi: 10.1002/aja.1002030302. [DOI] [PubMed] [Google Scholar]
- 27.Kalamidas SA, Kondomerkos DJ, Kotoulas OB, Hann AC. Electron microscopic and biochemical study of the effects of rapamycin on glycogen autophagy in the newborn rat liver. Microsc Res Tech. 2004;63:215–9. doi: 10.1002/jemt.20032. [DOI] [PubMed] [Google Scholar]
- 28.Rangaraju S, Verrier JD, Madorsky I, Nicks J, Dunn WA, Jr., Notterpek L. Rapamycin activates autophagy and improves myelination in explant cultures from neuropathic mice. J Neurosci. 2010;30:11388–97. doi: 10.1523/JNEUROSCI.1356-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yue Z, Jin S, Yang C, Levine AJ, Heintz N. Beclin 1, an autophagy gene essential for early embryonic development, is a haploinsufficient tumor suppressor. Proc Natl Acad Sci USA. 2003;100:15077–82. doi: 10.1073/pnas.2436255100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cecconi F, Levine B. The Role of Autophagy in Mammalian Development: Cell Makeover Rather than Cell Death. Dev Cell. 2008;15:344–57. doi: 10.1016/j.devcel.2008.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Heasman J. Morpholino Oligos: Making Sense of Antisense? Dev Biol. 2002;243:209–14. doi: 10.1006/dbio.2001.0565. [DOI] [PubMed] [Google Scholar]
- 32.Yokoyama T, Miyazawa K, Naito M, Toyotake J, Tauchi T, Itoh M, et al. Vitamin K2 induces autophagy and apoptosis simultaneously in leukemia cells. Autophagy. 2008;4:629–40. doi: 10.4161/auto.5941. [DOI] [PubMed] [Google Scholar]
- 33.Michiorri S, Gelmetti V, Giarda E, Lombardi F, Romano F, Marongiu R, et al. The Parkinson-associated protein PINK1 interacts with Beclin1 and promotes autophagy. Cell Death Differ. 2010;17:962–74. doi: 10.1038/cdd.2009.200. [DOI] [PubMed] [Google Scholar]
- 34.Levine B, Klionsky DJ. Development by self-digestion: molecular mechanisms and biological functions of autophagy. Dev Cell. 2004;6:463–77. doi: 10.1016/S1534-5807(04)00099-1. [DOI] [PubMed] [Google Scholar]
- 35.Ninkovic J, Stigloher C, Lillesaar C, Bally-Cuif L. Gsk3beta/PKA and Gli1 regulate the maintenance of neural progenitors at the midbrain-hindbrain boundary in concert with E(Spl) factor activity. Development. 2008;135:3137–48. doi: 10.1242/dev.020479. [DOI] [PubMed] [Google Scholar]
- 36.Vanderlaan G, Tyurina OV, Karlstrom RO, Chandrasekhar A. Gli function is essential for motor neuron induction in zebrafish. Dev Biol. 2005;282:550–70. doi: 10.1016/j.ydbio.2005.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Briscoe J, Pierani A, Jessell TM, Ericson J. A homeodomain protein code specifies progenitor cell identity and neuronal fate in the ventral neural tube. Cell. 2000;101:435–45. doi: 10.1016/S0092-8674(00)80853-3. [DOI] [PubMed] [Google Scholar]
- 38.Karlstrom RO, Tyurina OV, Kawakami A, Nishioka N, Talbot WS, Sasaki H, et al. Genetic analysis of zebrafish gli1 and gli2 reveals divergent requirements for gli genes in vertebrate development. Development. 2003;130:1549–64. doi: 10.1242/dev.00364. [DOI] [PubMed] [Google Scholar]
- 39.Nguyen V, Chokas AL, Stecca B, Ruiz i Altaba A. Cooperative requirement of the Gli proteins in neurogenesis. Development. 2005;132:3267–79. doi: 10.1242/dev.01905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bergeron SA, Milla LA, Villegas R, Shen MC, Burgess SM, Allende ML, et al. Expression profiling identifies novel Hh/Gli regulated genes in developing zebrafish embryos. Genomics. 2008;91:165–77. doi: 10.1016/j.ygeno.2007.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Grinblat Y, Sive H. Zic Gene Expression Marks Anteroposterior Pattern in the Presumptive Neurectoderm of the Zebrafish Gastrula. Dev Dyn. 2001;222:688–93. doi: 10.1002/dvdy.1221. [DOI] [PubMed] [Google Scholar]
- 42.Maurus D, Harris WA. Zic-associated holoprosencephaly: zebrafish Zic1 controls midline formation and forebrain patterning by regulating Nodal, Hedgehog, and retinoic acid signaling. Genes Dev. 2009;23:1461–73. doi: 10.1101/gad.517009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kim HT, Kim EH, Yoo KW, Lee MS, Choi JH, Park HC, et al. Isolation and Expression Analysis of Alzheimer’s Disease-Related Gene xb51 in Zebrafish. Dev Dyn. 2008;237:3921–6. doi: 10.1002/dvdy.21806. [DOI] [PubMed] [Google Scholar]
- 44.Park HC, Kim CH, Bae YK, Yeo SY, Kim SH, Hong SK, et al. Analysis of upstream elements in the HuC promoter leads to the establishment of transgenic zebrafish with fluorescent neurons. Dev Biol. 2000;227:279–93. doi: 10.1006/dbio.2000.9898. [DOI] [PubMed] [Google Scholar]
- 45.Petko JA, Kabbani N, Frey C, Woll M, Hickey K, Craig M, et al. Proteomic and functional analysis of NCS-1 binding proteins reveals novel signaling pathways required for inner ear development in zebrafish. BMC Neurosci. 2009;10:27. doi: 10.1186/1471-2202-10-27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Son OL, Kim HT, Ji MH, Yoo KW, Rhee M, Kim CH. Cloning and expression analysis of a Parkinson's disease gene, uch-L1, and its promoter in zebrafish. Biochem Biophys Res Commun. 2003;312:601–7. doi: 10.1016/j.bbrc.2003.10.163. [DOI] [PubMed] [Google Scholar]
- 47.Sun Z, Gitler AD. Discovery and characterization of three novel synuclein genes in zebrafish. Dev Dyn. 2008;237:2490–5. doi: 10.1002/dvdy.21569. [DOI] [PubMed] [Google Scholar]
- 48.Anken RH. On the role of the central nervous system in regulating the mineralisation of inner-ear otoliths of fish. Protoplasma. 2006;229:205–8. doi: 10.1007/s00709-006-0219-6. [DOI] [PubMed] [Google Scholar]
- 49.Makky K, Tekiela J, Mayer AN. Target of rapamycin (TOR) signaling controls epithelial morphogenesis in the vertebrate intestine. Dev Biol. 2007;303:501–13. doi: 10.1016/j.ydbio.2006.11.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Whitlock KE, Westerfield M. The olfactory placodes of the zebrafish form by convergence of cellular fields at the edge of the neural plate. Development. 2000;127:3645–53. doi: 10.1242/dev.127.17.3645. [DOI] [PubMed] [Google Scholar]
- 51.Higashijima S, Hotta Y, Okamoto H. Visualization of cranial motor neurons in live transgenic zebrafish expressing green fluorescent protein under the control of the islet-1 promoter/enhancer. J Neurosci. 2000;20:206–18. doi: 10.1523/JNEUROSCI.20-01-00206.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Lépine S, Allegood JC, Park M, Dent P, Milstien S, Spiegel S. Sphingosine-1-phosphate phosphohydrolase-1 regulates ER stress-induced autophagy. Cell Death Differ. 2011;18:350–61. doi: 10.1038/cdd.2010.104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Xia HG, Zhang L, Chen G, Zhang T, Liu J, Jin M, et al. Control of basal autophagy by calpain1 mediated cleavage of ATG5. Autophagy. 2010;6:61–6. doi: 10.4161/auto.6.1.10326. [DOI] [PMC free article] [PubMed] [Google Scholar]