Abstract
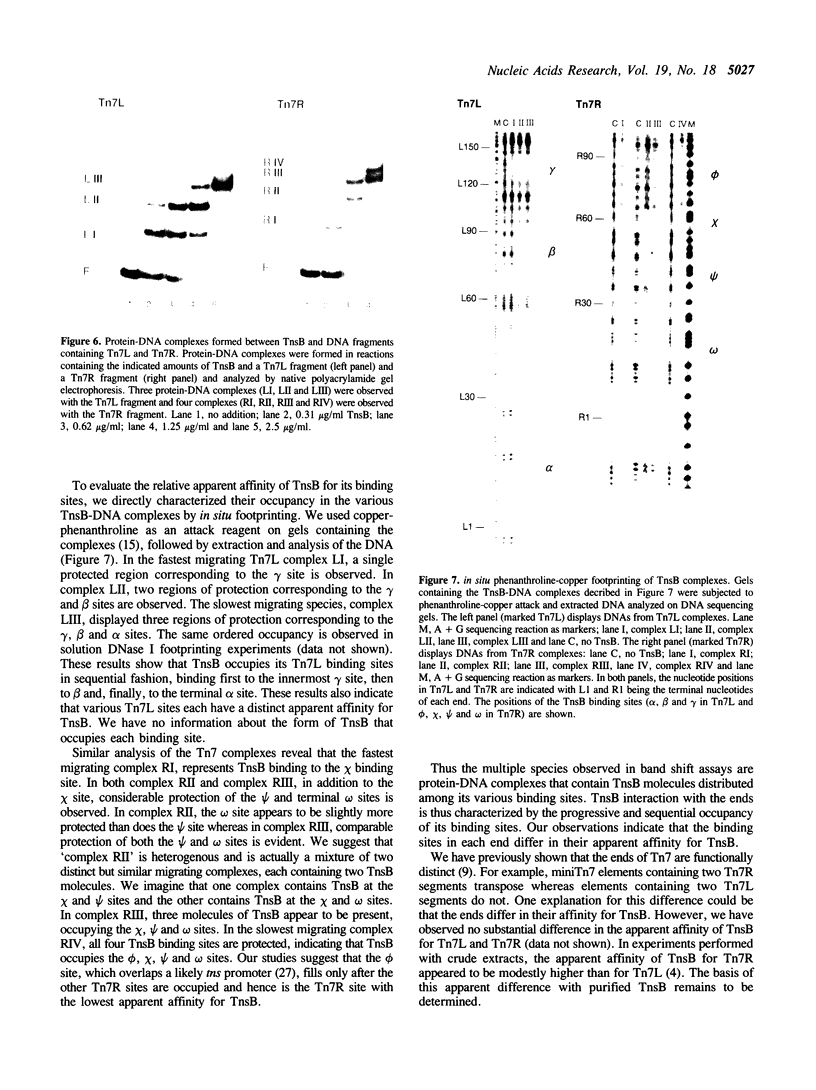
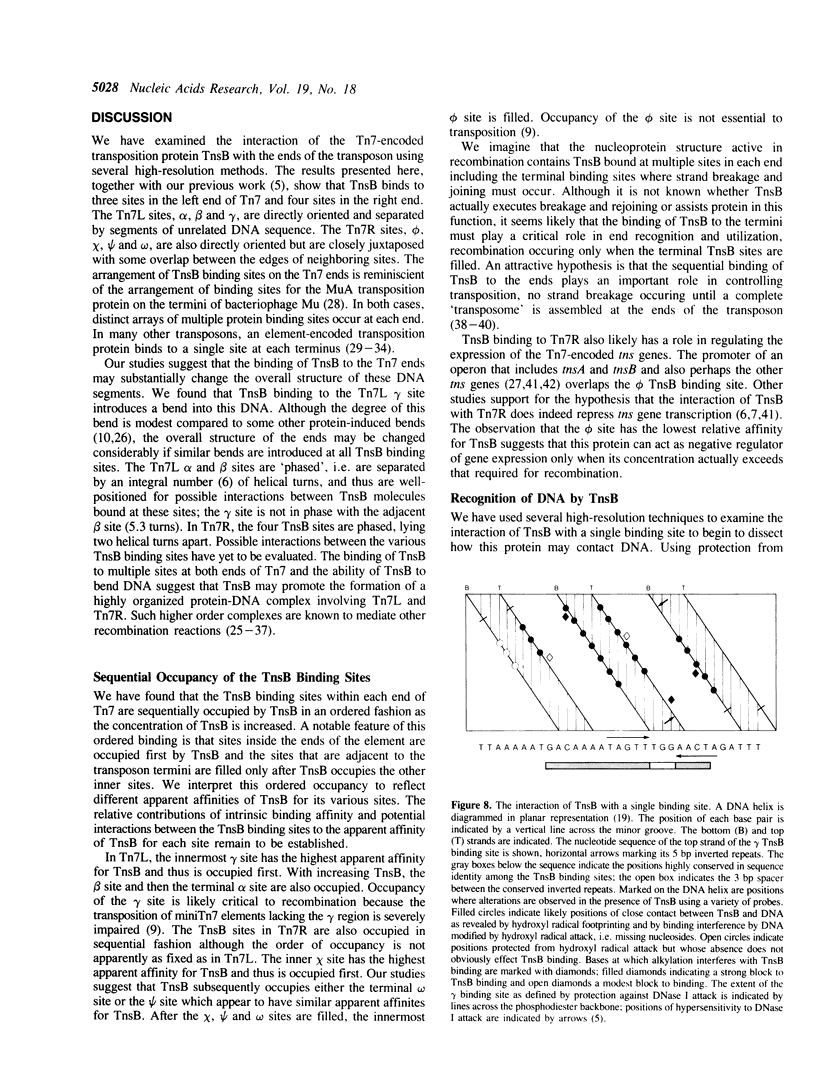
We have used several high resolution methods to examine the interaction of TnsB, a transposition protein encoded by the bacterial transposon Tn7, with its binding sites at the ends of the transposon. These binding sites lie within the DNA segments that are directly involved in transposition. We show that the binding of TnsB to DNA can promote DNA bending, suggesting that the interaction of TnsB with the ends may result in formation of a highly organized protein-DNA complex. We also identify likely positions of close contact between of TnsB and its binding sites. Analysis of the interaction of TnsB with intact Tn7 ends reveals TnsB occupies its binding sites in a particular order, the sites immediately adjacent to the transposon termini being occupied only after other inner sites are bound. Such ordered occupancy suggests that the various binding sites have differing apparent affinities for TnsB.
Full text
PDF

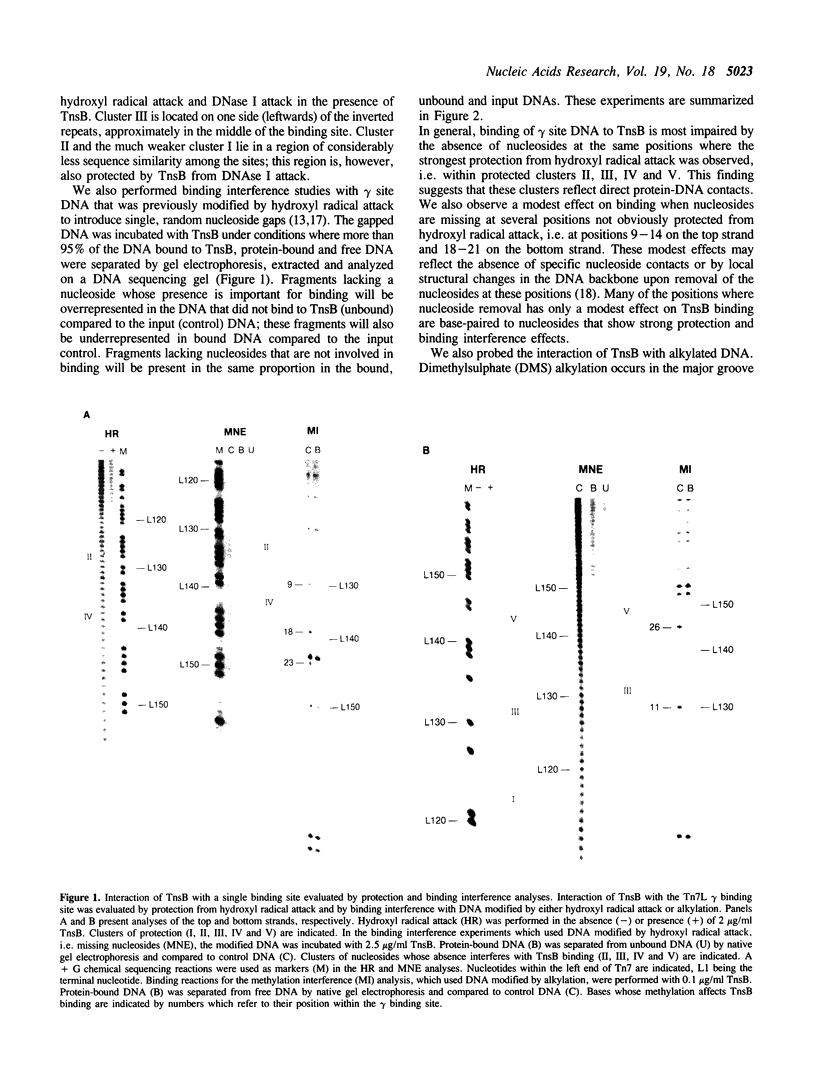

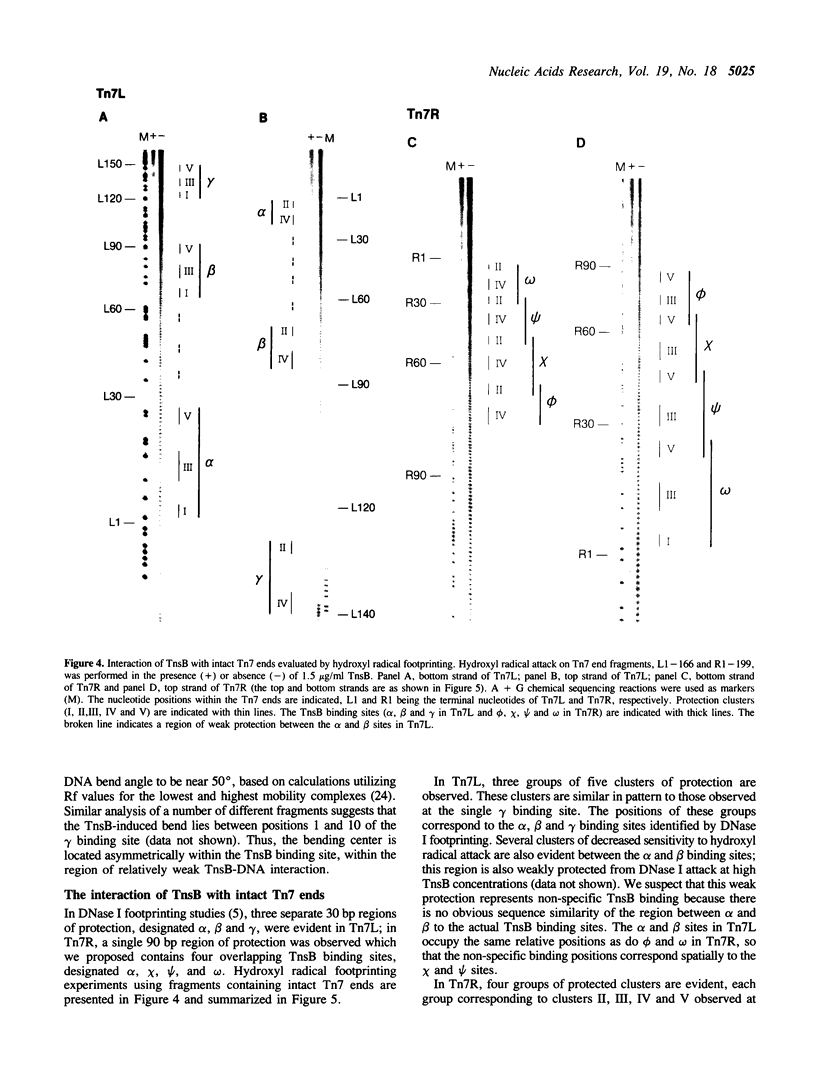
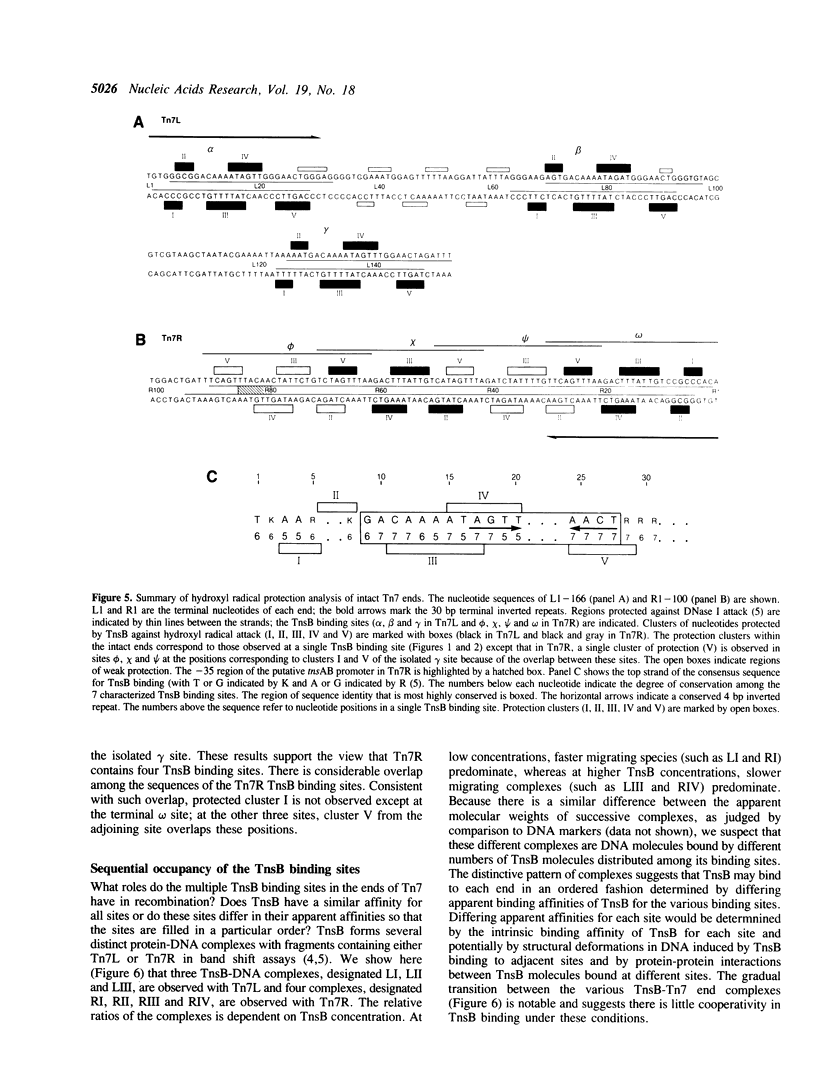

Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Arciszewska L. K., Drake D., Craig N. L. Transposon Tn7. cis-Acting sequences in transposition and transposition immunity. J Mol Biol. 1989 May 5;207(1):35–52. doi: 10.1016/0022-2836(89)90439-7. [DOI] [PubMed] [Google Scholar]
- Barth P. T., Datta N., Hedges R. W., Grinter N. J. Transposition of a deoxyribonucleic acid sequence encoding trimethoprim and streptomycin resistances from R483 to other replicons. J Bacteriol. 1976 Mar;125(3):800–810. doi: 10.1128/jb.125.3.800-810.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chalepakis G., Beato M. Hydroxyl radical interference: a new method for the study of protein-DNA interactions. Nucleic Acids Res. 1989 Feb 25;17(4):1783–1783. doi: 10.1093/nar/17.4.1783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Craigie R., Mizuuchi K. Role of DNA topology in Mu transposition: mechanism of sensing the relative orientation of two DNA segments. Cell. 1986 Jun 20;45(6):793–800. doi: 10.1016/0092-8674(86)90554-4. [DOI] [PubMed] [Google Scholar]
- Craigie R., Mizuuchi M., Mizuuchi K. Site-specific recognition of the bacteriophage Mu ends by the Mu A protein. Cell. 1984 Dec;39(2 Pt 1):387–394. doi: 10.1016/0092-8674(84)90017-5. [DOI] [PubMed] [Google Scholar]
- Derbyshire K. M., Hwang L., Grindley N. D. Genetic analysis of the interaction of the insertion sequence IS903 transposase with its terminal inverted repeats. Proc Natl Acad Sci U S A. 1987 Nov;84(22):8049–8053. doi: 10.1073/pnas.84.22.8049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flores C., Qadri M. I., Lichtenstein C. DNA sequence analysis of five genes; tnsA, B, C, D and E, required for Tn7 transposition. Nucleic Acids Res. 1990 Feb 25;18(4):901–911. doi: 10.1093/nar/18.4.901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gay N. J., Tybulewicz V. L., Walker J. E. Insertion of transposon Tn7 into the Escherichia coli glmS transcriptional terminator. Biochem J. 1986 Feb 15;234(1):111–117. doi: 10.1042/bj2340111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hatfull G. F., Noble S. M., Grindley N. D. The gamma delta resolvase induces an unusual DNA structure at the recombinational crossover point. Cell. 1987 Apr 10;49(1):103–110. doi: 10.1016/0092-8674(87)90760-4. [DOI] [PubMed] [Google Scholar]
- Hayes J. J., Tullius T. D. The missing nucleoside experiment: a new technique to study recognition of DNA by protein. Biochemistry. 1989 Nov 28;28(24):9521–9527. doi: 10.1021/bi00450a041. [DOI] [PubMed] [Google Scholar]
- Huisman O., Errada P. R., Signon L., Kleckner N. Mutational analysis of IS10's outside end. EMBO J. 1989 Jul;8(7):2101–2109. doi: 10.1002/j.1460-2075.1989.tb03619.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ichikawa H., Ikeda K., Wishart W. L., Ohtsubo E. Specific binding of transposase to terminal inverted repeats of transposable element Tn3. Proc Natl Acad Sci U S A. 1987 Dec;84(23):8220–8224. doi: 10.1073/pnas.84.23.8220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J., Zwieb C., Wu C., Adhya S. Bending of DNA by gene-regulatory proteins: construction and use of a DNA bending vector. Gene. 1989 Dec 21;85(1):15–23. doi: 10.1016/0378-1119(89)90459-9. [DOI] [PubMed] [Google Scholar]
- Koudelka G. B., Harrison S. C., Ptashne M. Effect of non-contacted bases on the affinity of 434 operator for 434 repressor and Cro. 1987 Apr 30-May 6Nature. 326(6116):886–888. doi: 10.1038/326886a0. [DOI] [PubMed] [Google Scholar]
- Kubo K. M., Craig N. L. Bacterial transposon Tn7 utilizes two different classes of target sites. J Bacteriol. 1990 May;172(5):2774–2778. doi: 10.1128/jb.172.5.2774-2778.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuo C. F., Zou A. H., Jayaram M., Getzoff E., Harshey R. DNA-protein complexes during attachment-site synapsis in Mu DNA transposition. EMBO J. 1991 Jun;10(6):1585–1591. doi: 10.1002/j.1460-2075.1991.tb07679.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuwabara M. D., Sigman D. S. Footprinting DNA-protein complexes in situ following gel retardation assays using 1,10-phenanthroline-copper ion: Escherichia coli RNA polymerase-lac promoter complexes. Biochemistry. 1987 Nov 17;26(23):7234–7238. doi: 10.1021/bi00397a006. [DOI] [PubMed] [Google Scholar]
- Makris J. C., Nordmann P. L., Reznikoff W. S. Mutational analysis of insertion sequence 50 (IS50) and transposon 5 (Tn5) ends. Proc Natl Acad Sci U S A. 1988 Apr;85(7):2224–2228. doi: 10.1073/pnas.85.7.2224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maxam A. M., Gilbert W. Sequencing end-labeled DNA with base-specific chemical cleavages. Methods Enzymol. 1980;65(1):499–560. doi: 10.1016/s0076-6879(80)65059-9. [DOI] [PubMed] [Google Scholar]
- McKown R. L., Waddell C. S., Arciszewska L. K., Craig N. L. Identification of a transposon Tn7-dependent DNA-binding activity that recognizes the ends of Tn7. Proc Natl Acad Sci U S A. 1987 Nov;84(22):7807–7811. doi: 10.1073/pnas.84.22.7807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mertens G., Klippel A., Fuss H., Blöcker H., Frank R., Kahmann R. Site-specific recombination in bacteriophage Mu: characterization of binding sites for the DNA invertase Gin. EMBO J. 1988 Apr;7(4):1219–1227. doi: 10.1002/j.1460-2075.1988.tb02934.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- New J. H., Eggleston A. K., Fennewald M. Binding of the Tn3 transposase to the inverted repeats of Tn3. J Mol Biol. 1988 Jun 5;201(3):589–599. doi: 10.1016/0022-2836(88)90640-7. [DOI] [PubMed] [Google Scholar]
- Prentki P., Chandler M., Galas D. J. Escherichia coli integration host factor bends the DNA at the ends of IS1 and in an insertion hotspot with multiple IHF binding sites. EMBO J. 1987 Aug;6(8):2479–2487. doi: 10.1002/j.1460-2075.1987.tb02529.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robertson C. A., Nash H. A. Bending of the bacteriophage lambda attachment site by Escherichia coli integration host factor. J Biol Chem. 1988 Mar 15;263(8):3554–3557. [PubMed] [Google Scholar]
- Rogers M., Ekaterinaki N., Nimmo E., Sherratt D. Analysis of Tn7 transposition. Mol Gen Genet. 1986 Dec;205(3):550–556. doi: 10.1007/BF00338097. [DOI] [PubMed] [Google Scholar]
- Salvo J. J., Grindley N. D. The gamma delta resolvase bends the res site into a recombinogenic complex. EMBO J. 1988 Nov;7(11):3609–3616. doi: 10.1002/j.1460-2075.1988.tb03239.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siebenlist U., Gilbert W. Contacts between Escherichia coli RNA polymerase and an early promoter of phage T7. Proc Natl Acad Sci U S A. 1980 Jan;77(1):122–126. doi: 10.1073/pnas.77.1.122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stark W. M., Boocock M. R., Sherratt D. J. Site-specific recombination by Tn3 resolvase. Trends Genet. 1989 Sep;5(9):304–309. doi: 10.1016/0168-9525(89)90113-3. [DOI] [PubMed] [Google Scholar]
- Surette M. G., Buch S. J., Chaconas G. Transpososomes: stable protein-DNA complexes involved in the in vitro transposition of bacteriophage Mu DNA. Cell. 1987 Apr 24;49(2):253–262. doi: 10.1016/0092-8674(87)90566-6. [DOI] [PubMed] [Google Scholar]
- Thompson J. F., Landy A. Empirical estimation of protein-induced DNA bending angles: applications to lambda site-specific recombination complexes. Nucleic Acids Res. 1988 Oct 25;16(20):9687–9705. doi: 10.1093/nar/16.20.9687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tullius T. D., Dombroski B. A. Hydroxyl radical "footprinting": high-resolution information about DNA-protein contacts and application to lambda repressor and Cro protein. Proc Natl Acad Sci U S A. 1986 Aug;83(15):5469–5473. doi: 10.1073/pnas.83.15.5469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waddell C. S., Craig N. L. Tn7 transposition: two transposition pathways directed by five Tn7-encoded genes. Genes Dev. 1988 Feb;2(2):137–149. doi: 10.1101/gad.2.2.137. [DOI] [PubMed] [Google Scholar]
- Wu H. M., Crothers D. M. The locus of sequence-directed and protein-induced DNA bending. Nature. 1984 Apr 5;308(5959):509–513. doi: 10.1038/308509a0. [DOI] [PubMed] [Google Scholar]
- Zerbib D., Jakowec M., Prentki P., Galas D. J., Chandler M. Expression of proteins essential for IS1 transposition: specific binding of InsA to the ends of IS1. EMBO J. 1987 Oct;6(10):3163–3169. doi: 10.1002/j.1460-2075.1987.tb02627.x. [DOI] [PMC free article] [PubMed] [Google Scholar]