Abstract
Disadvantageous hybridization favors the evolution of prezygotic isolating behaviors, generating a geographic pattern of interspecific mate discrimination where members of different species drawn from sympatric populations exhibit stronger preference for members of their own species than do individuals drawn from allopatric populations. Geographic shifts in species' boundaries can relax local selection against hybridization; under such scenarios the fate of enhanced species preference is unknown. Lineages established from populations in the region of sympatry that have been maintained as single-species laboratory cultures represent cases where allopatry has been produced experimentally. Using such cultures dating from the 1950s, we assess how Drosophila pseudoobscura and D. persimilis mate preferences respond to relaxed natural selection against hybridization. We found that the propensity to hybridize generally declines with increasing time in experimental allopatry, suggesting that maintaining enhanced preference for conspecifics may be costly. However, our data also suggest a strong role for drift in determining mating preferences once secondary allopatry has been established. Finally, we discuss the interplay between populations in establishing the presence or absence of patterns consistent with reinforcement.
Introduction
Reinforcement, the process by which natural selection against hybridization strengthens prezygotic reproductive isolation between species, enhances prezygotic isolating mechanisms including interspecific mate discrimination among sympatric taxa [1]–[6]. Reinforcement generates a geographic pattern in species discrimination where members of potentially hybridizing species from sympatric populations exhibit a lower propensity to mate interspecifically than do individuals drawn from allopatric populations [7]–[10]. Although enhanced species discrimination is beneficial in light of natural selection against hybridization [4], its evolutionary fate in the absence of interspecific interactions is not clear. In part, the outcome may hinge on what prevents enhanced species discrimination from initially spreading throughout a species' range. Below, we address two of the most likely possibilities and outline how these generate specific predictions regarding the evolutionary response to relaxation of selection against hybridization.
One reason why enhanced species discrimination may remain restricted to regions of sympatry is that the phenotypes favored by reinforcement can differ from those favored by intraspecific mate choice [11] in regions of allopatry, producing direct fitness costs to enhanced species recognition in areas where hybridization does not occur. In such cases, species-recognition systems, either the courtship signals, mating preferences, or both, will evolve at the expense of those typically involved in mate choice within species [12], [13], producing patterns of enhanced interspecific mate recognition in individuals from populations in regions of sympatry and perhaps more discriminating intraspecific mate preferences in individuals from regions of allopatry. A related possibility is that enhanced species recognition imposes fitness costs indirectly, which limits enhanced recognition to regions of sympatry [14]. For example, the evolution of other traits related to mate choice, including elaborate courtship behaviors (e.g. zig-zag and rolling dances [15]) or investment in specialized signaling or sensory structures (e.g. fin size [16]), could enable enhanced species discrimination and thus avoid hybridization in regions of sympatry. However, absent the risk of hybridization, fitness decrements associated with such traits would select against their evolution or maintenance in regions of allopatry. Under either scenario, relaxation or loss of selection against hybridization - for example through shifts in species ranges that alter or eliminate regions of sympatry [17] - would favor the loss of such species recognition traits and favor a return to the phenotype expressed in regions of allopatry.
Alternatively, costs associated with greater species-recognition need not be invoked to explain the geographic patterns of interspecific mate discrimination. Instead, the traits involved in prezygotic isolation could be effectively neutral in allopatry, and thus mate preference among conspecific populations may be free to diversify via genetic drift or population-specific patterns of sexual selection [18], [19]. This would produce a geographic pattern where individuals from populations in regions of sympatry would exhibit relatively strong, uniform species-discrimination abilities whereas individuals from regions of allopatry would possess weaker discrimination abilities and stronger, population-specific intraspecific mate preferences [18].
In both cases outlined above, interspecific mate discrimination may be reduced or even absent in regions of allopatry because alleles for species discrimination are not beneficial in the absence of selection to avoid hybridization [17]. Establishing which of these scenarios occur in nature can inform our understanding of the processes maintaining patterns of mate discrimination both in sympatry and in allopatry, processes which in turn contribute to the generation and maintenance of barriers between closely related species.
For a number of reasons, it is challenging to study the response of traits involved in enhanced species recognition to relaxed selection against hybridization. In nature, the degree of contact between species frequently grades from high in regions of true syntopy and lessens with distance as sympatry gives way to allopatry. Moreover, the size, location and intensity of this gradient may fluctuate over time. Together, this means that it can be difficult to establish precisely when and the degree to which evolutionarily relevant contact between taxa ceases and true allopatry begins. In addition, competition between sibling species within the region of sympatry can lead to divergent selection on other traits, particularly those associated with food acquisition [3], [20]–[22]. The resulting ecological character displacement can be confounded with, or even mistaken for, reproductive character displacement associated with selection for enhanced species discrimination [3], [12]. All this, combined with gene flow between regions of sympatry and allopatry, can complicate empirical investigation of the evolutionary fate of enhanced species recognition [6], [23], [24]. One solution to these problems is to study populations taken from regions of sympatry and maintained in experimental allopatry [25]. Because naturally occurring variation among populations can persist during routine care and maintenance of laboratory cultures [26], single-species collections from nature represent a rich resource for the experimental study of species-recognition systems and how they might affect the maintenance of species boundaries.
The behavior and genetics of mate choice, speciation, and reinforcement have been particularly well studied in Drosophila pseudoobscura and D. persimilis, and reinforcement in this sibling species pair has been well documented in several independent studies [27]–[34]. Starting with Dobzhansky's pioneering work [1], [27], [35], collections of these species have been made from geographic populations within regions of allopatry and sympatry and maintained as laboratory isolates. For collections made from within the region of sympatry, these isolates represent populations that have evolved heightened species recognition abilities in nature [2] that were placed into and maintained in experimental allopatry. Hence, these historical collections offer a unique resource that can be used to study the evolutionary fate of enhanced species recognition once selection against hybridization has been removed.
Here, we explore variation in the strength of species recognition between D. pseudoobscura and D. persimilis to uncover the evolutionary fate of enhanced species discrimination associated with reinforcement. Using historical collections maintained in experimental allopatry, we examine differences in propensity to hybridize between populations collected from within the regions of sympatry and allopatry. Our experimental design is powerful because we assay populations that have been maintained in experimental allopatry for 10 to 60 years (up to ∼880 generations), allowing inferences to be drawn about the fate of species discrimination across multiple time points. Additionally, our design enables us to examine changes in mate discrimination resulting from changes in traits related to male courtship and/or female preferences. If enhanced species discrimination is costly, either through direct selection against female discrimination or through selection on traits linked to female choice, we predict that D. pseudoobscura females originating from regions of sympatry with D. persimilis will become less discriminating with increasing time since isolation. However, if there is no cost to enhanced species recognition, we expect that populations collected from the regions of sympatry will have higher species discrimination abilities than their allopatric counterparts.
Methods
Study system
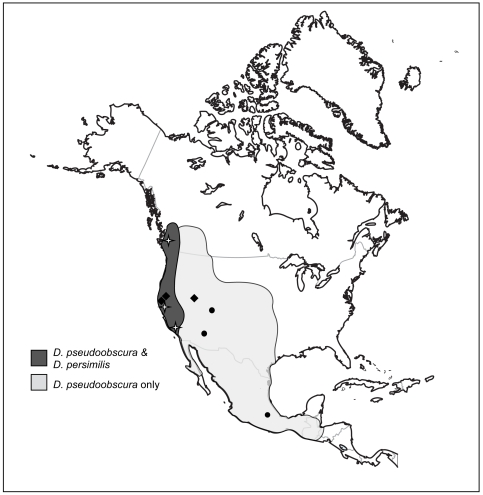
Drosophila pseudoobscura is a wide-ranging species, occurring from southwestern Canada, through the western United States, and into Mexico (Figure 1). The range of D. persimilis is contained exclusively within the northern Pacific coastal range of D. pseudoobscura (Figure 1). These sibling species diverged ∼850,000 to 500,000 years ago [36], [37]. Hybrid matings occur naturally at a low frequency in the wild [38], [39], resulting in hybrid male sterility [40], [41]. Males court females of both species indiscriminately and mate choice is driven largely by female preference for conspecific males [2], [42]. Documented patterns of species discrimination within D. pseudoobscura indicate that reinforcement is occurring; preference for conspecific males is greater in females from populations where D. pseudoobscura and D. persimilis are sympatric than in females from allopatric populations, and species discrimination ability varies in females among sympatric populations [31].
Figure 1. Species ranges and localities.
The distribution of Drosophila pseudoobscura and D. persimilis [57]. Localities for allopatric populations of D. pseudoobscura are shown in black circles and sympatric populations in black diamonds. Localities for D. persimilis are indicated by open stars.
Lineages and rearing conditions
Multiple lineages from both sympatric and allopatric populations of D. pseudoobscura and D. persimilis were obtained from the Drosophila Species Stock Center for each of three time intervals of collection: 1950s, 1990s, and 2000s (Table 1; strains Mather 17 and Mather 32 were kindly provided by M. Noor). These intervals were chosen because they contained D. persimilis and D. pseudoobscura population collections from within the regions of sympatry and allopatry. Given these criteria and culture availability, we were restricted in the final composition of our sample populations (Figure 1, Table 1, and see below). Where possible, isolates were chosen from similar geographic localities across time periods. We were able to use flies from the same sympatric population (Mather, California) at each of the three collection times. All lineages had been maintained in culture at least 1 year prior to acquisition. The experiments were conducted in late spring and early summer of 2009, thus the majority of 2000s lineages represent a virtually contemporary collection. Flies were raised on a sucrose-yeast-agar diet and maintained on a 12∶12 light cycle at 20°C and 75% humidity. Flies were reared in food vials at moderate larval density with relatively little variation in subsequent adult body size.
Table 1. Species lineages.
| Species | Lineage | Population | Location | Year of Collection |
| D. persimilis | 111.01 | - | Quesnel, British Columbia | 1958 |
| 111.46 | - | Mount St. Helena, California | 1997 | |
| 111.48 | - | Mount St. Helena, California | 1997 | |
| 111.5 | - | Santa Cruz Island, California | 2004 | |
| 111.51 | - | Santa Cruz Island, California | 2004 | |
| D. pseudoobscura | 121.88 | allopatric | Edo de Mexico, Mexico | 1958 |
| 121.89 | allopatric | Edo de Mexico, Mexico | 1958 | |
| 121.94 | allopatric | Mesa Verde, Colorado | 1996 | |
| 121.151 | allopatric | Flagstaff, Arizona | 1993 | |
| 121.15 | allopatric | Organ Pipe Monument, Arizona | 2007 | |
| 121.153 | allopatric | Wilcox, Arizona | 2007 | |
| 121.41 | sympatric | Mather, California | 1959 | |
| 121.42 | sympatric | Mather, California | 1959 | |
| 121.64 | sympatric | Mather, California | 1959 | |
| Mather 17 | sympatric | Mather, California | 1997 | |
| Mather 32 | sympatric | Mather, California | 1997 | |
| 121.103 | sympatric | Mather, California | 2001 | |
| 121.104 | sympatric | Mather, California | 2001 | |
| 121.148 | sympatric | San Francisco, California | 2006 | |
| 121.155 | sympatric | Eugene, Oregon | 2006 |
Fly lineages used in outcrossing and subsequent courtship trials. Lineages grouped in boxes were crossed to generate an outcrossed population from which flies were collected for experiment. 111.01 was not outcrossed as no additional population from that collection year was available. All lines were from the Drosophila Species Stock Center, except Mather 17 and 32 (provided by M. Noor).
Crosses and Mating Assays
To reduce potential inbreeding depression, lineages from the same species, time point, and where possible, populations were crossed in both directions (Table 1) [2], [25], [31], [32], [43]. When crosses from the same population were not possible, crosses were made using the nearest locality from that time period. Within 4 hours of eclosion, unmated progeny from these crosses were anesthetized with CO2, sorted by sex and placed into single-sex vials with food for 9 days. At least 1 day prior to the experiment, males and females were aspirated into individual food vials to reduce courtship inhibition resulting from crowding [44]. Mating assays were conducted 11 days post-eclosion, during the first three hours of light cycle. For the trials, an individual female was placed in the male vial and the cotton plug pushed down such that approximately 2.5 cm (∼10 cm3) of space remained in which flies could interact [45], (Noor pers. comm.). On any given day, all possible mating trial combinations (Table 2) were conducted and trials were randomized for time since collection and pair direction both within and across days. This “no-choice” design employs well-established protocols for this species pair [2], [30], [45], except that in our case flies were aged an additional 4 days to improve overall mating success (data not shown). All assays were conducted blind; the observer did not know whether crosses were conspecific vs. heterospecific, sympatric vs. allopatric, or the year of collection.
Table 2. Latency data.
| A | 1950s | 1990s | 2000s | |||
| N | Latency | N | Latency | N | Latency | |
| ♂ persimilis X ♀ persimilis | 86 | 54 (50) | 100 | 61 (60) | 95 | 73 (65) |
| ♂ pseudoobscura X ♀ pseudoobscura | 100 | 46 (51) | 99 | 40 (50) | 101 | 42 (43) |
| ♂ pseudoobscura(S) X ♀ persimilis | 202 | 46 (56) | 197 | 57 (59) | 198 | 48 (51) |
| 198 | 43 (51) | |||||
| ♂ pseudoobscura(A) X ♀ persimilis | 200 | 49 (54) | 201 | 60 (66) | 200 | 44 (52) |
| ♂ persimilis X ♀ pseudoobscura(S) | 197 | 62 (55) | 196 | 56 (51) | 198 | 77 (66) |
| 198 | 62 (54) | |||||
| ♂ persimilis X ♀ pseudoobscura(A) | 196 | 61 (58) | 199 | 72 (62) | 195 | 63 (54) |
Sample size and average courtship latency in seconds for each type of conspecific and heterospecific pairing for each of the three collection time points. Standard deviations shown in parentheses.
Trials were observed for up to 5 minutes to determine the time of the onset of courtship, typically identified by male wing vibration or rarely a copulation attempt [46], followed by an additional 5 minute observation. Time to initiate courtship (courtship latency), number of copulation attempts and time until successful copulation were recorded using a custom software program (FlyMate, available by request from EMM). Each fly was used only once [32]. Following mating trials, food vials were cleaned with ethanol and reused in subsequent mating trials on later days.
Analysis and Interpretation
We made multiple comparisons to evaluate changes in species discrimination patterns over time. We compared courtship and mating characteristics for conspecific versus heterospecific crosses to confirm that male court females of either species indiscriminately and that females preferentially mate with conspecifc partners over heterospecific males. For conspecific D. pseudoobscura trials, pairings were constructed equally from allopatric and sympatric populations. We also compared courtship and mating variation between sympatric and allopatric populations in the heterospecific crosses, as well as among the three collection times (1950s, 1990s, and 2000s). Analyses include only trials where at least one copulation attempt occurred; however, including males that performed courtship song but did not attempt copulation did not affect our results (analyses not shown). A contingency table chi-square test was used to assess two indices of pair mating success, the number of successful copulations (copulation that lasted more than 60 seconds) [32], and the Noor Score, a metric of mating success in which a score of 2 is assigned for successful copulation within two attempts, 1 is assigned to all other successful copulations, and 0 is assigned to unsuccessful pairings [30]. The effect of cross type (e.g. conspecific/heterospecific, allopatry/sympatry, collection time) on courtship latency and number of copulation attempts was analyzed by a series of six individual one-way ANOVAs. Subsequent post-hoc pair wise comparisons were made using Tukey-Kramer tests [47], [48]. For all crosses, Clopper-Pearson exact binomial confidence intervals were calculated using JavaStat [49], [50].
Results
We observed 3,949 courtship trials of which 3,356 (85%) contained at least one copulation attempt (Table 2). Neither time since isolation nor cross type (conspecific versus heterospecific) affected the occurrence of at least one copulation attempt (χ2 = 5.13, df = 2, p>0.05; χ2 = 0.85, df = 1, p>0.05 respectively). However, D. persimilis males were significantly less likely to attempt at least one copulation (regardless of the female they were paired with) than their congeneric counterparts (χ2 = 83.164, df = 1, p<0.0001) and copulation attempts were less frequent with D. pseudoobscura females, regardless of the identity of the courting male (χ2 = 17.668, df = 1, p<0.0001).
As predicted, there was no difference in courtship latency for crosses involving males courting conspecific or heterospecific females (F1,3354 = 3.21 p = 0.073; Table 2) although across all pairings, D. pseudoobscura males courted more quickly and had more copulation attempts than D. persimilis males (mean ± SE; Latency: D. persimilis 64.38±1.38 sec, D. pseudoobscura 48.44±1.36 sec, F1,3354 = 67.51, p<0.0001, Table 2; Copulation Attempts: D. persimilis 3.56±0.094, D. pseudoobscura 5.28±0.094, F1,3353 = 165.99, p<0.0001; Table 3). There was no change in this general pattern when each of the collection time points was examined individually (Table 2; Table 3).
Table 3. Copulation data.
| 1950s | 1990s | 2000s | |
| Copulation Attempts | Copulation Attempts | Copulation Attempts | |
| M persimilis X F persimilis | 1.81 (1.49) | 1.34 (0.79) | 1.44 (2.33) |
| M pseudoobscura X F pseudoobscura | 1.30 (0.81) | 1.61 (1.66) | 1.70 (1.55) |
| 6.52 (3.91) | 4.17 (3.67) | 6.97 (5.32) | |
| 8.78 (5.95) | |||
| M pseudoobscura(A) X F persimilis | 6.12 (3.99) | 4.32 (3.49) | 5.70 (4.29) |
| M persimilis X F pseudoobscura(S) | 3.18 (2.13) | 4.86 (3.78) | 2.78 (1.98) |
| 4.09 (2.82) | |||
| M persimilis X F pseudoobscura(A) | 3.54 (2.39) | 5.34 (3.19) | 4.02 (2.87) |
Number of copulation attempts for each type of conspecific and heterospecific pairing for each of the three collection time points. Standard deviations shown in parentheses.
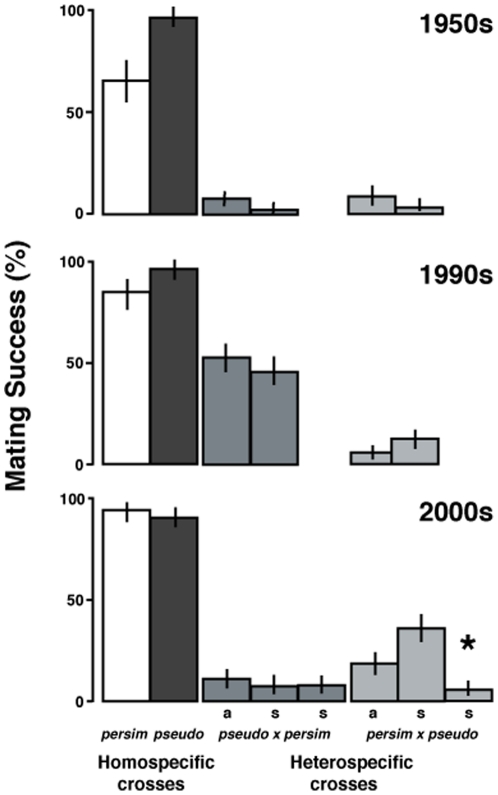
Mating success for conspecific pairings ranged from 72% to 95%. D. persimilis mated more readily with conspecifics as time since isolation decreased, whereas D. pseudoobscura exhibited the opposite pattern (Figure 2). Heterospecific pairings were less successful (2–51%). If enhanced species recognition is costly, then we expect species discrimination ability to be negatively correlated with time since isolation, however the pattern of successful copulations varied among collection time points and pairing type. Specifically, our data provide mixed support for the predicted pattern of stronger species discrimination in heterospecific pairings of D. pseudoobscura females from regions of sympatry relative to those from regions of allopatry (Figure 2, Table 4). For populations isolated in the 2000s, the results were split; the sympatric Mather population mated with D. persimilis at significantly higher levels than the allopatric population. In contrast, the non-Mather sympatric population exhibited a significant pattern consistent with a history of reinforcement. For the historically isolated populations, the 1990s sympatric population differed significantly from the allopatric population in the direction opposite of that expected under reinforcement, and the 1950s populations were indistinguishable in their species discrimination ability. Results were similar using the Noor Score alternative metric of mating success (Table 4).
Figure 2. Copulation success.
Copulation success rates for conspecific and both directions of sympatric (s) and allopatric (a) heterospecific pairings (male×female) between D. pseudoobscura (pseudo) and D. persimilis (persim). Panel is divided by the time of lineage collection. Clopper-Pearson exact binomial confidence intervals are given for each cross type and statistically significant pattern consistent with reinforcement is indicated with a star.
Table 4. Analysis of mating success across collection times.
| A | |||||
| Collection | N | df | -Log Likelihood | Chi Square | P-value |
| 1950s | 393 | 1 | 2.137 | 4.274 | 0.0387 |
| 1990s | 395 | 1 | 3.187 | 6.373 | 0.0116* |
| 2000s | |||||
| Mather | 393 | 1 | 7.216 | 14.433 | 0.0001* |
| Non-Mather | 393 | 1 | 7.325 | 14.65 | 0.0001* |
Analysis of variation in (A) mating success and (B) Noor Score between sympatric and allopatric populations of D. pseudoobscura females paired with D. persimilis males from each of the three collection times. P-values in bold indicate values significant after sequential bonferroni correction, asterisk indicates pattern in the opposite direction from that expected under reinforcement.
Discussion
Studying the evolutionary fate of enhanced species discrimination provides insight into the processes and mate choice behaviors generating and maintaining species barriers, as well as the pliability of the barriers themselves. Freed from the constraints imposed by strong selection against hybridization, it is unclear how species-level mating preferences will respond to relaxed selection. Historical, single-species collections provide a potentially powerful experimental system with which to track changes in mating preference patterns over time, perhaps particularly so when applied to studies of the evolutionary fate of enhanced species discrimination following the loss of natural selection against hybridization. Using historical collections maintained in experimental allopatry for hundreds of generations, we found considerable variation in several aspects of mating propensity within and across collection times and cross types. Despite this variation, however, some important trends are evident. In general, we found lower levels of successful mating across all pairing types relative to other studies with this system [2], [45]. And, although heterospecific matings occurred at relatively low frequency regardless of collection time or the populations paired, only the lineages experimentally isolated most recently exhibited enhanced mate discrimination in a pattern consistent with being formed by reinforcement. This suggests that enhanced species discrimination may decay readily once selection against hybridization is removed. Below we attempt to determine what might be responsible for these patterns.
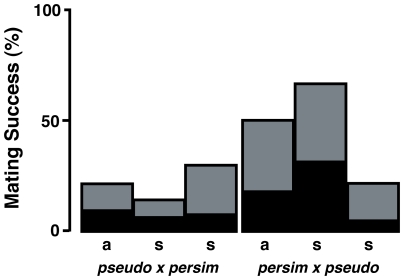
Larval diet can influence the degree of species discrimination in other Drosophilids [51], and so we performed mating trials to determine if our observed pattern of generally lower levels of mating success across all time points could be explained by diet. To explore fully this possibility, we would ideally conduct all mating trials using larvae from all populations reared on diets used in earlier studies and test for effects of larval diet on species discrimination. While the exact diet used in the earliest research is not known for certain [27], two media types were prevalent including Kalmus media [52] and Spassky Cream of wheat media [53]. We did not explore the effects of these potential media. However, we reared approximately 25 crosses of each pair type for the 2000s time point on a food recipe used in several other recent studies [32], [54], (Noor pers. comm.), which differed from our recipe in that it contained dextrose in addition to sucrose (Table S1). All pairings exhibited higher mating success on this enriched diet (Figure 3). Although statistically significant differences in the relative proportions of successful matings are seen compared to a standard average increase (χ2 = 17.29, df = 5, p = 0.00398), much of this was driven by the Mather population (Mather 2000s discussed further below); excluding the Mather population upheld the relative levels of species discrimination among groups regardless of larval diet (χ2 = 6.739, df = 3, p = 0.0807). Thus, while larval diet was responsible for the pattern of overall reduced mating success in the current study relative to earlier work with this system [2], [32], [45], we reject diet as being responsible for the observed variation in mating success among populations and time points.
Figure 3. Diet effects on mating success.
Copulation success rates for both directions of sympatric (s) and allopatric (a) heterospecific pairings (male×female) between D. pseudoobscura (pseudo) and D. persimilis (persim) from the 2000s time point reared on sucrose only diet (black bars) and sucrose+dextrose diet (gray bars).
Differences in the number of successful copulations (i.e., copulations lasting at least 60 seconds) between heterospecific and conspecific crosses were not a function of variation in male courtship intensity. Courtship latency was similar across cross types and time periods and was consistent with other studies [2], [42]. Drosophila pseudoobscura males court on average 16 seconds faster and have 2 more copulation attempts than males of D. persimilis. Although such interspecific differences in courtship intensity could have important evolutionary consequences if females select between directly competing males, such scenarios are likely limited in nature [31].
The absence of a role for male courtship intensity or persistent enhanced mate discrimination following the establishment of experimental allopatry indicates that enhanced female choosiness for conspecific males may carry some cost and thus be selected against in the absence of reinforcement. This likely suggests that, in this case selection for enhanced interspecific mate discrimination, selection may have seized upon the same traits involved in intraspecific sexual selection. Although the exact targets of selection are unknown, possibilities include cuticular hydrocarbon profiles [13] or courtship song pattern [29].
Our results suggest a role for selection against enhanced species discrimination in experimental allopatry, however, the observed variation in species discrimination among experimental lineages indicates that drift may also be an important factor. Within collection time periods, variation in mating success among the sympatric populations of D. pseudoobscura reflects a genetic basis for variation in species discrimination; considerable standing variation in species discrimination exists within both sympatric and allopatric populations [32], [45]. The high levels of mating success exhibited by the Mather 2000 lineages is unusual in that these flies mated more readily than even the allopatric populations of D. pseudoobscura, and at a higher frequency than any other heterospecific cross type in our study. Indeed, the mating propensity we observed exceeds the levels reported for this population in other studies, particularly after accounting for the effects of diet (e.g., 67% success on comparable sucrose-dextrose food recipe, Figure 3; compared to maximum values of 10–45% [45]; 16.67% [43]; 15.6–36.5% [2]; 30% [30]; 0–4.26% [31]; 8.6–48% [32]). As inbred lineages demonstrate higher variation in mating success than outbred populations [32], the strong propensity to mate with heterospecifics exhibited by the Mather 2000 lineage may indicate that it suffers from low genetic diversity. Preliminary genetic analysis of the two lineages that were crossed to create the Mather 2000 lineage for this experiment found that each possessed the same rare allele at a locus on the XL chromosome (Noor pers. comm.), suggesting that the lineages used in this study established from those stocks had lower genetic diversity than other lineages used in the study. Thus, low genetic diversity at the loci contributing to species discrimination may help explain the relatively indiscriminant mating pattern exhibited by this lineage.
Our research highlights the interplay between populations in studying mate preferences generally, and establishing the presence or absence of patterns consistent with reinforcement. Studies assessing the presence or absence of reinforcement do so based on enhanced mate discrimination in sympatric populations relative to allopatric ones. This comparison inherently ties experimental outcomes to not just the sympatric population of interest, but also to the allopatric reference population. Thus, changes in the relative choosiness of an allopatric population can determine whether or not a pattern consistent with reinforcement is seen, regardless of choosiness of the sympatric population.
Although our experimental approach offers some advantages over traditional comparative methods that draw on extant natural populations, it brings some complications of its own. First, while being maintained in single-population isolates in the laboratory, flies experience relaxed selection for enhanced mate discrimination and are simultaneously subjected to novel selection on other traits related to lab adaptation [25]. Consequently, selection may occur on secondary traits correlated with the focal traits, changing the focal trait value even in the absence of direct selection on species discrimination. In addition, founder effects and small population sizes coupled with occasional bottlenecks may increase the importance of drift, affecting mating preferences and levels of discrimination between lineages within and between species in unpredictable ways as similar effects have been seen in other traits [26]. Cycles of founder-flush-bottlenecking can produce ethological isolation between populations and inbreeding depression can reduce the mating propensity even with members of the same population [55], [56]. Hence, negative results discovered using our approach must be interpreted with caution.
In conclusion, historical laboratory collections offer a unique and powerful experimental tool to address questions regarding the evolutionary fate of reinforcement-driven enhanced interspecific mate discrimination once selection against hybridization is relaxed. While our experimental design is powerful, limited numbers of stocks limited the number of populations available to test our hypotheses. More independent populations, for example, would allow for a more rigorous testing of the role of drift or selection in producing our observed patterns; unfortunately, such additional replicated populations are not available. This highlights one of the key values of preserving historical cultures and calls for regular and repeated sampling from the same localities; deposition and culture of lineages in this manner will preserve biological resources – time capsules – which are likely to be of increased utility as species distributions change. Our finding of generally reduced species discrimination with time spent in experimental allopatry likely suggests that intraspecific sexual selection may alter mating preferences in the absence of selection against hybridization; however, drift likely moderates mating preferences as well. The patterns of mating preferences observed for any population allow us to glimpse the many selective forces acting on the pliability of the species barrier across both space and time.
Supporting Information
Food recipes. Food recipes used in examining mating success based on diet based on a 1-liter recipe.
(DOC)
Acknowledgments
E. Bakota wrote the FlyMate program for data collection. The following students assisted in observing courtship trials: E. Bakota, A. Serna, M. Vila, E. Joyce, M. White, and S. Onopa. M. Noor provided the Mather 32 and Mather 17 lines of D. pseudoobscura, as well as feedback on methodology and an earlier draft of this manuscript. K. Pfennig also provided constructive comments on an earlier draft of this manuscript. Toby Liss provided valuable assistance in figure and manuscript editing.
Footnotes
Competing Interests: The authors have declared that no competing interests exist.
Funding: EMM was supported by a National Science Foundation Bioinformatics Postdoctoral Fellowship. This research was supported by National Science Foundation grants Division of Environmental Biology #0805818, Integrative Organismal Systems #0920720, and a University of Houston Grants to Enhance and Advance Research Award to WAF. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Dobzhansky T. Genetics and the Origin of Species. New York: Columbia University Press; 1937. 364 [Google Scholar]
- 2.Noor MAF. Speciation driven by natural selection in Drosophila. Nature. 1995;375:674–675. doi: 10.1038/375674a0. [DOI] [PubMed] [Google Scholar]
- 3.Noor MAF. Reinforcement and other consequences of sympatry. Heredity. 1999;83:503–508. doi: 10.1038/sj.hdy.6886320. [DOI] [PubMed] [Google Scholar]
- 4.Servedio MR, Noor MAF. The role of reinforcement in speciation: Theory and data. Annu Rev Ecol Evol S. 2003;34:339–364. [Google Scholar]
- 5.Geyer LB, Palumbi SR. Reproductive character displacement and the genetics of gamete recognition in tropical sea urchins. Evolution. 2003;57:1049–1060. doi: 10.1111/j.0014-3820.2003.tb00315.x. [DOI] [PubMed] [Google Scholar]
- 6.Matute DR. Reinforcement of gametic isolation in Drosophila. PLoS Biol. 2010;2010 23;8(3):e1000341. doi: 10.1371/journal.pbio.1000341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Coyne JA, Orr HA. Patterns of speciation in Drosophila. Evolution. 1989;43:362–381. doi: 10.1111/j.1558-5646.1989.tb04233.x. [DOI] [PubMed] [Google Scholar]
- 8.Sætre G-P, Moum T, Bures S, Kral M, Adamjan M, et al. A sexually selected character displacement in flycatchers reinforces premating isolation. Nature. 1997;387:589–592. [Google Scholar]
- 9.Rundle HD, Schluter D. Reinforcement of stickleback mate preferences: Sympatry breeds contempt. Evolution. 1998;52:200–208. doi: 10.1111/j.1558-5646.1998.tb05153.x. [DOI] [PubMed] [Google Scholar]
- 10.Kronfrost MR, Young LG, Gilbert LE. Reinforcement of mate preferences among hybridizing Heliconius butterflies. J Evol Biol. 2007;20:278–285. doi: 10.1111/j.1420-9101.2006.01198.x. [DOI] [PubMed] [Google Scholar]
- 11.Blows MW. Interaction between natural and sexual selection during the evolution of mate recognition. P Roy Soc B-Biol Sci. 2002;269:1113–1118. doi: 10.1098/rspb.2002.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Higgie M, Blows MW. Are traits that experience reinforcement also under sexual selection? Am Nat. 2007;170:409–420. doi: 10.1086/519401. [DOI] [PubMed] [Google Scholar]
- 13.Higgie M, Blows MW. The evolution of reproductive character displacement conflicts with how sexual selection operates within a species. Evolution. 2008;62:1192–1203. doi: 10.1111/j.1558-5646.2008.00357.x. [DOI] [PubMed] [Google Scholar]
- 14.Pfennig KS, Pfennig DW. Character displacement: ecological and reproductive responses to a common evolutionary problem. Q Rev Biol. 2009;84:253–276. doi: 10.1086/605079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kitano J, Mori S, Peichel CL. Divergence of male courtship displays between sympatric forms of anadromous threespine stickleback. Behaviour. 2008;145:443–461. [Google Scholar]
- 16.Ptacek MB. Interspecific mate choice in sailfin and shortfin species of mollies. Anim Behav. 1998;56:1145–1154. doi: 10.1006/anbe.1998.0909. [DOI] [PubMed] [Google Scholar]
- 17.Wellenreuther M, Tynkkynen K, Svensson EI. Simulating range expansion: male species recognition and loss of premating isolation in damselflies. Evolution. 2010;64:242–252. doi: 10.1111/j.1558-5646.2009.00815.x. [DOI] [PubMed] [Google Scholar]
- 18.McPeek MA, Gavrilets S. The evolution of female mating preferences: differentiation from species with promiscuous males can promote speciation. Evolution. 2006;60:1967–1980. [PubMed] [Google Scholar]
- 19.Ryan MJ, Rand AS. Species recognition and sexual selection as a unitary problem in animal communication. Evolution. 1993;47:647–657. doi: 10.1111/j.1558-5646.1993.tb02118.x. [DOI] [PubMed] [Google Scholar]
- 20.Adams DC, Rohlf FJ. Ecological character displacement in Plethodon: biomechanical differences found from a geometric morphometric study. P Natl Acad Sci USA. 2000;97:4106–4111. doi: 10.1073/pnas.97.8.4106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Grant PR, Grant BR. Speciation and hybridization in island birds. Philos T Roy Soc B. 1996;351:765–772. [Google Scholar]
- 22.Pfennig DW, Pfennig KS. Character displacement and the origins of diversity. Am Nat. 2010;176:S22–S44. doi: 10.1086/657056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Liou LW, Price TD. Speciation by reinforcement of premating isolation. Evolution. 1994;48:1451–1459. doi: 10.1111/j.1558-5646.1994.tb02187.x. [DOI] [PubMed] [Google Scholar]
- 24.Nosil P, Crespi BJ, Sandoval CP. Reproductive isolation driven by the combined effects of ecological adaptation and reinforcement. Proc R Soc Lond B. 2003;270:1911–1918. doi: 10.1098/rspb.2003.2457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Harshman LG, Hoffmann AA. Laboratory selection experiments using Drosophila: what do they really tell us? Trends Ecol Evol. 2000;15:32–36. doi: 10.1016/s0169-5347(99)01756-5. [DOI] [PubMed] [Google Scholar]
- 26.Krebs RA, Roberts SP, Bettencourt BR, Feder ME. Changes in thermotolerance and Hsp70 expression with domestication in Drosophila melanogaster. J Evol Biol. 2001;14:75–82. doi: 10.1046/j.1420-9101.2001.00256.x. [DOI] [PubMed] [Google Scholar]
- 27.Lewontin RC, Moore JA, Provine WB, Wallace B. Dobzhansky's Genetics of Natural Populations I–XLIII. New York: Columbia University Press; 1981. [Google Scholar]
- 28.Noor MAF, Coyne JA. Genetics of a difference in cuticular hydrocarbons between Drosophila pseudoobscura and D. persimilis. Genet Res. 1996;68:117–123. doi: 10.1017/s0016672300034005. [DOI] [PubMed] [Google Scholar]
- 29.Noor MAF, Aquadro CF. Courtship songs of Drosophila pseudoobscura and D. persimilis: analysis of variation. Anim Behav. 1998;56:115–125. doi: 10.1006/anbe.1998.0779. [DOI] [PubMed] [Google Scholar]
- 30.Noor MAF, Grams KL, Bertucci LA, Reiland J. Chromosomal inversions and the reproductive isolation of species. P Natl Acad Sci USA. 2001;98:12084–12088. doi: 10.1073/pnas.221274498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Noor MAF, Ortiz-Barrientos D. Simulating natural conditions in the laboratory: A re-examination of sexual isolation between sympatric and allopatric populations of Drosophila pseudoobscura and Drosophila persimilis. Behav Genet. 2006;36:322–327. doi: 10.1007/s10519-005-9033-8. [DOI] [PubMed] [Google Scholar]
- 32.Ortiz-Barrientos D, Counterman BA, Noor MAF. The genetics of speciation by reinforcement. PLoS Biol. 2004;2:2256–2263. doi: 10.1371/journal.pbio.0020416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ortiz-Barrientos D, Noor MAF. Evidence for a one-allele assortative mating locus. Science. 2005;310:1467. doi: 10.1126/science.1121260. [DOI] [PubMed] [Google Scholar]
- 34.Williams MA, Blouin AG, Noor MAF. Courtship songs of Drosophila pseudoobscura and D. persimilis. II. Genetics of species differences. Heredity. 2001;86:68–77. doi: 10.1046/j.1365-2540.2001.00811.x. [DOI] [PubMed] [Google Scholar]
- 35.Dobzhansky T. Genetic nature of species differences. Am Nat. 1937;71:404–420. [Google Scholar]
- 36.Hey J, Neilsen R. Multilocus methods for estimating population sizes, migration rates and divergence time, with applications to the divergence of Drosophila pseudoobscura and D. persimilis. Genetics. 2004;167:747–760. doi: 10.1534/genetics.103.024182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tamura K, Subramanian S, Kumar S. Temporal patterns of fruit fly (Drosophila) evolution revealed by mutation clocks. Mol Bio Evol. 2004;21:36–44. doi: 10.1093/molbev/msg236. [DOI] [PubMed] [Google Scholar]
- 38.Dobzhansky T. Is there gene exchange between Drosophila pseudoobscura and D. persimilis in their natural habitats? Am Nat. 1973;107:312–314. [Google Scholar]
- 39.Powell JR. Interspecific cytoplasmic gene flow in the absence of nuclear gene flow: Evidence from Drosophila. P Natl Acad Sci USA. 1983;80:492–495. doi: 10.1073/pnas.80.2.492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Dobzhansky T. Studies on hybrid sterility. II. Localization of sterility factors in Drosophila pseudoobscura hybrids. Genetics. 1936;21:113–135. doi: 10.1093/genetics/21.2.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Prakash S. Origin of reproductive isolation in the absence of apparent genic differentiation in a geographic isolate of Drosophila pseudoobscura. Genetics. 1972;72:143–155. doi: 10.1093/genetics/72.1.143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mayr E. Experiments on sexual isolation in Drosophila. VII. The nature of the isolating mechanisms between Drosophila pseudoobscura and Drosophila persimilis. P Natl Acad Sci USA. 1946;32:128–137. doi: 10.1073/pnas.32.5.128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lorch PD, Servedio MR. Postmating-prezygotic isolation is not an important source of selection for reinforcement within and between species in Drosophila pseudoobscura and D. persimilis. Evolution. 2005;59:1039–1045. [PubMed] [Google Scholar]
- 44.Noor MAF. Environmental effects on male courtship intensity in Drosophila pseudoobscura (Diptera: Drosophilidae). J Insect Behav. 1997;10:305–312. [Google Scholar]
- 45.Barnwell CV, Noor MAF. Failure to replicate two mate preference QTLs across multiple strains of Drosophila pseudoobscura. J Hered. 2008;99:653–656. doi: 10.1093/jhered/esn069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Brown RGB. Courtship behaviour in the Drosophila obscura group. I.: D. pseudoobscura. Behaviour. 1964;23:61–106. doi: 10.1163/156853965x00174. [DOI] [PubMed] [Google Scholar]
- 47.Kramer CY. Extension of multiple range tests to group means with unequal numbers of replications. Biometrics. 1956;12:309–310. [Google Scholar]
- 48.Tukey J. A problem of multiple comparisons. 1953. In Dittoed manuscript of 396 pages. Princeton University.
- 49.Clopper CJ, Pearson ES. The use of confidence or fiducial limits illustrated in the case of the binomial. Biometrika. 1934;26:404–413. [Google Scholar]
- 50.Pezzullo JC. 2010. JavaStat– Binomial and Poisson Confidence Intervals.
- 51.Jennings JH, Etges WJ. Species hybrids in the laboratory but not in nature: A reanalysis of premating isolation between Drosophila arizonae and D. mojavensis. Evolution. 2010;64:587–598. doi: 10.1111/j.1558-5646.2009.00834.x. [DOI] [PubMed] [Google Scholar]
- 52.Kalmus H. Skimmed milk as a culture media for Drosophila. Drosophila Information Service. 1941;14:56. [Google Scholar]
- 53.Spassky B. Cream of wheat-molasses fly medium. Drosophila Information Service. 1943;17:67–68. [Google Scholar]
- 54.Ortiz-Barrientos D, Chang AS, Noor MAF. A recombinational portrait of the Drosophila pseudoobscura genome. Genet Res. 2006;87:23–31. doi: 10.1017/S0016672306007932. [DOI] [PubMed] [Google Scholar]
- 55.Galiana A, Moya A, Ayala FJ. Founder-flush speciation in Drosophila pseudoobscura: A large-scale experiment. Evolution. 1993;47:432–444. doi: 10.1111/j.1558-5646.1993.tb02104.x. [DOI] [PubMed] [Google Scholar]
- 56.Meffert LM, Bryant EH. Divergent ambulatory and grooming behavior in serially bottlenecked lines of the housefly. Evolution. 1992;46:1399–1407. doi: 10.1111/j.1558-5646.1992.tb01132.x. [DOI] [PubMed] [Google Scholar]
- 57.Machado CA, Kliman RM, Markert JA, Hey J. Inferring the History of Speciation from Multilocus DNA Sequence Data: The Case of Drosophila pseudoobscura and Close Relatives. Mol Biol Evol. 2002;19(4):472–488. doi: 10.1093/oxfordjournals.molbev.a004103. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Food recipes. Food recipes used in examining mating success based on diet based on a 1-liter recipe.
(DOC)