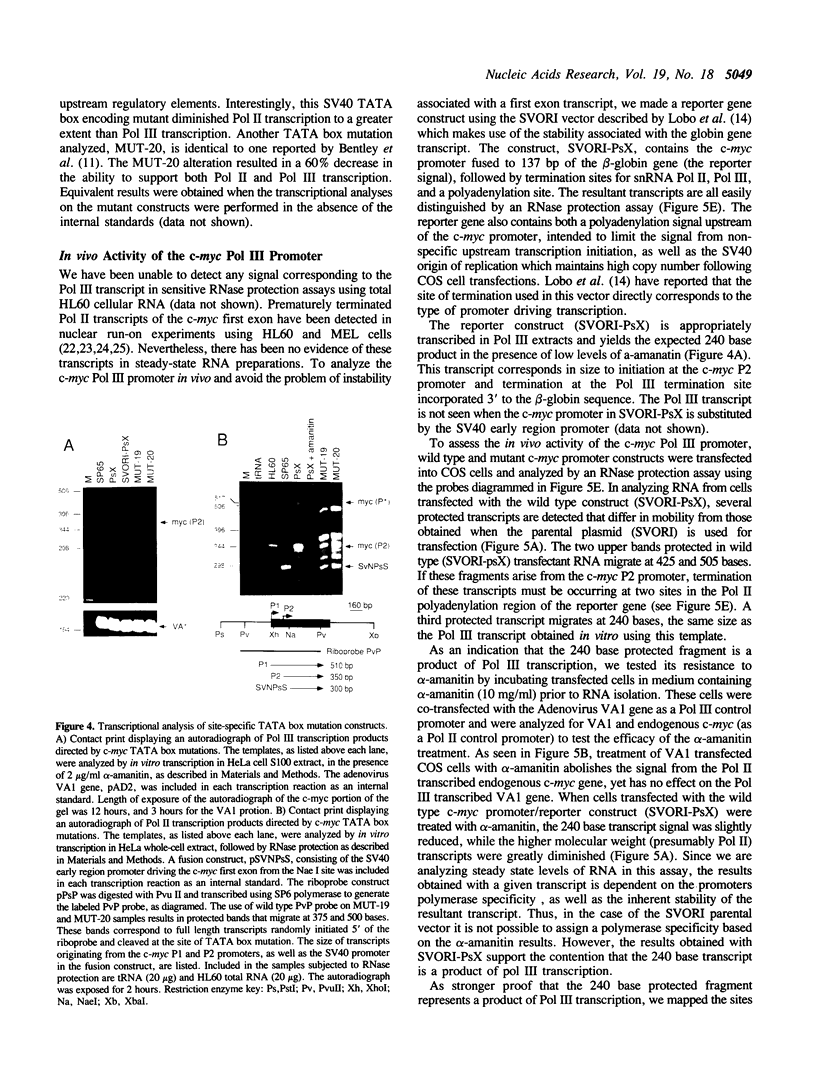
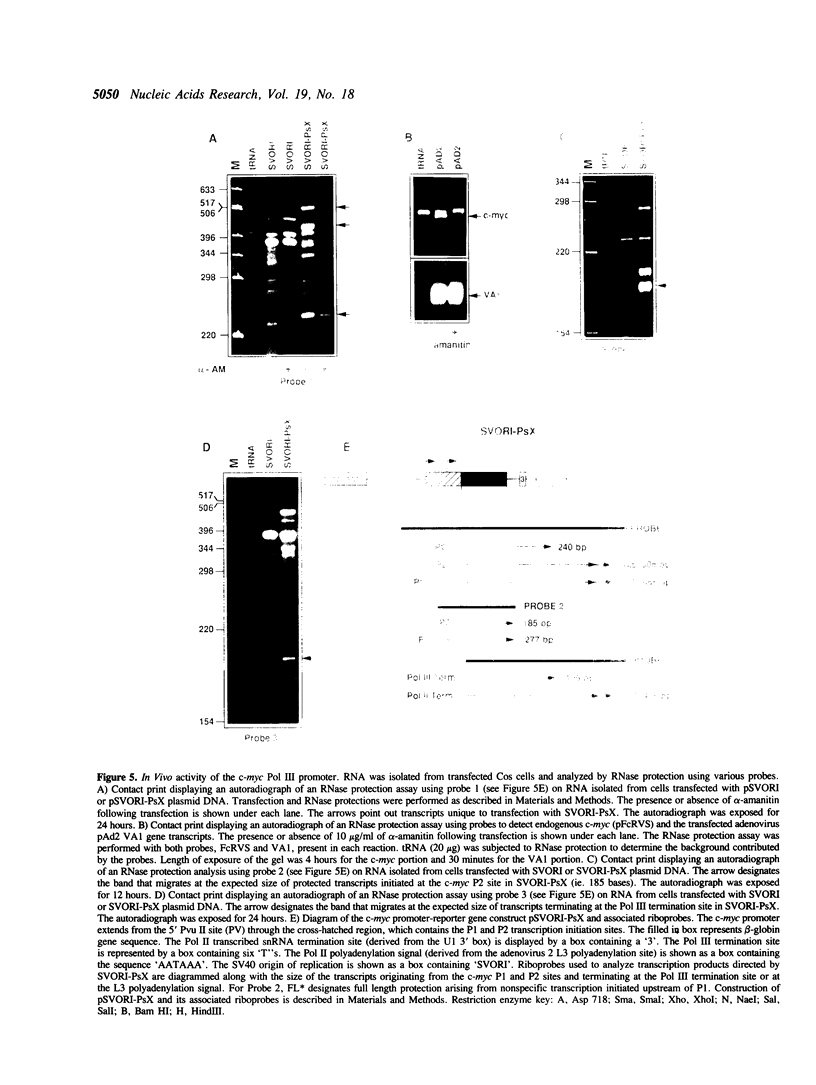
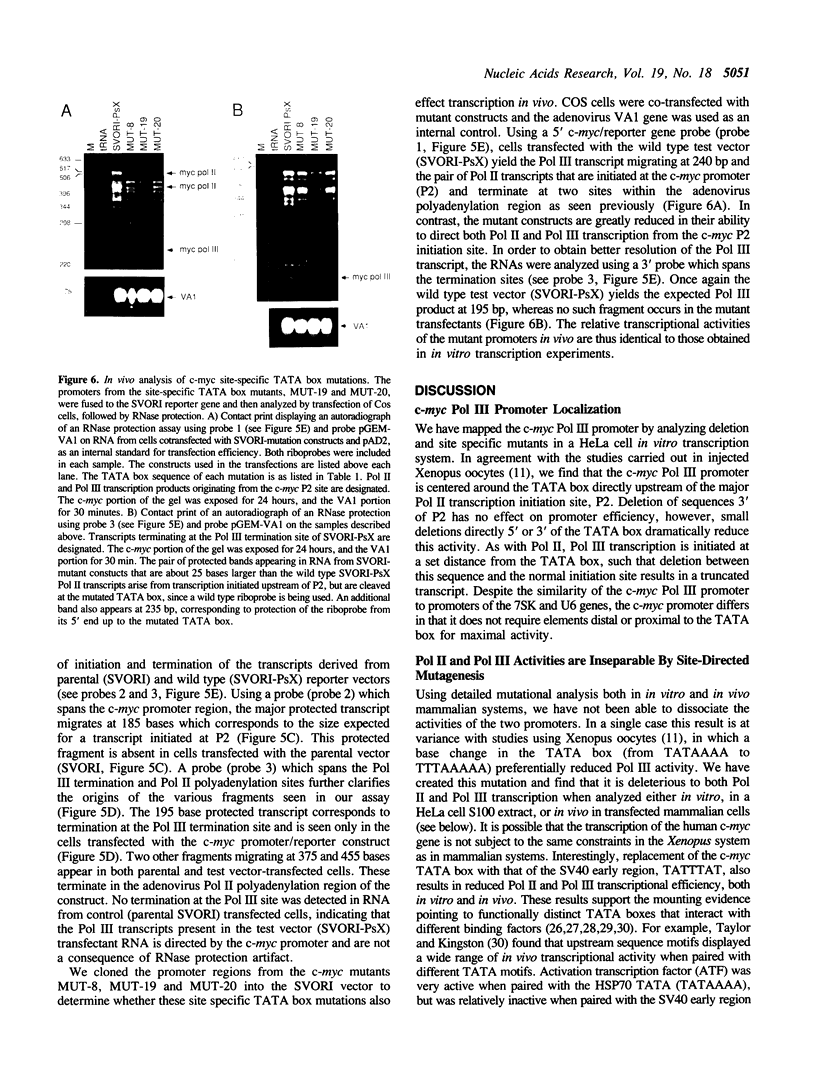
Abstract
The c-myc promoter has the unusual property of displaying both RNA polymerase II (Pol II) and RNA polymerase III (Pol III) activities. Both Pol II and Pol III utilize the same transcription initiation site. We have now examined the effects of mutations in crucial regions of the c-myc promoter to assess their effects on both transcriptional activities. In doing this we show that both Pol II and Pol III activities require sequences that are located within the stronger of the two principal c-myc promoter regions (P2). Further, we show that the Pol III activity using this initiation site does not require an A box or distal upstream sequences. Like the Pol II activity, it does require an intact TATA sequence and alterations at this site result in the simultaneous loss of both Pol II and Pol III activities. The superimposition of two apparently inseparable promoter activities makes it possible to consider common features, possible common protein elements in each holoenzyme complex, as well as a potential role for each enzyme in the regulated expression of the c-myc gene.
Full text
PDF


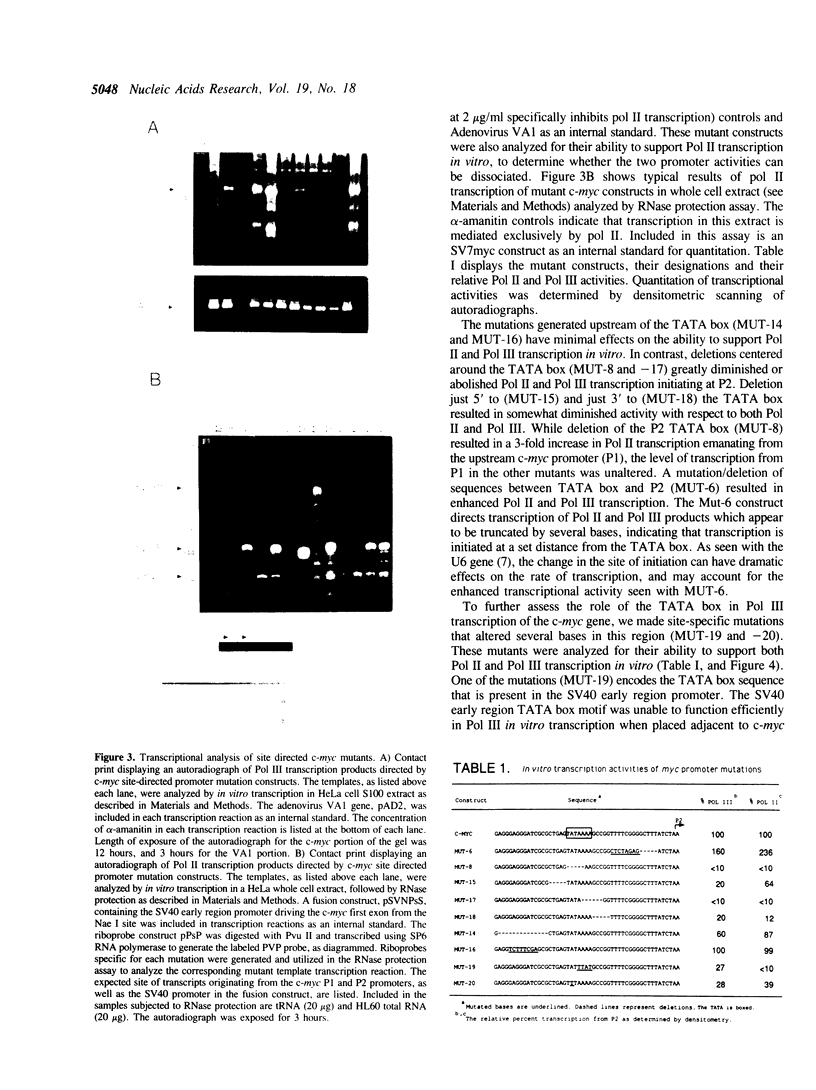

Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Bark C., Weller P., Zabielski J., Janson L., Pettersson U. A distant enhancer element is required for polymerase III transcription of a U6 RNA gene. Nature. 1987 Jul 23;328(6128):356–359. doi: 10.1038/328356a0. [DOI] [PubMed] [Google Scholar]
- Bentley D. L., Brown W. L., Groudine M. Accurate, TATA box-dependent polymerase III transcription from promoters of the c-myc gene in injected Xenopus oocytes. Genes Dev. 1989 Aug;3(8):1179–1189. doi: 10.1101/gad.3.8.1179. [DOI] [PubMed] [Google Scholar]
- Bentley D. L., Groudine M. A block to elongation is largely responsible for decreased transcription of c-myc in differentiated HL60 cells. Nature. 1986 Jun 12;321(6071):702–706. doi: 10.1038/321702a0. [DOI] [PubMed] [Google Scholar]
- Bentley D. L., Groudine M. Sequence requirements for premature termination of transcription in the human c-myc gene. Cell. 1988 Apr 22;53(2):245–256. doi: 10.1016/0092-8674(88)90386-8. [DOI] [PubMed] [Google Scholar]
- Berk A. J. Adenovirus promoters and E1A transactivation. Annu Rev Genet. 1986;20:45–79. doi: 10.1146/annurev.ge.20.120186.000401. [DOI] [PubMed] [Google Scholar]
- Campisi J., Gray H. E., Pardee A. B., Dean M., Sonenshein G. E. Cell-cycle control of c-myc but not c-ras expression is lost following chemical transformation. Cell. 1984 Feb;36(2):241–247. doi: 10.1016/0092-8674(84)90217-4. [DOI] [PubMed] [Google Scholar]
- Carbon P., Murgo S., Ebel J. P., Krol A., Tebb G., Mattaj L. W. A common octamer motif binding protein is involved in the transcription of U6 snRNA by RNA polymerase III and U2 snRNA by RNA polymerase II. Cell. 1987 Oct 9;51(1):71–79. doi: 10.1016/0092-8674(87)90011-0. [DOI] [PubMed] [Google Scholar]
- Chen W., Struhl K. Saturation mutagenesis of a yeast his3 "TATA element": genetic evidence for a specific TATA-binding protein. Proc Natl Acad Sci U S A. 1988 Apr;85(8):2691–2695. doi: 10.1073/pnas.85.8.2691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chirgwin J. M., Przybyla A. E., MacDonald R. J., Rutter W. J. Isolation of biologically active ribonucleic acid from sources enriched in ribonuclease. Biochemistry. 1979 Nov 27;18(24):5294–5299. doi: 10.1021/bi00591a005. [DOI] [PubMed] [Google Scholar]
- Chung J., Sussman D. J., Zeller R., Leder P. The c-myc gene encodes superimposed RNA polymerase II and III promoters. Cell. 1987 Dec 24;51(6):1001–1008. doi: 10.1016/0092-8674(87)90586-1. [DOI] [PubMed] [Google Scholar]
- Das G., Henning D., Wright D., Reddy R. Upstream regulatory elements are necessary and sufficient for transcription of a U6 RNA gene by RNA polymerase III. EMBO J. 1988 Feb;7(2):503–512. doi: 10.1002/j.1460-2075.1988.tb02838.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dignam J. D., Lebovitz R. M., Roeder R. G. Accurate transcription initiation by RNA polymerase II in a soluble extract from isolated mammalian nuclei. Nucleic Acids Res. 1983 Mar 11;11(5):1475–1489. doi: 10.1093/nar/11.5.1475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eick D., Bornkamm G. W. Transcriptional arrest within the first exon is a fast control mechanism in c-myc gene expression. Nucleic Acids Res. 1986 Nov 11;14(21):8331–8346. doi: 10.1093/nar/14.21.8331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Folk W. R. Changing directions in Pol III transcription. Genes Dev. 1988 Apr;2(4):373–375. doi: 10.1101/gad.2.4.373. [DOI] [PubMed] [Google Scholar]
- Fowlkes D. M., Shenk T. Transcriptional control regions of the adenovirus VAI RNA gene. Cell. 1980 Nov;22(2 Pt 2):405–413. doi: 10.1016/0092-8674(80)90351-7. [DOI] [PubMed] [Google Scholar]
- Gallant J. W., Kurz J., Krause M. O. Increased rates of specific RNA polymerase III transcription precede overexpression of cellular oncogenes upon induction of transformation. Oncogene Res. 1989;4(1):39–46. [PubMed] [Google Scholar]
- Harbury P. A., Struhl K. Functional distinctions between yeast TATA elements. Mol Cell Biol. 1989 Dec;9(12):5298–5304. doi: 10.1128/mcb.9.12.5298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hernandez N., Lucito R. Elements required for transcription initiation of the human U2 snRNA gene coincide with elements required for snRNA 3' end formation. EMBO J. 1988 Oct;7(10):3125–3134. doi: 10.1002/j.1460-2075.1988.tb03179.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelly K., Cochran B. H., Stiles C. D., Leder P. Cell-specific regulation of the c-myc gene by lymphocyte mitogens and platelet-derived growth factor. Cell. 1983 Dec;35(3 Pt 2):603–610. doi: 10.1016/0092-8674(83)90092-2. [DOI] [PubMed] [Google Scholar]
- Kerppola T. K., Kane C. M. Intrinsic sites of transcription termination and pausing in the c-myc gene. Mol Cell Biol. 1988 Oct;8(10):4389–4394. doi: 10.1128/mcb.8.10.4389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lobo S. M., Hernandez N. A 7 bp mutation converts a human RNA polymerase II snRNA promoter into an RNA polymerase III promoter. Cell. 1989 Jul 14;58(1):55–67. doi: 10.1016/0092-8674(89)90402-9. [DOI] [PubMed] [Google Scholar]
- Loeken M., Bikel I., Livingston D. M., Brady J. trans-activation of RNA polymerase II and III promoters by SV40 small t antigen. Cell. 1988 Dec 23;55(6):1171–1177. doi: 10.1016/0092-8674(88)90261-9. [DOI] [PubMed] [Google Scholar]
- Manley J. L., Fire A., Cano A., Sharp P. A., Gefter M. L. DNA-dependent transcription of adenovirus genes in a soluble whole-cell extract. Proc Natl Acad Sci U S A. 1980 Jul;77(7):3855–3859. doi: 10.1073/pnas.77.7.3855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mattaj I. W., Dathan N. A., Parry H. D., Carbon P., Krol A. Changing the RNA polymerase specificity of U snRNA gene promoters. Cell. 1988 Nov 4;55(3):435–442. doi: 10.1016/0092-8674(88)90029-3. [DOI] [PubMed] [Google Scholar]
- Mechti N., Piechaczyk M., Blanchard J. M., Marty L., Bonnieu A., Jeanteur P., Lebleu B. Transcriptional and post-transcriptional regulation of c-myc expression during the differentiation of murine erythroleukemia Friend cells. Nucleic Acids Res. 1986 Dec 22;14(24):9653–9666. doi: 10.1093/nar/14.24.9653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melton D. A., Krieg P. A., Rebagliati M. R., Maniatis T., Zinn K., Green M. R. Efficient in vitro synthesis of biologically active RNA and RNA hybridization probes from plasmids containing a bacteriophage SP6 promoter. Nucleic Acids Res. 1984 Sep 25;12(18):7035–7056. doi: 10.1093/nar/12.18.7035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy S., Di Liegro C., Melli M. The in vitro transcription of the 7SK RNA gene by RNA polymerase III is dependent only on the presence of an upstream promoter. Cell. 1987 Oct 9;51(1):81–87. doi: 10.1016/0092-8674(87)90012-2. [DOI] [PubMed] [Google Scholar]
- Murphy S., Moorefield B., Pieler T. Common mechanisms of promoter recognition by RNA polymerases II and III. Trends Genet. 1989 Apr;5(4):122–126. doi: 10.1016/0168-9525(89)90043-7. [DOI] [PubMed] [Google Scholar]
- Murphy S., Pierani A., Scheidereit C., Melli M., Roeder R. G. Purified octamer binding transcription factors stimulate RNA polymerase III--mediated transcription of the 7SK RNA gene. Cell. 1989 Dec 22;59(6):1071–1080. doi: 10.1016/0092-8674(89)90763-0. [DOI] [PubMed] [Google Scholar]
- Nepveu A., Marcu K. B. Intragenic pausing and anti-sense transcription within the murine c-myc locus. EMBO J. 1986 Nov;5(11):2859–2865. doi: 10.1002/j.1460-2075.1986.tb04580.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nevins J. R. Induction of the synthesis of a 70,000 dalton mammalian heat shock protein by the adenovirus E1A gene product. Cell. 1982 Jul;29(3):913–919. doi: 10.1016/0092-8674(82)90453-6. [DOI] [PubMed] [Google Scholar]
- Potter H., Weir L., Leder P. Enhancer-dependent expression of human kappa immunoglobulin genes introduced into mouse pre-B lymphocytes by electroporation. Proc Natl Acad Sci U S A. 1984 Nov;81(22):7161–7165. doi: 10.1073/pnas.81.22.7161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sassone-Corsi P., Borrelli E. Promoter trans-activation of protooncogenes c-fos and c-myc, but not c-Ha-ras, by products of adenovirus early region 1A. Proc Natl Acad Sci U S A. 1987 Sep;84(18):6430–6433. doi: 10.1073/pnas.84.18.6430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott M. R., Westphal K. H., Rigby P. W. Activation of mouse genes in transformed cells. Cell. 1983 Sep;34(2):557–567. doi: 10.1016/0092-8674(83)90388-4. [DOI] [PubMed] [Google Scholar]
- Simon M. C., Fisch T. M., Benecke B. J., Nevins J. R., Heintz N. Definition of multiple, functionally distinct TATA elements, one of which is a target in the hsp70 promoter for E1A regulation. Cell. 1988 Mar 11;52(5):723–729. doi: 10.1016/0092-8674(88)90410-2. [DOI] [PubMed] [Google Scholar]
- Singh K., Carey M., Saragosti S., Botchan M. Expression of enhanced levels of small RNA polymerase III transcripts encoded by the B2 repeats in simian virus 40-transformed mouse cells. Nature. 1985 Apr 11;314(6011):553–556. doi: 10.1038/314553a0. [DOI] [PubMed] [Google Scholar]
- Sollner-Webb B. Surprises in polymerase III transcription. Cell. 1988 Jan 29;52(2):153–154. doi: 10.1016/0092-8674(88)90500-4. [DOI] [PubMed] [Google Scholar]
- Struhl K. Constitutive and inducible Saccharomyces cerevisiae promoters: evidence for two distinct molecular mechanisms. Mol Cell Biol. 1986 Nov;6(11):3847–3853. doi: 10.1128/mcb.6.11.3847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor I. C., Kingston R. E. Factor substitution in a human HSP70 gene promoter: TATA-dependent and TATA-independent interactions. Mol Cell Biol. 1990 Jan;10(1):165–175. doi: 10.1128/mcb.10.1.165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weil P. A., Segall J., Harris B., Ng S. Y., Roeder R. G. Faithful transcription of eukaryotic genes by RNA polymerase III in systems reconstituted with purified DNA templates. J Biol Chem. 1979 Jul 10;254(13):6163–6173. [PubMed] [Google Scholar]
- White R. J., Stott D., Rigby P. W. Regulation of RNA polymerase III transcription in response to F9 embryonal carcinoma stem cell differentiation. Cell. 1989 Dec 22;59(6):1081–1092. doi: 10.1016/0092-8674(89)90764-2. [DOI] [PubMed] [Google Scholar]